Abstract
ICP4 of herpes simplex virus (HSV) is essential for productive infection due to its central role in the regulation of HSV transcription. This study identified a region of ICP4 that is not required for viral growth in culture or at the periphery of experimentally inoculated mice but is critical for productive growth in the trigeminal ganglia. This region of ICP4 encompasses amino acids 184 to 198 and contains 13 nearly contiguous serine residues that are highly conserved among the alphaherpesviruses. A mutant in which this region is deleted (ΔSER) was able to grow on the corneas of mice and be transported back to the trigeminal ganglia. ΔSER did not grow in the trigeminal ganglia but did express low levels of several immediate-early (ICP4 and ICP27) and early (thymidine kinase [tk] and UL42) genes. It expressed very low levels of the late gC gene and did not appear to replicate DNA. This pattern of gene expression was similar to that observed for a tk mutant, dlsptk. Both ΔSER and dlsptk expressed higher levels of the latency-associated transcript (LAT) per genome earlier in infected ganglia than did the wild-type virus, KOS. However, infected ganglia from all three viruses accumulated the same level of LAT per genome at 30 days postinfection (during latency). The data suggest that the polyserine tract of ICP4 provides an activity that is required for lytic infection in ganglia to progress to viral DNA synthesis and full lytic gene expression. In the absence of this activity, higher levels of LAT per genome accumulate earlier in infection than with wild-type virus.
Herpes simplex virus type 1 (HSV-1), a member of the Alphaherpesvirinae, is recognized by its rapid lytic growth characteristics, variable host range, and propensity to establish and reactivate from latent infection in neurons of sensory ganglia. Lytic HSV infection progresses with the regulated expression of viral genes in three temporal phases, immediate-early (IE; α), early (E; β), and late (L; γ) (37), coordinating the events required for assembly of infectious progeny virus with the inevitable destruction of the infected cell. In neurons, the virus can either initiate a productive infection or establish a latent infection in which transcription from the viral genome is relatively silent (44, 45, 72), with the exception of the latency-associated transcript (LAT) (13, 14, 60, 64, 72). In vivo, the latent genomes persist in the sensory ganglia (11, 23, 64), where certain stresses can trigger reactivation of the lytic cycle, resulting in recurrent infection at mucocutaneous sites and shedding of progeny virus (for reviews, see references 28 and 73). The transition between lytic and latent infection presumably requires an elaborate array of viral functions which is reflected in the number and complexity of accessory proteins encoded by the HSV genome. While there are probably several critical checkpoints that determine the fate of HSV infection in the neuron, one of the most obvious targets for regulating lytic growth is the viral protein ICP4.
ICP4 is a large (175-kDa) DNA-binding (21, 26) phosphoprotein (12, 61) that exists as a homodimer (55, 68) and is localized primarily in the nucleus of the infected cell (12, 61). During viral infection, ICP4 is required for the transcriptional activation of most of the essential E and L genes (19, 24, 30, 31, 57). ICP4 also acts as a repressor of its own expression (19, 58, 59) as well as that of LAT (3, 4, 62) and L/ST (long/short transcripts) (5, 78). ICP4 contains discrete functional domains which determine DNA binding, dimerization, nuclear localization, and transcriptional activation (18, 59, 60, 68). The DNA-binding activity is important for transcriptional activation by ICP4 (60, 68) and essential to its function as a repressor of activated transcription (35, 36, 63). Two additional regions within ICP4 contribute to transcriptional activation, a large domain defined by the carboxy-terminal region of the protein (amino acids [aa] 775 to 1298) (17, 18, 68) and a small serine-rich region near the amino terminus (aa 143 to 210) (68). The carboxy-terminal domain is required for high-level activation by ICP4 and, hence viral growth, because it supports the interaction of ICP4 with TFIID (6). ICP4 proteins lacking the carboxy-terminal region activate transcription poorly (6, 17, 18, 34). Further removal of the amino acids between residues 143 and 210 completely abrogates the activation function (65, 68, 69).
Deletion of only the serine-rich region (aa 143 to 210) results in a virus that is marginally impaired for growth in culture and in the eyes of infected mice but is completely impaired for viral growth in the trigeminal ganglia (77). Besides an unusual polyserine tract (aa 184 to 198), this region contains consensus sites for the cellular kinases protein kinase A (PKA), PKC, and casein kinase II (CKII), as well as a target of in vitro autophosphorylation (76, 77). Given the abundance of potential kinase targets, it follows that removal of this domain results in a significantly hypophosphorylated ICP4 (20, 76). Despite the apparent interaction of this region with several kinases, mutants lacking this region are qualitatively functional by all in vitro assessments (6, 34, 65, 69). The entire region between aa 143 and 210 demonstrates relatively little conservation among the alphaherpesvirus ICP4 homologs. However, similar stretches of polyserine residues corresponding to residues 184 to 198 of HSV-1 ICP4 are found in region 1 of the ICP4 homologs of HSV-2 (24), varicella-zoster virus, simian varicella herpesvirus, pseudorabies virus, Marek’s disease virus, equine herpesvirus 1, and region 5 of bovine herpesvirus 1 (1, 8, 25, 32, 33, 54, 66). A similar polyserine tract also appears in a small cellular protein, P15 (29, 47), which supports the activation function of VP16 and possibly other activators (29, 40). While the functional significance of the polyserine motif is not known, the conservation of these residues among these neurotropic viruses may indicate that it imparts to ICP4 and its homologs an activity required for gene regulation or function in neurons, despite its dispensability in cell culture.
In the classically defined temporal cascade of HSV gene expression, activation of only one temporal phase, γ2 or L, is dependent on viral replication (39, 42, 52). In neurons, viral DNA synthesis is similarly required for induction of the γ2 genes, but in contrast to lytic infection, it has also been reported to enhance expression of the α and β genes (43, 56). During acute infection, reduced expression of α and β genes has been observed in mouse ganglia infected with a thymidine kinase (tk) mutant (43) or a mutant that blocks replication in neurons, as well as in HSV-infected primary rat cervical ganglia neuron cultures treated with inhibitors of viral DNA replication (56). It has been hypothesized that in neurons, viral replication may enhance transcription of the IE genes directly by altering the template (43, 56). The enhanced expression of ICP4 following replication may also be reflective of alleviation from repression by LAT (3, 4, 62) a transcript of unknown function which has been reported to down-regulate productive cycle genes (7, 27). By this scenario, viral DNA replication is the rate-limiting step, pivotal in determining whether lytic infection proceeds or is suspended to the latent state. Assuming that gene expression may differ between neuronal and nonneuronal cells, it follows that some of the viral activities regulating gene expression may also differ.
