Abstract

Development of new targeted inhibitors for oncogenic KRAS mutants may benefit from insight into how a given mutation influences the accessibility of protein residues and how compounds interact with mutant or wild-type KRAS proteins. Targeted proteomic analysis, a key validation step in the KRAS inhibitor development process, typically involves both intact mass- and peptide-based methods to confirm compound localization or quantify binding. However, these methods may not always provide a clear picture of the compound binding affinity for KRAS, how specific the compound is to the target KRAS residue, and how experimental conditions may impact these factors. To address this, we have developed a novel top-down proteomic assay to evaluate invitro KRAS4B-compound engagement while assessing relative quantitation in parallel. We present two applications to demonstrate the capabilities of our assay: maleimide-biotin labeling of a KRAS4BG12D cysteine mutant panel and treatment of three KRAS4B proteins (WT, G12C, and G13C) with small molecule compounds. Our results show the time- or concentration-dependence of KRAS4B-compound engagement in context of the intact protein molecule while directly mapping the compound binding site.
The RAS family of GTPases (HRAS, KRAS, and NRAS) behave as molecular switches through their association with GDP (inactive) or GTP (active) to activate multiple signaling cascades (e.g., MAPK and PI3K pathways) driving cell growth and differentiation, cell cycle regulation, and proliferation.1−4 When the RAS genes are mutated, primarily at codons 12, 13, and 61, GTPase-activating proteins (GAPs)5 are unable to hydrolyze the nucleotide, thereby preventing the deactivation of RAS and ultimately leading to oncogenesis.6,7
Prior RAS drug discovery efforts have led to the development of covalent inhibitors specific to KRASG12C, commonly found in non-small cell lung tumors.7,8 Ostrem et al. first demonstrated allosteric inhibition and illuminated the switch II pocket using a disulfide-fragment tethering screen, producing compounds specifically targeting the GDP-bound state of KRASG12C.9 This vital discovery was followed by the discovery of two FDA approved KRASG12C inhibitors: AMG510 (Sotorasib)10−12 and MRTX849 (Adagrasib).13−15 More recently, new inhibitor design efforts have focused on compounds specifically targeting KRAS4B G12D,16,17 G12S,18 and G12R19 mutants, along with a novel class of pan-(KRAS) mutant inhibitors20 and a bifunctional inhibitor that alters KRAS-membrane interactions.21,22 However, many oncogenic mutations and other RAS isoforms remain to be effectively targeted for therapeutic use, presenting a formidable challenge to the field.
Targeted proteomic analyses comprise a valuable component of the inhibitor development process, enabling quantification of compound engagement, determination of binding kinetics, and evaluation of compound specificity.11,14,23−26 These experiments typically employ “bottom-up” proteomics,14,26 which involves the proteolytic digestion of a sample into peptides,27 and/or intact mass analysis,9,26,28 which identifies the molecular weight of proteins in a sample.29 While peptide-based strategies maximize analytical sensitivity, digestion eliminates the connectivity of any protein modifications and can potentially introduce chemical artifacts or compound side reactions.30−32 Thus, it can be challenging to accurately quantify a specific modification or determine the total number of modifications to a single protein molecule. Conversely, intact mass analysis maintains the integrity of the protein sequence while identifying all intact and modified protein forms, also known as proteoforms, within the sample.33,34 However, intact mass alone cannot provide precise compound localization or the order in which multiple compound engagement events may have occurred.
Therefore, an alternative proteomic strategy is needed to fully understand the modification profile of compound-treated proteins. Top-down proteomic analysis provides the sequence specificity of bottom-up analysis while extending the information gathered from intact mass analysis (MS1) by subjecting each identified proteoform to subsequent targeted fragmentation (MS2).29 This enables the direct characterization of each proteoform to determine the protein sequence while localizing any post-translational modifications.33,35 Employing the top-down approach can thereby greatly enhance the information obtained for a target protein following compound treatment.
Here, we present a novel top-down proteomic assay to evaluate KRAS4B compound engagement invitro. By investigating labeled or compound-treated recombinant KRAS4B proteins (rKRAS4B) without disrupting the protein primary structure, we can directly visualize and accurately identify each proteoform within a sample. Our method enables the determination of binding stoichiometry, identification of off-target compound engagement, and elucidation of unanticipated compound reaction products. We validated this method by two proof-of-concept experiments, maleimide-biotin (MBtn) labeling of a rKRAS4BG12D cysteine mutant panel and treatment of three rKRAS4B proteins (WT, G12C, and G13C) with selected compounds from the UCSF Small Molecule Discovery Center (SMDC) library and FDA-approved inhibitors. We further generated fractional relative quantitation values to fully characterize compound engagement. Our results show the time- or concentration-dependence of KRAS4B-compound engagement in context of the intact protein molecule while directly mapping the compound binding site.
Experimental Section
Detailed experimental methods, instrument parameters, and data analysis parameters are provided in the Supporting Information.
Production of Recombinant KRAS4B Proteins
DNA for all rKRAS4B (1-169) proteins used were created and validated by ATUM (Newark, CA) as Gateway Entry clones optimized forEscherichia coliexpression. Final expression clones were created by Gateway recombination using methods described by Esposito et al.36 and expressed using protocols outlined by Taylor et al.37
Maleimide-Biotin Labeling of Engineered Cysteines in KRAS4BG12D
Purified proteins were buffer exchanged with 20 mM HEPES/150 mM NaCl (pH 7.3) and labeled with a 50-fold molar excess of EZ-Link Maleimide-PEG2-Biotin (MBtn, Thermo Fisher Scientific) in 1X phosphate-buffered saline (PBS, pH 7.4). The labeling reaction occurred at room temperature across five aliquoted protein tubes, while 1X PBS was added to a sixth tube as a control. The reaction was quenched with 500 mM tris(2-carboxyethyl)phosphine (TCEP) at time points 0 (control), 0.5, 1, 2, 4, and 24 h.
