Summary
Primary hyperoxaluria type 1 (PH1) is a rare inherited metabolic disorder characterized by oxalate overproduction in the liver, resulting in renal damage. It is caused by mutations in the AGXT gene. Combined liver and kidney transplantation is currently the only permanent curative treatment. We combined locus-specific gene correction and hepatic direct cell reprogramming to generate autologous healthy induced hepatocytes (iHeps) from PH1 patient-derived fibroblasts. First, site-specific AGXT corrected cells were obtained by homology directed repair (HDR) assisted by CRISPR-Cas9, following two different strategies: accurate point mutation (c.731T>C) correction or knockin of an enhanced version of AGXT cDNA. Then, iHeps were generated, by overexpression of hepatic transcription factors. Generated AGXT-corrected iHeps showed hepatic gene expression profile and exhibited in vitro reversion of oxalate accumulation compared to non-edited PH1-derived iHeps. This strategy set up a potential alternative cellular source for liver cell replacement therapy and a personalized PH1 in vitro disease model.
Subject areas: Biological sciences, Cell biology, Molecular genetics
Graphical abstract

Highlights
-
•
AGXT p.Ile244Thr mutation was precisely corrected in PH1-derived fibroblasts by HDR
-
•
AGXT targeted knockin was generated in PH1-derived fibroblasts
-
•
iHeps were derived from PH1 and AGXT-corrected fibroblasts by direct reprogramming
-
•
AGXT-corrected iHeps metabolize glyoxylate, reverting oxalate accumulation in vitro
Biological sciences; Cell biology; Molecular genetics
Introduction
Primary hyperoxalurias (PH) are rare inborn errors of glyoxylate metabolism that result in hepatic oxalate overproduction.1,2,3,4,5 Depending on the affected enzyme, three types have been described, being primary hyperoxaluria type 1 (PH1) the most frequent and severe one.4 PH1 is a rare autosomal recessive inherited metabolic liver disorder caused by the deficiency in the hepatic peroxisomal enzyme alanine-glyoxylate aminotransferase (AGT), codified by the AGXT gene, which catalyzes the conversion of glyoxylate to glycine in the liver.6,7 Due to the deficiency in AGT activity, the accumulated glyoxylate is converted into oxalate by the enzyme lactate dehydrogenase (LDH). Oxalate is an end-product metabolite that cannot be processed by mammals and must be excreted by urine. In excess, it forms nonsoluble calcium oxalate crystals that accumulate in different tissues, especially in kidneys, resulting in progressive renal damage (urolithiasis, nephrocalcinosis) and, ultimately, end stage renal disease (ESRD)4 but also in other tissues, leading to systemic oxalosis.8 PH1 has an estimated prevalence of 1–3 per million inhabitants,9,10,11 however, higher rates have been described in historically isolated populations, such as the Canary Islands in Spain, due to a founder effect.12 Recent population genomic studies have pointed to a much higher prevalence,13,14 since milder forms of PH can be wrongly diagnosed as idiopathic kidney stone disease. p.I244T (c.731T>C) mutation at AGXT exon 7,15 is the second most common mutation in PH1 patients worldwide and the most frequent one in the Canary Islands, appearing in the context of the AGT minor allele, a polymorphic variant with reduced catalytic activity and more susceptible to the effect of pathogenic missense mutations.16,17
Classical treatments of this disorder aim to preserve renal function, decreasing urinary calcium oxalate saturation, through the increase of fluid intake, the use of crystallization inhibitors and the administration of pyridoxine.18,19 Recently, a substrate reduction therapy, consisting of the periodic administration of a therapeutic siRNA for silencing the upstream enzyme glycolate oxidase (GO), responsible for glyoxylate production, has been approved.20,21 Once kidney damage is established, patients need to be subjected to intensive dialysis as a temporary treatment while awaiting for liver-kidney transplantation (LKTx).18 Combined or sequential LKTx is currently the only definitive curative treatment, since the genetic defect in the liver is responsible for oxalate overproduction.22,23 In addition to the shortage of donor livers and the potential complications of the transplantation, transplanted patient must be under immunosuppressive treatment for their entire life. Furthermore, although transplantation outcomes in PH1 patients have improved in the last years, significant challenges remain,24 making LKTx a suboptimal solution. Other therapeutics are now being tested in clinical trials, including substrate reduction therapy targeting either GO or LDH enzymes, chaperone therapy to restore the functional enzyme conformation and intestinal oxalate degradation using probiotics or oral enzyme administration.19 Nevertheless, these approaches do not represent a persistent cure for the disorder. Thus, the development of new therapeutic approaches is an urgent necessity to achieve an effective and definitive cure for PH, and in particular for PH1.
Gene addition therapy has positioned as a real alternative for many genetic diseases, being hematologic and hepatic diseases the most successfully addressed.25,26,27,28 Promising results have also been obtained in preclinical studies in PH1 animal models using different viral vectors.29,30,31 Most recently, targeted gene editing, which consists in accurate site-specific genome modifications, has emerged as a precise alternative to the classical gene therapy in order to avoid insertional mutagenesis risks, associated to random transgene integration, and to take advantage of endogenous gene regulation. In particular, the use of nucleases to trigger the repair mechanisms of the cells that can drive the editing of the genome, and more specifically the use of the CRISPR/Cas system, has spread the clinical applications of gene editing technology.32,33 In this work, we have developed two alternative gene editing strategies, point mutation editing using single-stranded oligodeoxynucleotides (ssODN) or targeted knockin of an enhanced AGXT cDNA, assisted by the CRIPSR-Cas9 nuclease system34 to specifically correct the AGXT gene in PH1 patient-derived cells by homology directed repair (HDR) in the presence of exogenous repair templates.35,36,37
On the other hand, hepatocyte transplantation has been reported as a promising alternative to whole liver transplant in many inherited metabolic liver disorders.38,39,40 Hepatocyte cell transplantation has shown favorable results in PH1 animal models in preclinical studies,41,42 and there is even a PH1 case report where hepatocyte transplantation was used as a bridging procedure to liver transplantation.43 To overcome the limitations associated to the in vitro culture of primary human hepatocytes, that rapidly dedifferentiate in vitro, and the allogenic transplantation, alternative cell sources are being investigated. Hepatocyte-like cells have been obtained from human embryonic stem cells (hESC) and human induced pluripotent stem cells (hiPSC).44,45,46 In this context, Estève et al. differentiated hepatocyte-like cells from PH1 patient-derived iPSCs.47 Most recently, direct cell reprogramming of differentiated somatic cells into induced hepatocytes (iHeps) by ectopic expression of different liver-enriched transcription factors48,49 has appeared as a promising alternative that would reduce the tumorigenic risk associated with the undifferentiated subpopulations that remain in differentiation protocols from iPSCs. So, we decided to investigate the generation of autologous phenotypically healthy induced hepatocytes as an alternative cell source for liver cell replacement therapy, avoiding intermediate pluripotent cell stages.
Here we propose the combination of gene editing and hepatic direct cell reprogramming for the generation of autologous AGXT-corrected induced-hepatocytes from PH1 patient-derived dermal fibroblasts as a potential therapeutic approach for the treatment of PH1 through liver cell replacement therapy. In addition, generated PH1-derived iHeps will constitute a useful patient-specific in vitro disease model (Graphical abstract).
Results
In this work, we explored the combination of locus-specific AGXT gene editing and hepatic direct cell reprogramming to generate phenotypically healthy induced hepatocytes from PH1 patient-derived fibroblasts.
Specific correction of AGXT gene was addressed in human fibroblasts derived from skin biopsy of PH1 patients carrying the c.731T>C point mutation at exon 7, a founder mutation prevalent in the Canary Islands. Two different homology directed repair (HDR) strategies were tested: the precise correction of the point mutation c.731T>C using single-stranded oligodeoxynucleotides (ssODN) and a more universal strategy by knocking in a full cDNA version of the AGXT gene at the endogenous locus (Figure 1A).
Figure 1.
Gene editing strategies for locus specific AGXT gene correction of PH1 patient-derived fibroblasts
(A) Two gene editing strategies for PH1 causing mutation correction: precise correction of the most frequent mutation in PH1 patient from the Canary Islands and targeted AGXT knockin of an enhanced version of AGXT cDNA. Scheme of the genome structure and locations where the gene editing was conducted is represented. Gray squares represent AGXT exons.
(B) Gene editing workflow. (1) PH1 patient-derived fibroblasts were synchronized with aphidicolin for cell-cycle arrest and then released to increase the percentage of cells in S phase. (2) Synchronized PH1 fibroblasts were electroporated to deliver the corresponding gene editing tools: gRNA and Cas9 as ribonucleoprotein particle and ssODN, for point-mutation correction at exon 7, or plasmid DNA, containing an enhanced version of AGXT cDNA for knockin strategy at exon 1, as HDR template. (3) Electroporated cells were isolated by limiting dilution, in the case of point mutation correction strategy, or by puromycin resistant selection, in the knockin strategy. (4) Expanded isolated clones were analyzed by RFLP and Sanger sequencing for HDR allele characterization, in the case of point mutation correction, or by in-out PCR amplification and Sanger sequencing for knockin integration confirmation.
Human fibroblast cultures were synchronized before delivering the gene editing tools by electroporation to increase the percentage of cells in S phase when HDR occurred. Monoclonal cell populations were isolated from the total pool of electroporated cells by limiting dilution or puromycin resistant selection, depending on the gene editing strategy, and HDR analysis was conducted in the isolated clones (Figure 1B).
Precise correction of one of the most prevalent AGXT missense mutations in PH1 patient-derived fibroblasts
For the precise correction of the c.731T>C point mutation at exon 7, the most prevalent AGXT mutation in the Canary Islands, first, two different guide RNAs (gRNA) were tested to induce double-strand breaks (DSBs) close to the point mutation. As HDR templates, two ssODN with the AGXT-corrected sequence and some silent changes, to avoid re-cutting of the corrected sequence, and including a restriction site to facilitate the identification of the corrected integration, were designed (Figure 2A). Gene editing outcomes for the different combinations of the gene editing tools were analyzed in PH1 patient-derived fibroblasts (Figure 2B). Most resulted alleles corresponded to non-edited ones (40.9%–58.9%), followed by deletions of less than 10bp (26.5%–41.4%) or more than 10bp (3.7%–19.3%). In addition to the precise correction of the point mutation (0%–4.8%), other ssODN aberrant insertions, that hampered recombinant analysis, appeared (0%–7.2%). Accurate correction efficiencies increased with E7-G1 single gRNA (4.8% with ssODN-100 or 3.3% with ssODN-150 with respect to 0% or 1.2% for E7-G2 dual gRNA, respectively), whose cleavage site was closer to the point mutation and to the nucleotide changes included in the ssODNs. Moreover, ssODN aberrant insertions decreased when the longest ssODN (ssODN-150) was used (3.3% with respect to 7.2% for ssODN-100), both in combination with synthetic E7-G1 sgRNA (Figure 2B).
Figure 2.
Precise correction of the c.731T>C point mutation in exon 7 in PH1 patient-derived fibroblasts
(A) Schematic representation of gene editing tools: gRNAs (E7-G1 and E7-G2) and ssODNs. Two sense ssODNs of 100nt and 150nt, respectively, with the AGXT-corrected sequence and silent changes, including an EcoRV restriction site close to the point mutation. E7-G1 gRNA induced DSBs closer to the missense point mutation.
(B) Gene editing outcomes of the point mutation correction experiments conducted with E7-G1 synthetic single gRNA or E7-G2 crRNA:tracrRNA dual gRNA, in combination with the ssODN-100 or ssODN-150. Targeted AGXT exon 7 sequences were PCR amplified, subcloned, and analyzed by RFLP and Sanger sequencing. In addition to the precise point mutation correction (purple percentages), other INDELs and aberrant ssODN insertions were detected (black percentages).
(C) Specific AGXT point mutation correction outcomes of gene editing isolated clones obtained from three different PH1-patient derived fibroblasts edited with E7-G1 sgRNA and ssODN-150. Bars represent the number of accurate point mutation corrected (purple) and RFLP positive (light pink) clones among isolated clones (n = 74 for PH1-1, n = 84 for PH1-2 and n = 73 for PH1-3). Percentage of clones with the precise correction is shown.
(D) Allele sequence characterization of AGXT-corrected clones obtained from the three different PH1 donors. All clones resulted in the correction of one allele (light red squares highlighting C>T substitutions). The second allele presented small deletions (3 or 5 bp, light red squares marking missing nucleotides with dashes) in the fully characterized clones. (Pink arrows represent the E7-G1 sgRNA directed close to the point mutation at AGXT exon 7).