In this study, we examined the contribution of the conserved polyserine tract in the HSV-1 regulatory protein ICP4 to viral growth in tissue culture and in neuronal and nonneuronal cells in vivo. The fate of the viral genomes containing mutations in the polyserine tract in murine trigeminal ganglia was also determined as infection proceeded from the acute to the latent phase. Last, gene expression from a polyserine tract mutant in acutely and latently infected ganglia was compared to that from wild-type (wt) virus and a tk mutant virus to assess the point where infection of ganglia is blocked. The results suggest that the polyserine tract supports an activity of ICP4 specifically required for growth in the sensory ganglia. In the absence of this function, lytic infection of ganglia is blocked at a point similar to that for tk mutants.
MATERIALS AND METHODS
Virus and cells.
Vero cells were used for the isolation of viral mutants, yield assays, and labeling of viral proteins. E5 cells (17, 62), which express complementing levels of the wild-type ICP4 upon infection, were used to assay and propagate viral stocks. The wt virus used in these studies was KOS (16). The mutants n12 (17), d8-10 (68), and dlsptk (10) were previously described. dlsptk was kindly provided by Donald Coen, Harvard Medical School, Boston, Mass. The procedures for propagating and assaying viral stocks were as previously described (22).
Virus construction.
The construction of plasmid p[i8-i10], containing only the ICP4 coding sequences corresponding to aa 143 to 210 (Fig. 1A), consisted of cloning the 78-bp PstI-AatII fragment from plasmid pi8 (68) and the 135-bp AatII-PstI fragment from plasmid pi10 (68) into the PstI site of pUC18. The previously described plasmids pi8 and pi10 contain in-frame PstI oligonucleotide insertions encoding three alanines after aa 142 and 210, respectively (68). The ΔSER mutation was constructed in p[i8-i10] by using a Transformer site-directed mutagenesis kit (Clontech) and the accompanying protocol. The mutagenic primer, 5′-GTCGTCGTCCTCGTCCTCGTCCAACGCGTTCCCGGAGTCCGACGAGGTCGA-3′, had a deletion of amino acids 184 to 198 and insertion of a unique MluI site and three amino acids, asparagine-alanine-leucine (Fig. 1A). This manipulation was confirmed by DNA sequencing. To generate the full-length ICP4 coding sequences with the ΔSER mutation, the 168-bp PstI fragment of p[i8-i10] containing the ΔSER mutation was cloned into PstI-digested pdi8-i10 (68), replacing the wild-type sequences between residues 143 and 210 with the analogous region containing the ΔSER mutation.
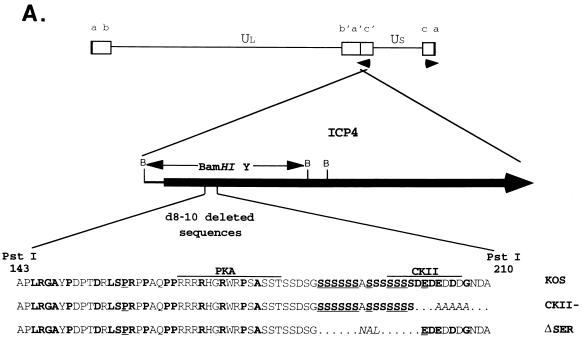
FIG. 1.
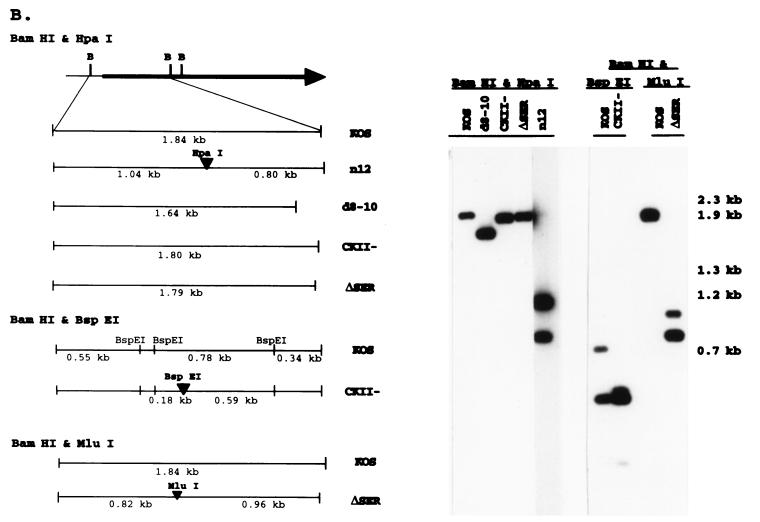
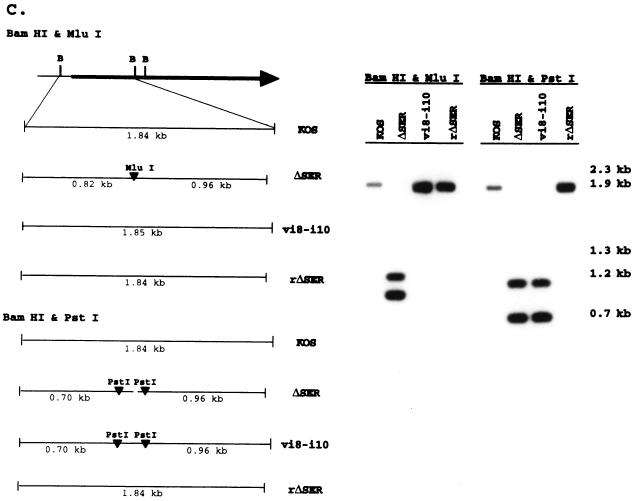
ICP4 serine tract mutations. (A) The genomic location and direction of transcription (arrowheads) of ICP4 are shown along with the BamHI restriction sites in reference to the ICP4 coding region. The BamHI Y fragment and the serine tract region (aa 143 to 210) deleted in mutant d8-10 (68, 77) are indicated. The corresponding wt (KOS) and mutant (CKII− and ΔSER) sequences are compared, showing the deleted (dots) and substituted (italics) residues in reference to the consensus sites for the cellular kinases PKA and CKII. Amino acids that are conserved in the HSV-1 and pseudorabies virus (8) or varicella-zoster virus (54) ICP4 homologs are designated in boldface; underlined residues are conserved in all three. (B) Fine map of the BamHI Y fragment in the wt (KOS) and mutant viruses is shown with the relevant restriction sites for determining the presence of the CKII− and ΔSER mutations. Southern blots of viral DNA digested with the indicated restriction enzymes and hybridized to labeled BamHI-Y probe are shown on the right along with the sizes of the expected fragments. (C) Fine map of the BamHI Y fragment is shown with the relevant restriction sites for determining the structures of the rΔSER and vi8-i10 viruses. Southern blots of viral DNA digested with the indicated restriction enzymes and hybridized to labeled BamHI-Y probe are shown on the right along with the sizes of the expected fragments.