Sample Preparation for Bottom-Up Mass Spectrometry
Labeled rKRAS4B proteins were subjected to in-solution tryptic digestion at pH 7 to minimize hydrolysis of the maleimide ring.32
Briefly, proteins were precipitated with ice-cold acetone, resuspended in 8 M urea in 25 mM Tris (pH 7), reduced with dithiothreitol (DTT), alkylated with iodoacetamide, quenched with DTT, diluted to 1 M urea, digested overnight with trypsin (Promega), quenched with formic acid (FA), desalted via a C18 spin column (Thermo Fisher Scientific), and dried via SpeedVac. Dried peptides were resuspended in Buffer A (0.2% MS-grade FA in 5% Optima-grade acetonitrile [ACN, Thermo Fisher Scientific]) for analysis by liquid chromatography–tandem mass spectrometry (LC–MS/MS).
Bottom-Up LC–MS/MS and Data Analysis
Peptides were separated by reverse-phase chromatography on an UltiMate 3000 system (Thermo Fisher Scientific) using Acclaim PepMap 100 C18 HPLC trap and analytical columns (Thermo Fisher Scientific) maintained at 60 °C. Peptides were loaded onto the trap column for 10 min followed by gradient separation of 5 to 50% Buffer B (0.2% MS-grade FA in 95% Optima-grade ACN) over 90 min. Peptides were ionized by a Nanospray Flex Ion Source (Thermo Fisher Scientific) with a stainless-steel emitter coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Each digested sample was injected two times for LC–MS/MS analysis by either higher-energy collisional dissociation (HCD) or electron transfer dissociation with supplemental HCD (EThcD) using data-dependent acquisition. The resulting files were searched against their respective custom sequence-specific database and a common contaminants database in Proteome Discoverer (Thermo Fisher Scientific) using SEQUEST and MSAmanda nodes.
Sample Preparation for Top-Down Mass Spectrometry
rKRAS4B proteins were first concentrated and desalted via solid phase extraction using C4 ZipTips (Millipore), followed by dilution in Buffer A for label or compound localization by top-down LC–MS/MS. An aliquot from each concentrated and desalted sample was diluted 1:1 with Buffer A for relative abundance quantitation by LC–MS.
Top-Down LC–MS/MS and Data Analysis
Proteins were separated by reversed-phase chromatography on an UltiMate 3000 system using self-packed PLRP-s (Agilent) trap and analytical columns maintained at 45 °C. Proteins were loaded onto the trap column for 10 min and then separated by first ramping to 25% Buffer B in 1 min followed by gradient separation of 25–45% B over 39 min. Proteins were ionized using fused-silica emitters (New Objective) packed in house with 2–3 mm of PLRP-s resin and a self-built University of Washington’s Proteomics Resource nanospray source coupled to an Orbitrap Fusion Lumos mass spectrometer.
Identified precursor m/z values corresponding to labeled rKRAS4B proteoforms were used to generate label-specific methods for targeted MS2 fragmentation by either HCD or electron transfer dissociation (ETD). Manual analysis of targeted fragmentation scans was performed using Xtract deconvolution in FreeStyle (Thermo Fisher Scientific). The list of identified fragment ions was then imported into ProSight Lite for comparison against the appropriate rKRAS4B proteoform sequence.38 The monoisotopic elemental composition mass of each compound was used for modification localization.
Compound Library Screening for KRAS4BG13C Specificity
The UCSF SMDC screened their library of ∼1600 disulfide tethering compounds for reactivity against rKRAS4BG13C in the active state, as described in Hallenbeck et al.39 Compounds displaying greater than 3 σ from the average labeling were further evaluated at 8 compound concentrations via 3-fold serial dilutions of the compound against rKRAS4B or rKRAS4BG13C.
Compound Concentration Course
Three selected UCSF SMDC hit compounds were evaluated for their compound localization, specificity to G13C in the active state, and relative quantitation. First, rKRAS4BC118S, rKRAS4BCG12C/C118S, or rKRAS4BG13C/C118S were nucleotide exchanged from the native GDP to either GppNHp (nonhydrolyzable GTP analog) or GDPβS (nonhydrolyzable GDP analog), as described in Agamasu et al.40 Compound labeling was carried out by incubating rKRAS4B loaded with GppNHp or GDPβS at 4 μM with 55.6 μM, 166.7 μM, or 500.0 μM compounds in DMSO for 1 h at room temperature in 100 mM ammonium acetate (pH 7.5) and 500 μM β-mercaptoethanol. A control was also prepared by adding DMSO in the place of the compound. The reaction was quenched by adding Buffer A. Labeling rKRAS4B with AMG510 and MRTX849 was performed as above except the reaction was carried out with 5 μM rKRAS4B in a buffer of 20 mM HEPES/150 mM NaCl/2 mM MgCl2 (pH 7.3).
Relative Quantitation LC–MS and Data Analysis
Proteins were resolved by reverse phase separation with a Vanquish Flex chromatographic system (Thermo Fisher Scientific) using a MabPac RP analytical column (Thermo Fisher Scientific) maintained at 50 °C over an 8 min gradient of Buffer B (47.5% Optima ACN, 47.5% Optima isopropanol [IPA, Thermo Fisher Scientific], 5% Optima H2O, and 0.2% FA) from 2 to 100%. Proteins were ionized using a heated electrospray ionization (HESI) source connected to an Exactive Plus EMR mass spectrometer (Thermo Fisher Scientific) collecting only intact protein (MS1) spectra. Quintuplicate injections were performed on each sample.