Once the most efficient combination of gRNA and ssODN (E7-G1 gRNA and the ssODN-150) was selected, point mutation correction experiments were conducted in PH1 patient-derived fibroblasts obtained from three different donors (PH1-1, PH1-2, and PH1-3). Synchronized fibroblasts (Figure S1A) were electroporated with the selected gene editing tools. Cleavage efficiency was close to 80% for all the three PH1 donors (Figure S1C). Monoclonal cell populations were isolated and analyzed for accurate HDR correction by restriction fragment length polymorphism (RFLP) and Sanger sequencing. On average, 12.8% of RFLP positive clones resulted in precise point mutation correction, representing an overall AGXT point mutation correction efficiency in PH1 patient-derived fibroblasts of 2.16% (4.05%, 1.19%, and 1.37% for PH1-1, PH1-2, and PH1-3, respectively) (Figure 2C). All AGXT-corrected clones obtained were heterozygous for the specific correction (Figure 2D). Small deletions of 3 bp or 5 bp were found in the second allele, according to the most prevalent gene editing outcomes found in the screening of the editing tools (29% small deletions, light blue in Figure 2B).
When we analyzed potential off-target activity, eight off-target sites for the E7-G1 gRNA, from the top 5 in silico predicted by two different web tools (Figure S2A), were characterized in the five AGXT-corrected clones obtained. Sequence alterations were detected at one of the predicted sites in two of the five AGXT-corrected clones analyzed (Figure S2B). To further investigate other undesirable side effects of Cas9 nuclease, an array-CGH was performed in the GC-1A and GC-2A corrected clones, in comparison to their corresponding non-edited PH1-derived cells. The results from a-CGH clearly demonstrate that CRISPR/Cas9 nuclease does not lead to significant chromosomal truncation or any detectable adverse effects on the integrity of specific gene-corrected clones’ genomes. Our data indicate that the corrected clones exhibited stable and intact karyotypes, supporting the safety and accuracy of this gene-editing approach. (Figure S3).
AGXT targeted knockin of an enhanced full cDNA to correct most AGXT gene mutations
The second proposed strategy, the targeted knockin of an enhanced version of the AGXT cDNA near the ATG start codon at the endogenous gene exon 1, was addressed in order to correct AGT deficiency in human fibroblasts derived from several PH1 patients (Figure 1A) and assure a more universal correction platform. A single guide RNA (E1-G1) to induce DSBs close to the translation start codon at AGXT exon 1 (7 bp downstream ATG codon) and four different plasmid DNA donors with an enhanced version of AGXT cDNA (hAGXT-RHEAM),50 a puromycin-resistance cassette (PuroR), and 600 bp homology arms flanking the previous sequences, to promote homologous recombination at AGXT endogenous locus, were designed (Figure 3A).
Figure 3.
AGXT targeted knockin of an enhanced cDNA at exon 1 in PH1 patient-derived fibroblasts
(A) Gene editing tools designed. E1-G1 sgRNA (blue arrow) induced double-strand breaks at AGXT exon 1, near ATG starting codon. Four different DNA plasmid donors with an enhanced version of AGXT cDNA (hAGXT-RHEAM) and a puromycin-resistance cassette (PuroR), flanked by 600 bp homology arms were tested. Left homology arm (LHA) were the same for all of them. Right homology arm 1 (RHA1), in pDonor_A and pDonor_B, was positioned 5 bp upstream to the E1-G1 genome cleavage site, symmetrically to LHA. Right homology arm 2 (RHA2), in pDonor_C and pDonor_D, was positioned 155 bp downstream to the cleavage site, at the end of AGXT exon 1. pDonor_B and pDonor_D included the E1-G1 target sequence at the end of each homology arm.
(B) Knockin integration efficiencies obtained for PH1 patient-derived fibroblasts from three different donors. Bars show the number of positive (blue) and negative (white) clones for the precise knock-in integration among puromycin resistant (PuroR) clones. Only clones properly amplified by PCR and Sanger-sequenced for the two homology arm junctions were considered as correct knock-in clones. The percentage of clones with the accurate knock-in integration is shown in blue.
Targeted knockin experiments were conducted in PH1 patient-derived fibroblasts obtained from three different donors. Synchronized cultures (Figure S1B) were electroporated with the designed gene editing tools, obtaining cleavage efficiencies ranging from 24% to 69% depending on the donor (Figure S1D). Puromycin resistant monoclonal cell populations were isolated and analyzed to confirm specific knockin integration at the target locus by PCR amplification and Sanger sequencing (Figure S8B). Only those clones in which both homology arms were verified for accurate integration were considered as correct knockin clones (Figure 3B). AGXT knockin integration efficiency in PH1 patient-derived human fibroblasts varied from 0% to 50% of puromycin resistant isolated clones, depending on the PH1 patient donor and the plasmid used. The addition of the gRNA target sequence at the end of each homology arm in the plasmid donor DNA, increased specific knockin efficiency (25.9% pDonor_B in comparison to 7.7% pDonor_A; and 6.5% pDonor_D compared to 0% pDonor_C). Homology arms symmetrically positioned close to the cleavage site improved knockin generation (25.9% pDonor_B in comparison to 6.5% pDonor_D and 7.7% pDonor_A with respect to 0% pDonor_C). Overall, the most efficient plasmid donor for knockin integration was pDonor_B (25.9%) which included the gRNA target at the end of the homology arms, to be linearized inside the cell by the CRISPR/Cas9 system, and both homology arms symmetrically close to the genome cleavage site of the gRNA. Heterozygous integration resulted more frequent (80%) than homozygous clones, among confirmed knockin clones.
Eight different off-targets sites for the E1-G1 gRNA, from the top 5 in silico predicted by two different web tools (Figure S2C), were analyzed in the ten confirmed AGXT knockin clones. Only heterozygous alterations were found in one of the AGXT knockin clone in OT4 predicted site, but these modifications were far from the predicted cleavage point (Figure S2D). In the same way, we have analyzed the potential undesirable side effects of the nuclease in clones specifically corrected, an array-CGH analysis was performed in 4 KI clones (pA-1A, pD-1A, pD-2A, and pB-3A), in comparison to their corresponding non-edited PH1-derived cells. The results from a-CGH clearly demonstrate that CRISPR/Cas9 nuclease does not lead to significant chromosomal truncation or any detectable adverse effects on the integrity of 3 out of these 4 clones’ genomes. Our data indicate that pA-1A, pD-2A, and pB-3A corrected clones exhibited stable and intact karyotypes. However, it is worth noting that clone pD-1A showed some interstitial deletions in chromosomes 2, 8, and 16. These findings suggest that rare off-target events may have occurred in clone pD-1A (Figure S3).
Generation of induced hepatocytes from PH1 and AGXT-corrected fibroblasts
Corrected fibroblasts must be converted into hepatic cells prior to confirm the metabolic correction of PH1 patient-derived cells, as AGXT is specifically expressed in the liver. PH1 patient-derived and AGXT gene-corrected fibroblasts obtained by the two different gene editing strategies were directly reprogrammed into induced hepatocytes (iHeps) by overexpression of four hepatic transcription factors: FOXA2, HNF1α, HNF4α, and TBX3, through lentiviral transduction, in combination with specific culture media (Figure 4A). Transduced cells were analyzed at different time points during the reprogramming protocol (Figure 4B), to confirm the lentiviral transduction and the acquisition of the hepatic profile. Previous to patient-derived samples, healthy donor human fibroblasts were reprogrammed to establish the best protocol for direct conversion of fibroblasts to hepatic cells. Flow cytometry analysis of the mCherry fluorescent tracking protein, also present in the hepatic transcription factor lentiviruses, confirmed the initial transduction of around 80% of starting fibroblasts (Figures S4A and S4B). The expression of the exogenous hepatic transcription factors FOXA2, HNF1a, and HNF4a was confirmed at different time points during the reprogramming process (Figures S4C and S4D), as well as the induction of the corresponding endogenous transcription factors (Figures S4E and S4F). Cell morphology changes were observed during the direct hepatic reprogramming process (Figure 4C): starting fibroblastic shape went through a transient phase of cytoplasm elongation becoming very thin with prominent nuclei and, once cells were transferred to the maturation medium, their cytoplasm swelled and they acquired a polyhedral morphology forming a confluent cobblestone, showing an epithelial-like morphology.
Figure 4.
Generation of induced-hepatocytes (iHeps) from human fibroblasts by hepatic direct cell reprogramming
(A) Human fibroblasts were transduced with lentiviruses coding for four hepatic transcription factors: FOXA2, HNF1α, HNF4α, and TBX3. Transduced cells were cultured in endoderm induction medium from day 1 post-transduction to day 3–5 and then in hepatic maturation medium until day 13–15.
(B) Time course of direct reprogramming protocol.
(C) Cell morphology evolution during hepatic direct cell reprogramming protocol. Representative phase contrast images of PH1 patient-derived cells at different time points are shown. All images were taken with 400× magnification. Scale bar: 25 μm.
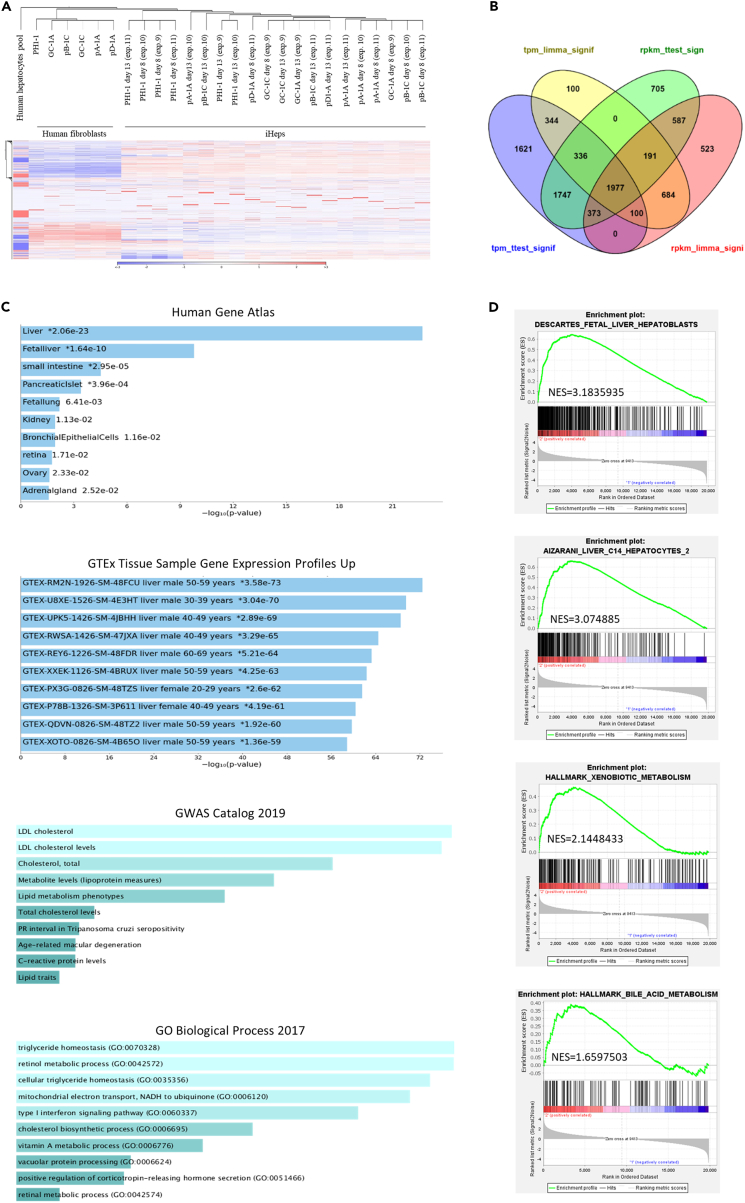
To further characterize the generated iHeps, a whole transcriptome analysis was conducted, comparing the starting fibroblasts with their generated iHeps at day 8 and 13 post-transduction. Clustering analysis on RNA sequencing demonstrated that generated iHeps separated clearly from the original fibroblasts (Figures 5A and S5A). Applying differential gene expression analyses of RNA-seq data from PH1-derived samples, corrected and non-corrected, we found 1,977 protein-coding genes with significant differential expression between the generated iHeps and the starting fibroblasts (Figure 5B; and 328 differentially expressed genes in the case of healthy donor samples (Figure S5B)). Common protein-coding genes significantly up-regulated and down-regulated in generated iHeps from both healthy and PH1-derived donors are listed in Tables S1 and S2. The hepatic fate acquisition by the reprogrammed cells was confirmed by the analysis of their 1,097 up-regulated genes, that demonstrated enrichment in genes involved in different liver functions (Figure 5C; 251 up-regulated genes in healthy donor iHeps (Figure S5C)), such as cholesterol, LDL, or triglyceride metabolism, among others. This hepatic profile was also corroborated by the gene set enrichment analysis (GSEA) analysis (Figures 5D and S5D), which demonstrated the enrichment in hepatocyte and hepatoblast characteristic genes, as well as in specific liver pathways, such as xenobiotic and bile acid metabolism.
Figure 5.
Transcriptome analysis of PH1-derived and AGXT-corrected iHeps generated by direct cell reprogramming and their starting fibroblasts
RNA-seq analysis of 10 independent experiments, conducted with one PH1 donor and five different AGXT-corrected clones, two of them with the precise point mutation correction and the other three with the AGXT targeted knockin.