The CKII− mutant was made by combining the HindIII-EarI fragment and the HindIII-PstI fragment from pi10 (68) with the oligonucleotides 5′-TCCGGAGCCGCAGCTGCA-3′ and 5′-GCTGCGGCTCC-3′, which replace the acidic amino acids of the CKII consensus site with the amino acid sequence SAAAAA and insert a new BspE1 restriction site (Fig. 1A).
The ΔSER and the CKII− mutations were introduced into the virus by cotransfecting 106 E5 cells with 3 μg of the 4.2-kb EcoRI fragment of either the ΔSER or the CKII− plasmid and 3 μg of n12 viral DNA, using the calcium phosphate coprecipitation method (17). n12 is a previously described ICP4-deficient virus containing an oligonucleotide specifying a nonsense mutation and an HpaI restriction site at aa 250 (17). Viruses carrying these mutations were selected for rescued growth on Vero cells, and the presence of the mutations in both copies of ICP4 was confirmed by several rounds of Southern blot analysis and plaque purification.
As a control, a virus containing the same i8 and i10 insertion mutations as were present in ΔSER was also constructed. Individually, the i8 and i10 insertions have been shown not to affect the activity of ICP4 (68). The full-length i8-i10 mutant ICP4 was constructed by inserting the 204-bp PstI fragment of p[i8-i10] into the PstI site of plasmid pdi8-i10 (68) to generate pi8-i10. Using the protocol described above for construction of the ΔSER virus, the 4.2-kb EcoRI fragment of pi8-i10 was cotransfected with n12 DNA on E5 cells, and subsequently the desired construct was selected for growth on Vero cells. The presence of the i8-i10 insertions in both copies of ICP4 was confirmed by Southern blot analysis.
To rescue the ΔSER viral mutation, 106 E5 cells were cotransfected with 3 μg of the 1.84-kb BamHI Y fragment from wild-type ICP4 plasmid pK1-2 (17) and 3 μg of ΔSER viral DNA, using calcium phosphate coprecipitation (17). Plaque isolates from the progeny of the transfection were amplified, and rescued viruses were identified by the presence of the full-length 1.84-kb BamHI Y fragment determined by Southern blot analysis. Such isolates were further plaque purified and rechecked by Southern blot analysis.
Virus yield assays.
The assay to determine virus yield was done as previously described (77). Burst size for each virus was expressed as the total PFU yield per infected cell.
Analysis of viral proteins.
Viral polypeptides were radiolabeled by incubating 6 × 105 cells infected at a multiplicity of infection (MOI) of 10 PFU/cell with 100 μCi of [35S]methionine per ml of methionine-deficient medium for 1 h at the indicated times postinfection. The polypeptides were extracted from cells, using sodium dodecyl sulfate (SDS) sample buffer as previously described (77), and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (48).
Animal studies.
Five- to six-week-old male and female CD-1 mice (Charles River Laboratories) were inoculated with 2 × 106 PFU/eye via corneal scarification (51). The assays monitoring acute growth at the site of inoculation and in the trigeminal ganglia, and that for in vitro reactivation, were done as previously described (51). For the PCR and reverse transcriptase (RT)-mediated PCR (RT-PCR) analysis of viral genome copy number and gene expression in the trigeminal ganglia, ganglia were removed at the indicated times postinfection and frozen in liquid nitrogen as previously described (44–46).
PCR analysis of viral DNA.
PCR analysis was used to quantify the average number of viral genomes present in ganglia DNA from infected mice as previously described (41), with minor modifications. The primers specific for the viral gene gC (5′-GGGTCCGTCCCCCCCAAT-3′ and 5′-CGTTAGGTTGGGGGCGCT-3′) and cellular β-actin (5′-AACCCTAAGGCCAACCGTGAAAAGATGACC-3′ and 5′-CCAGGGAGGAAGAGGATGCGGC-3′) (46, 49) were used to amplify the target sequences without additives included in the PCR amplification of β-actin. The PCR products were separated on native 8% polyacrylamide gels containing 0.5× Tris-borate-EDTA (TBE), transferred to GeneScreen Plus (NEN), and then probed with oligonucleotides specific for the PCR products that were 32P end labeled as previously described (45). The probed filters were exposed to X-ray film to obtain an autoradiographic image and also quantified with an Ambis Radioanalytic Imager. The number of viral genomes per cell equivalent was interpolated from linear regression curves (r ≥ 0.990) generated from the standards prepared as previously described (41), using Kaleidagraph software.
RT-PCR analysis.
Total RNA was prepared from pooled ganglia by equilibrium centrifugation as previously described (45), with some modifications. Following centrifugation, approximately 10 to 20 μg of total RNA was incubated with 4 U of RQ-DNase (Promega) and 20 U of RNasin (Promega) in a 40-μl standard reaction mixture for 1 h at 37°C and then extracted with Ultraspec (Biotecx) according to the manufacturer’s protocol. Prior to synthesis of cDNA, 3 μg of total RNA was annealed for 2 h at 65°C in a 12-μl reaction mixture containing reverse primers specific for the viral mRNAs encoding ICP4 (5′-GATCCCCCTCCCGCGCTTCGTCCG-3′), gC (5′-CGTTAGGTTGGGGGCGCT-3′), LAT (5′-ACGAGGGAAAACAATAAGGG-3′), and the cellular β-actin (5′-CCAGGGAGGAAGAGGATGCGGC-3′) (44, 45). Reverse primers for the viral mRNAs encoding ICP27 (5′-TGTGGGGCGCTGGTTGAGGAT-3′), TK (5′-AGGGGGTACGAAGCCATACGCGCTT-3′), and UL42 (5′-CGTGATCGCCAACTCCA-3′) were also included in the 12-μl annealing mix. Half of the annealed RNA was reverse transcribed in a 20-μl reaction mixture containing 1 mM deoxynucleoside triphosphates (dNTPs) and avian myeloblastosis virus RT (Promega); the other half was incubated in an identical reaction mixture containing no RT. One-tenth of each mixture (with and without RT) was subjected to PCR for ICP4, TK, and gC, and 1/200 was used for the PCR amplification of LAT and β-actin. The primer sets and optimal PCR conditions for ICP4, TK, gC, LAT, and β-actin were as previously described (45), with minor changes to the PCRs for TK and β-actin. The PCR amplification of the 103-bp TK-specific product was optimal in the presence of 1.5 mM Mg2+, 10% glycerol, and 5% dimethyl sulfoxide, and the PCR for β-actin contained no additives. The PCRs for ICP27 and UL42 included the reverse primers listed above with the forward primers for ICP27 (5′GCCGCGACGACCTGGAAT3′) and UL42 (5′GGAATCCTACAGGCGTTTGC3′). These reactions were optimized by the addition of 10% glycerol and of 10% glycerol plus 5% dimethyl sulfoxide, respectively. To amplify the 218-bp ICP27- and the 263-bp UL42-specific products, 1/10 of the cDNA was subjected to 30 cycles of 1-min denaturing at 95°C, 1-min annealing at 55°C, and 1-min extension at 72°C. All PCRs were done in a 100-μl volume containing 1× PCR buffer (Boehringer Mannheim Biochemicals), 200 μM dNTPs, and 2.5 U of Taq polymerase (Boehringer Mannheim Biochemicals). For the ICP4 PCR, 2.5 U of Amplitaq Gold (Perkin-Elmer) was substituted for Taq polymerase (Boehringer Mannheim Biochemicals).
