Abstract
Dyslipidemia has long been implicated in elevating mortality risk; yet, the precise associations between lipid traits and mortality remained undisclosed. Our study aimed to explore the causal effects of lipid traits on both all-cause and cause-specific mortality. One-sample Mendelian randomization (MR) with linear and nonlinear assumptions was conducted in a cohort of 407,951 European participants from the UK Biobank. Six lipid traits, consisting of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides, apolipoprotein A1 (ApoA1), apolipoprotein B (ApoB), and lipoprotein(a), were included to investigate the causal associations with mortality. Two-sample MR was performed to replicate the association between each lipid trait and all-cause mortality. Univariable MR results showed that genetically predicted higher ApoA1 was significantly associated with a decreased all-cause mortality risk (HR[95% CI]:0.93 [0.89–0.97], P value = 0.001), which was validated by the two-sample MR analysis. Higher lipoprotein(a) was associated with an increased risk of all-cause mortality (1.03 [1.01–1.04], P value = 0.002). Multivariable MR confirmed the direct causal effects of ApoA1 and lipoprotein(a) on all-cause mortality. Meanwhile, nonlinear MR found no evidence for nonlinearity between lipids and all-cause mortality. Our examination into cause-specific mortality revealed a suggestive inverse association between ApoA1 and cancer mortality, a significant positive association between lipoprotein(a) and cardiovascular disease mortality, and a suggestive positive association between lipoprotein(a) and digestive disease mortality. High LDL-C was associated with an increased risk of cardiovascular disease mortality but a decreased risk of neurodegenerative disease mortality. The findings suggest that implementing interventions to raise ApoA1 and decrease lipoprotein(a) levels may improve overall health outcomes and mitigate cancer and digestive disease mortality.
Supplementary key words: all-cause mortality, apolipoproteins, cause-specific mortality, lipid profiles, Mendelian randomization, UK Biobank
Dyslipidemia is abnormal lipid metabolism commonly characterized by a prevalent pattern of elevated levels of serum low-density lipoprotein cholesterol (LDL-C), triglyceride (TG), and/or reduced high-density lipoprotein cholesterol (HDL-C) (1). This metabolic imbalance predisposes individuals to the development of atherosclerosis, which in turn contributes to various vascular diseases and premature mortality (2). According to estimates from the World Health Organization, dyslipidemia is highly prevalent worldwide, affecting 40% of the population and accounting for one-third of ischemic heart disease cases, resulting in approximately 2.6 million deaths annually (3). While LDL-C has historically been the primary focus of intensive lipid management strategies, recent research has shown that apolipoprotein B (ApoB), apolipoprotein A-1 (ApoA1), and lipoprotein(a) (Lp(a)) similarly hold significant predictive value for both cardiovascular disease (CVD) and all-cause mortality (4, 5, 6). Therefore, it is imperative to conduct a comprehensive exploration of lipid profiles to evaluate their impacts on mortality risk and devise a rational approach to the management of lipids.
Current evidence on the relationship between lipid traits and all-cause mortality is ambiguous. Some studies reported a counterintuitively negative correlation between LDL-C levels and all-cause mortality (7, 8), while others showed no significant association (9, 10). Recent observational research suggested the potential for a nonlinear relationship between LDL-C and all-cause mortality (11). but a subsequent Mendelian randomization (MR) study did not detect such a nonlinear causal relationship (12). Moreover, the causal nature of other related lipid traits with all-cause mortality remains unclear. Furthermore, emerging evidence indicates that the role of lipids varies among specific deaths. As an example, a prospective study reported that higher levels of HDL-C, ApoA1, and Lp(a) were associated with reduced cancer mortality, but not with CVD mortality (13). Despite extensive research implicating lipid metabolism in the development of various diseases, research on the causal role of lipids in cause-specific mortalities, apart from CVD and cancer mortality, is still lacking.
In this study, we systematically explored the causal associations of six lipid traits, specifically LDL-C, HDL-C, TG, ApoA1, ApoB and Lp(a), with all-cause mortality and cause-specific mortality in a large-scale prospective cohort of the UK Biobank (UKB). A better understanding of the relationships between lipid traits and mortality could indicate a better management strategy for lipids.
Materials and methods
Study population
The prospective cohort of the UKB comprises approximately 500,000 participants recruited from 2006 to 2010 through 22 health assessment centers scattered across the United Kingdom (14). To minimize population stratification bias in our MR analysis, we restricted the study population to genetically verified white British individuals (15). In detail, we implemented individual-level quality control to exclude 92,859 non-European ancestries, 731 participants with excessive or low heterozygosity, 556 with sex chromosome aneuploidy, 161 with excessive genetic relatedness (more than 10 putative third-degree relatives in the kinship), 154 with sex mismatch, resulting in a final study population of 407,951 participants. All participants provided informed consent, and the study was approved by the Research Ethics Committee (REC; approval number: 11/NW/0382) and conducted in accordance with the principles of the Declaration of Helsinki.
Ascertainment of exposure and outcomes
Serum lipids were quantified using the Beckman Coulter AU5800 by enzymatic protective selection analysis for LDL-C (mmol/L), enzyme immunoinhibition analysis for HDL-C (mmol/L), glycerol-3-phosphate-peroxidase analysis for TG (mmol/L) and immunoturbidimetric analysis for ApoA1 (g/L), ApoB (g/L), and Lp(a) (nmol/L). All lipid traits were inversely rank-normalized before genetic analysis, allowing comparison of effect estimates across traits.
Mortality data were obtained from the National Health Service Information Centre and the National Health Service Central Register Scotland. The primary causes of death were classified using codes from the International Classification of Diseases, Tenth Revision. The main outcome of interest was all-cause mortality, and secondary outcomes included mortality due to specific causes, including CVD, cancer, respiratory disease, digestive disease, and neurodegenerative disease. Follow-up time was defined as the period from the date of recruitment to the date of death or 12th November 2021, whichever came first. Details on the coding of mortality causes and the number of deaths per cause are shown in supplemental Table S1.
Single-nucleotide polymorphisms and genetic risk score as instrumental variables
Genetic variants associated with LDL-C, HDL-C, and TG were obtained from a recent large-scale, exome-wide association study of lipid traits in approximately 300,000 Europeans (16). Genetic variants associated with ApoB and ApoA1 were extracted from a genome-wide association study (GWAS) in the UKB, which included a total of 441,016 participants (17). Genetic variants related to Lp(a) were obtained from a GWAS in the Precocious Coronary Artery Disease cohort of European descent (18).