(A) Hierarchical clustering of RNA-Seq analyzed samples and heatmap of the gene expression analysis. Red color represents overexpression, while blue color indicates underexpression.
(B) Differential gene expression analysis of PH1 and AGXT-corrected fibroblasts and their generated iHeps. Differentially expressed protein coding genes in the generated iHeps at day 8 and 13 of the reprogramming protocol with respect to the starting fibroblasts, analyzed by four different tests, is represented.
(C) Enrichment analysis of the 1097 significative up-regulated genes in the generated iHeps with the web tool Enrichr indicated enrichment in liver genes, including different hepatic metabolic pathways.
(D) Gene set enrichment analysis (GSEA) of generated iHeps compared to human fibroblasts. GSEA demonstrated enrichment in hepatoblast and hepatocyte characteristic genes, as well as in genes involved in different liver functions, in the iHeps compared to the starting fibroblasts. NES = normalized enrichment score.
Based on the differential expression analysis, a set of the most expressed hepatic genes among the up-regulated ones in healthy donor-derived iHeps was analyzed by RT-qPCR (Figure S6). Four of these hepatic up-regulated genes were finally selected to validate the hepatic reprogramming outcomes by RT-qPCR in experiments conducted with 2 PH1 patient-derived and their corresponding AGXT-corrected fibroblasts. Consistent and reproducible induction of these hepatic genes in the generated iHeps in comparison with the starting fibroblasts was demonstrated in a set of independent experiments (Figure 6A). Remarkably, the expression of ABCC2 in the generated iHeps was very close to that of primary hepatocytes (93.9%) (Figure S6).
Figure 6.
Characterization of iHeps generated from PH1 patient-derived fibroblasts and AGXT gene corrected clones
(A) Expression of hepatocyte specific genes in iHeps generated by direct cell reprogramming from PH1 patient-derived fibroblasts and AGXT gene corrected fibroblasts. The expression of genes involved in different liver functions was analyzed by RT-qPCR. Data from twenty-six independent experiments, conducted with two different PH1-donor fibroblasts (PH1-1 (n = 8) and PH1-2 (n = 4)), three AGXT point mutation corrected clones from the two PH1 donors (GC-1A (n = 4) and GC-1C (n = 1) derived from PH1-1. GC-2A (n = 2) derived from PH1-2) and four AGXT targeted knockin clones (pA-1A (n = 2), pB-1C (n = 2), and pD-1A (n = 2)), derived from PH1-1. pD-2A (n = 1) derived from PH1-2). Violin plot of data show median (bold dashed line) and interquartile range (light dashed line). Normal distribution of the data was assessed by a Kolmogorov-Smirnov test. Nonparametric Kruskal-Wallis with Dunn’s multiple comparisons test was used to compare, for each analyzed gene, iHeps with hepatocytes ($) or with fibroblasts (#). Significant differences between cell populations are marked as follows: $/#: p < 0.05; $$/##: p < 0.01; $$$/###: p < 0.001; $$$$/####: p < 0.0001. ABCC2: ATP binding cassette subfamily C member 2, CP: ceruloplasmin, FGL1: fibrinogen-like protein 1 and HP: haptoglobin.
(B) Glycogen storage in the generated iHeps. Microscope images of PAS staining on starting human fibroblasts and generated iHeps, from healthy donor, PH1 patients and AGXT-corrected clones (GC-1A from PH1-1 and GC-2A from PH1-2). HepG2 cell line was used as positive control. All images were taken with 100× magnification. Scale bar: 75 μm.
In addition, all generated iHeps, independently from their origin, acquired the hepatocyte-specific capacity of storing glycogen, as demonstrated by PAS staining (Figure 6B), proving its hepatic functionality.
In vitro reversion of oxalate accumulation in AGXT-corrected iHeps
AGT protein induction was confirmed by western blot in healthy donor, PH1 patient-derived and AGXT-corrected induced hepatocytes at the end of the reprogramming process for one of the PH1 donors (Figure 7A).
Figure 7.
AGT protein induction in generated iHeps and in vitro reversion of oxalate accumulation in AGXT-corrected iHeps with respect to non-corrected PH1-derived iHeps
(A) Western-blot analysis of AGT protein in PH1-derived, AGXT-corrected, and healthy donor-derived iHeps.
(B) Determination of oxalate accumulation in PH1-derived, AGXT-corrected, and healthy donor-derived iHeps cultured in the presence of 1 mM of glyoxylate for 48 h, expressed as nmol of produced oxalate in culture media. Each graph represents the average of 3 or 6 independent experiments conducted with a PH1 donor and their corresponding AGXT-corrected clones, generated by accurate point mutation correction (GC-1A and GC-2A) and AGXT targeted knockin (pA-1A, pD-1A, pD-2A, and pB-3A), for each PH1 donor. Mean ± SD of three or six biological replicates is represented for each cell line. Shapiro-Wilk test for n = 3 or Kolmogorov-Smirnov test for n = 6 confirmed normal distribution. Significant differences between cell populations were analyzed by one-way ANOVA with Tukey’s multiple comparisons test (∗∗: p < 0.01; ∗∗∗∗: p < 0.0001).
(C) Gene expression of LDH-A in the generated iHeps indicated as the percentage of expression referred to human hepatocytes (100%), analyzed by RT-qPCR. Mean ± SD of 1–4 biological replicates is represented.
(D) Activity of LDH-A in the generated iHeps analyzed by colorimetric assay in protein extracted from cell cultures prior glyoxylate addition. Activity was expressed as nmol of NADH produced per minute and per μg of protein.
Oxalate concentration in the culture supernatants of generated iHeps was determined after 48 h of culture with the AGT enzyme substrates, glyoxylate and alanine, and the co-enzyme, pyridoxal-phosphate. Optimal glyoxylate concentration in culture media was previously defined to assure an optimal cell survival and oxalate production. Oxalate accumulation decreased in all the AGXT-corrected iHeps, generated by the two different gene editing strategies, after hepatic direct cell reprogramming, in comparison with the corresponding non-edited PH1-derived iHeps (Figure 7B). The expression and the activity of LDH-A were assessed to assure that the changes in the oxalate accumulation observed in the iHep culture is clearly determined by the reactivation of the final step in the glyoxylate pathway. Differences in the expression of LDH-A or in the activity of LDH-A between the corrected and uncorrected iHeps do not justify that the reduction in oxalate production observed in the corrected clones could be caused by a mechanism other than restoration of AGT activity derived from gene correction. Such gene correction is what ultimately would lead to the restoration of glyoxylate metabolism (Figures 7C and 7D). These results, confirmed for the three PH1 donors and their AGXT-corrected iHeps, validate the therapeutic potential of this combined gene editing and hepatic direct reprogramming approach, as well as the potential of the direct reprogramming protocol to generate PH1 patient-specific in vitro models.
Discussion
We generated AGXT gene-corrected PH1 patient-derived induced hepatocytes with glyoxylate metabolizing capacity as a relevant cellular source to be evaluated as a potential therapeutic approach for PH1. These AGXT-corrected iHeps could represent an alternative accessible autologous cell source for liver cell replacement therapy or a bridging procedure to whole liver transplantation. This approach could be applied to several other inherited hepatic metabolic disorders.
Different ex vivo gene therapy attempts have been previously conducted for the treatment of PH1. Estève et al. developed a conventional gene addition therapy with a lentiviral vector coding the AGXT cDNA,47 and also targeted gene therapy into the AAVS1 safe harbor locus assisted by CRISPR/Cas9 system,51 in PH1 patient-derived iPSCs, achieving the restoration of AGT expression in the hepatocyte-like cells differentiated from its genetically modified PH1-iPSCs. However, the specific correction of AGXT gene has remained elusive so far. Here, we demonstrate the locus-specific AGXT gene correction in PH1 patient-derived fibroblasts by two different gene editing strategies maintaining the endogenous gene regulation: an accurate correction of p.I244T mutation at exon 7, with minimal genome sequence alteration, and a knockin of an enhanced AGXT cDNA at exon 1, as a universal strategy for PH1 patients.
Regarding precise point mutation correction, we found that accurate gene editing efficiency depends deeply on the gRNA and ssODN design, with the best results obtained using a gRNA with its cleavage site the closest to the target base to be modified, in combination with a ssODN carrying the nucleotide modifications also as close as possible to the point mutation and cleavage site. In the case of AGXT targeted knockin strategy, we found that homology arms symmetrically close to the cleavage site, in combination with the addition of the gRNA target sequence at the end of each homology arm increased the knockin efficiency, as it has been previously described.52 Applying the optimized gene editing tools, we managed to isolate fibroblast clones from three different PH1 patients with the precise point mutation correction in heterozygosity, as well as heterozygous, but also, homozygous AGXT targeted knockin clones, demonstrating the feasibility of specific AGXT locus editing in PH1 patient-derived cells. Considering that PH1 is a recessive inherited disease, the restored expression of AGXT gene from the corrected allele in the case of heterozygous clones will be sufficient for the proper glyoxylate metabolism to occur. Nevertheless, considering that the knockin of an enhanced AGXT cDNA has render homozygous clones for the correction and that almost all mutations in AGXT gene could be addressed with this strategy, this would be the gene editing approach to pursue.
One important aspect of ex vivo gene editing approaches is the possibility of a deep characterization of the potential therapeutic product. On-target and off-target effects derived from the use of CRISPR/Cas9 system can be screened out before the eventual administration of the cellular product to the patient, discarding the edited clones that do not meet the safety requirements. With the objective of reducing nuclease side effects, such as unspecific DSB in genomic sequence other than the target locus, a dual system of Cas9 nickases could be used, although the necessity of getting two different gRNA with proper score and in the precise genomic location could make the design more complicated and it would not be always possible. In addition to gRNA off-target screening, we also suggest to perform a deep on-target analysis to discard large deletions that can be caused by CRISPR/Cas9 system53 and check for random integration of repair templates, as it has been reported, although in a low frequency.54 In this sense, we found aberrant insertions of the ssODN used as repair template at the target site. This kind of on-target imperfect insertions has been previously reported,55 and highlights the importance of carefully analyzing the edited cell population. In this regard, we observed that ssODN length affects the percentage of these aberrant insertions, as these events decreased by half when using the longest ssODN. The a-CGH analysis conducted in gene-corrected clones revealed stable and intact karyotypes. However, one of the knockin corrected clones showed interstitial deletions in different chromosomes, including chromosome 2 where AGXT locus is placed, highlighting the importance of carefully monitoring and characterizing gene-edited clones for any potential unintended genomic alterations. In any case, the effect of extended periods of cell culture for clone isolation and expansion may also have an impact on chromosomal stability, and observed alterations in clone pD-1A cannot unambiguously be attributed to gene editing process.
Ex vivo gene editing, not only allows previous analyses of the cell product, but it also will overcome the pre-existing humoral and cell-mediated SpCas9 immunogenicity reported in humans,56,57,58 which may affect in vivo editing efficiency.
Traditionally, gene editing therapy was only viable for accessible cells that could be maintained and expanded in vitro, such as blood cells. However, advances in hiPSCs and direct lineage reprogramming have opened the possibility of editing an easy-to-access cell sample from a patient and then convert this into the desired final cell type. Hepatocyte-like cells have been previously generated from PH1-derived hiPSCs.47 Here, we demonstrate the hepatic direct cell reprogramming of healthy donor, PH1 patient-derived and AGXT-corrected fibroblasts into induced hepatocytes. This strategy opens the possibility of generating an alternative cell source for liver cell replacement therapy, using autologous corrected cells, without going through an undifferentiated step, avoiding the concerns related to the tumorigenic risk of iPSC-derived products.
The comparison of iPSC-derived hepatocyte-like cells and/or directly reprogrammed iHeps generated in different studies is complex since there are not stablished standards and because of the wide interindividual variability in the gene expression, that has been reported in livers and primary human hepatocytes.59,60 This donor-dependent variability would be expected to influence the reprogramming outcomes too. Currently, the main goal is to generate fully mature functional hepatocytes, as any of them have demonstrated functionality at the same levels as adult primary human hepatocytes with respect to a variety of hepatic functions. Despite this fact, differential gene expression analysis of our generated iHeps demonstrated the induction of specific hepatic genes involved in different liver functions, although at a low activation level. Even with a general low activation of hepatic genes, we detected AGT protein in the generated iHeps and, most remarkably, we have demonstrated a consistent and reproducible cell reprogramming to glyoxylate-metabolizing iHeps derived from three different PH1 patients, which is an essential factor to consider this protocol as a useful tool for research in PH1 field. AGT protein detection together with LDH-A expression and activity in the generated induced-hepatocytes, confirm that the reversion of oxalate accumulation in AGXT-corrected iHeps compared to non-corrected PH1-derived iHeps is due to the final steps in the glyoxylate pathway (conversion of glyoxylate to glycine by the AGT enzyme or to oxalate by the LDH enzyme). Our next goal will be to enhance the maturity of generated iHeps, which can be achieved by means of chemical compounds, genetic manipulation, 3D structures, and/or addition of supporting cells.61,62,63,64,65,66
Another optimization, with respect to the direct reprogramming protocol, would be to replace the use of individual lentiviral vectors to express the hepatic transcription factors with an all-in-one polycistronic vector,67 in order to reduce the presumable heterogeneity of the transduced cell population with variable hepatic conversion outcomes. However, considering its clinical application, it would be more suitable to use non-viral methods, such as mRNA transfection,68 activation of the endogenous transcription factors using a dead-Cas9 fused to transcriptional activators69 or, ideally, avoid gene transfer using small molecules.64,70,71 Before considering the corrected iHeps with regenerative purposes, as they constitute a potent tool to explore gene editing combined with cellular reprogramming as a therapeutic alternative, an in vivo reversion of PH1 phenotype after transplantation of gene-edited iHeps should be assessed. Unfortunately, immunodeficient mouse models of the disease are not available so far.