Intact mass intensities of all proteoforms were generated using multiple software packages to demonstrate the comparability between methods including manual analysis of an averaged scan using Xtract, sliding window Xtract using BioPharma Finder (Thermo Fisher Scientific), and deconvolution of an averaged scan using UniDec.41 The percent compound engagement was calculated for each injection using the following equation
Averages and standard deviations of the resulting percentages were generated in Microsoft Excel.
Results and Discussion
Evaluation of KRAS4B Engineered Cysteine Reactivity
A rKRAS4B library was developed at the NCI RAS Initiative for new lead compound evaluation by disulfide tethering screen in the context of the G12D mutation.42 Each rKRAS4B protein contained G12D and C118S mutations, along with a single point mutation to cysteine based on predicted solvent exposure from available KRAS4B crystal structures.43−45 The reactivity of “WT” (rKRAS4BG12D/C118S) and three cysteine substitutions (rKRAS4BD33C, rKRAS4BK42C, and rKRAS4BD47C) were evaluated using MBtn labeling over 6 time points ranging from 0 to 24 h. Traditional peptide-based proteomic methods were then employed to determine the location and relative abundance of the resulting MBtn labels.
We hypothesized that the engineered cysteine residues would be preferentially labeled, while the native cysteine residues (C51 and C80) would be unavailable for labeling. Instead, all cysteine containing peptides across all time points were observed to be labeled by MBtn (Table S1; Figures S1–S5), with labeled D33C and C80 observed at high abundance. We also observed the doubly labeled peptides of K42C or D47C and C51, suggesting that all cysteines could be labeled on a single rKRAS4B molecule. However, trypsin digestion prevented us from retaining connectivity between all cysteine residues or accurately determining the labeling status of each rKRAS4B protein.
Due to these limitations, we developed a top-down proteomic assay to evaluate the relative abundance of labeled rKRAS4B, localize MBtn within each proteoform, and enable the detection of multiply labeled rKRAS4B proteoforms (Figure S6). rKRAS4B proteoforms collected at each labeling time point were concentrated, desalted, and subsequently analyzed by top-down LC–MS. Figure S7 shows the 915–1015 m/z region of the intact protein (MS1) spectra of all detected proteoforms, while Figure S8 shows an example of the full scan MS1 spectrum at 0 and 24 h for rKRAS4BD47C. Differences in the relative abundance of each proteoform can be observed across all time points. While the control rKRAS4B is minimally labeled with a single MBtn after 24 h (Figure S7a), rKRAS4BD33C and rKRAS4BD47C are rapidly labeled with a single MBtn after 0.5 h (Figures S7b,d), and rKRAS4BK42C is gradually labeled with a single MBtn by 24 h (Figure S7c). Likewise, a second MBtn label is visible on rKRAS4BD47C at 0.5 h (Figure S7d), on rKRAS4BK42C at 4 h (Figure S7c), and on rKRAS4BD33C at 24 h (Figure S7b). A third MBtn peak is also visible on rKRAS4BD47C at 4 h (Figure S7d). Visualizing these rKRAS4B proteoforms by intact mass provided clarity to the rate of MBtn labeling and confirmed off-target binding, thus demonstrating the reactivity of available rKRAS4B cysteine residues.
Each observed proteoform was then targeted for top-down MS2 fragmentation to both localize each MBtn label to a specific rKRAS4B cysteine residue and determine the order of cysteine labeling for each rKRAS4B protein. Figure S9 shows the MS2 fragmentation coverage maps of each proteoform at 24 h. The first cysteine labeled in the control was C51 (Figure S9a), while the engineered cysteine was preferentially labeled in the mutated proteins (Figure S9b–d). The second label in the mutated proteins was C51, matching the availability of this residue on the control sample at extended time points (Figure S9b–d). For rKRAS4BD47C specifically, it was confirmed that all three cysteine residues were labeled with MBtn (Figure S9d).
The results obtained by top-down proteomic analysis contrasted markedly from the initial peptide-based analysis, which identified labeling of all rKRAS4B cysteines. Eliminating the trypsin digestion step therefore provided greater context for the reactivity of native and engineered rKRAS4B cysteine residues. While other intact mass analysis methods can identify the number of labels on each protein molecule, our ability to directly isolate and efficiently fragment labeled rKRAS4B proteoforms is unparalleled, enabling our top-down assay to localize each label to a specific cysteine, determine the binding order, and confirm our initial hypothesis: engineered cysteine residues predicted to be solvent exposed (D33C, K42C, and D47C) were observed to be preferentially labeled by MBtn over native cysteine residues (C51, C80). We further hypothesize that the addition of MBtn to D47C destabilized the rKRAS4B structure, exposing C51 and C80 to labeling.
Determining Small Molecule Reactivity to KRAS4BG13C
To demonstrate the capabilities of our top-down assay in the context of drug discovery workflows, we selected three compounds (Table S2) from a UCSF SMDC library covalently targeting GppNHp-bound KRAS4BG13C based on their initial dose–response curves, all of which suggested high specificity to active KRAS4BG13C (Figure S10).46 rKRAS4BC118S and rKRAS4BG12C/C118S nucleotide exchanged with GppNHp were included as controls in order to evaluate whether these compounds exhibited nonspecific binding. All three proteins were treated with each compound at a final concentration of 0, 55.5, 166.7, or 500 μM for 1 h.