All single nucleotide polymorphisms (SNPs) for lipid traits were selected at the genome-wide significance level (P < 5×10−8) and filtered by excluding SNPs with imputation r2 <0.9, minor allele frequency <0.01, Hardy-Weinberg equilibrium P <1×10−4, and high linkage disequilibrium coefficient r2 >0.1. Ultimately, a total of 147, 195, 163, 440, 255, and 16 SNPs were retained for LDL-C, HDL-C, TG, ApoA1, ApoB, and Lp(a), respectively. We removed those SNPs associated with potential confounding at a Bonferroni corrected level (P < 0.05/number of SNPs), including smoking, drinking, Townsend deprivation index, physical activity, body mass index, Hemoglobin A1c, Diastolic Blood Pressure, and Systolic Blood Pressure. As a result, we used 65 SNPs as instrumental variables for LDL-C, 69 SNPs for HDL-C, 43 SNPs for TG, 269 SNPs for ApoA1, 161 SNPs for ApoB, and 4 SNPs for Lp(a) in univariable MR (UVMR) analysis (supplemental Tables S2–S7). For each lipid trait, we generated a weighted genetic risk score (GRS) by using the allelic effect of the SNPs as reported in previous public GWAS. For the multivariable MR (MVMR) analysis, the SNPs of all lipid traits were clumped by r2 <0.1 within 1 Mb distance. A total of 254 SNPs were finally involved in the MVMR analysis of LDL-C, TG, and ApoB, 290 SNPs in the analysis of HDL-C and ApoA1, and 387 SNPs in the analysis of Lp(a), ApoA1 and ApoB (supplemental Tables S8–S10). Using the phenotypic correlation matrix from UKB data, we calculated the conditional F-statistics for exposures. The F-statistic for each instrument-exposure association ranged from 8111.5 to 199,475.7, indicating a lower risk of weak instrument bias in MR analysis (19). Detailed strength of GRSs associated with blood lipids in UVMR and MVMR for the overall population and subgroups are shown in supplemental Tables S11 and S12.
Linear MR analysis
We applied the two-stage method as the primary approach to estimate the causal effect of lipid traits on mortality in linear MR analyses. In the first stage, a linear regression of the lipid traits on the GRS was conducted to generate predicted values of the exposure, with adjustment of age at recruitment, sex, assessment center, genotyping array, and top 10 genetic principal components. In the second stage, a Cox regression analysis was conducted to examine the association between the predicted values of the lipids on mortality, with adjustment for the same covariates (20).
As a replication analysis, we performed a two-sample MR using the aforementioned GWAS summary-statistic data of lipids from UKB and summary data of all-cause mortality from FinnGen Consortium (Round 6) (21). FinnGen is a public-private partnership project combining genotype data from Finnish biobanks and digital health record data from Finnish health registries, including up to 135,638 women and men of Finnish ancestry. The GWAS of all-cause mortality involved 21,649 individuals with mortality and 238,711 controls of European ancestry from FinnGen consortium. We employed the inverse-variance weighted (IVW) method as the primary MR analysis, and used the methods of weighted median, MR-Egger, and MR-pleiotropy residual sum and outliers (MR-PRESSO) to assess the robustness of our results.
To account for correlations among different lipid traits, Pearson correlation coefficients were calculated and graphically represented by a heat map (supplemental Fig. S1). We then performed an MVMR analysis that jointly estimated the causal association of highly related traits with three models. In model 1, LDL-C, TG, and ApoB were included to identify which one or more traits appeared to be responsible for the effect of LDL-related traits on the risk of mortality. In model 2, HDL-C and ApoA1 were incorporated to illustrate the potential causal roles of HDL-related phenotypes in mortality. Model 3 was developed to examine the independent influence of Lp(a) while minimizing collinearity issues. Given the strong correlations between ApoA1 and HDL-C (Pearson R = 0.92), as well as between ApoB and LDL-C (Pearson R = 0.96), only Lp(a), ApoA1 and ApoB were included in this model for adjustment (supplemental Fig. S1).
Finally, five complementary MR methods with different assumptions about horizontal pleiotropy, including IVW, MR-Egger, MR-PRESSO for UVMR, and MVMR-IVW and MVMR-Egger for MVMR, were performed to assess the robustness of the findings. Cochran Q test and I2 index were used to evaluate the presence of heterogeneity.
Nonlinear MR analysis
To characterize the shape of the causal association between each lipid trait and all-cause mortality, we performed nonlinear MR analyses using a fractional polynomial method (22). We employed a strategy of conditioning on centiles of instrumental variables-free exposure to generate localized average causal effect (LACE) estimates in each stratum. Then we performed a meta-regression of the LACE estimates against the mean of the exposure in each stratum and visualized the best fitting using the derivative of fractional polynomial models of degree 1 and degree 2. Finally, we reported two results: 1) P-values from the fractional polynomial nonlinear test, which is used to evaluate the applicability of the nonlinear model compared with the linear model in the estimation of different strata LACE; and 2) P-values from a trend test based on meta-regression to evaluate the presence of non-linear trends.
All statistical tests are 2-tailed, a Bonferroni corrected P-value <8.3×10−3 (0.05/6 lipid traits) was considered a statistical significance for multiple testing, and results between P-value ≥8.3×10−3 and P-value <0.05 was reported as suggestively significant. All MR analyses were performed using the “MendelianRandomization” (version 0.5.1), “MRPRESSO” (version 0.3), and “MVMR” (version 0.3) packages in R software (version 3.6.3).
Results
Basic characteristics of participants
Of 407,982 individuals included in the analysis (mean age 56.9 ± 8.0 years; 45.9% men), 25,147 (6.2%) cases of CVD, 21,907 (5.4%) cases of respiratory disease, 16,645 (4.1%) cases of cancer, 57,334 (14.1%) cases of digestive diseases, 414 (0.1%) cases of neurodegenerative diseases have been documented at baseline. During a median follow-up period of 12.4 (IQR: 12.0–13.4) years, 30,991 (7.5%) participants were dead. Summary demographic information of individuals studied is available in Table 1.
Table 1.