This work demonstrates that ex vivo gene editing in combination with hepatic direct cell reprogramming is a feasible strategy to generate AGXT-corrected induced-hepatocytes from PH1 dermal fibroblasts with the ability to reverse the PH1 pathological oxalate accumulation in vitro, in comparison to their corresponding non-corrected PH1-derived iHeps, demonstrating the therapeutic potential of our proposed innovative treatment. In addition, PH1 patient-derived iHeps could be employed as a personalized in vitro disease model for pathophysiology studies and therapeutic screenings.
Limitations of the study
This work demonstrates the feasibility of editing human AGXT locus to correct one of the most common mutations or to generate a knockin of AGXT under its physiological regulation. However, homologous-directed repair assisted by the CRISPR/Cas9 system achieved is still inefficient, and we are now exploring new designs of homologous-directed repair templates that could enhance the specific recombination process. This efficacy improvement would allow to increase the generation of homozygous clones too, that although is not strictly necessary for the treatment of a recessive inherited disease, would reduce the heterogeneity of the corrected clones.
The genome safety studies carried out show that there is indeed a risk for nuclease side effects. Alternative editing systems are discussed that could improve the security of the gene correction. This improvement would be recommended at least when aiming to obtain an alternative cell source for regenerative purposes.
After direct reprogramming protocol, induced hepatocytes show a general, although low, activation of hepatic genes. Enhancement of hepatic gene expression in a more physiological environment is expected, and testing the improved functionality of reprogrammed cells after transplanting is a clear next step to continue with.
Considering these patient-derived induced hepatocytes as a relevant disease model that can be generated for every specific patient, other enzymes involved in glyoxylate metabolism, different from AGT, should be characterized. This would allow to apply this technology to patients with other type of hyperoxaluria.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Recombinant Anti-AGXT antibody [EPR13232] | Abcam | Cat#ab178708 |
| HRP-conjugated Donkey anti-rabbit IgG | Amersham | Cat# NA934; RRID: AB_772206 |
| HRP mouse Anti-Rabbit native IgG antibody | Sigma-Aldrich | Cat# R3155; RRID:AB_1079117 |
| Bacterial and virus strains | ||
| Competent E. coli One Shot® TOP10 cells | Invitrogen | Cat#C404003 |
| Biological samples | ||
| Primary human hepatocytes | Dr. Josep M Canals (Hospital Clinic de Barcelona) and Dra. Trinidad Serrano (Hospital de Zaragoza) | N/A |
| 20-donor pooled human cryopreserved hepatocytes | Quimigen | Liver Pool #X008000 |
| Primary human fibroblasts from skin biopsy | Dr. Eduardo Salido (University Hospital of La Laguna, Tenerife, Sapin) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Aphidicolin | Sigma-Aldrich | Cat#A0781 |
| Puromycin | Sigma-Aldrich | Cat#P8833 |
| Protamine sulfate | Sigma-Aldrich | Cat#P3369 |
| Collagen solution (95 - 98% Type I bovine collagen) | Stem Cell | Cat#04902 |
| Nicotinamide | Sigma-Aldrich | Cat#N0636 |
| Recombinant Human FGF-basic (154 a.a.) | Peprotech | Cat#100-18B |
| Recombinant Human HGF (Insect derived) | Peprotech | Cat#100-39 |
| A-83-01 (≥98% (HPLC)) | Sigma-Aldrich | Cat#SML0788 |
| CHIR99021 (≥98% (HPLC)) | Sigma-Aldrich | Cat#SML1046 |
| Dexamethasone powder (γ-irradiated, BioXtra, suitable for cell culture, ≥80% (HPLC)) | Sigma-Aldrich | Cat#D8893 |
| ITS Liquid Media Supplement (100×) | Sigma-Aldrich | Cat#I3146 |
| L-NAME (Nω-Nitro-L-arginine methyl ester hydrochloride) (≥97% (TLC), powder) | Sigma-Aldrich | Cat#N5751 |
| Recombinant Human Oncostatin M (227 a.a.) | Peprotech | Cat#300-10 |
| TRIzol™ Reagent | Invitrogen | Cat#15596-018 |
| Protein A/G PLUS-Agarose | Santa Cruz Biotechnology | Cat#sc-2003 |
| Critical commercial assays | ||
| P2 Primary Cell 4D-Nucleofector X Kit | Lonza | Cat#V4XP-2032, Cat#V4XP-2024 |
| Zero Blunt® PCR Cloning Kit | Invitrogen | Cat#K2700 |
| RETROscript Reverse Transcription Kit | Invitrogen | Cat#AM1710 |
| FastStart Universal SYBR Green Master (Rox) | Roche | Cat#4913850001 |
| Sodium glyoxylate monohydrate (≥93%) | Sigma-Aldrich | Cat#G4502 |
| Pyridoxal 5′-phosphate hydrate powder (BioReagent, suitable for cell culture) | Sigma-Aldrich | Cat#P3657 |
| Oxalate Kit | Trinity Biotech | Cat#591D |
| LDH Assay Kit | Abcam | Cat#ab102526 |
| SurePrint G3 Human CGH Array 8 × 60 K | Agilent | Cat#G4450A |
| Deposited data | ||
| RNA-Seq raw and analyzed data | This paper | GEO: GSE226019 |
| Experimental models: Cell lines | ||
| HEK 293T/17 | ATCC | CRL-11268 |
| HepG2 | ATCC | HB-8065 |
| Oligonucleotides | ||
| Primers for AGXT sequence characterization: see Table S3 | This paper | N/A |
| ssODN-100nt: GAAAGAAGATGTACTCCCGCAAG ACGAAGCCCTTCTCCTTCTACCTGGATATCAAGT GGCTGGCCAACTTCTGGGGCTGTGACGACCAG CCCAGGATGTG |
This paper | N/A |
| ssODN-150nt: TGCTCAGCCTGCTTCTTTCTCCCCA GAAAGAAGATGTACTCCCGCAAGACGAAGCCCT TCTCCTTCTACCTGGATATCAAGTGGCTGGCCAA CTTCTGGGGCTGTGACGACCAGCCCAGGATGTG AGGCCTGGCAGGGATGGGAAGGTGG |
This paper | N/A |
| Primers for the generation of knock-in plasmid templates: see Table S4 | This paper | N/A |
| Primers for gene editing analyses: see Table S5 | This paper | N/A |
| Primers for PCR amplification of E7-G1 and E1-G1 off-target sites: see Table S6 | This paper | N/A |
| Primers for gene expression analysis by RT-qPCR: See Table S7 | This paper | N/A |
| Primer and probes for VCN determination: see Table S8 | This paper | N/A |
| Recombinant DNA | ||
| pLOX-CMV:hTERT-IRES-TK was a gift from Didier Trono | Salmon et al., 2000 (https://doi.org/10.1006/mthe.2000.0141)72 | Addgene plasmid #12245 |
| Knock-in plasmid templates: (E1-G1)-LHA-hAGXT-RHEAM-SV40polyA-LoxP-PGKpromoter-PuroR/ΔTK-bGHpolyA-LoxP-RHA1/2-(E1-G1) | This paper | N/A |
| SSFV:HepaTF-E2A-mCherry-wpre∗ | This paper | N/A |
| Software and algorithms | ||
| Breaking-Cas | Oliveros et al., 2016 (https://doi.org/10.1093/NAR/GKW407)73 | https://bioinfogp.cnb.csic.es/tools/breakingcas/ |
| CRISPOR | Concordet and Haeussler, 2018 (https://doi.org/10.1093/NAR/GKY354)74 | http://crispor.tefor.net/ |
| FlowJo software v.10.5 | BD | https://www.flowjo.com/solutions/flowjo |
| TIDE (Tracking of Indels by DEcomposition) software | Brinkman et al., 2014 (https://doi.org/10.1093/NAR/GKU936)75 | https://tide.nki.nl/ |
| Multi Experiment Viewer (MeV) (version 4.9.0) software | Saeed et al., 2003 (https://doi.org/10.2144/03342MT01)76 | https://webmev.tm4.org/about |
| Gene Set Enrichment Analysis (GSEA) (version 4.1.0) software | Subramanian et al., 2005 (https://doi.org/10.1073/PNAS.0506580102)77 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Enrichr | Xie et al., 2021 (https://doi.org/10.1002/CPZ1.90)78 | https://maayanlab.cloud/Enrichr/ |
| GraphPad Prism (version 9 for Windows) | GraphPad Software | https://www.graphpad.com/ |
| Feature Extraction Software v10.7 | Agilent Technologies | https://www.agilent.com/en/product/mirna-microarray-platform/mirna-microarray-software/feature-extraction-software-228496#productdetails |
| Agilent Genomic Workbench v7.0.4 | Agilent Technologies | https://www.agilent.com/en/product/cgh-cgh-snp-microarray-platform/cgh-cgh-snp-microarray-software/agilent-genomic-workbench-228497 |
| Other | ||
| Alt-R® S.p.Cas9 Nuclease V3 | IDT (Integrated DNA Technologies) | Cat# 1081058 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, María García-Bravo (maria.garciabravo@ciemat.es).
Materials availability
Any additional information related to materials generated in this study is available from the lead contact upon request.
Data and code availability
-
•
Cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Human fibroblasts
PH1 patient-derived fibroblasts, homozygous for the c.731T>C (p.Ile244Thr) mutation at AGXT exon 7, were obtained from three different donors of both sexes, two women of 38 and 52 years old (PH1-1 and PH1-2) and a 54 year-old man (PH1-3). This ensure the study’s generalizability, although differences based on the gender are not expected in the results of this work. Informed consent was obtained from the patients for the biopsies obtention, and the procedure was approved by the Comité Ético de Investigación Clínico del Hospital Universitario de Canarias (Spain). Experiments described in this work were authorized by Centro de Investigaciones Energéticas, Medio Ambientales y Tecnológicas (CIEMAT).
Human fibroblasts were cultured in DMEM (1X) with GlutaMAX, 10% HyClone Research Grade Fetal Bovine Serum and 1% penicillin/streptomycin solution, under normoxic conditions (37°C, 21% O2, 5% CO2 and 95% relative humidity).
Method details
Cells and culture conditions
Isolation of primary human fibroblasts
A 3-mm skin biopsy was collected from the forearm of healthy donors or PH1 patients at University Hospital of La Laguna (Tenerife, Spain) for fibroblast isolation. All donors signed an informed consent, and the study was approved by the Ethical Committee of the University Hospital of La Laguna (Tenerife, Spain). Biopsies were washed with DMEM (1X) with GlutaMAX (Gibco, Cat#61965) and incubated in this medium with 2mg/ml type IV collagenase (Sigma, Cat#C5138) and 1% penicillin/streptomycin solution (Sigma, Cat#P4333) overnight at 37°C. The epidermis layer was separated from the dermis and discarded. The piece of dermis was maintained in the medium with collagenase until completely dissociated. Cell suspension was centrifuged, and cell pellet was seeded with DMEM (1X) with GlutaMAX, supplemented with 10% HyClone Research Grade Fetal Bovine Serum (GE healthcare, Cat#SV30160.03) and 1% penicillin/streptomycin solution. Fibroblast growth appeared 7-14 days after initial plating and medium was replaced as necessary during this incubation period.
Human fibroblast culture
Human fibroblasts were cultured in Dulbecco’s Modified DMEM (1X) with GlutaMAX (Gibco, Cat#61965), 10% HyClone Research Grade Fetal Bovine Serum (GE healthcare, Cat#SV30160.03) and 1% penicillin/streptomycin solution (Sigma, Cat#P4333), from now on called cDMEM, under normoxic conditions (37°C, 21% O2, 5% CO2 and 95% relative humidity). Fibroblasts were seeded at a density of 9x103cell/cm2 and were serially passaged at a split ratio 1:2, using 0.25% trypsin-EDTA 1X (Gibco, Cat#25200072), when confluence was reached.
Primary fibroblasts were immortalized by lentiviral transduction with a vector coding for the telomerase reverse transcriptase, hTERT, the catalytic subunit of the telomerase enzyme (pLOX-TERT-iresTK, Addgene plasmid Cat#12245)72 (see lentiviral transduction at lentivirus procedures section). Transduced fibroblasts were expanded and, 1-2 months afterward, the vector copy number (VCN, quantification of proviral vector genomes in the cell genomic DNA) were checked to assure the integration of the vector with a VCN around 1 (see lentivirus titration at lentivirus procedures section).