Upon initial visualization of all proteoforms from each sample by top-down LC–MS, each compound was observed to exhibit varying degrees of specificity. Figure 1 displays a zoomed-in view of the MS1 spectra (21+ charge states) from the 500 μM samples, while Figures S11–S13 plot the extracted ion chromatogram (XIC) for all identified proteoforms at each concentration. 917101 and 917178 labeled all cysteines present across all proteins, as evident by the double compound addition on rKRAS4BC118S and the triple compound addition on rKRAS4BG12C/C118S and rKRAS4BG13C/C118S (Figure 1a,c), suggesting substantial off-target binding. Conversely, 917969 targeted only a single cysteine residue (Figure 1b), suggesting specific engagement. Additionally, the combined relative abundance of all compound-labeled proteoforms was the highest in the rKRAS4BG13C/C118S samples in agreement with the initial high throughput screens (Figure S10).
Figure 1.

Top-down characterization of compound engagement to the active state of KRAS4B. MS1 region containing the 21+ charge state following incubation with (a) 917101, (b) 917969, and (c) 917178 at 500 μM for 1 h. Compound engagement for rKRAS4BC118S (top); rKRAS4BG12C/C118S (middle); and rKRAS4BG13C/C118S (bottom). Peaks corresponding to each compound addition are labeled (cyan: 1 compound addition; orange: 2 compound additions; and green: 3 compound additions).
For all samples, each observed proteoform was then targeted for top-down MS2 fragmentation to localize each compound addition to a specific cysteine residue. Figures S14–S16 show the MS2 fragmentation coverage maps obtained from the 500 μM samples. For 917101, the two sequentially eluting single compound additions observed for rKRAS4BC118S (Figure S11a) were determined to be compound-labeled C80 and C51, respectively, and the double compound addition was determined to be both C51 and C80 labeled (Figure S14a). The single compound additions observed for both rKRAS4BG12C/C118S (Figure S11b) and rKRAS4BG13C/C118S (Figure S11c) were determined to be compound-labeled C12 (Figure S14b) and C13 (Figure S14c), respectively, while the sequentially eluting double compound additions observed for both samples (Figures S11b,c) were determined to be compound-labeled C12/C13 and C80, followed by C12/C13 and C51 (Figures S14b,c), and the triple compound additions were determined to be compound-labeled C12/13, C51, and C80 (Figures S14b,c). For 917969, compound binding was not detected for rKRAS4BC118S, and only a single compound addition was observed for both rKRAS4BG12C/C118S and rKRAS4BG13C/C118S (Figure S12). Targeted MS2 fragmentation confirmed that each single compound addition was to C12 or C13 (Figures S15b,c).
For 917178, the two sequentially eluting single compound additions observed for rKRAS4BC118S (Figure S13a) were determined to be compound-labeled C80 or C51, while the double compound addition was determined to be both C51 and C80 (Figure S16a). Interestingly, for the two sequentially eluting single compound additions observed for rKRAS4BG12C/C118S and rKRAS4BG13C/C118S (Figures S13b,c), 917178 was observed to label C12, followed by C51, on the former (Figure S16b), and C51, followed by C13, on the latter (Figure S16c), although the C13 labeled proteoform was the most abundant (Figure S13c). Differential patterns of double compound additions were also observed, with rKRAS4BG12C/C118S determined to be labeled at C12 and C80 or C12 and C51 (Figure S16b), and rKRAS4BG13C/C118S determined to be labeled at C13 and C80, C51 and C80, or C13 and C51 (Figure S16c). Both observed triple compound additions were confirmed to be C12/13, C51, and C80 labeled (Figures S16b,c).
While these initial results suggested that the selected compounds preferentially targeted C13, we also evaluated how the degree of compound engagement changed when rKRAS4B was in the inactive state by repeating our top-down assay after exchanging the native nucleotide with the nonhydrolyzable GDP analog GDPβS. Figure 2 shows the zoomed-in view of the 21+ charge states from the 500 μM samples, while Figures S17–S19 plot the XIC for all identified proteoforms at each concentration. We then employed targeted top-down MS2 fragmentation to localize each compound addition to a specific cysteine residue. Figures S20–S22 contain the MS2 fragment maps obtained from the 500 μM samples.
Figure 2.

Top-down characterization of compound engagement to the inactive state of KRAS4B. MS1 region containing the 21+ charge state following incubation with (a) 917101, (b) 917969, and (c) 917178 at 500 μM for 1 h. Compound engagement for rKRAS4BC118S (top); rKRAS4BG12C/C118S (middle); and rKRAS4BG13C/C118S (bottom). Peaks corresponding to each compound addition are labeled (cyan: 1 compound addition; orange: 2 compound additions; and green: 3 compound additions).
Each compound displayed analogous characteristics when targeting the inactive form of rKRAS4B. For example, 917969 maintained specificity to the mutated cysteine residues of C12 and C13 (Figure S21), although the single compound additions exhibited higher relative abundances (Figure S18). 917101 and 917178 again exhibited off-target binding to the native cysteine residues across all proteins, albeit at a lower relative abundance in comparison to that of the active state (Figures S17, S19, S20, and S22). For the single compound addition to rKRAS4BC118S, the compound labeled C80 proteoform eluted first, while the compound labeled C51 proteoform eluted second at a higher abundance (Figures S17a, S19a, S20a, and S22a). For the double compound additions to rKRAS4BG12C/C118S and rKRAS4BG13C/C118S, C12/C13 and C80 labeled eluted first, while C12/C13 and C51 labeled eluted second (Figures S17b,c, S19b,c, S20b,c, and S22b,c).