Baseline characteristics of participants in the UK Biobanka
| Variable | Baseline Characteristics |
|---|---|
| No. | 407,982 |
| Follow-up time, years, median (IQR) | 12.73 (23.00–13.42) |
| Enrollment age, years, mean (SD) | 56.91 (8.00) |
| Male sex, N (%) | 187,417 (45.9%) |
| BMI, kg/m2, mean (SD) | 27.41 (4.76) |
| Smoking status, N (%) | |
| Prefer not to answer | 1,427 (0.3%) |
| Never | 221,914 (54.4%) |
| Previous | 143,433 (35.2%) |
| Current | 41,176 (10.1%) |
| Drinking status, N (%) | |
| Prefer not to answer | 350 (0.1%) |
| Never | 12,790 (3.1%) |
| Previous | 13,959 (3.4%) |
| Current | 380,851 (93.4%) |
| Inactive physical activity, N (%) | 64,976 (19.7%) |
| Lipid-lowering drug users, N (%) | 71,251 (17.6%) |
| Disease at enrollment | |
| Cardiovascular disease, N (%) | 25,147 (6.2%) |
| Respiratory disease, N (%) | 21,907 (5.4%) |
| Cancer, N (%) | 16,645 (4.1%) |
| Digestive disease, N (%) | 57,334 (14.1%) |
| Neurodegenerative disease, N (%) | 414 (0.1%) |
| HbA1c%, mean (SD) | 5.44 (0.59) |
| SBP, mmHg, mean (SD) | 140.24 (19.66) |
| DBP, mmHg, mean (SD) | 82.30 (10.67) |
| LDL-C, mmol/L, mean (SD) | 3.57 (0.87) |
| HDL-C, mmol/L, mean (SD) | 1.45 (0.38) |
| ApoA1, g/L, mean (SD) | 1.54 (0.27) |
| ApoB, g/L, mean (SD) | 1.03 (0.24) |
| TG, mmol/L, mean (SD) | 1.76 (1.02) |
| Lp(a), mg/dl, median [IQR] | 20.10 (9.32–60.11) |
| Death during follow-up, N (%) | 30,991 (7.5%) |
ApoA1, Apolipoprotein A; ApoB, Apolipoprotein B; BMI, Body mass index; DBP, Diastolic blood pressure; HbA1c, Glycated haemoglobin; HDL-C, High-density lipoprotein cholesterol; IQR, Interquartile range; LDL-C, Low-density lipoprotein cholesterol; Lp(a), Lipoprotein(a); SBP, Systolic blood pressure; SD, standard derivation; TG, Triglyceride.
Data are expressed as mean ± standard deviation, number (percentage), or median (interquartile range).
Effects of genetically predicted lipid traits on mortality in UVMR
In UVMR, ApoA1 was found to be inversely associated with all-cause mortality, with a 1-standard deviation (SD) increase in the level of ApoA1 causing an approximately 7% lower risk of all-cause mortality (HR [95% CI] per SD: 0.93 [0.89–0.97], P = 0.001) (Fig. 1). On the other hand, evidence of positive association was observed in ApoB (1.07 [1.02–1.12], P = 0.002) and Lp(a) (1.03 [1.01–1.04], P = 0.002) with all-cause mortality (Fig. 1). The MR estimates obtained from the weighted median, MR-Egger, IVW, and MR-PRESSO analyses were consistent with the findings, as presented in supplemental Table S13. MR-Egger intercept test suggested pleiotropic SNPs may be present for ApoB, but MR-PRESSO did not identify any pleiotropic SNPs. Besides, there was no evidence of heterogeneity or directional pleiotropy for other lipid traits with all-cause mortality (supplemental Table S23).
Fig. 1.
Associations of lipid profiles with all-cause and cause-specific mortality in univariable Mendelian randomization analysis. ApoA1, Apolipoprotein A1; ApoB, Apolipoprotein B; CI, confidence interval; HDL-C, high-density lipoprotein-cholesterol; HR, hazard ratio; LDL-C, low-density lipoprotein-cholesterol; Lp(a), lipoprotein (a); TG, triglyceride.
The replication analyses of two-sample MR also revealed a suggestively negative effect of ApoA1 (OR [95% CI]: 0.93 [0.87–0.99], P = 0.032) and a significantly positive effect of ApoB (1.13 [1.05–1.23], P = 0.001) on all-cause mortality risk, with no evidence of pleiotropy or heterogeneity. No causal evidence for other lipid traits and all-cause mortality was observed (supplemental Table S26).
As for cause-specific mortality, ApoA1 was found to be suggestively associated with CVD mortality (HR [95% CI]: 0.88 [0.80–0.97], P = 0.011) and cancer mortality (0.94 [0.88–0.99], P = 0.035) (Fig. 1). Higher Lp(a) was found to be significantly associated with increased CVD mortality risk (1.11 [1.07–1.15], P < 0.001) and suggestively associated with increased digestive disease mortality risk (1.08 [1.01–1.17], P = 0.045). Similarly, ApoB showed a positive association with CVD mortality (1.39 [1.26–1.54], P < 0.001) and neurodegenerative disease mortality (1.32 [1.11–1.57], P = 0.001). Finally, it is worth noting that LDL-C played an opposite causal role in CVD mortality (1.46 [1.29–1.64], P < 0.001) and neurodegenerative disease mortality (0.80 [0.66–0.98], P = 0.027) (Fig. 1). Other MR methods did not give appreciable differences from the two-stage method (supplemental Tables S13 and S23). In addition to these results, we did not observe any significant association between genetically predicted TG and the risk of cause-specific mortality.
Direct causal effect of genetically predicted lipid traits on mortality in MVMR
In MVMR, ApoA1 showed an inverse and direct causal effect on all-cause mortality after controlling for HDL-C (HR [95% CI]: 0.92 [0.87–0.97], P = 0.002), while Lp(a) presented a positive direct effect after controlling for ApoA1 and ApoB (1,02 [1.01–1.04], P = 0.005). However, the association for ApoB was attenuated after controlling for LDL-C (1.05 [1.00–1.12], P = 0.076) (Fig. 2). The findings were consistent across MVMR-IVW and MVMR-Egger analyses (supplemental Table S20–S22), and no pleiotropy or heterogeneity was detected (supplemental Table S24).
Fig. 2.
Associations of lipid traits with all-cause mortality in multivariable Mendelian randomization analysis. ApoA1, Apolipoprotein A1; ApoB, Apolipoprotein B; CI, Confidence interval; HDL-C, High-density lipoprotein-cholesterol; HR, Hazard Ratio; LDL-C, Low-density lipoprotein-cholesterol; Lp(a), Lipoprotein (a); TG, Triglyceride.