AGXT sequence characterization
The AGXT gene editing target sequences of PH1 patient-derived fibroblast lines obtained from three different donors were characterized. Genomic DNA were extracted using DNeasy Blood & Tissue Kit (Qiagen, Cat#695004), following the manufacture instructions. In the case of exon 7, 768bp surrounding the described c.731T>C point mutation were PCR amplified and Sanger sequenced, confirming the presence of this point mutation in all the PH1 fibroblast samples (Figure S8A). For the knock-in strategy, a region of 2630bp at 5’AGXT was characterized throughout 4 PCR amplifications that were then sequenced (Figure S8B). A set of changes, including a 74bp duplication in intron 1, with respect to NCBI Reference Sequence NG_008005.1, were detected. This set of changes detected (reflected in Figure S8B), had been previously described as part of AGXT minor allele.17 Primers for PCR amplification and sequencing are summarized in Table S3.
Gene editing tools
CRISPR/Cas9 system components
Guide RNAs (gRNAs) to induce double strand breaks (DSBs) at the target site of the genome were designed based on the specific PH1 patient sequences (including the point mutation c.731T>C) (Figure S8), using different web tools available: Dr. Zhang's Lab (https://zlab.bio/guide-design-resources), BreakingCas73 (https://bioinfogp.cnb.csic.es/tools/breakingcas/) or CRISPOR.org 74 (http://crispor.tefor.net/). E1-G1 (Target sequence+PAM: GGTCACCAGCAGCTTGTGAG-AGG) and E7-G1 (Target sequence+PAM: TGGCCAGCCACTTGGTGTCC-AGG) chemically modified synthetic sgRNAs supplied by Synthego (Menlo Park, California, USA) combines the crRNA and tracrRNA elements into a single RNA molecule. E7-G2 gRNA (Target sequence+PAM: CCAGAAGTTGGCCAGCCACT-TGG), consisting of a duplex system: Alt-R® CRISPR-Cas9 crRNA + Alt-R® CRISPR-Cas9 tracrRNA in equimolar concentration, were obtained from IDT (Integrated DNA Technologies, Coralville, Iowa, USA). To form the crRNA:tracrRNA duplex, 2μl crRNA (200μM) and 2μl tracrRNA (200μM) were mixed with 6μl of buffer solution and incubated at 95°C for 5 minutes to anneal.
Recombinant Alt-R® S.pyogenes Cas9 Nuclease V3, purchased from IDT (Integrated DNA Technologies, Cat#1081058), was used in the different experiments.
gRNA and Cas9 nuclease were delivery into PH1 patient-derived fibroblasts by electroporation as ribonucleoprotein (RNP) complex. To form these RNP complexes, 10μl of prepared 40μM IDT duplex crRNA:tracrRNA or 4μl of 100μM Synthego sgRNA were mixed with 1μl of 60μM Cas9 nuclease purchased from IDT and incubated 10 minutes at room temperature.
ssODN templates for point mutation correction
Single-stranded oligodeoxynucleotide (ssODN) templates were designed based on exon 7 AGXT consensus sequence (NCBI Reference Sequence: NG_008005.1), to reverse the point mutation c.731T>C and to introduce some useful silent changes (GACACC > GATATC). Two different sense-ssODN were designed, one of 100nt and the other one of 150nt (key resources table). In addition to the point mutation correction (GACACC > GATATC), ssODN incorporated synonymous substitutions to avoid re-cutting of the corrected sequence and a EcoRV restriction site to facilitate HDR identification by Restriction Fragment Length Polymorphism (RFLP) analysis (GACACC > GATATC). These silent changes were positioned in the middle of the ssODN. ssODN synthesis was ordered to SIGMA-Aldrich (St. Louis, Missouri, USA).
Plasmid templates for knock-in generation
Four different plasmid donors were designed as DNA templates for the AGXT knock-in strategy (Figure 3). All of them include an enhanced version of the AGXT cDNA, hAGXT-RHEAM, that consists of five amino acid changes that improve protein kinetic stability and increase enzyme activity,50 followed by a SV40 polyadenylation (polyA) signal and a selection cassette based on the puromycin resistance and thymidine kinase fusion gene, driven by the PGK promoter and ending in the bGH-polyA signal (Figure S9A). All these elements were flanked by 600bp homology arms with the sequences around the E1-G1 gRNA target site, to facilitate homologous recombination. The four plasmid donors share the same left homology arm (LHA) but with two different right homology arms (RHA-1 and RHA-2). Furthermore, two of them incorporated the target sequence of the E1-G1 gRNA at the end of each homology arm. Primers used to generate the four different pDonors are summarized in Table S4.
Gene editing procedures
Fibroblast culture synchronization
Confluent PH1 patient-derived fibroblasts were culture in 2μg/ml aphidicolin (Sigma, Cat#A0781) in cDMEM for 17 hours (over-night) (cell cycle arrest). These fibroblast cultures were then washed with 1X PBS and cultured in cDMEM for 3 hours to release the cell cycle arrest. Cell cycle synchronization was analyzed by flow cytometry based on DNA cell content. 3x105 cells of synchronized culture and non-synchronized culture as control were fixed with 500μl of 70% ethanol at 4°C for 1 hour. Then, cell samples were centrifuged, and cell pellets were resuspended in 300μl PBA (PBS with 0.1% BSA and 0.02% sodium azide) with 50μg/ml RNAse for 20 minutes at room-temperature. Finally, 1X propidium iodide (Sigma, Cat#P4170) was added and incubated for 15 minutes at room-temperature protected from light. Proportion of cells in the different cell cycle phases was determined by flow cytometry measuring propidium iodide signal in LSR Fortessa Cell Analyser (BD, Becton Dickinson). Acquired data were analyzed with FlowJo software v.10.5 (BD).
Electroporation of PH1-derived fibroblasts
Gene editing tools were delivery into synchronized PH1 patient-derived fibroblasts by electroporation. Fibroblasts were nucleofected using P2 Primary Cell 4D-Nucleofector X Kit (Lonza, Cat#V4XP-2032, Cat#V4XP-2024) in Amaxa 4D-Nucleofector System (Lonza, Cat#AAF-1002X). Cells were harvested by trypsinization and 1.5x10⁵ for point mutation correction or 7.5x10⁵ cells for knock-in generation were resuspended in 20μl or 100μl nucleofection solution, respectively. RNP complex, previously prepared (see RNP complex at CRISPR/Cas9 section), and HDR templates, if applicable, were added to the cell suspension: 2μl of IDT duplex crRNA:tracrRNA + Cas9 or 1μl of Synthego sgRNA + Cas9 for 20μl 16-well nucleocuvette strips, with the corresponding amount of ssODN in the case of point mutation correction experiments; or 5μl of Synthego sgRNA + Cas9 RNP complex with 2.5μg plasmid donor DNA for 100μl nucleocuvette in the case of knock-in experiments. Human fibroblasts were electroporated using DT-130 electroporation program. After pulse completed, cells were incubated in the nucleocuvette for 10 minutes before resuspended in pre-warmed cDMEM medium and transferred to the corresponding cell culture plates. Figure S10 summarizes the different electroporation conditions.
Puromycin-resistant clone isolation
In the case of knock-in experiments, cells electroporated with the corresponding knock-in gene editing tools were expanded for 5 days. Then, gene edited cells were selected with 1μg/ml puromycin (Sigma, Cat#P8833) for 7 days to obtain isolated knock-in clones.
Gene editing analyses
INDELs frequency analysis by TIDE assay
Genomic DNA from the pool of edited fibroblasts was extracted using DNeasy Blood & Tissue Kit (Qiagen, Cat#695004) and AGXT exon 7 or exon 1 target were PCR amplified and sequenced (primers summarized at Table S5). INDELs generated by the CRISPR/Cas9 RNP complex were quantified using TIDE (Tracking of Indels by DEcomposition) software75 (https://tide.nki.nl/). Unedited PH1 patient-derived fibroblasts were used as negative controls for INDEL frequency determination.
Point mutation correction characterization
Restriction Fragment Length Polymorphism (RFLP) analysis was conducted in the pool of electroporated fibroblasts or in isolated clones to detect HDR sequences. Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, Cat#695004) and exon 7 AGXT gene editing target site was amplified by PCR with primers listed in Table S5, resulting in a 650bp amplicon. Gene edited alleles were analyzed by RFLP through EcoRV digestion of exon 7 AGXT target PCR. EcoRV-HF restriction enzyme from New England Biolabs (Cat#R3195S) were used following manufacturer’s instructions. Digested products were analyzed by electrophoresis in 1% agarose gels.
AGXT exon 7 target PCR from the pool of electroporated fibroblasts were subcloned into bacteria vectors using the Zero Blunt® PCR Cloning Kit (Invitrogen, Cat#K2700), following the manufacturer’s instructions. Subcloned vectors were transformed into competent E. coli One Shot® TOP10 cells (Invitrogen, Cat#C404003) following manufacturer’s instructions. Individual transformant colonies were picked from the LB plates and grew overnight in 3mL LB containing 50μg/mL kanamycin. Then, plasmid DNA was purified using NucleoSpin Plasmid kit (Macherey-Nagel, Cat#740499). Finally, isolated plasmids with the AGXT exon 7 gene editing targets were analyzed by EcoRV-HF and BamHI (New England Biolabs, Cat#R3195S, Cat#R0136S) digestion. To further characterize edited alleles, the PCR products inserted into the PCR-Blunt vector were sequenced using the M13 forward and/or reverse primers.
AGXT targeted knock-in characterization
Two primers (Table S5) were used to specifically amplify the integrated left homology arm (LHA): primer Pre-LHA_Fw hybridized at the genome sequence while primer RHEAM(Ex1-Ex2)_Rv hybridized only at hAGXT-RHEAM between exon 1 and 2 (Figure S8B). Three primers (Table S5) were used to specifically amplify accurate integrated right homology arms: primer bGH-polyA_Fw only hybridized at the puromycin resistant cassette, and primers Post-RHA1_Rv and Post-RHA2_Rv hybridized at genomic DNA (Figure S9B). All PCR amplicons were characterized by Sanger sequencing using the primers listed in Table S5.
Homozygosity of knock-in integration was analyzed in the confirmed knock-in clones by PCR amplification based on different amplicon sizes (500bp for knock-in integration allele versus 713bp for non-edited PH1 allele; Figure S9B). Primers KI.Ex1_Fw_2 and AGXT_Ex2_Rv listed in Table S5 were used.
Off-target analysis in fibroblast clones
Two different web tools were used for the in-silico prediction of the off-target sites of the different gRNAs employed: CRISPOR74 (http://crispor.tefor.net/crispor.py) and BreakingCas73 (https://bioinfogp.cnb.csic.es/tools/breakingcas/). The top 5 predicted off-targets identified by each web tool were selected for analysis. As some of the predicted off-targets were common to both web tools, a total of 8 potential off-target sites were analyzed for each gene editing strategy. This analysis was conducted in the genomic DNA isolated using DNeasy Blood & Tissue Kit (Qiagen, Cat#695004) from the AGXT-corrected clones. Predicted off-target sites were amplified by PCR, using the primers listed in Table S6. These PCR products were characterized by Sanger sequencing. In case of detecting any sequence abnormality with respect to the reference sequences, the PCR amplicon was subcloned using the Zero Blunt® PCR Cloning Kit (Invitrogen, Cat#K2700) and transformed into competent E. coli One Shot® TOP10 cells (Invitrogen, Cat#C404003). Individual colonies were grown, and plasmid DNA was purified using NucleoSpin Plasmid kit (Macherey-Nagel, Cat#740499). Finally, PCR products inserted in the PCR-Blunt vector were sequenced using the same primers used for PCR amplification (Table S6).
Comparative Genomic Hybridization analysis
For the Comparative Genomic Hybridization (aCGH) analysis, we used a dual-color oligonucleotide-based array (SurePrint G3 Human CGH Array 8 × 60 K; Agilent, Cat#G4450A) with an overall median probe spacing of 41 kb. Genomic DNA samples from AGXT-corrected fibroblast clones after gene editing, along with the corresponding PH1 patient-derived reference DNA samples before gene editing, were labeled with Cy5 and Cy3 dyes (Agilent), respectively. The hybridization was performed following the manufacturer's protocols.
Subsequently, the arrays were scanned using the G2565BA DNA Microarray Scanner (Agilent Technologies), and the data were extracted using Feature Extraction Software v10.7 (Agilent Technologies). To identify contiguous genomic regions corresponding to copy number variations, we used the Aberration Detection Method 2 (ADM-2) algorithm in Agilent Genomic Workbench v7.0.4 (Agilent Technologies). The genomic regions with potential alterations were scored based on the average quality weighted log ratio of the sample and reference channels.