The combination of intact mass analysis and targeted MS2 fragmentation provided by our top-down assay generated a level of molecular detail inaccessible to alternative proteomic methods. Our results provided precise localization of each compound addition while maintaining the integrity of the protein sequence for each proteoform analyzed. Our inclusion of rKRAS4B native cysteine residues C51 and C80 allowed a more comprehensive evaluation of compound binding sites while simultaneously revealing the order and extent of nonspecific binding. Additionally, assessing compound engagement with both nucleotide states of rKRAS4B provided a holistic understanding of how these small molecules interact with the full nucleotide cycle of rKRAS4B.
Relative Quantitation of Compound Engagement
To complement our top-down assay, we also created an LC–MS relative quantitation workflow to evaluate the concentration dependence of KRAS4B C13-targeting compound engagement. For this higher-throughput experiment, we performed the LC–MS analysis using a larger column and a faster gradient (Figure S23) to rapidly obtain quintuplicate injections while maintaining high chromatographic reproducibility (Figures S24–S41). Consequently, this resulted in the coelution of all proteoforms of the same mass. While we could no longer distinguish between proteoforms (e.g., rKRAS4BC118S with C51 or C80 labeled by compound) by MS1 alone, this further simplified our analysis of off-target binding by solely accounting for a single chromatographic peak.
Relative fractional abundances of each compound addition were calculated using MS1 intensity values. Two methods were first evaluated using the rKRAS4BG13C/C118S + 917101 data set for comparison against manual analysis using the Xtract algorithm: BioPharma Finder, where deconvolution is performed via sliding window averaged scan across the chromatogram, and the freely available UniDec, which performs deconvolution of a user-created averaged scan.41Figure S26b provides the fractional abundance of each compound addition calculated using each data analysis method, demonstrating that all methods provided similar results. BioPharma Finder was used to automate the remainder of the data analysis presented here, but alternate software could also be used.
Figure 3 depicts the fractional abundance of all rKRAS4B compound addition proteoforms, while Tables S3–S20 provide the values used to generate this figure. When rKRAS4B was in the active state (Figure 3a,c,e), the single compound addition to rKRAS4BG13C/C118S, which was localized to C13 (Figures S14–S16), was the most abundant proteoform across all samples except for 917178 at 500 μM. Compound 917178 exhibited extensive off-target binding at high concentrations (Figure 3e), which diminished the abundance of the rKRAS4BG13C/C118S single compound addition proteoform.
Figure 3.
Relative quantitation of compound engagement by intact mass. Fractional abundances of all proteoforms were plotted as a function of compound concentration for 917101 (a,b), 917969 (c,d), and 917178 (e,f) with rKRAS4B in the active state (upper row) and inactive state (lower row). rKRAS4BC118S proteins are in purple, rKRAS4BG12C/C118S proteins are in blue, and rKRAS4BGG13C/C118S proteins are in black.
When rKRAS4B was in the inactive state (Figure 3b,d,f), the single compound addition proteoforms of rKRAS4BG12C/C118S and rKRAS4BG13C/C118S increased in abundance in comparison to the active state. Interestingly, the single compound addition to rKRAS4BG12C/C118S, which was localized to C12 (Figures S20–S22), was the most abundant proteoform across all samples at high concentrations. Additionally, the nonspecificity of 917178 that was observed in the active state (Figure 3e) was not reciprocated in the inactive state (Figure 3f) as noted by the lower relative abundance of rKRAS4BC118S proteoforms and double or triple compound additions to rKRAS4BG12C/C118S and rKRAS4BG13C/C118S. In total, these data suggest that the selected compounds have different mutant residue selectivities and reactivities depending on the nucleotide state of rKRAS4B.
For the three example compounds analyzed, our data led to the classification of 917969 as slow but highly specific, 917101 as rapid but slightly nonspecific, and 917178 as rapid but highly nonspecific for rKRAS4BG13C, all at proteoform-resolved detail that simply would not have been accessible by standard proteomic methods employed in drug discovery workflows. Moreover, we could confidently monitor the increase in covalent modification as concentration increased, even for low abundance proteoforms such as the multiple off-target binding populations for all compounds. Combining proteoform fractional abundance values with the localization data obtained by targeted top-down MS2 fragmentation thus provided a complete understanding of the specificity and selectivity of these compounds to rKRAS4BG13C that was previously unknown from the initial high throughput screen (Figure S10).
Evaluation of Clinical Compounds
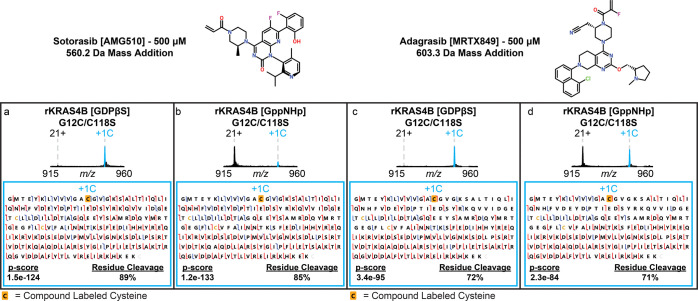
To further validate our top-down assay, we incorporated FDA-approved inhibitors Sotorasib (AMG510) and Adagrasib (MRTX849), which specifically target C12 when KRAS is in the GDP-bound inactive state.7,15,47 Since these compounds have been extensively characterized for use in human patients, we hypothesized that we would observe Sotorasib and Adagrasib rapidly labeling C12 with no off-target binding when rKRAS4B is bound to GDPβS. We performed the top-down assay in a manner similar to evaluating the UCSF SMDC compounds to confirm that these compounds specifically targeted C12 (Figure 4). No off-target labeling of C13 or the native cysteine residues occurred on any protein evaluated (Figure S42). Interestingly, we did observe minimal labeling on C12 from both compounds when rKRAS4BG12C/C118S was bound to GppNHp, suggesting that C12 can still be targeted when rKRAS4B is in the active state.