As for cause-specific mortality, both LDL-C and ApoB retained a causal association with CVD mortality (HR [95% CI]: 1.24 [1.07–1.44], P = 0.004 for LDL-C; 1.26 [1.11–1.44], P < 0.001 for ApoB), and with neurodegenerative disease mortality (0.58 [0.45–0.74], P = 0.004 for LDL-C; 1.75 [1.40–2.19], P < 0.001 for ApoB) (supplemental Fig. S2). The effect size of ApoA1 on cancer mortality was similar to UVMR results (0.92 [0.85–0.99], P = 0.036) (supplemental Fig. S3), and remained significant across different MR methods. Elevated Lp(a) was still significantly associated with a higher risk of CVD mortality (1.09 [1.06–1.13], P < 0.001) and suggestively associated with a higher risk of digestive disease mortality (1.09 [1.01–1.18], P = 0.032) (supplemental Fig. S4).
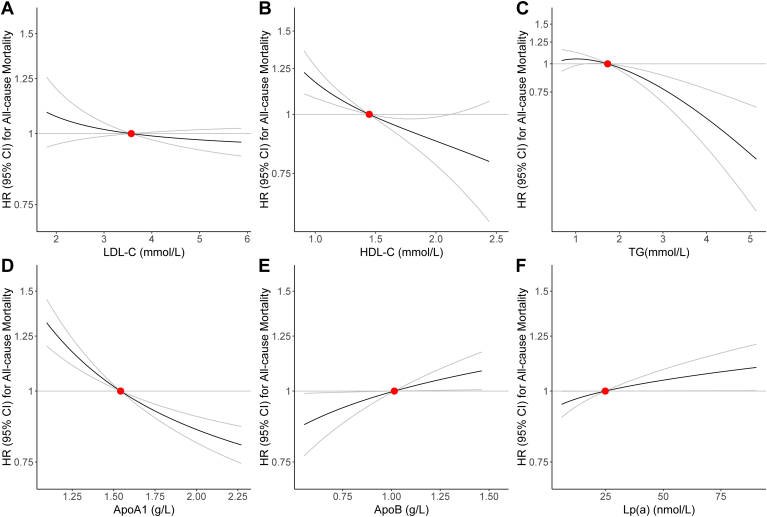
Nonlinear MR analysis of the association between lipid traits and all-cause mortality
To further examine the shape of the genetic association between lipid traits and all-cause mortality, we performed nonlinear MR analyses by calculating LACE estimates for each centile of instrumental variable-free exposure. Our results suggested that there were no significant nonlinear causal associations between lipid traits and all-cause mortality (Fig. 3; P for nonlinearity all >0.05 in supplemental Table S25). Nevertheless, the shape of the relationship suggested that genetically predicted higher ApoA1 was associated with a decreased risk of all-cause mortality, whilst higher Lp(a) was associated with an increased risk of all-cause mortality, which further confirmed the associations from the linear MR analysis (Fig. 3).
Fig. 3.
Nonlinear Mendelian randomization for the causal effects of (A) LDL-C, (B) HDL-C, (C) TG, (D) ApoA1, (E) ApoB, (F) Lp(a) on the risk of all-cause mortality. Red point indicates the reference point, which is the median of lipid trait. Grey lines represent 95% CI. Gradient at each point of the curve is the localized average causal effect. ApoA1, Apolipoprotein A1; ApoB, Apolipoprotein B; CI, Confidence interval; HDL-C, High-density lipoprotein-cholesterol; HR, Hazard Ratio; LDL-C, Low-density lipoprotein-cholesterol; Lp(a), Lipoprotein (a); TG, Triglyceride.
Sensitivity analyses
A set of sensitivity analyses was performed to assess the robustness of our findings. First, in the stratified analyses by sex and age groups (younger individuals <65 years old vs. older individuals ≥65 years old), we observed a causal association between ApoA1 and all-cause mortality in both males (HR [95% CI]: 0.92 [0.87–0.98], P = 0.008) and females (0.94 [0.88–0.99], P = 0.043) as well as in younger individuals (0.93 [0.88–0.99], P = 0.014), as supported by UVMR (supplemental Tables S14–S17). Correspondingly, MVMR analysis yielded consistent associations across the three groups, providing robustness to our findings (supplemental Table S21). Meanwhile, Lp(a) also showed a positive association with all-cause mortality in subgroup analyses by age and sex, especially in the males (1.04 [1.02–1.06], P < 0.001 for UVMR; 1.04 [1.01–1.06], P = 0.001 for MVMR) (supplemental Tables S14, S15, S16, and S22). The disparity between age groups could potentially be ascribed to the relatively low representation of the elderly population, which accounts for only around 20% of the total population.
Second, we excluded individuals with baseline CVD, cancer, respiratory, digestive, or neurodegenerative diseases to minimize reverse causation bias. UVMR analysis demonstrated a significant causal effect of ApoA1 and Lp(a) on all-cause mortality using a two-stage method (HR [95% CI]:0.93 [0.89–0.99], P = 0.001 for ApoA1; 1.03 [1.01–1.04], P = 0.002 for Lp(a)), with consistent effects across other MR methods (supplemental Table S18). MVMR results supported the strong independent effect of ApoA1 and Lp(a) on all-cause mortality risk, in accordance with our primary analyses (supplemental Tables S20–S22).
Third, we repeated the MR analyses in individuals without using lipid-lowering drugs, to address potential confounding factors such as medication adherence, unknown time to treatment, and differences in the type and dosage of lipid-lowering drugs. We observed a significant negative association between ApoA1 and all-cause mortality (HR [95% CI]: 0.93 [0.88–0.98], P = 0.006 for UVMR; 0.90 [0.84–0.97], P = 0.003 for MVMR), and a roughly significant association for Lp(a) (1.02 [0.99–1.04], P = 0.115 for UVMR; 1.02 [0.99–1.04], P = 0.127 for MVMR) (supplemental Tables S19 and S21).
Fourth, as suggested by the reviewers, we accessed recently published GWAS datasets for LDL-C, HDL-C, TG (23), and Lp(a) (24) to obtain more SNPs. The SNP selection process precisely adhered to the previously described method. We identified 158, 187, 178, and 10 SNPs to proxy LDL-C, HDL-C, TG, and Lp(a), respectively (supplemental Table S27). Utilizing these SNPs, we constructed weighted GRSs and repeated the UVMR and MVMR analyses. UVMR analysis revealed positive associations of Lp(a) with the risk of all-cause mortality (1.09[1.03,1.15], P = 0.002), CVD mortality (1.17[1.07, 1.27], P < 0.001), and digestive-specific mortality (1.26[1.04,1.52], P = 0.019). Genetically predicted higher LDL-C levels were associated with an increased risk of CVD mortality (HR [95% CI]: 1.54 [1.36, 1.75], P < 0.001) while genetically predicted lower HDL-C levels were associated with a decreased risk of CVD mortality (0.83[0.76, 0.92], P < 0.001) (supplemental Tables S28). MVMR outcomes corroborated the direct causal impacts of Lp(a) on all-cause, CVD, and digestive disease mortality, with control for ApoA1 and ApoB. Additionally, the direct causal effect of LDL-C on CVD mortality was affirmed after accounting for TG and ApoB (supplemental Table S29).