Direct cell reprogramming protocol
Fibroblasts were transduced with four lentiviral vectors coding for selected human hepatic transcription factors (HepaTFs): FOXA2, HNF1α, HNF4α and TBX3 (for lentiviral vectors and transduction see lentivirus procedures section) and cultured in hepatic-specific culture media (Figure 4). Two defined culture media were employed sequentially, endoderm induction medium for 3-5 days (cDMEM supplemented with 4.99mM nicotinamide [Sigma, Cat#N0636], 10ng/ml recombinant human FGF-basic [Peprotech, Cat#100-18B], 20ng/ml recombinant human HGF [Peprotech, Cat#100-39], 0.10% DMSO [Sigma, Cat#D2438], 2μM A-83-01 [Sigma, Cat#SML0788] and 3μM CHIR99021 [Sigma, Cat#SML1046]) followed by hepatic maturation medium until day 13 (cDMEM supplemented with 0.1μM dexamethasone [Sigma, Cat#D8893], ITS (insulin, transferrin, sodium selenite) liquid media supplement 1X [Sigma, Cat#I3146], 640μM N-nitro-Larginine methyl ester hydrochloride [L-NAME; Sigma, Cat#N5751], 20ng/ml recombinant human HGF [Peprotech, Cat#100-39], 20ng/ml recombinant human oncostatin M [Peprotech, Cat#300-10], 0.10% DMSO [Sigma, Cat#D2438], 2μM A-83-01 [Sigma, Cat#SML0788] and 3μM CHIR99021 [Sigma, Cat#SML1046]). Medium was replaced every two days.
Cell morphology was observed along the direct reprogramming protocol with the Nikon ECLIPSE Ts2R-FL microscope and images were acquired using Leica DFC70000T camera.
iHeps characterization
mCherry flow cytometry analysis
Transduced cells were trypsinized and centrifuged. Then, cell pellet was resuspended in PBA (PBS with 0.1% BSA and 0.02% sodium azide) and 1μg/ml DAPI (Sigma) was added as death cell marker. Flow cytometry analyses were conducted in LSR Fortessa Cell Analyser (BD, Becton Dickinson) and acquired data were then analyzed with FlowJo software v.10.5 (BD).
RNA-sequencing analysis
RNA was extracted from fibroblasts and iHeps samples at days 8 and 13 of the reprogramming protocol using TRIzol reagent (Invitrogen, Cat#15596-018) following manufacturer’s instructions. A sample of cryopreserved primary human hepatocytes consisted of a pool of cells from 20 different donors (Quimigen) was included in one of the two RNA-sequencing analysis. Extracted RNA was purified with RNeasy MinElute Cleanup Kit (Qiagen, Cat#74204). Purified RNA samples were quantified with Qubit™ RNA BR Assay Kit (Invitrogen, Cat#Q10210) and their quality and integrity were determined with Qubit RNA IQ Assay (Invitrogen, cat#Q33221) using Qubit 4 Fluorometer (Invitrogen, Cat#Q33238). Only samples with RNA IQ ≥ 7 (RNA quality score) were sequenced.
Human transcriptome sequencing and differential expression analysis were ordered to STABvida (Madan Parque, Caparica, Portugal). cDNA library of RNA samples was constructed using Kapa Stranded mRNA Library Preparation Kit and sequenced in the lllumina Novaseq platform, using 150bp paired-end sequencing reads. The generated sequence raw data was analyzed using CLC Genomics Workbench 12.0.3 (Qiagen). Reads were trimmed and then mapped against the reference genome: Human genome hg38. The result of mapping served to determine the gene expression levels based on the TPM (Trancripts Per Kilobase Million per Million) and RPKM (Reads Per Kilobase MillionReads per Kilobase Million) methods.
Further differential gene expression analyses with TPM and RPKM values comparing human fibroblast starting samples with generated iHeps were performed using Multi Experiment Viewer (MeV) (version 4.9.0) software76 (https://webmev.tm4.org/about). T-test (p<0.001) and LIMMA-test (p<0.001) were applied to both TPM and RPKM values of coding genes. Gene set enrichment analyses were conducted with GSEA (version 4.1.0) software77 (https://www.gsea-msigdb.org/gsea/index.jsp), using the TPM values of protein coding genes, comparing the generated iHeps with the starting fibroblast samples. Additional enrichment analysis was performed with the Enrichr utility78 (https://maayanlab.cloud/Enrichr/).
Gene expression analysis by RT-qPCR
Gene expression analyses for specific genes were performed by real time-quantitative PCR (RT-qPCR). First, RNA from cells was purified using TRIzol reagent (Invitrogen, Cat#15596-018) following manufacturer’s instructions. Next, reverse transcription reaction was conducted using RETROscript Reverse Transcription Kit (Invitrogen, Cat#AM1710), employing random decamers according with manufacturer’s instruction, to generate cDNA. Finally, RT-qPCR was conducted using FastStart Universal SYBR Green Master (Rox) (Roche, Cat#4913850001) in 7500 Fast Real-Time PCR device (Applied Biosystems). Primers for amplification of gene targets are listed in Table S7. GAPDH was used as housekeeping gene, to normalize the target gene expression by ΔCt method.
Different samples of cryopreserved primary human hepatocytes from different donors, kindly provided by Dr. Josep M Canals (Hospital Clinic de Barcelona, Spain) and Dra. Trinidad Serrano (Hospital de Zaragoza, Spain), and a mixed gender 20-donor pooled human cryopreserved hepatocytes (Quimigen) were used as controls to determine the hepatic gene expression levels of the generated iHeps by means of ΔΔCt method.
Periodic Acid-Schiff (PAS) staining
For fibroblasts or HepG2 cells staining, round coverslips Ø12mm (0.13-016mm) (ThermoScientific-Menzel) were previously coated with 0.1% gelatin, and for iHeps staining coverslips were previously coated with collagen solution 1:45 (Type I bovine collagen, Stem Cell Cat#04902). Samples were fixed with 4% paraformaldehyde (PFA) solution (Paraformaldehyde 16%; AlfaAesar, Cat#43368) in PBS for 15 minutes at room temperature. Fixed cells samples were staining for the identification of glycogen accumulation using Periodic Acid-Schiff Kit (Sigma, Cat#395B), according to manufacturer's indications. Briefly, fixed cells were rinsed with water for 1 minute and incubated in Periodic Acid for 5 minutes at room temperature. After several changes of distilled water to rinse the slides, they were incubated with Schiff’s reagent for 15 minutes at room temperature. Finally, samples were washed with water and visualized under light microscope (Nikon ECLIPSE Ts2R-FL). Images were taken with a Leica DFC70000T camera.
Immunoprecipitation and western blot
Total cellular protein was extracted from iHeps and HepG2 cell samples by sonication in lysis buffer (100 mM K3PO4, 250 mM sucrose, 10 μM Pyridoxal-P) containing a protease inhibitor (Complete Mini Protease Inhibitor Cocktail) (Roche, Cat#04693124001) and a phosphatase inhibitor (PhosSTOP) (Sigma, Cat#4906845001) mixture. Sonication was performed using a Fisherbrand Sonic Dismembrator (Fisher Scientific) with 3 cycles of 10 seconds and 30 seconds between cycles, and a 50% amplitude.
Protein concentration was determined using the Bio-Rad Protein Assay Dye Reagent (Bio-Rad, Cat#5000006) and a BSA standard curve and the resulted absorbance was measured at 595nm. Three hundred micrograms of iHeps lysates and 5μg of HepG2 cell lysates were pre-incubated first with horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG 1:5000 (Amersham, Cat#NA934), and second with the Protein A/G PLUS reagent (Santa Cruz Technologies, Cat#sc-2003). After centrifugation at maximal speed, supernatant was incubated with rabbit monoclonal anti-AGXT antibody 1:1000 (Abcam, Cat#ab178708) at 4°C for 1 hour and afterwards with Protein A/G PLUS reagent (Santa Cruz Technologies, Cat#sc-2003) again at 4°C for 1 hour. Immunoprecipitated samples were prepared in 4x Laemmli Sample Buffer (Bio-Rad, Cat#1610747) and β-mercaptoethanol 1:10 (Sigma, Cat#8.05740), and warm at 75°C for 10 minutes. Prepared samples were resolved on 4–15% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, Cat#4561084) by electrophoresis in Tris/Glycine/SDS running buffer (Bio-Rad, Cat#1610732) and transferred to nitrocellulose membrane (Trans-Blot Turbo transfer pack, Bio-Rad, Cat#1704158) using Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, California, USA). The membrane was blocked with 5% non-fat dry milk in 0.1% Tween-20 diluted in phosphate-buffered saline (PBS-T) for 30 minutes at room temperature and then incubated overnight at 4°C with rabbit monoclonal anti-AGXT antibody 1:1000 (Abcam, Cat#ab178708) diluted in blocking solution. Next day, the membrane was washed three times for 10 minutes in PBS 1X and then incubated with horseradish peroxidase (HRP)-conjugated mouse anti-rabbit native IgG 1:5000 (Sigma-Aldrich, Cat#R3155) in the blocking solution for 1 hour at room temperature. After washing three times for 10 minutes with PBS 1X, the immunoblot was revealed using the Clarity Western ECL Substrate (Bio-Rad, Cat#1705061) and visualized in ChemiDoc XRS+ Gel Imaging System using ImageLab software (Bio-Rad, Hercules, California, USA).
In vitro oxalate accumulation assay
To assay the oxalate accumulation in generated iHeps, from day 8 of the reprogramming protocol onwards we change the DMEM used to prepare the maturation medium for one without alanine (Gibco, Cat#41966), supplemented with NEAA (Non-essential Amino Acid Solution, Lonza Cat#13-114E), resulting in a final concentration of 0.1mM L-alanine. Then at day 13, 1mM sodium glyoxylate monohydrate (Sigma, Cat#G4502) were added to the maturation medium in combination with 0.3μM pyridoxal 5'-phosphate hydrate (Sigma, Cat#P3657). Culture supernatants were collected after 48 hours. These samples were acidified with HCl 4N to adjust pH between 5 and 7, and oxalate concentration were determined using Oxalate Kit (Trinity Biotech, Cat#591D), adjusting the manufacturer's protocol. Briefly, 400μl of culture supernatant were mixed with 200μl of 80mg/ml activated charcoal, followed by centrifugation. Twenty microliters of purified samples were then used for the enzymatic reaction, adding 100μl of oxalate reagent A and 10μl of oxalate reagent B. After 5 minutes at room temperature, oxalate was measure by absorbance at 590nm in Tecan Infinite 200 Pro Reader using Tecan i-control 2.0 software. Oxalate concentration was determined by interpolation on the oxalate standard curve performed in parallel.
LDH-A activity assay
LDH-A acitivity in the generated iHeps was assessed by a colorimetric assay (LDH Assay Kit Cat#ab102526) in the sonicated samples once protein concentration was determined, following manufacturer's protocol. Briefly, LDH substrate mix was added to the samples diluted in LDH assay buffer. Immediately, absorbance was measure at 450 nm in Tecan Infinite 200 Pro Reader using Tecan i-control 2.0 software every 2 minutes for a total period of 30 minutes. NADH generated was determined by interpolation on the NADH standard curve performed in parallel, and finally considering the time interval in which all the samples fit in the linear range of the reaction. Activity was expressed as nmol of NADH produced per minute and per μg of protein.
Lentivirus procedures
Lentiviral vectors
pLOX-CMV:hTERT-IRES-TK is a lentiviral vector coding for the telomerase reverse transcriptase, hTERT, the catalytic subunit of the telomerase enzyme, under the control of the CMV promoter (Addgene plasmid, Cat#12245),72 used for the immortalization of primary human fibroblasts.
For the direct reprogramming experiments, we used four self-inactivating lentiviral vectors, SSFV:HepaTF-E2A-mCherry-wpre∗ (Figure S7), coding for the four different hepatic transcription factors (HepaTFs). They were based on pLVX lentiviral backbone (derived from pLVX-IRES-ZsGreen1 vector, Clontech #632187). The corresponding hepatic transcription factor: FOXA2, HNF1α, HNF4α or TBX3, were expressed under the SFFV promoter as a bicistronic mRNA together with the mCherry fluorescent protein, joined by an E2A peptide element to the HepaTF. HepaTFs were chemically synthesized by GeneService company, based on consensus human mRNA sequences from GenBank (FOXA2: BC011780.2A; HNF1α: BC104908.1; HNF4α: BC137540.1; TBX3: BC025258.1). mCherry fluorescent protein (p.mCherry, Clontech Cat#632522) was then joined to the HepaTF with the E2A element by overlapping PCR. Mutated version of Wpre (woodchuck hepatitis virus post-transcriptional regulatory element) was included at the end of the cDNA to stabilize transgene RNA.