Figure 4.
Top-down characterization of FDA-approved compounds. MS1 region containing the 21+ charge state following incubation with Sotorasib (AMG510; a-b) or Adagrasib (MRTX849; c-d) at 500 μM for 1 h. Complete engagement was observed with rKRAS4B in the inactive GDPβS state with targeted MS2 fragmentation confirming the localization of each compound to C12 (a and c). Compound engagement was also observed with rKRAS4B in the active, GppNHp state to C12 as confirmed with targeted MS2 fragmentation (b and d).
After performing relative quantitation analysis of AMG510 and MRTX849-treated KRAS4B proteoforms (Figures S43–S55 and Tables S21–S32), we produced the expected result of near complete engagement to C12 by both compounds at the lowest concentration used when rKRAS4BG12C/C118S was bound to GDPβS and no binding was observed to C13 or native cysteines (Figure S43). When calculating the engagement of these compounds to G12C in the active state, we observed AMG510 gradually reaching approximately 15% engagement and MRTX849 approaching 50% engagement by 500 μM, demonstrating that these compounds can still engage their target residue at elevated concentrations (Figure S43). These results, while unexpected, demonstrate the novel insights that our top-down assay can provide while exhaustively characterizing how compounds interact with rKRAS4B invitro during development.
Conclusions
By directly investigating labeled or compound-treated rKRAS4B proteins without disrupting the protein primary structure, we can directly visualize and accurately identify each proteoform within a sample. The novel method described here enables determination of binding stoichiometry, identification of off-target compound engagement, and elucidation of unanticipated compound reaction products, all facilitated by top-down LC–MS and targeted MS2 fragmentation. Additionally, we incorporated complementary methods for the relative quantitation of labeled and unlabeled proteins to rapidly quantify compound engagement. Our results show the time- or concentration-dependence of rKRAS4B-compound engagement in the context of the intact protein molecule while directly mapping the compound binding site. This adaptable methodology can be used to extensively evaluate rKRAS4B-compound engagement and facilitate new inhibitor development. Our hope is that our method may help accelerate the identification of new successful targeted inhibitors for KRAS4B and other RAS isoforms.
Acknowledgments
Special thanks to Elizabeth Donahue Vo, Michelle Arkin, and the UCSF SMDC for running the initial rKRAS4BG13C high-throughput screen, and to Jessica Baker for performing follow-up validation of selected compounds.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.3c05626.
Detailed experimental methods and supporting figues depicting XICs of MBtn labeled tryptic peptides, pictorial representation of the top-down assay, rKRAS4B intact mass of MBtn labeling over time, full scan MS1 of rKRAS4B following MBtn labeling, MBtn labeled rKRAS4B fragmentation maps, UCSF SMDC initial high-throughput screen for selected compounds, XICs of compound engagement to rKRAS4B in the GppNHp state, fragmentation maps of compounds bound to rKRAS4B in the GppNHp state, XICs of compound engagement to rKRAS4B in the GDPβS state, fragmentation maps of compounds to rKRAS4B in the GDPβS state, pictorial representation of the relative quantitation assay, relative quantitation replicate MS1 data for UCSF compounds, treatment of rKRAS4BC118S and rKRAS4BG13C/C118S with AMG510 or MRTX849, relative quantitation plots of AMG510 and MRTX849, relative quantitation replicate MS1 data for AMG510 or MRTX849, tabular observation of MBtn-labeled peptides across all time points, table of UCSF compounds, and relative quantitation values for compound engagement (PDF)
This project has been funded with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The authors declare no competing financial interest.
Notes
Raw files, BioPharma Finder reports, Proteome Discoverer search results, ProSight Lite exports, and relative quantitation results are available to download from the MassIVE repository with identifier MSV000093278.
This paper was published on March 18, 2024. The title was revised slightly, and the paper was reposted on March 18, 2024.