Discussion
In this study, we comprehensively investigated the causal effects of blood lipids on all-cause mortality and cause-specific mortality using data on 407,951 individuals from the UKB cohort. Our main findings were three-fold. First, both linear and nonlinear MR suggested a linear relationship between genetically predicted ApoA1 and Lp(a) with all-cause mortality. Second, genetically predicted higher ApoA1 was suggestively associated with a decreased risk of cancer mortality, while genetically predicted higher Lp(a) was significantly associated with an increased risk of CVD and suggestively associated with an increased risk of digestive disease mortality. Third, LDL-C exhibited an opposite causal effect in CVD and neurodegenerative disease mortality, despite no significant causal association with all-cause mortality. These findings shed light on the complex relationships between lipid biomarkers and mortality, highlighting the importance of appropriate management of lipids in mortality, especially of ApoA1 and Lp(a).
ApoA1 is a prominent protein of high-density lipoprotein particles, and it facilitates the efflux of cholesterol from cells while also possessing strong anti-inflammatory and anti-atherogenic properties (25). Prior studies showed that elevated serum ApoA1 was a strong beneficial prognostic marker for CVD and mortality in different populations (26, 27). However, a recent observational study reported a U-shaped association between ApoA1 levels and CVD as well as all-cause mortality, indicating that both low and high ApoA1 may lead to an increased risk of death (28). Nonetheless, our study revealed a robust inverse causal association between ApoA1 levels and all-cause mortality, which persisted in various sensitivity analyses and was confirmed in the replication analysis. Both linear and nonlinear MR supported a negative correlation between ApoA1 and all-cause mortality. This inconsistency with the previous study can potentially be attributed to the nature of observational studies, as they are more susceptible to residual confounding or reverse causation than MR analyses (29). In addition, our UVMR results showed a total effect of ApoA1 on CVD mortality, whilst MVMR presented no evidence for a direct effect of ApoA1 after controlling for HDL-C, suggesting that the causal effect of ApoA1 on CVD mortality may be mediated by HDL-C. Finally, our study uncovered a novel causal relationship between ApoA1 and cancer mortality. This finding is in line with a previous retrospective study conducted on patients undergoing percutaneous coronary intervention, which showed that lower ApoA1 levels increased the risk of gastrointestinal cancer mortalities (30). One possible explanation for the protective role of high ApoA1 in cancer mortality is its beneficial properties, including antioxidative and anti-inflammatory capacities, that are linked to improved vascular function (31). Animal experimental study also addressed that dysregulation of ApoA1 is implicated in systemic alterations in lipid and cholesterol metabolism, potentially impacting immune cell homeostasis and inflammatory responses associated with malignancy (32). On the other hand, in-vitro studies also disclosed more direct tumor suppressive roles of ApoA1, such as altering sustained proliferative signal mechanisms, reducing resistance to apoptotic cell death, inhibiting angiogenesis, and modulating activation of invasion and metastasis behaviors (33).
In contrast to ApoA1, Lp(a) is a highly heritable biomarker that has been extensively studied in relation to CVDs due to its promotion of atherosclerosis and thrombosis (34). Numerous studies have established a causal link between Lp(a) and CVDs, but studies on the association between Lp(a) and mortality remain scarce and inconclusive. For example, an early meta-analysis of 36 prospective studies displayed that elevated Lp(a) was exclusively associated with vascular outcomes like stroke and coronary heart disease, but not with increased cancer mortality or other non-vascular death (35). A recent observational study in the general U.S. population demonstrated that higher Lp(a) levels were significantly associated with higher all-cause and CVD mortality (36). Our study corroborated and extended previous research by showing a linear and positive causal association between Lp(a) with the risk of all-cause and CVD mortality. Concordant with our results, a dose-response meta-analysis has further supported that higher Lp(a) levels are linearly associated with elevated risks of all-cause mortality and CVD-related death both in the general population and among patients with pre-existing CVDs (37). Two other genetic association studies by Arsenault et al. and Langsted et al. also reported similar findings with a strong association between high Lp(a) and increased mortality risk, lending support to our conclusions (38, 39). In addition to those addressed above, we discovered a causal relationship between Lp(a) and digestive disease mortality for the first time. Several lines of evidence from research on its role in liver fibrosis, non-alcoholic fatty liver disease, and inflammatory bowel disease support the biological plausibility of this finding (40, 41, 42). The exact mechanism behind these associations is unclear, but it has been proposed that Lp(a) mediates clinical events primarily through three mechanisms. First, Lp(a) contains a moiety similar to low-density lipoprotein, with one ApoB molecule that can accelerate atherosclerosis (43). Second, the high homology of its apolipoprotein(a) component to the fibrinolytic proenzyme plasminogen suggests a potential antifibrinolytic role for Lp(a), which can interfere with the binding of plasminogen to fibrin and ultimately contribute to the formation of atherogenic plaques (44). Third, Lp(a) is the major lipoprotein carrier of oxidized phospholipids, a highly reactive endogenous stimulant of pain receptors, which may trigger sterile inflammation and calcification and eventually lead to various diseases (45).