Lentivirus production and titration
Lentiviruses were produced using second-generation lentiviral system with HEK293T cell line as packaging cells, following the protocol previously described by Tiscornia and colleagues79 with some modifications. Briefly, calcium phosphate transient transfection was used to transfect the plasmids into the cells. One million HEK293T cells were seeded in a 150 mm TC-treated culture dish (Corning) 24 hours before transfection in DMEM (1X) with GlutaMAX (Gibco, #61965), 10% HyClone (GE healthcare, Cat#SV30160.03). Transfer plasmid, envelop plasmid pMD2.G-VSVG (Addgene plasmid, Cat#12259) and packaging plasmid pCMV-ΔR8.7 (Addgene plasmid, Cat#12263) (22.5μg, 12μg and 27.5μg, respectively) were mixed with 457μl CaCl2 2.5M and 3.2ml milli-Q dH2O. Then, 3.6ml HBS 2X (pH=7) was added dropwise to form Ca2+/DNA precipitates that were then added over HEK293T cells. After 5-6 hours, medium was removed, and fresh medium was added for 48 hours. Culture supernatant was harvested, centrifuged and filtered (Millex PVDF filter unit 0.45μm, Millipore Cat#SLHV033N) to eliminate cellular debris. Finally, virus supernatant was concentrated by ultracentrifugation (7000g, 2.5 hours at 16°C; Optima L-100 XP Ultracentrifugue, Beckman-Coulter) and resuspended in saline solution (NaCl 0.9%). Virus stocks were stored at -80°C.
Virus production was titered in HEK293T cells by serial dilution. In the case of lentiviral vectors carrying a fluorescent protein (mCherry), transducing units (TU) were calculated based on flow cytometry measures 48-72 hours after transduction, applying this formula:
Lentiviral vectors that did not expressed a fluorescent protein were tittered by vector copy number (VCN) determination of serial dilutions of transduced cells, with the following formula:
For VCN determination or quantification of proviral vector genomes in the cell genomic DNA, DNA was extracted from transduced cells using DNeasy Blood & Tissue Kit (Qiagen) and quantitative PCR was conducted to evaluate VCN, based on the simultaneous amplification of Ψ (psi) viral region and human albumin gene as endogenous genome reference, using ABsolute QPCR Mix, low ROX (ThermoFisher, Cat#AB1318). A synthetic dsDNA fragment (gBlock) containing Ψ and albumin sequences, was used as standard curve. Primers and probes used are showed on Table S8.
Lentiviral transduction
For pLOX-CMV:hTERT-IRES-TK lentiviral transduction, human fibroblasts were seeded in 6-well cell culture plate at a density of 1.05x104cell/cm2 24 hours before transduction in cDMEM. Cells were transduced with the appropriate amount of lentiviral vectors, MOI=1.5 (multiplicity of infection, the ratio of transducing lentiviral particles per cell), at a final volume of 400μl cDMEM/well with 8μg/ml of protamine sulfate (Sigma, Cat#P3369) for 6 hours. Afterwards, 600μl/well of cDMEM were added overnight and transduction medium was replaced with fresh culture medium the next day.
For HepaTFs lentiviral transduction for direct cell reprogramming protocol, 6-well cell culture plates were treated with collagen solution 1:45 (Type I bovine collagen, Stem Cell, Cat#04902) for 1 hour and washed with PBS 1X twice before seeding the cells. Human fibroblasts were seeded at a density of 1.05x104cell/cm2, 24 hours before transduction. Cells were transduced with the appropriate mix of lentiviral vectors, at a final volume of 400μl cDMEM/well with 8μg/ml of protamine sulfate (Sigma, Cat#P3369) for 6 hours: MOI=5 for FOXA2, HNF1α and HNF4α and MOI=3 for TBX3 (total MOI=18) in the case of healthy donor fibroblasts; and MOI=10 for FOXA2, HNF1α and HNF4α and MOI=3 for TBX3 (total MOI=33) in the case of PH1 and AGXT-corrected fibroblasts. Afterwards, 600μl/well of cDMEM were added overnight and transduction medium was replaced to fresh culture medium next day.
Quantification and statistical analysis
Statistical analyses were conducted with GraphPad Prism software (version 9 for Windows) (https://www.graphpad.com/). For experiments with n<5, nonparametric two-tailed Mann-Whitney test was used to compare two variable and nonparametric Kruskal-Wallis with Dunn’s multiple comparisons test was used to compare multiple variables. For experiments with n≥5, a Kolmogorov-Smirnov test was conducted first to assess the normal distribution of the data. In case of normal distribution, parametric two-tailed unpaired T test, for two variables comparison, or one-way ANOVA with Tukey’s multiple comparisons test, for more than two variables comparison, were used. If data did not follow normal distribution, previously mentioned nonparametric tests were used. Statistical details of specific experiments, such as the exact number of n, can be found at the corresponding figure legend.
Acknowledgments
The authors thank Aurora de la Cal, María del Carmen Sánchez, Soledad Moreno, Nadia Abu-sabha, Montserrat Aldea, and Sergio Losada for dedicated administrative help. Also, the contribution of PH1 patients and their health care provider (Dr. Armando Torres) is deeply appreciated.
This work was supported by grants from “Ministerio de Economía, Comercio y Competitividad” (EPISEVI 2.0 [RETOS RTC-2015-3393-1] to J.-C.S.), “Ministerio de Ciencia, Innovación y Universidades” (PKDefin [SAF2017-84248-P] and Hemagen [PID2020-119637RB-I00] to J.-C.S.), “Spanish National Research and Development Plan”, Instituto de Salud Carlos III and FEDER (PI20/01837 to S.R.-P., CIBERER (“Acciones Cooperativas y Complementarias Intramurales” 2014 and 2016), to J.-C.S., the Oxalosis and Hyperoxaluria Foundation to M.G.-B., the Asociación Española Contra el Cáncer (LABAE20049RODR to S.R.-P.) and the Fundación Eugenio Rodríguez Pascual to M.G.-B.
The authors also received funds from Instituto de Investigación Sanitaria ‘‘Fundación Jiménez Díaz’’, and CIBERER (CB06/07/0014) and TerCel (RD16/0011/0011), initiatives of the ‘‘Instituto de Salud Carlos III’’.
V.N.R. and A.M.V. contracts were funded by "Ayuda para contratos predoctorales para la Formación de Profesorado Universitario" from “Ministerio de Educación, Cultura y Deporte”: FPU16/02228 and FPU17/02179.
Author contributions
Conceptualization, M.G.-B. and J.-C.S.; methodology, M.G.-B. and V.N.-R.; software, R.G.-E., S.R.-P., and V.N.-R.; investigation, V.N.-R., A.G.-T., A.M.-V., F.J.M., S.R.-P., and M.G.-B.; resources, E.S.; writing – original draft, V.N.-R.; writing – review and editing, M.G.-B. and V.N.-R.; supervision, J.-C.S.;funding acquisition, J.-C.S. and M.G.-B.
Declaration of interests
The authors declare no competing interests.
Published: March 21, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109530.
Contributor Information
José-Carlos Segovia, Email: jc.segovia@ciemat.es.
María García-Bravo, Email: maria.garciabravo@ciemat.es.
Supplemental information
References
- 1.Salido E., Pey A.L., Rodriguez R., Lorenzo V. Primary hyperoxalurias: Disorders of glyoxylate detoxification. Biochim. Biophys. Acta. 2012;1822:1453–1464. doi: 10.1016/j.bbadis.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Harambat J., Fargue S., Bacchetta J., Acquaviva C., Cochat P. Primary hyperoxaluria. Internet J. Nephrol. 2011;2011 doi: 10.4061/2011/864580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cochat P., Rumsby G. Primary hyperoxaluria. N. Engl. J. Med. 2013;369:649–658. doi: 10.1056/NEJMRA1301564. [DOI] [PubMed] [Google Scholar]
- 4.Hoppe B. An update on primary hyperoxaluria. Nat. Rev. Nephrol. 2012;8:467–475. doi: 10.1038/NRNEPH.2012.113. [DOI] [PubMed] [Google Scholar]
- 5.Archer H.E., Dormer A.E., Scowen E.F., Watts R.W. Primary hyperoxaluria. Lancet. 1957;273:320–322. doi: 10.1016/s0140-6736(57)92210-9. [DOI] [PubMed] [Google Scholar]
- 6.Danpure C.J., Jennings P.R. Peroxisomal alanine:glyoxylate aminotransferase deficiency in primary hyperoxaluria type I. FEBS Lett. 1986;201:20–24. doi: 10.1016/0014-5793(86)80563-4. [DOI] [PubMed] [Google Scholar]
- 7.Oppici E., Montioli R., Cellini B. Liver peroxisomal alanine:glyoxylate aminotransferase and the effects of mutations associated with Primary Hyperoxaluria Type I: An overview. Biochim. Biophys. Acta. 2015;1854:1212–1219. doi: 10.1016/j.bbapap.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 8.Sas D.J., Harris P.C., Milliner D.S. Recent advances in the identification and management of inherited hyperoxalurias. Urolithiasis. 2019;47:79–89. doi: 10.1007/s00240-018-1093-3. [DOI] [PubMed] [Google Scholar]
- 9.Cochat P., Deloraine A., Rotily M., Olive F., Liponski I., Deries N. Epidemiology of primary hyperoxaluria type 1. Nephrol. Dial. Transplant. 1995;10:3–7. doi: 10.1093/ndt/10.supp8.3. [DOI] [PubMed] [Google Scholar]
- 10.van Woerden C.S., Groothoff J.W., Wanders R.J.A., Davin J.C., Wijburg F.A. Primary hyperoxaluria type 1 in The Netherlands: Prevalence and outcome. Nephrol. Dial. Transplant. 2003;18:273–279. doi: 10.1093/ndt/18.2.273. [DOI] [PubMed] [Google Scholar]
- 11.Groothoff J.W., Metry E., Deesker L., Garrelfs S., Acquaviva C., Almardini R., Beck B.B., Boyer O., Cerkauskiene R., Ferraro P.M., et al. Clinical practice recommendations for primary hyperoxaluria: an expert consensus statement from ERKNet and OxalEurope. Nat. Rev. Nephrol. 2023;193:194–211. doi: 10.1038/s41581-022-00661-1. [DOI] [PubMed] [Google Scholar]
- 12.Lorenzo V., Alvarez A., Torres A., Torregrosa V., Hernández D., Salido E. Presentation and role of transplantation in adult patients with type 1 primary hyperoxaluria and the I244T AGXT mutation: Single-center experience. Kidney Int. 2006;70:1115–1119. doi: 10.1038/sj.ki.5001758. [DOI] [PubMed] [Google Scholar]
- 13.Bastarache L., Hughey J.J., Hebbring S., Marlo J., Zhao W., Ho W.T., Van Driest S.L., McGregor T.L., Mosley J.D., Wells Q.S., et al. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science. 2018;359:1233–1239. doi: 10.1126/SCIENCE.AAL4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopp K., Cogal A.G., Bergstralh E.J., Seide B.M., Olson J.B., Meek A.M., Lieske J.C., Milliner D.S., Harris P.C., Rare Kidney Stone Consortium, et al. Phenotype-Genotype Correlations and Estimated Carrier Frequencies of Primary Hyperoxaluria. J. Am. Soc. Nephrol. 2015;26:2559–2570. doi: 10.1681/ASN.2014070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Von Schnakenburg C., Rumsby G. Primary hyperoxaluria type 1: A cluster of new mutations in exon 7 of the AGXT gene. J. Med. Genet. 1997;34:489–492. doi: 10.1136/jmg.34.6.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santana A., Salido E., Torres A., Shapiro L.J. Primary hyperoxaluria type 1 in the Canary Islands: A conformational disease due to I244T mutation in the P11L-containing alanine:glyoxylate aminotransferase. Proc. Natl. Acad. Sci. USA. 2003;100:7277–7282. doi: 10.1073/pnas.1131968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams E.L., Acquaviva C., Amoroso A., Chevalier F., Coulter-Mackie M., Monico C.G., Giachino D., Owen T., Robbiano A., Salido E., et al. Primary hyperoxaluria type 1: Update and additional mutation analysis of the AGXT gene. Hum. Mutat. 2009;30:910–917. doi: 10.1002/humu.21021. [DOI] [PubMed] [Google Scholar]
- 18.Cochat P., Hulton S.A., Acquaviva C., Danpure C.J., Daudon M., De Marchi M., Fargue S., Groothoff J., Harambat J., Hoppe B., et al. Primary hyperoxaluria Type 1: indications for screening and guidance for diagnosis and treatment. Nephrol. Dial. Transplant. 2012;27:1729–1736. doi: 10.1093/ndt/gfs078. [DOI] [PubMed] [Google Scholar]
- 19.Shee K., Stoller M.L. Perspectives in primary hyperoxaluria — historical, current and future clinical interventions. Nat. Rev. Urol. 2022;19:137–146. doi: 10.1038/s41585-021-00543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrelfs S.F., Frishberg Y., Hulton S.A., Koren M.J., O’Riordan W.D., Cochat P., Deschênes G., Shasha-Lavsky H., Saland J.M., Van’t Hoff W.G., et al. Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1. N. Engl. J. Med. 2021;384:1216–1226. doi: 10.1056/NEJMOA2021712. [DOI] [PubMed] [Google Scholar]