The PCR products were separated on native 8% polyacrylamide–TBE gels, electrophoretically transferred to nylon membranes (GeneScreen Plus; NEN), UV cross-linked, and subsequently probed with 32P-end-labeled internal probes specific for ICP4, TK, gC, LAT, and β-actin as previously described (44, 45). The internal probes used to detect the ICP27 and UL42 PCR products were 5′-AGCACCCAGACGCCTCGTCCGACGGA-3′ and 5′-TCCATAACACGATCTTTGGGGAGCAGGTG-3′, respectively. The resulting blots were exposed to X-ray film and quantitatively analyzed with an Ambis Radioanalytic Imager. Because the LAT RT-PCR signal varied greatly between different viruses over time, the following experiment was used to demonstrate the linearity of the LAT RT-PCR conducted in the context of this study. cDNA from KOS-infected ganglia at 30 days postinfection was serially diluted in cDNA from mock-infected ganglia by 0.5-log dilutions over 3 orders of magnitude. This preparation was subjected to PCR, analyzed by Southern blotting, quantified, and plotted as counts versus dilution. Over the 1,000-fold range tested, a linear relationship was obtained.
RESULTS
The acute growth phenotypes of mutants within the serine-rich region of ICP4.
Identification of the residues responsible for the neuron-specific growth defect of the mutant d8-10 (77) required making finer mutations within this region of ICP4. As illustrated in Fig. 1A, the sequences most conserved among the alphaherpesviruses are the polyserine tract and several residues constituting a consensus site for CKII. To address the contributions of these sites to viral growth, mutations were introduced at each site as described in Materials and Methods. The ΔSER mutation deleted the polyserine tract, and the CKII− mutation deleted the CKII consensus site by replacing the acidic residues of the consensus site with alanines (Fig. 1A). Two additional viruses were constructed as controls. Mutant vi8-i10 contained the wt sequences along with the same i8 and i10 insertions exploited in the construction of the ΔSER mutant. The virus rΔSER was isolated following transfection of ΔSER viral DNA with the 1.84-kb BamHI Y fragment encoding the first 521 aa of ICP4 (Fig. 1A) and screening for repair of the ΔSER mutation.
Southern blot analysis (Fig. 1B) confirmed the introduction of the ΔSER and CKII− mutations in both copies of ICP4 by demonstrating the insertion of new restriction sites (MluI and BspE1, respectively) as well as the loss of the HpaI site present in the n12 (parental) virus (17). The vi8-i10 mutant contains the same i8 and i10 mutations as ΔSER, as indicated by the corresponding PstI restriction sites (68). The loss of the MluI and PstI sites from rΔSER (Fig. 1C) confirms the rescue of the serine tract mutation.
The ΔSER and CKII− mutants and control viruses were analyzed, along with the wt virus KOS and the mutant d8-10, for the ability to grow in Vero cells and for effects on viral gene expression. As previously described, d8-10 grows in tissue culture with reduced viral yields (68, 69, 77). To compare the growth deficiency of d8-10 with any imparted by the ΔSER and CKII− mutations, Vero cells were infected with each virus at an MOI of 5 PFU per cell, and viral yields were determined by plaque assay on E5 cells. The ΔSER and CKII− viruses demonstrated greater viral yields than d8-10 (Table 1). The ranges of viral yields (burst sizes) over four experiments were as follows; 45 to 72 for d8-10, 130 to 360 for ΔSER, 300 to 700 for CKII−, and 600 to 840 for KOS. The yields for vi8-i10 and rΔSER were similar to those for KOS, indicating that the i8 and i10 mutations or mutations outside the 1.84-kb BamHI-Y loci of ΔSER did not significantly impair viral growth.
TABLE 1.
Growth of ICP4 mutants in Vero cells
| Virus | Burst sizea |
|---|---|
| KOS | 820 |
| d8-10 | 45 |
| CKII− | 480 |
| ΔSER | 170 |
| i8-i10 | 690 |
| rΔSER | 1,100 |
Yield of virus harvested at 18 h postinfection divided by number of infected cells.
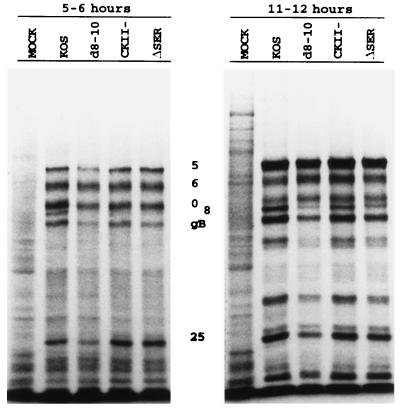
The ΔSER and CKII− mutants exhibited only slightly reduced growth in tissue culture compared to KOS. Accordingly, both viruses demonstrated polypeptide profiles (Fig. 2) at early and late times postinfection that were very similar to those of KOS, while the more growth-deficient d8-10 virus resulted in a delayed pattern of gene expression, consistent with previous findings (68, 69, 77).
FIG. 2.
Polypeptide profiles of the serine tract mutants. Vero cells were infected (MOI of 10) with the indicated viruses, pulsed with [35S]methionine at the indicated times postinfection, and processed for SDS-PAGE analysis on an SDS–9% polyacrylamide gel. The positions of selected viral proteins are designated.
The main focus of this study is to identify the elements within the region deleted in d8-10 that are responsible for the growth deficiency in the trigeminal ganglia. The murine eye model was used to assess the growth phenotypes in vivo of viruses with mutations in the serine-rich region. Mice were infected via corneal scarification with either ΔSER, CKII−, d8-10, or KOS as described in Materials and Methods. Growth in the eye was quantified from eye swab samples collected at 1 day postinfection, and growth in the trigeminal ganglia was quantified from ganglion homogenate samples collected and prepared 3 days postinfection. Table 2 summarizes the growth phenotypes of the viruses in the eye and in the trigeminal ganglia from several experiments. In the eye, the viruses had a growth profile similar to that observed in Vero cells (Table 1), in that ΔSER and CKII− were not as deficient for growth as d8-10 but not as vigorous as KOS. However, the growth characteristics of the viruses in the trigeminal ganglia differed dramatically. While the CKII− virus yields in the ganglia were approximately 30-fold lower than the wt yields, the ΔSER yields were very similar to that of d8-10 in that very few harvested ganglia yielded any infectious virus, with the average yield being more than 103-fold lower than that of KOS (Table 2). These results indicated that the polyserine tract was the element within the serine-rich region contributing specifically to growth in the trigeminal ganglia. Additional support of this conclusion comes from the growth phenotypes of vi8-i10 and rΔSER. Both vi8-i10 and rΔSER demonstrated a growth phenotype similar to that of KOS in the eye and in the trigeminal ganglia, indicating again that the ΔSER mutation was responsible for the severe growth defect observed in ganglia.