Supplementary Material
References
- Bourne H. R.; Sanders D. A.; McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 1991, 349 (6305), 117–127. 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Simanshu D. K.; Nissley D. V.; McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell 2017, 170 (1), 17–33. 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten M.; Barbacid M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020, 37 (4), 543–550. 10.1016/j.ccell.2020.03.013. [DOI] [PubMed] [Google Scholar]
- Krygowska A. A.; Castellano E. PI3K: A Crucial Piece in the RAS Signaling Puzzle. Cold Spring Harbor Perspect. Med. 2018, 8 (6), a031450. 10.1101/cshperspect.a031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J.; Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93 (1), 269–309. 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Prior I. A.; Lewis P. D.; Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72 (10), 2457–2467. 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. R.; Rosenberg S. C.; McCormick F.; Malek S. RAS-targeted therapies: is the undruggable drugged?. Nat. Rev. Drug Discovery 2020, 19 (8), 533–552. 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punekar S. R.; Velcheti V.; Neel B. G.; Wong K. K. The current state of the art and future trends in RAS-targeted cancer therapies. Nat. Rev. Clin. Oncol. 2022, 19 (10), 637–655. 10.1038/s41571-022-00671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem J. M.; Peters U.; Sos M. L.; Wells J. A.; Shokat K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503 (7477), 548–551. 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J.; Rex K.; Saiki A. Y.; Mohr C.; Cooke K.; Bagal D.; Gaida K.; Holt T.; Knutson C. G.; Koppada N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575 (7781), 217–223. 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- Lanman B. A.; Allen J. R.; Allen J. G.; Amegadzie A. K.; Ashton K. S.; Booker S. K.; Chen J. J.; Chen N.; Frohn M. J.; Goodman G.; et al. Discovery of a Covalent Inhibitor of KRAS(G12C) (AMG 510) for the Treatment of Solid Tumors. J. Med. Chem. 2020, 63 (1), 52–65. 10.1021/acs.jmedchem.9b01180. [DOI] [PubMed] [Google Scholar]
- Nakajima E. C.; Drezner N.; Li X.; Mishra-Kalyani P. S.; Liu Y.; Zhao H.; Bi Y.; Liu J.; Rahman A.; Wearne E.; Ojofeitimi I.; Hotaki L. T.; Spillman D.; Pazdur R.; Beaver J. A.; Singh H. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2022, 28 (8), 1482–1486. 10.1158/1078-0432.CCR-21-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin J.; Engstrom L. D.; Hargis L.; Calinisan A.; Aranda R.; Briere D. M.; Sudhakar N.; Bowcut V.; Baer B. R.; Ballard J. A.; et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discovery 2020, 10 (1), 54–71. 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell J. B.; Fischer J. P.; Baer B. R.; Blake J. F.; Bouhana K.; Briere D. M.; Brown K. D.; Burgess L. E.; Burns A. C.; Burkard M. R.; et al. Identification of the Clinical Development Candidate MRTX849, a Covalent KRAS(G12C) Inhibitor for the Treatment of Cancer. J. Med. Chem. 2020, 63 (13), 6679–6693. 10.1021/acs.jmedchem.9b02052. [DOI] [PubMed] [Google Scholar]
- Dhillon S. Adagrasib: First Approval. Drugs 2023, 83 (3), 275–285. 10.1007/s40265-023-01839-y. [DOI] [PubMed] [Google Scholar]
- Wang X.; Allen S.; Blake J. F.; Bowcut V.; Briere D. M.; Calinisan A.; Dahlke J. R.; Fell J. B.; Fischer J. P.; Gunn R. J.; et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J. Med. Chem. 2022, 65 (4), 3123–3133. 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- Hallin J.; Bowcut V.; Calinisan A.; Briere D. M.; Hargis L.; Engstrom L. D.; Laguer J.; Medwid J.; Vanderpool D.; Lifset E.; et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat. Med. 2022, 28 (10), 2171–2182. 10.1038/s41591-022-02007-7. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Guiley K. Z.; Shokat K. M. Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S). Nat. Chem. Biol. 2022, 18 (11), 1177–1183. 10.1038/s41589-022-01065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Morstein J.; Ecker A. K.; Guiley K. Z.; Shokat K. M. Chemoselective Covalent Modification of K-Ras(G12R) with a Small Molecule Electrophile. J. Am. Chem. Soc. 2022, 144 (35), 15916–15921. 10.1021/jacs.2c05377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.; Herdeis L.; Rudolph D.; Zhao Y.; Böttcher J.; Vides A.; Ayala-Santos C. I.; Pourfarjam Y.; Cuevas-Navarro A.; Xue J. Y.; et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature 2023, 619 (7968), 160–166. 10.1038/s41586-023-06123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z.; Marshal C. B.; Nishikawa T.; Gossert A. D.; Jansen J. M.; Jahnke W.; Ikura M. Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site. Cell Chem. Biol. 2018, 25 (11), 1327. 10.1016/j.chembiol.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Morstein J.; Shrestha R.; Van Q. N.; Lopez C. A.; Arora N.; Tonelli M.; Liang H.; Chen D.; Zhou Y.; Hancock J. F.; Stephen A. G.; Turbyville T. J.; Shokat K. M. Direct Modulators of K-Ras-Membrane Interactions. ACS Chem. Biol. 2023, 18 (9), 2082–2093. 10.1021/acschembio.3c00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah R.; Thomas J. R.; Shafer C. M. Drug discovery considerations in the development of covalent inhibitors. Bioorg. Med. Chem. Lett. 2014, 24 (1), 33–39. 10.1016/j.bmcl.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Shi T.; Song E.; Nie S.; Rodland K. D.; Liu T.; Qian W. J.; Smith R. D. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16 (15–16), 2160–2182. 10.1002/pmic.201500449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebler D. C.; Zimmerman L. J. Targeted quantitation of proteins by mass spectrometry. Biochemistry 2013, 52 (22), 3797–3806. 