Several interesting observations emerged from the current study. Unlike conventional observational studies, our investigation did not reveal any significant linear or non-linear causal relationship between LDL-C and all-cause mortality. However, we observed an opposite pattern of association of LDL-C with CVD mortality and neurodegenerative disease mortality in the main analysis. Different patterns of association between LDL-C and cause-specific mortality may help explain the overall insignificant association between LDL-C and all-cause mortality. Given that high levels of LDL-C are a crucial factor in atherosclerosis, it is reasonable that LDL-C is positively associated with CVD mortality. Recent studies have also reported that there might be an inverse association between LDL-C and Parkinson’s disease (46, 47), which is similar to the association between LDL-C and neurodegenerative disease mortality observed in our study. Based on this evidence, higher LDL-C levels may potentially contribute to a reduction in neurodegenerative diseases by minimizing neuronal impairments, facilitating compensatory repair of injured neurons, and ultimately lowering the risk of neurodegenerative mortality (48, 49). Concurrently, ApoB exhibited a positive correlation with both CVD and neurodegenerative mortality. This implies that although ApoB and LDL-C share common roles in CVDs, the mass of cholesterol carried by LDL particles as measured by LDL-C rather than ApoB may have distinct roles in neurodegenerative disease mortality. Finally, although high TG levels have traditionally been associated with increased atherogenic risk, our analysis did not reveal any statistically significant association between TG levels and the risk of all-cause or cause-specific mortality. This observation aligns with a body of prior research, including randomized controlled trials, which also failed to demonstrate a substantial impact of TG reduction on cardiovascular events or mortality (50, 51, 52, 53). One plausible hypothesis posits that the metabolic rate of TG or its metabolites might genuinely influence the risk of mortality independently of the conventionally measured plasma TG levels (54). However, Thomsen et al., in a study involving 10,208 subjects in Denmark, obtained inconsistent results by employing four genetic variants within the LPL gene region as IVs for genetic prediction of TG levels. Their study suggested that lower non-fasting TG levels were associated with a reduced risk of all-cause mortality (55). These discrepancies might be attributed to variations in study populations, especially among individuals with multimorbidity or specific conditions such as diabetes (56). In this view, future investigations can conduct a more granular examination of the association between lipid levels and mortality risk in diverse populations with varying disease profiles.
Strengths and limitations
To our knowledge, this study is the first to systematically investigate the causal relationships between blood lipids and all-cause mortality, utilizing both linear and nonlinear MR methods. By employing MR analysis, we were able to minimize the potential influence of confounding factors and reverse causation, thereby increasing the robustness of our findings. The use of a large sample size and the construction of a GRS rather than a single SNP also enhanced our statistical power to detect the causation. Moreover, taking advantage of the individual data, we thoroughly examined the potential causal relationships between different lipid traits with cause-specific mortality, providing a comprehensive insight to reconcile discrepancies from previous studies. Finally, consistent findings from both primary and sensitivity analyses, along with similar findings from validation datasets, provide support for our conclusions.
Several limitations should also be considered in interpreting our findings. First, MR relies on several strict assumptions. Despite implementing various strategies to mitigate potential pleiotropy, such as excluding pleiotropic SNPs and using MVMR estimates and different MR methods, our findings may still be subject to bias due to residual and unmeasured confounding factors. Second, genetic variants reflect the effect of lifelong changes in blood lipids on mortality, while clinical interventions tend to have a greater impact on lipid level changes later in life. Although consistent results were obtained for ApoA1 after excluding participants using lipid-lowering agents, the true shape and strength of the association might be more intricately influenced by real-world environments than currently portrayed. Third, the UKB cohort may not be fully representative of the general UK population, with a low response rate of only 5% and a potential health-worker effect. Regarding this, earlier publications from the UKB have shown that studies using MR Methods are less affected by selection bias, indicating only a minor impact on our findings. Finally, due to constraints in accessing independent individual-level data, we were unable to conduct formal statistical replication of nonlinear MR analysis. However, we utilized a two-sample MR approach with all-cause mortality data from FinnGen to validate the robustness of our primary outcome.
Conclusions
Our results suggested that genetically predicted elevated levels of ApoA1 might confer a protective effect on the risk of all-cause and cancer mortality, whereas elevated levels of Lp(a) were associated with a higher risk of all-cause, CVD, and digestive disease mortality. It is noteworthy that LDL-C exhibited an opposite causal effect on CVD and neurodegenerative disease mortality, but no causal effect on all-cause mortality. These findings emphasize the potential for targeted interventions to modulate lipid levels and reduce mortality risk in a more personalized and tailored manner, with implications for public health policies and clinical practice.
Data availability
Data from the UK Biobank are available on application (https://www.ukbiobank.ac.uk). Summary-level data for all-cause mortality are publicly available to researchers from FinnGen consortium (https://www.finngen.fi/en/access_results). For statistical code relating to the individual level data analysis in UK Biobank, please contact the corresponding author at jianggzh5@mail.sysu.edu.cn.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
This research has been conducted using the UK Biobank Resource (application No. 78619). We thank the participants and researchers from the UK Biobank who contributed or collected data.
Author contributions
J. L., Z. W., and G. J. conceptualization; J. L., Z. W., J. Z, and G. J. data curation; J. L., Z. W., and C. Z. formal analyses; J. L., Z. W., and G. J. investigation; J. L., Z. W., J. Z., F. J., C. Z., and G. J. validation; J. L., Z. W., L. H., F. J., C. Z., and G. J. methodology; J. L., Z. W., L. H., F. J., C. Z., and G. J. project administration; J. L., Z. W., and G. J. Supervision; J. L., and J. Z. visualization; J. L. writing–original draft; J. L., Z. W., G. J., J. Z., F. J., C. Z., and L. H. writing–review and editing. G. J. funding
Funding and additional information
This work was supported by the National Key R&D Program of China [2022YFC2502402], Shenzhen Science and Technology Program [JCYJ20220530145210024], and the Guangdong Basic and Applied Basic Research Foundation [2023A1515012624].