TABLE 2.
Growth of ICP4 mutants in mice
| Virus | Avg titer (PFU/ml)
|
|
|---|---|---|
| Eye | Ganglia | |
| KOS | 7.4 × 105 (18/20a) | 5.5 × 104 (18/22) |
| d8-10 | 1.4 × 104 (15/16) | 17 (3/16) |
| CKII− | 4.5 × 104 (4/4) | 1.5 × 103 (4/4) |
| ΔSER | 9.0 × 104 (12/12) | 20 (1/12) |
| rΔSER | 1.1 × 106 (6/6) | 7.5 × 104 (6/6) |
| vi8-i10 | 8.4 × 105 (6/6) | 1.3 × 105 (4/4) |
Number of samples yielding PFU over the total number of samples used in calculating the numerical average.
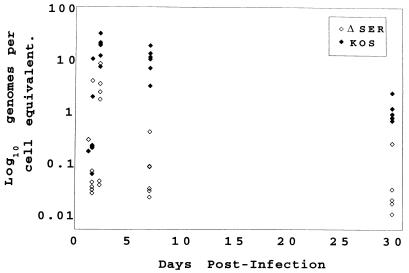
ΔSER DNA replicates poorly in the trigeminal ganglia yet establishes latent infection.
To address the block in lytic growth of the ΔSER mutant in the trigeminal ganglia, a more detailed study of viral replication was initiated. Mice were infected with ΔSER or the wt virus, and ganglia were harvested at selected time points over a 30-day period postinfection. Viral DNA content (load) at each time interval was determined by quantitative PCR (41). Figure 3 graphically illustrates the data, each data point representing the viral load of an individual ganglion. As shown, ΔSER DNA was detected in the ganglia as early as 33 h postinfection, with the viral load peaking at 56 h (Fig. 3). By 7 days, the viral load had generally decreased in the ΔSER-infected ganglia, approaching the levels observed at 30 days postinfection. In contrast, the wt-infected ganglia demonstrated at 56 h a viral load greater than that observed for ΔSER, and this increased viral load was relatively unchanged at 7 days postinfection.
FIG. 3.
Accumulation of viral DNA in ΔSER- and KOS-infected ganglia. Viral DNA content at the designated time points in KOS- or ΔSER-infected trigeminal ganglia was determined by PCR as described in Materials and Methods (41). Each data point was determined from an individual ganglion.
Collectively, these data indicate that the ΔSER virus probably did not replicate in the trigeminal ganglia. While it cannot be ruled out that the increased viral load at 56 h postinfection in the ΔSER-infected ganglia represents low levels of viral replication, it may simply reflect increased load due to the retrograde transport of virus still growing at peripheral sites of infection. By 7 days postinfection, with the peripheral infection resolving, the viral load in the ganglia may be a better representation of viral replication occurring at that site.
It has been shown that replication in the trigeminal ganglia is not required for establishment of latency (10, 23, 41, 51) and that d8-10 establishes latent infection (77). The ability of the viruses constructed for this study to establish latent infection was determined by two methods; the first measured the viral DNA content directly by determining the genome copy number at 30 days postinfection, and the second determined the efficiency of reactivation as an indicator of previously latent virus. Quantitative PCR (41) was used to determine the DNA copy number in latently infected trigeminal ganglia. As summarized in Table 3, the average viral load in ΔSER-infected ganglia was about 10-fold lower than the viral load in wt-infected ganglia, while the viral load in d8-10-infected ganglia was further reduced. As expected, the viral load in rΔSER-infected ganglia was more similar to that in KOS-infected ganglia. Explant reactivation assays (51) were also performed at 30 days postinfection for mice infected with d8-10, ΔSER, CKII−, rΔSER, and KOS; percentages of explanted ganglia that yielded infectious virus are shown in Table 3. These results are noteworthy only in that the ability of the mutants to reactivate confirms the establishment of latency and the presence of intact genomes in ganglia at 30 days postinfection. The frequency of reactivation for d8-10 agrees with that previously reported for this mutant (77).
TABLE 3.
Viral DNA content and reactivatable virus in ganglia at 30 days postinfection
| Virus | Genomes/cell equivalenta | % Reactivationb (no. yielding infectious virus/total no. of samples) |
|---|---|---|
| KOS | 0.54 | 100 (4/4) |
| d8-10 | <0.01 | 12 (1/8) |
| CKII− | ND | 50 (3/6) |
| ΔSER | 0.06 | 12 (1/8) |
| rΔSER | 0.19 | 100 (4/4) |
Determined by quantitative PCR as described by Katz et al. (41). Each viral sample represents six pooled ganglia. ND, not determined.
Percentage of explanted ganglia that yield infectious virus.
Viral gene expression in ΔSER-infected ganglia.
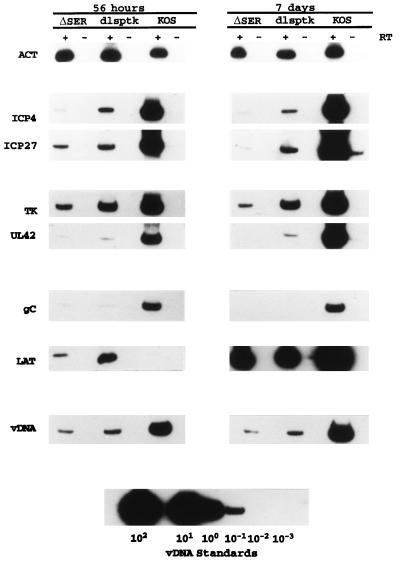
To determine where in the lytic cycle the ΔSER mutation affected viral growth in neurons, viral gene expression in the trigeminal ganglia was analyzed by quantitative RT-PCR (45). Included in the analyses were KOS and a tk mutant, dlsptk (10), a virus with a well-characterized defect for growth in the trigeminal ganglia (10, 41, 43, 45). In this set of experiments, ganglia from infected mice were harvested at 56 h, 7 days, and 30 days postinfection. Six ganglia from each time point were pooled, an aliquot of each pooled sample was processed for DNA to determine viral DNA content (41), and the remainder was processed for RT-PCR. Primers specific for the viral genes encoding ICP4, ICP27, TK, UL42, gC, and LAT and for the cellular β-actin gene (44, 45) were used for the synthesis and amplification of cDNA. The PCR products were separated on native polyacrylamide gels and transferred to Nytran membranes. Gene-specific products were identified by Southern blot analysis using radiolabeled oligonucleotide probes.