10.1021/bi400110b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. S.; Quinn J. G.; Saabye M. J.; Guerrero J. F. S.; Nonomiya J.; Lian Q.; Phung W.; Izrayelit Y.; Walters B. T.; Gustafson A.; Endres N. F.; Beresini M. H.; Mulvihill M. M. High-Throughput Kinetic Characterization of Irreversible Covalent Inhibitors of KRAS(G12C) by Intact Protein MS and Targeted MRM. Anal. Chem. 2022, 94 (2), 1230–1239. 10.1021/acs.analchem.1c04463. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Fonslow B. R.; Shan B.; Baek M. C.; Yates J. R. 3rd Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113 (4), 2343–2394. 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli M. P.; Janes M. R.; Li L. S.; Hansen R.; Peters U.; Kessler L. V.; Chen Y.; Kucharski J. M.; Feng J.; Ely T.; Chen J. H.; Firdaus S. J.; Babbar A.; Ren P.; Liu Y. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discovery 2016, 6 (3), 316–329. 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- Lermyte F.; Tsybin Y. O.; O’Connor P. B.; Loo J. A. Top or Middle? Up or Down? Toward a Standard Lexicon for Protein Top-Down and Allied Mass Spectrometry Approaches. J. Am. Soc. Mass Spectrom. 2019, 30 (7), 1149–1157. 10.1007/s13361-019-02201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal M. S.; Liang Y.; Phinney K. W.; Stein S. E. Quantitative bottom-up proteomics depends on digestion conditions. Anal. Chem. 2014, 86 (1), 551–558. 10.1021/ac4027274. [DOI] [PubMed] [Google Scholar]
- Muller T.; Winter D. Systematic Evaluation of Protein Reduction and Alkylation Reveals Massive Unspecific Side Effects by Iodine-containing Reagents. Mol. Cell. Proteomics 2017, 16 (7), 1173–1187. 10.1074/mcp.M116.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyatzis A. E.; Bringans S. D.; Piggott M. J.; Duong M. N.; Lipscombe R. J.; Arthur P. G. Limiting the Hydrolysis and Oxidation of Maleimide-Peptide Adducts Improves Detection of Protein Thiol Oxidation. J. Proteome Res. 2017, 16 (5), 2004–2015. 10.1021/acs.jproteome.6b01060. [DOI] [PubMed] [Google Scholar]
- Smith L. M.; Kelleher N. L.; Consortium for Top Down P. Proteoform: a single term describing protein complexity. Nat. Methods 2013, 10 (3), 186–187. 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M.; Kelleher N. L. Proteoforms as the next proteomics currency. Science 2018, 359 (6380), 1106–1107. 10.1126/science.aat1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catherman A. D.; Skinner O. S.; Kelleher N. L. Top Down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445 (4), 683–693. 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D.; Garvey L. A.; Chakiath C. S. Gateway cloning for protein expression. Methods Mol. Biol. 2009, 498, 31–54. 10.1007/978-1-59745-196-3_3. [DOI] [PubMed] [Google Scholar]
- Taylor T.; Denson J. P.; Esposito D. Optimizing Expression and Solubility of Proteins in E. coli Using Modified Media and Induction Parameters. Methods Mol. Biol. 2017, 1586, 65–82. 10.1007/978-1-4939-6887-9_5. [DOI] [PubMed] [Google Scholar]
- Fellers R. T.; Greer J. B.; Early B. P.; Yu X.; LeDuc R. D.; Kelleher N. L.; Thomas P. M. ProSight Lite: graphical software to analyze top-down mass spectrometry data. Proteomics 2015, 15 (7), 1235–1238. 10.1002/pmic.201400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenbeck K. K.; Davies J. L.; Merron C.; Ogden P.; Sijbesma E.; Ottmann C.; Renslo A. R.; Wilson C.; Arkin M. R. A Liquid Chromatography/Mass Spectrometry Method for Screening Disulfide Tethering Fragments. SLAS Discovery 2018, 23 (2), 183–192. 10.1177/2472555217732072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agamasu C.; Ghirlando R.; Taylor T.; Messing S.; Tran T. H.; Bindu L.; Tonelli M.; Nissley D. V.; McCormick F.; Stephen A. G. KRAS Prenylation Is Required for Bivalent Binding with Calmodulin in a Nucleotide-Independent Manner. Biophys. J. 2019, 116 (6), 1049–1063. 10.1016/j.bpj.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty M. T.; Baldwin A. J.; Marklund E. G.; Hochberg G. K.; Benesch J. L.; Robinson C. V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015, 87 (8), 4370–4376. 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenbeck K. K.; Turner D. M.; Renslo A. R.; Arkin M. R. Targeting Non-Catalytic Cysteine Residues Through Structure-Guided Drug Discovery. Curr. Top. Med. Chem. 2016, 17 (1), 4–15. 10.2174/1568026616666160719163839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaiah S.; Bindu L.; Tran T. H.; Gillette W. K.; Frank P. H.; Ghirlando R.; Nissley D. V.; Esposito D.; McCormick F.; Stephen A. G.; Simanshu D. K. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. U.S.A. 2016, 113 (44), E6766–E6775. 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaiah S.; Tran T. H.; Messing S.; Agamasu C.; Gillette W. K.; Yan W.; Waybright T.; Alexander P.; Esposito D.; Nissley D. V.; McCormick F.; Stephen A. G.; Simanshu D. K. Structures of N-terminally processed KRAS provide insight into the role of N-acetylation. Sci. Rep. 2019, 9 (1), 10512. 10.1038/s41598-019-46846-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogabe S.; Kamada Y.; Miwa M.; Niida A.; Sameshima T.; Kamaura M.; Yonemori K.; Sasaki S.; Sakamoto J. I.; Sakamoto K. Crystal Structure of a Human K-Ras G12D Mutant in Complex with GDP and the Cyclic Inhibitory Peptide KRpep-2d. ACS Med. Chem. Lett. 2017, 8 (7), 732–736. 10.1021/acsmedchemlett.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin M. R.; Ang K. K.; Chen S.; Davies J.; Merron C.; Tang Y.; Wilson C. G.; Renslo A. R. UCSF Small Molecule Discovery Center: innovation, collaboration and chemical biology in the Bay Area. Comb. Chem. High Throughput Screening 2014, 17 (4), 333–342. 10.2174/1386207317666140323133841. [DOI] [PubMed] [Google Scholar]
- Nakajima E. C.; Drezner N.; Li X.; Mishra-Kalyani P. S.; Liu Y.; Zhao H.; Bi Y.; Liu J.; Rahman A.; Wearne E.; Ojofeitimi I.; Hotaki L. T.; Spillman D.; Pazdur R.; Beaver J. A.; Singh H. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2022, 28 (8), 1482–1486. 10.1158/1078-0432.CCR-21-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.