Supplemental data
References
- 1.Klop B., Elte J.W., Cabezas M.C. Dyslipidemia in obesity: mechanisms and potential targets. Nutrients. 2013;5:1218–1240. doi: 10.3390/nu5041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P., Buring J.E., Badimon L., Hansson G.K., Deanfield J., Bittencourt M.S., et al. Atheroscler. Nat. Rev. Dis. Primers. 2019;5:56. doi: 10.1038/s41572-019-0106-z. [DOI] [PubMed] [Google Scholar]
- 3.Pirillo A., Casula M., Olmastroni E., Norata G.D., Catapano A.L. Global epidemiology of dyslipidaemias. Nat. Rev. Cardiol. 2021;18:689–700. doi: 10.1038/s41569-021-00541-4. [DOI] [PubMed] [Google Scholar]
- 4.Dong J., Yang S., Zhuang Q., Sun J., Wei P., Zhao X., et al. The associations of lipid profiles with cardiovascular diseases and death in a 10-year prospective cohort study. Front. Cardiovasc. Med. 2021;8 doi: 10.3389/fcvm.2021.745539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin G., Chen W., Dai C., Xu K. The significance of apolipoprotein-A in the long-term death of patients with STEMI. J. Healthc. Eng. 2022;2022 doi: 10.1155/2022/5941117. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Obisesan O.H., Kou M., Wang F.M., Boakye E., Honda Y., Uddin S.M.I., et al. Lipoprotein(a) and subclinical vascular and valvular calcification on cardiac computed tomography: the atherosclerosis risk in communities study. J. Am. Heart Assoc. 2022;11 doi: 10.1161/JAHA.121.024870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaysen G.A., Ye X., Raimann J.G., Wang Y., Topping A., Usvyat L.A., et al. Lipid levels are inversely associated with infectious and all-cause mortality: international MONDO study results. J. Lipid Res. 2018;59:1519–1528. doi: 10.1194/jlr.P084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang J.J., Katsanos A.H., Khorchid Y., Dillard K., Kerro A., Burgess L.G., et al. Higher low-density lipoprotein cholesterol levels are associated with decreased mortality in patients with intracerebral hemorrhage. Atherosclerosis. 2018;269:14–20. doi: 10.1016/j.atherosclerosis.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Psaty B.M., Anderson M., Kronmal R.A., Tracy R.P., Orchard T., Fried L.P., et al. The association between lipid levels and the risks of incident myocardial infarction, stroke, and total mortality: the Cardiovascular Health Study. J. Am. Geriatr. Soc. 2004;52:1639–1647. doi: 10.1111/j.1532-5415.2004.52455.x. [DOI] [PubMed] [Google Scholar]
- 10.Fried L.P., Kronmal R.A., Newman A.B., Bild D.E., Mittelmark M.B., Polak J.F., et al. Risk factors for 5-year mortality in older adults: the Cardiovascular Health Study. JAMA. 1998;279:585–592. doi: 10.1001/jama.279.8.585. [DOI] [PubMed] [Google Scholar]
- 11.Johannesen C.D.L., Langsted A., Mortensen M.B., Nordestgaard B.G. Association between low density lipoprotein and all cause and cause specific mortality in Denmark: prospective cohort study. BMJ. 2020;371 doi: 10.1136/bmj.m4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Yang X., Huang W., Zou C., Lu J., Zhang J., et al. Investigating linear and nonlinear associations of LDL cholesterol with chronic kidney disease, atherosclerotic cardiovascular disease and all-cause mortality: a prospective and Mendelian Randomization Study. Atherosclerosis. 2023;87 doi: 10.1016/j.atherosclerosis.2023.117394. [DOI] [PubMed] [Google Scholar]
- 13.Katzke V.A., Sookthai D., Johnson T., Kühn T., Kaaks R. Blood lipids and lipoproteins in relation to incidence and mortality risks for CVD and cancer in the prospective EPIC-Heidelberg cohort. BMC Med. 2017;15:218. doi: 10.1186/s12916-017-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai S.Z., Zhang J.H., Zhao G.A., Wang N.J., Li G.H., So H.C., et al. Causal associations of short and long sleep durations with 12 cardiovascular diseases: linear and nonlinear Mendelian randomization analyses in UK Biobank. Eur. Heart J. 2021;42:3349–3357. doi: 10.1093/eurheartj/ehab170. [DOI] [PubMed] [Google Scholar]
- 16.Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson T.G., Sanderson E., Palmer T.M., Ala-Korpela M., Ference B.A., Smith G.D., et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke R., Peden J.F., Hopewell J.C., Kyriakou T., Goel A., Heath S.C., et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. New Engl. J. Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 19.Burgess S., Thompson S.G., Collaboration C.C.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011;40:755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 20.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1546–1718. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurki M.I., Karjalainen J., Palta P., Sipilä T.P., Kristiansson K., Donner K.M., et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staley J.R., Burgess S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet. Epidemiol. 2017;41:341–352. doi: 10.1002/gepi.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham S.E., Clarke S.L., Wu K.-H.H., Kanoni S., Zajac G.J.M., Ramdas S., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley D., Blaszczak A., Yin Z., Liu J., Joseph J.J., Wright V., et al. Clusterin impairs hepatic insulin sensitivity and adipocyte clusterin associates with cardiometabolic risk. Diabetes Care. 2019;42:466–475. doi: 10.2337/dc18-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walldius G., Jungner I., Holme I., Aastveit A.H., Kolar W., Steiner E. High apolipoprotein B, low apolipoprotein A-I, and improvement in the prediction of fatal myocardial infarction (AMORIS study): a prospective study. Lancet. 2001;358:2026–2033. doi: 10.1016/S0140-6736(01)07098-2. [DOI] [PubMed] [Google Scholar]
- 27.Florvall G., Basu S., Larsson A. Apolipoprotein A1 is a stronger prognostic marker than are HDL and LDL cholesterol for cardiovascular disease and mortality in elderly men. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:1262–1266. doi: 10.1093/gerona/61.12.1262. [DOI] [PubMed] [Google Scholar]
- 28.Faaborg-Andersen C.C., Liu C., Subramaniyam V., Desai S.R., Sun Y.V., Wilson P.W.F., et al. U-shaped relationship between apolipoprotein A1 levels and mortality risk in men and women. Eur. J. Prev. Cardiol. 2022;30:293–304. doi: 10.1093/eurjpc/zwac263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishiyama H., Funamizu T., Iwata H., Endo H., Chikata Y., Doi S., et al. Low apolipoprotein A1 was associated with increased risk of cancer mortality in patients following percutaneous coronary intervention: a 10-year follow-up study. Int. J. Cancer. 2022;151:1482–1490. doi: 10.1002/ijc.34164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohatgi A., Westerterp M., von Eckardstein A., Remaley A., Rye K.-A. HDL in the 21st century: a multifunctional roadmap for future HDL research. Circulation. 2021;143:2293–2309. doi: 10.1161/CIRCULATIONAHA.120.044221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tall A.R., Yvan-Charvet L. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 2015;15:104–116. doi: 10.1038/nri3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgila K., Vyrla D., Drakos E. Apolipoprotein A-I (ApoA-I), immunity, inflammation and cancer. Cancers (Basel) 2019;11:1097. doi: 10.3390/cancers11081097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mach F., Baigent C., Catapano A.L., Koskinas K.C., Casula M., Badimon L., et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk: the Task Force for the Management of Dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS) Eur. Heart J. 2019;41:111–188. doi: 10.1093/eurheartj/ehz455. [DOI] [PubMed] [Google Scholar]