The RT-PCR analysis of viral gene expression during the acute infection is shown in Fig. 4. Signals for all amplified messages were readily detected for the replication-deficient mutants ΔSER and dlsptk, except for those of UL42 and gC, which approached the level of detection, especially at 7 days postinfection. Consistent with observations made by Kosz-Vnenchak et al. (43) and Nichol et al. (56), the levels of ICP4 and ICP27 signals detected from the infected ganglia relative to viral DNA content indicate that expression of ICP4 and ICP27 in dlsptk was lower per genome than that in the wt virus. The level of ICP4 per genome in ΔSER was lower than that observed for the dlsptk mutant at 56 h and 7 days postinfection. In contrast, ICP27 expression in ΔSER was similar to that in dlsptk at 56 h postinfection; however, it was reduced compared to that in dlsptk at 7 days postinfection.
FIG. 4.
Viral gene expression in the trigeminal ganglia during acute infection. RT-PCR products of viral and cellular transcripts from ganglia infected with ΔSER, KOS, or the tk mutant dlsptk (10) from pooled ganglia harvested at 56 h and 7 days postinfection are shown. PCR products of viral DNA (vDNA) for each sample are also shown along with the PCR signals used to establish a viral DNA standard curve. ACT, β-actin.
The reduced abundance of ICP4 mRNA during the acute infection in ΔSER did not appear to greatly affect the level of tk per genome relative to dlsptk. Although the tk signal is lower in ΔSER than in dlsptk, the two mutants demonstrated similar ratios of tk to ICP4 signal, indicating that the ΔSER mutation in ICP4 does not abrogate its ability to activate tk, a gene whose expression stringently depends on ICP4 (16, 19, 30, 38, 57). Expression of another E gene, UL42, in ΔSER and dlsptk during the acute infection appears to be more restricted than the expression of tk. While UL42 was easily detected in KOS at 56 h and 7 days, very little UL42 was detected in the ΔSER and dlsptk samples. Again, consistent with their ICP4 RT-PCR profiles, the ΔSER UL42 signal was lower than that from the dlsptk-infected ganglia and hence does not reveal any obvious difference in UL42 expression between the mutants.
RT-PCR analysis of L gene expression supports the observation that ΔSER and dlsptk are not replicating. As a true late gene, expression of the gC gene is dependent on viral replication (39, 42, 52). It might be expected that if ΔSER and dlsptk were not replicating, no gC would be detected. Barely detectable levels of gC were detected in the ΔSER- and dlsptk-infected ganglia at 56 h postinfection, and no gC was detected at 7 days postinfection. It may be that the small amount of gC detected is due to read-through transcription from a 5′ E gene as previously hypothesized by Kramer et al. to explain the presence of small amounts of gC signal in dlsptk-infected ganglia (44, 46). Alternatively, the low expression of gC may indicate that there is very low level viral DNA synthesis in the ΔSER- and dlsptk-infected ganglia.
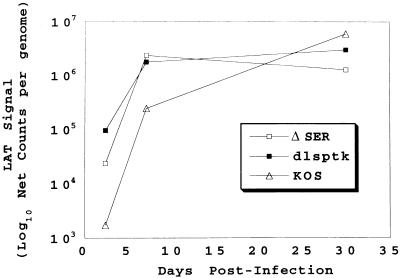
Perhaps the most interesting observation regarding viral gene expression in the trigeminal ganglia relates to the expression of LAT. At 56 h postinfection, the LAT signal per genome in the ΔSER- and dlsptk-infected ganglia was greater than that observed in the wt-infected ganglia (see Fig. 6). It is also interesting that despite the 10- to 30-fold increase in LAT expression in the mutant-infected ganglia, LAT expression at 56 h was lower in ΔSER than in dlsptk. By 7 days, there was no difference in LAT expression in the ΔSER- and dlsptk-infected ganglia. These levels were still greater on a per-genome basis than the amount of LAT per viral DNA content observed in wt-infected ganglia (8- to 10-fold).
FIG. 6.
LAT expression per genome during acute and latent infection. The 56-h- and 7-day-postinfection LAT RT-PCR and viral DNA PCR data from Fig. 4 were combined with the corresponding 30-day-postinfection LAT RT-PCR and viral DNA PCR data to illustrate LAT expression per genome as a function of time. As the absolute amount of LAT was not quantitatively determined with standards, the LAT RT-PCR signal per genome is represented simply as net counts per genome.
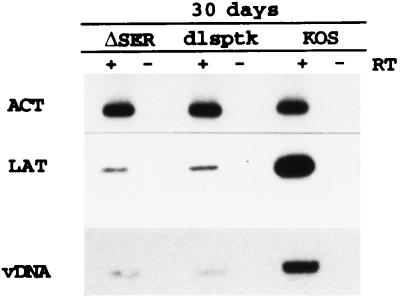
During latency, while expression of the acute viral genes is difficult to detect (45, 72), the LAT transcript is abundantly expressed (13, 14, 64, 70, 72). At 30 days postinfection, the levels of LAT detected in the mutant- and wt-infected ganglia per genome were very similar (Fig. 5). Therefore, as illustrated in Fig. 6, LAT expression per genome was initially greater and peaked earlier in the ΔSER- and dlsptk-infected ganglia than in the wt-infected ganglia. By 30 days postinfection, the difference was no longer apparent.
FIG. 5.
LAT expression at 30 days postinfection. RT-PCR products of LAT and β-actin (ACT) prepared from ganglia infected with the designated viruses and harvested at 30 days postinfection are shown in the top two panels. The bottom panel shows the PCR products of viral DNA (vDNA) for each virus. These data represent an animal experiment different from that used for Fig. 4 but are typical of previous results.
DISCUSSION
Viral mutants in which the serine-rich region (aa 143 to 210) of ICP4 is deleted exhibit reduced viral yields and delayed E and L gene synthesis in tissue culture but demonstrate a total block for replication in the trigeminal ganglia of infected mice. This result suggested that this region of ICP4 is necessary for the progression of lytic infection in neurons. In this study, we conducted a finer mutational analysis of the region and isolated a mutant whose phenotype strengthens and confirms this hypothesis. Specifically, a mutant in which only the highly conserved polyserine residues (aa 184 to 198) have been deleted, ΔSER, exhibited a more restored growth phenotype in tissue culture and at peripheral sites of infection in mice but continued to exhibit the restricted lytic growth in the mouse trigeminal ganglia characteristic of a mutant with the entire region (aa 143 to 210) deleted (68, 69, 77). This phenotype suggested that the polyserine residues comprise an element embedded within ICP4 required for the protein to function properly during lytic infection of neuronal tissue.