- 21.Sas D.J., Magen D., Hayes W., Shasha-Lavsky H., Michael M., Schulte I., Sellier-Leclerc A.L., Lu J., Seddighzadeh A., Habtemariam B., et al. Phase 3 trial of lumasiran for primary hyperoxaluria type 1: A new RNAi therapeutic in infants and young children. Genet. Med. 2022;24:654–662. doi: 10.1016/J.GIM.2021.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Cochat P., Fargue S., Harambat J. Primary hyperoxaluria type 1: Strategy for organ transplantation. Curr. Opin. Organ Transplant. 2010;15:590–593. doi: 10.1097/MOT.0b013e32833e35f5. [DOI] [PubMed] [Google Scholar]
- 23.Grenda R., Kaliciński P. Combined and sequential liver–kidney transplantation in children. Pediatr. Nephrol. 2018;33:2227–2237. doi: 10.1007/s00467-017-3880-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstralh E.J., Monico C.G., Lieske J.C., Herges R.M., Langman C.B., Hoppe B., Milliner D.S., IPHR Investigators Transplantation Outcomes in Primary Hyperoxaluria. Am. J. Transplant. 2010;10:2493–2501. doi: 10.1111/j.1600-6143.2010.03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathwani A.C., Reiss U.M., Tuddenham E.G.D., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D., et al. Long-Term Safety and Efficacy of Factor IX Gene Therapy in Hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/nejmoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K., et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C., et al. Lentiviral hematopoietic stem cell gene therapy in patients with wiskott-aldrich syndrome. Science. 2013;341 doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson A.A., Walters M.C., Kwiatkowski J., Rasko J.E.J., Ribeil J.-A., Hongeng S., Magrin E., Schiller G.J., Payen E., Semeraro M., et al. Gene Therapy in Patients with Transfusion-Dependent β-Thalassemia. N. Engl. J. Med. 2018;378:1479–1493. doi: 10.1056/nejmoa1705342. [DOI] [PubMed] [Google Scholar]
- 29.Salido E.C., Li X.M., Lu Y., Wang X., Santana A., Roy-Chowdhury N., Torres A., Shapiro L.J., Roy-Chowdhury J. Alanine-glyoxylate aminotransferase-deficient mice, a model for primary hyperoxaluria that responds to adenoviral gene transfer. Proc. Natl. Acad. Sci. USA. 2006;103:18249–18254. doi: 10.1073/pnas.0607218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salido E., Rodriguez-Pena M., Santana A., Beattie S.G., Petry H., Torres A. Phenotypic correction of a mouse model for primary hyperoxaluria with adeno-associated virus gene transfer. Mol. Ther. 2011;19:870–875. doi: 10.1038/mt.2010.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castello R., Borzone R., D’Aria S., Annunziata P., Piccolo P., Brunetti-Pierri N. Helper-dependent adenoviral vectors for liver-directed gene therapy of primary hyperoxaluria type 1. Gene Ther. 2016;23:129–134. doi: 10.1038/gt.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porteus M.H. A New Class of Medicines through DNA Editing. N. Engl. J. Med. 2019;380:947–959. doi: 10.1056/nejmra1800729. [DOI] [PubMed] [Google Scholar]
- 33.Sharma G., Sharma A.R., Bhattacharya M., Lee S.S., Chakraborty C. CRISPR-Cas9: A Preclinical and Clinical Perspective for the Treatment of Human Diseases. Mol. Ther. 2021;29:571–586. doi: 10.1016/J.YMTHE.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 36.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher R.A., Strom S.C. Human hepatocyte transplantation: Worldwide results. Transplantation. 2006;82:441–449. doi: 10.1097/01.tp.0000231689.44266.ac. [DOI] [PubMed] [Google Scholar]
- 39.Fitzpatrick E., Mitry R.R., Dhawan A. Human hepatocyte transplantation: State of the art. J. Intern. Med. 2009;266:339–357. doi: 10.1111/j.1365-2796.2009.02152.x. [DOI] [PubMed] [Google Scholar]
- 40.Fox I.J., Roy-Chowdhury J. Hepatocyte transplantation. J. Hepatol. 2004;40:878–886. doi: 10.5005/jp/books/12112_10. [DOI] [PubMed] [Google Scholar]
- 41.Guha C., Yamanouchi K., Jiang J., Wang X., Roy Chowdhury N., Santana A., Shapiro L.J., Salido E., Roy-Chowdhury J. Feasibility of hepatocyte transplantation-based therapies for primary hyperoxalurias. Am. J. Nephrol. 2005;25:161–170. doi: 10.1159/000085408. [DOI] [PubMed] [Google Scholar]
- 42.Jiang J., Salido E.C., Guha C., Wang X., Moitra R., Liu L., Roy-Chowdhury J., Roy-Chowdhury N. Correction of hyperoxaluria by liver repopulation with hepatocytes in a mouse model of primary hyperoxaluria type-1. Transplantation. 2008;85:1253–1260. doi: 10.1097/TP.0b013e31816de49e. [DOI] [PubMed] [Google Scholar]
- 43.Beck B.B., Habbig S., Dittrich K., Stippel D., Kaul I., Koerber F., Goebel H., Salido E.C., Kemper M., Meyburg J., Hoppe B. Liver cell transplantation in severe infantile oxalosisa potential bridging procedure to orthotopic liver transplantation? Nephrol. Dial. Transplant. 2012;27:2984–2989. doi: 10.1093/ndt/gfr776. [DOI] [PubMed] [Google Scholar]
- 44.Hu C., Li L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell. 2015;6:562–574. doi: 10.1007/s13238-015-0180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito Y., Ikemoto T., Morine Y., Shimada M. Current status of hepatocyte-like cell therapy from stem cells. Surg. Today. 2021;51:340–349. doi: 10.1007/s00595-020-02092-6. [DOI] [PubMed] [Google Scholar]
- 46.Roy-Chowdhury N., Wang X., Guha C., Roy-Chowdhury J. Hepatocyte-like cells derived from induced pluripotent stem cells. Hepatol. Int. 2017;11:54–69. doi: 10.1007/s12072-016-9757-y. [DOI] [PubMed] [Google Scholar]
- 47.Estève J., Blouin J.M., Lalanne M., Azzi-Martin L., Dubus P., Bidet A., Harambat J., Llanas B., Moranvillier I., Bedel A., et al. Generation of induced pluripotent stem cells-derived hepatocyte-like cells for ex vivo gene therapy of primary hyperoxaluria type 1. Stem Cell Res. 2019;38 doi: 10.1016/j.scr.2019.101467. [DOI] [PubMed] [Google Scholar]
- 48.Kogiso T., Nagahara H., Otsuka M., Shiratori K., Dowdy S.F. Transdifferentiation of human fibroblasts into hepatocyte-like cells by defined transcriptional factors. Hepatol. Int. 2013;7:937–944. doi: 10.1007/s12072-013-9432-5. [DOI] [PubMed] [Google Scholar]
- 49.Rombaut M., Boeckmans J., Rodrigues R.M., van Grunsven L.A., Vanhaecke T., De Kock J. Direct reprogramming of somatic cells into induced hepatocytes: Cracking the Enigma code. J. Hepatol. 2021;75:690–705. doi: 10.1016/j.jhep.2021.04.048. [DOI] [PubMed] [Google Scholar]
- 50.Mesa-Torres N., Yunta C., Fabelo-Rosa I., Gonzalez-Rubio J.M., Sánchez-Ruiz J.M., Salido E., Albert A., Pey A.L. The consensus-Based approach for gene/enzyme replacement therapies and crystallization strategies: The case of human alanine-Glyoxylate aminotransferase. Biochem. J. 2014;462:453–463. doi: 10.1042/BJ20140250. [DOI] [PubMed] [Google Scholar]
- 51.Estève J., Blouin J.M., Lalanne M., Azzi-Martin L., Dubus P., Bidet A., Harambat J., Llanas B., Moranvillier I., Bedel A., et al. Targeted gene therapy in human-induced pluripotent stem cells from a patient with primary hyperoxaluria type 1 using CRISPR/Cas9 technology. Biochem. Biophys. Res. Commun. 2019;517:677–683. doi: 10.1016/j.bbrc.2019.07.109. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.W., et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V., et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019;10:1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanza D.G., Gaspero A., Lorenzo I., Liao L., Zheng P., Wang Y., Deng Y., Cheng C., Zhang C., Seavitt J.R., et al. Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles. BMC Biol. 2018;16:69. doi: 10.1186/s12915-018-0529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivera-Torres N., Banas K., Bialk P., Bloh K.M., Kmiec E.B. Insertional mutagenesis by CRISPR/Cas9 ribonucleoprotein gene editing in cells targeted for point mutation repair directed by short single-stranded DNA oligonucleotides. PLoS One. 2017;12:e0169350. doi: 10.1371/journal.pone.0169350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M., et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferdosi S.R., Ewaisha R., Moghadam F., Krishna S., Park J.G., Ebrahimkhani M.R., Kiani S., Anderson K.S. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019;10:1842. doi: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner D.L., Amini L., Wendering D.J., Burkhardt L.M., Akyüz L., Reinke P., Volk H.D., Schmueck-Henneresse M. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 2019;25:242–248. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 59.Goyak K.M.O., Johnson M.C., Strom S.C., Omiecinski C.J. Expression profiling of interindividual variability following xenobiotic exposures in primary human hepatocyte cultures. Toxicol. Appl. Pharmacol. 2008;231:216–224. doi: 10.1016/j.taap.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gramignoli R., Dorko K., Tahan V., Skvorak K.J., Ellis E., Jorns C., Ericzon B.G., Fox I.J., Strom S.C. Hypothermic storage of human hepatocytes for transplantation. Cell Transplant. 2014;23:1143–1151. doi: 10.3727/096368913X668627. [DOI] [PubMed] [Google Scholar]
- 61.Chen C., Soto-Gutierrez A., Baptista P.M., Spee B. Biotechnology Challenges to In Vitro Maturation of Hepatic Stem Cells. Gastroenterology. 2018;154:1258–1272. doi: 10.1053/j.gastro.2018.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du Y., Wang J., Jia J., Song N., Xiang C., Xu J., Hou Z., Su X., Liu B., Jiang T., et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 63.Shan J., Schwartz R.E., Ross N.T., Logan D.J., Thomas D., Duncan S.A., North T.E., Goessling W., Carpenter A.E., Bhatia S.N. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W., Ding S. Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol. Sci. 2010;31:36–45. doi: 10.1016/j.tips.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Pettinato G., Lehoux S., Ramanathan R., Salem M.M., He L.X., Muse O., Flaumenhaft R., Thompson M.T., Rouse E.A., Cummings R.D., et al. Generation of fully functional hepatocyte-like organoids from human induced pluripotent stem cells mixed with Endothelial Cells. Sci. Rep. 2019;9:8920. doi: 10.1038/s41598-019-45514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L.Y., Liu L.P., Ge J.Y., Yuan Y.Y., Sun L.L., Xu H., Huang P.Y., Hui L.J., Isoda H., Ohkohchi N., et al. A Multiple-Cell Microenvironment in a 3-Dimensional System Enhances Direct Cellular Reprogramming Into Hepatic Organoids. Transplant. Proc. 2018;50:2864–2867. doi: 10.1016/j.transproceed.2018.03.076. [DOI] [PubMed] [Google Scholar]
- 67.Ballester M., Bolonio M., Santamaria R., Castell J.V., Ribes-Koninckx C., Bort R. Direct conversion of human fibroblast to hepatocytes using a single inducible polycistronic vector. Stem Cell Res. Ther. 2019;10:317. doi: 10.1186/s13287-019-1416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simeonov K.P., Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakraborty S., Ji H., Kabadi A.M., Gersbach C.A., Christoforou N., Leong K.W. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim K.T., Lee S.C., Gao Y., Kim K.P., Song G., An S.Y., Adachi K., Jang Y.J., Kim J., Oh K.J., et al. Small Molecules Facilitate Single Factor-Mediated Hepatic Reprogramming. Cell Rep. 2016;15:814–829. doi: 10.1016/j.celrep.2016.03.071. [DOI] [PubMed] [Google Scholar]
- 71.Tang W., Guo R., Shen S.-J., Zheng Y., Lu Y.-T., Jiang M.-M., Cui X., Jiang C.-Z., Xie X. Chemical cocktails enable hepatic reprogramming of human urine-derived cells with a single transcription factor. Acta Pharmacol. Sin. 2019;40:620–629. doi: 10.1038/s41401-018-0170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salmon P., Oberholzer J., Occhiodoro T., Morel P., Lou J., Trono D. Reversible Immortalization of Human Primary Cells by Lentivector-Mediated Transfer of Specific Genes. Mol. Ther. 2000;2:404–414. doi: 10.1006/MTHE.2000.0141. [DOI] [PubMed] [Google Scholar]
- 73.Oliveros J.C., Franch M., Tabas-Madrid D., San-León D., Montoliu L., Cubas P., Pazos F. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016;44:W267–W271. doi: 10.1093/NAR/GKW407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/NAR/GKY354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brinkman E.K., Chen T., Amendola M., Van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342MT01. [DOI] [PubMed] [Google Scholar]
- 77.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021;1 doi: 10.1002/CPZ1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tiscornia G., Singer O., Verma I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.