- 35.Erqou S., Kaptoge S., Perry P.L., Di Angelantonio E., Thompson A., White I.R., et al. Lipoprotein(a) concentration and the risk of coronary heart disease, stroke, and nonvascular mortality. JAMA. 2009;302:412–423. doi: 10.1001/jama.2009.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z.-W., Li M., Li J.-J., Liu N.-F. Association of lipoprotein(a) with all-cause and cause-specific mortality: a prospective cohort study. Eur. J. Intern. Med. 2022;106:63–70. doi: 10.1016/j.ejim.2022.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Amiri M., Raeisi-Dehkordi H., Verkaar A.J.C.F., Wu Y., van Westing A.C., Berk K.A., et al. Circulating lipoprotein (a) and all-cause and cause-specific mortality: a systematic review and dose-response meta-analysis. Eur. J. Epidemiol. 2023;38:485–499. doi: 10.1007/s10654-022-00956-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langsted A., Kamstrup P.R., Nordestgaard B.G. High lipoprotein(a) and high risk of mortality. Eur. Heart J. 2019;40:2760–2770. doi: 10.1093/eurheartj/ehy902. [DOI] [PubMed] [Google Scholar]
- 39.Arsenault B.J., Pelletier W., Kaiser Y., Perrot N., Couture C., Khaw K.-T., et al. Association of long-term exposure to elevated lipoprotein(a) levels with parental Life span, chronic disease-free survival, and mortality risk: a mendelian randomization analysis. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meroni M., Longo M., Lombardi R., Paolini E., Macchi C., Corsini A., et al. Low lipoprotein(a) levels predict hepatic fibrosis in patients with nonalcoholic fatty liver disease. Hepatol. Commun. 2022;6:535–549. doi: 10.1002/hep4.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam J.S., Jo S., Kang S., Ahn C.W., Kim K.R., Park J.S. Association between lipoprotein(a) and nonalcoholic fatty liver disease among Korean adults. Clin. Chim. Acta. 2016;461:14–18. doi: 10.1016/j.cca.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Katsiki N., Al-Rasadi K., Mikhailidis D.P. Lipoprotein (a) and cardiovascular risk: the show must go on. Curr. Med. Chem. 2017;24:989–1006. doi: 10.2174/0929867324666170112111948. [DOI] [PubMed] [Google Scholar]
- 43.Ward N.C., Kostner K.M., Sullivan D.R., Nestel P., Watts G.F. Molecular, population, and clinical aspects of lipoprotein(a): a bridge too far? J. Clin. Med. 2019;8:2073. doi: 10.3390/jcm8122073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boffa M.B. Beyond fibrinolysis: the confounding role of Lp(a) in thrombosis. Atherosclerosis. 2022;349:72–81. doi: 10.1016/j.atherosclerosis.2022.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Leibundgut G., Scipione C., Yin H., Schneider M., Boffa M.B., Green S., et al. Determinants of binding of oxidized phospholipids on apolipoprotein (a) and lipoprotein (a) J. Lipid Res. 2013;54:2815–2830. doi: 10.1194/jlr.M040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang F., Zhan Y., Hammar N., Shen X., Wirdefeldt K., Walldius G., et al. Lipids, apolipoproteins, and the risk of Parkinson disease. Circ. Res. 2019;125:643–652. doi: 10.1161/CIRCRESAHA.119.314929. [DOI] [PubMed] [Google Scholar]
- 47.Jin U., Park S.J., Park S.M. Cholesterol metabolism in the brain and its association with Parkinson's disease. Exp. Neurobiol. 2019;28:554–567. doi: 10.5607/en.2019.28.5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv Y.B., Yin Z.X., Chei C.L., Brasher M.S., Zhang J., Kraus V.B., et al. Serum cholesterol levels within the high normal range are associated with better cognitive performance among Chinese elderly. J. Nutr. Health Aging. 2016;20:280–287. doi: 10.1007/s12603-016-0701-6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou F., Deng W., Ding D., Zhao Q., Liang X., Wang F., et al. High low-density lipoprotein cholesterol inversely relates to dementia in community-dwelling older adults: the Shanghai aging study. Front. Neurol. 2018;9:952. doi: 10.3389/fneur.2018.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rauch B., Schiele R., Schneider S., Diller F., Victor N., Gohlke H., et al. OMEGA, a randomized, placebo-controlled trial to test the effect of highly purified omega-3 fatty acids on top of modern guideline-adjusted therapy after myocardial infarction. Circulation. 2010;122:2152–2159. doi: 10.1161/CIRCULATIONAHA.110.948562. [DOI] [PubMed] [Google Scholar]
- 51.Kromhout D., Giltay E.J., Geleijnse J.M. n-3 fatty acids and cardiovascular events after myocardial infarction. New Engl. J. Med. 2010;363:2015–2026. doi: 10.1056/NEJMoa1003603. [DOI] [PubMed] [Google Scholar]
- 52.Galan P., Kesse-Guyot E., Czernichow S., Briancon S., Blacher J., Hercberg S. Effects of B vitamins and omega 3 fatty acids on cardiovascular diseases: a randomised placebo controlled trial. BMJ. 2010;341 doi: 10.1136/bmj.c6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicholls S.J., Lincoff A.M., Garcia M., Bash D., Ballantyne C.M., Barter P.J., et al. Effect of high-dose omega-3 fatty acids vs corn oil on major adverse cardiovascular events in patients at high cardiovascular risk: the STRENGTH randomized clinical trial. JAMA. 2020;324:2268–2280. doi: 10.1001/jama.2020.22258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das Pradhan A., Glynn R.J., Fruchart J.-C., MacFadyen J.G., Zaharris E.S., Everett B.M., et al. Triglyceride lowering with pemafibrate to reduce cardiovascular risk. New Engl. J. Med. 2022;387:1923–1934. doi: 10.1056/NEJMoa2210645. [DOI] [PubMed] [Google Scholar]
- 55.Thomsen M., Varbo A., Tybjærg-Hansen A., Nordestgaard B.G. Low nonfasting triglycerides and reduced all-cause mortality: a mendelian randomization study. Clin. Chem. 2014;60:737–746. doi: 10.1373/clinchem.2013.219881. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y., Fang Y., Magliano D.J., Charchar F.J., Sobey C.G., Drummond G.R., et al. Fasting triglycerides are positively associated with cardiovascular mortality risk in people with diabetes. Cardiovasc. Res. 2023;119:826–834. doi: 10.1093/cvr/cvac124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the UK Biobank are available on application (https://www.ukbiobank.ac.uk). Summary-level data for all-cause mortality are publicly available to researchers from FinnGen consortium (https://www.finngen.fi/en/access_results). For statistical code relating to the individual level data analysis in UK Biobank, please contact the corresponding author at jianggzh5@mail.sysu.edu.cn.