The ΔSER mutant demonstrated a defective growth phenotype in the trigeminal ganglia of mice. The gene expression profile of ΔSER in the trigeminal ganglia was similar to, if not somewhat more impaired than, that of a tk mutant, dlsptk. This finding indicates that ΔSER infection was blocked at a similar point in lytic growth as dlsptk. While the dlsptk mutant does not replicate in the ganglia, presumably because it cannot supplement the limiting pools of NTPs, it is not clear why ΔSER is blocked for viral replication.
The RT-PCR data of ΔSER-infected ganglia indicated that both ICP4 and ICP27 were present. Furthermore, the ΔSER mutation did not appear to impair the ability of ICP4 to activate tk. UL42 was also present at low levels. In fact, tk and UL42 expression in ΔSER-infected ganglia was similar to that observed in dlsptk-infected ganglia, considering their corresponding ICP4 RT-PCR signals. In light of the similar phenotypes of these mutants, it is noteworthy that the wt phenotype of the rescued ΔSER virus as well as the acyclovir sensitivity of ΔSER confirmed that the defective phenotype was due to a mutation in ICP4, not in tk. While the tk and UL42 expression profiles of ΔSER did not explain the block in replication, the extremely low expression of gC and the PCR analysis of viral genomes in the infected ganglia indicated that somewhere between E gene expression and viral replication, the ΔSER mutation inhibited the progression of lytic growth in neurons.
While tk expression is considered an excellent indicator of ICP4 function, there are ICP4 mutants that are capable of activating tk in the context of viral infection yet fail to replicate (68). n208, a mutant in which the carboxy-terminal activation domain of the protein has been deleted (68), exhibits activated levels of some early genes like tk but not others (67). Consequently, E genes are not homogeneous with respect to activation by ICP4. Additionally, activation of some E genes, like UL9 and UL30, may be further complicated by their unusual sensitivity to the status of DNA replication (4, 74). There is also the possibility that some E genes are differentially regulated in neurons; for example, the UL9 promoter has been reported to contain a cyclic AMP response element and to be differentially induced in PC12 cells (14). Perhaps in neurons, the requirements for ICP4 activation are more stringent for a particular subset of E genes, requiring an activity which is lacking in the ΔSER ICP4 protein. Thus, while differential activation of the E genes is one hypothesis for the block in lytic growth in ΔSER-infected ganglia, the available RT-PCR data (as well as in vitro data) suggest that the ΔSER ICP4 retains some activation function. Last, it may be that the ΔSER mutation simply reduces the activation function of ICP4 such that it is not readily noticeable in cell culture beyond a slight reduction in viral yield. However, in neurons, some specific promoters may not be as responsive to ICP4, and thus the defect imparted by the ΔSER mutation would have greater consequences for the expression of such genes.
Another function of ICP4 which the RT-PCR data suggest is intact in the ΔSER protein is its ability to repress ICP4 and LAT. In fact, the 56-h postinfection data (Fig. 4) might suggest that the ΔSER mutation enhanced repression of these two genes, given their reduced abundance in the ΔSER-infected ganglia. Another viral product known to be repressed by ICP4 is the L/ST (open reading frame [ORF] P) transcript (5, 49, 78). We were unsuccessful in detecting the ORF P transcript by RT-PCR. The level of ORF P transcript may be relevant in that it has been shown to repress expression of the neurovirulence factor γ34.5 (50). However, in this case less neurovirulence is associated with the derepression of ORF P (50). As is the case for activation, the serine tract region does not appear to greatly affect repression in tissue culture or in vitro (35, 36, 65). Thus, if repression was affected by the ΔSER mutation, it would have to be an effect specific to function in neurons.
A less likely possibility is that the ΔSER mutation does not permit ICP4 to act more directly at the level of DNA replication. Although ICP4 does not participate in origin-dependent DNA synthesis in cell culture (75), perhaps its function as a transcriptional activator induces local changes in the viral genome that result in more efficient replication in neurons. In neurons, the converse seems to be true; viral replication enhances the transcriptional activation of the lytic genes (43, 56).
An interesting result from this study relates to the expression profile of LAT. LAT expression in ΔSER and dlsptk during the acute infection in ganglia was substantially greater (per genome) than in KOS, but by 30 days postinfection this difference was no longer apparent. The increased LAT expression during acute times in ΔSER- and dlsptk-infected ganglia may partly be a function of less ICP4 being expressed, as ICP4 has been shown to repress expression of LAT (3, 4, 62). Another possibility may be that LAT is expressed only from genomes in which lytic infection had either never initiated or already been aborted. This scenario predicts a population of ΔSER and dlsptk viruses that were no longer expressing the lytic genes and were essentially latent.
The robust expression of LAT in the replication-deficient mutants appears to be coincident with the reduced expression or silencing of lytic gene expression and has been hypothesized to be one mechanism by which to silence expression of the lytic genes (7, 27). As described by Garber et al. (27) and Chen et al. (7), LAT expression is associated with a decreased accumulation of ICP4 and tk transcripts. Down-regulating the lytic cycle genes would be a mechanism to slow the progression of productive infection. In the case of ΔSER and dlsptk, the block in replication prevents full expression of the IE and E genes. This, in combination with the earlier and more abundant expression of LAT, may expedite the process of achieving latency. In the KOS-infected ganglia, achieving this state is a more protracted process, perhaps because viral replication allows the IE genes to be fully induced, leading to lower LAT expression (3, 4, 62). Although replication-deficient mutants such as ΔSER and dlsptk establish latent infection with a lower viral genome burden, the numbers change very little during the acute and latent infection (44), whereas during acute times, wt-infected ganglia typically contain 10 to 100 times the number of genomes at latency. Therefore, if we define the efficiency of establishing latency as the percentage of genomes retained in the trigeminal ganglia, then ΔSER and dlsptk are more efficient at establishing latent infection.
The polyserine tract of ICP4 is conserved in many of the neurotropic herpesvirus ICP4 homologs and is required for growth in the trigeminal ganglia but is otherwise unremarkable for its effect on ICP4 function in Vero cells or in vitro. One possible explanation for the viral growth advantage specified by these residues may be that the polyserine residues directly complement a coactivator function that is missing or limiting in neuronal cells but is abundantly expressed in nonneuronal cells. It is also possible that these residues influence the activities of ICP4 in neurons as a function of phosphorylation of the ICP4 protein via modification by neuronal kinases and/or phosphatases. Both scenarios predict that this region is important in regulating the activity of ICP4 in neurons and in part helps determine whether the virus replicates or assumes a state that favors the establishment of latency.
ACKNOWLEDGMENTS
We thank Colton Smith and Lorna Samaniego for helpful discussions and comments on the manuscript.
This work was supported by NIH grant AI27431.
REFERENCES
- 1.Anderson A S, Francesconi A, Morgan R W. Complete nucleotide sequence of the Marek’s disease virus ICP4 gene. Virology. 1992;189:657–667. doi: 10.1016/0042-6822(92)90589-h. [DOI] [PubMed] [Google Scholar]
- 2.Baradaran K, Dabrowski C E, Schaffer P A. Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9, and UL10 genes and identification of a novel delayed-early gene product, OBPC. J Virol. 1994;68:4251–4261. doi: 10.1128/jvi.68.7.4251-4261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor A H, O’Hare P. Regulation and cell-type-specific activity of a promoter located upstream of the latency associated transcript of herpes simplex virus type 1. J Virol. 1990;64:3269–3279. doi: 10.1128/jvi.64.7.3269-3279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor A H, O’Hare P. Localization of cis-acting sequence requirements in the promoter of the latency-associated transcript of herpes simplex virus type 1 required for cell-type-specific activity. J Virol. 1992;66:3573–3582. doi: 10.1128/jvi.66.6.3573-3582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohenzky R A, Papavassiliou A G, Gelman I H, Silverstein S. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. J Virol. 1993;67:632–642. doi: 10.1128/jvi.67.2.632-642.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrozza M J, DeLuca N A. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol Cell Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, Kramer M F, Schaffer P A, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A K. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 1989;17:4637–4646. doi: 10.1093/nar/17.12.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 10.Coen D M, Kosz-Vnenchak M, Jacobson J G, Leib D A, Bogard C L, Schaffer P A, Tyler K L, Knipe D M. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci USA. 1989;86:4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook M L, Basatone V B, Stevens J G. Evidence that neurons harbor latent herpes virus. Infect Immun. 1974;9:946–951. doi: 10.1128/iai.9.5.946-951.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtney R J, Benyesh-Melnick M. Isolation and characterization of a large molecular weight polypeptide of herpes simplex virus type 1. Virology. 1974;62:539–551. doi: 10.1016/0042-6822(74)90414-0. [DOI] [PubMed] [Google Scholar]
- 13.Croen D D, Ostrove J M, Dragovic L J, Smialec J E, Straus S E. Latent herpes simplex virus in human trigeminal ganglia: detection of an immediate early gene “anti-sense” transcript by in situ hybridization. N Engl J Med. 1987;317:1427–1431. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 14.Deatly A M, Spivack J G, Lavi E, Fraser N W. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia in latently infected mice. Proc Natl Acad Sci USA. 1987;84:3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deb S P, Deb S, Brown D R. Cell-type-specific induction of the UL9 gene of HSV-1 by cell signaling pathway. Biochem Biophys Res Commun. 1994;205:44–41. doi: 10.1006/bbrc.1994.2627. [DOI] [PubMed] [Google Scholar]
- 16.DeLuca N A, Courtney M A, Schaffer P A. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J Virol. 1984;52:767–776. doi: 10.1128/jvi.52.3.767-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLuca N A, McCarthy A, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca N A, Schaffer P A. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985;5:1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLuca N A, Schaffer P A. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 1987;15:4491–4511. doi: 10.1093/nar/15.11.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLuca N A, Schaffer P A. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J Virol. 1988;62:732–743. doi: 10.1128/jvi.62.3.732-743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiDonato J A, Spitzner J R, Muller M R. A predictive model of DNA recognition by herpes simplex virus protein ICP4. J Mol Biol. 1991;219:451–470. doi: 10.1016/0022-2836(91)90186-a. [DOI] [PubMed] [Google Scholar]
- 22.Dixon R A F, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the early gene encoding the herpes simplex virus type 1 immediate-early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efstathiou S, Minson A C, Field H J, Anderson J R, Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986;57:446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett R D. Transactivation of transcription by herpes virus products: requirement for two HSV-1 immediate early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett R D, Fenwick M L. Comparative DNA sequence of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J Gen Virol. 1990;73:2167–2171. doi: 10.1099/0022-1317-71-6-1387. [DOI] [PubMed] [Google Scholar]
- 26.Faber S W, Wilcox K W. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences. Nucleic Acids Res. 1986;14:6067–6083. doi: 10.1093/nar/14.15.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber D A, Schaffer P A, Knipe D M. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71:5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Blanco M A, Cullen B R. Molecular basis of latency in pathogenic human viruses. Science. 1991;254:815–820. doi: 10.1126/science.1658933. [DOI] [PubMed] [Google Scholar]
- 29.Ge J, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 30.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;83:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godowski P J, Knipe D M. Transcriptional control of herpes virus gene expression: gene functions required for positive and negative regulation. Proc Natl Acad Sci USA. 1986;83:256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray W L, Gusick N J, Ek-Kommonen C, Kempson S E, Fletcher T M., III The inverted repeat regions of the simian varicella virus and varicella-zoster virus genomes have a similar genetic organization. Virus Res. 1995;39:181–193. doi: 10.1016/0168-1702(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 33.Grundy F J, Baumann R P, O’Callaghan D J. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology. 1989;172:223–236. doi: 10.1016/0042-6822(89)90124-4. [DOI] [PubMed] [Google Scholar]
- 34.Gu B, DeLuca N A. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J Virol. 1994;68:7953–7965. doi: 10.1128/jvi.68.12.7953-7965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu B, Kuddus R, DeLuca N A. Repression of activator-mediated transcription by herpes simplex virus via a mechanism involving interactions with the basal transcription factors TATA-binding protein and TFIIB. Mol Cell Biol. 1995;15:3618–3626. doi: 10.1128/mcb.15.7.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu B, Rivera-Gonzalez R, Smith C A, DeLuca N A. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc Natl Acad Sci USA. 1995;90:9528–9532. doi: 10.1073/pnas.90.20.9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honess R W, Roizman B. Regulation of herpes virus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imbalzano A N, Coen D M, DeLuca N A. Herpes simplex virus transactivator ICP4 operationally substitutes for the cellular transcription factor Sp1 for efficient expression of the viral thymidine kinase gene. J Virol. 1991;65:565–574. doi: 10.1128/jvi.65.2.565-574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson P A, Everett R D. DNA replication is required for abundant expression of a plasmid-borne late US11 gene of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:3609–3625. doi: 10.1093/nar/14.9.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser K, Steizer G, Meisterernst M. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz J P, Bodin E T, Coen D M. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J Virol. 1990;64:4288–4295. doi: 10.1128/jvi.64.9.4288-4295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kibbler P K, Duncan J, Keith B D, Hupel T, Smiley J R. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J Virol. 1991;65:6749–6760. doi: 10.1128/jvi.65.12.6749-6760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer M F, Chen S, Knipe D M, Coen D M. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J Virol. 1998;72:1177–1185. doi: 10.1128/jvi.72.2.1177-1185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer M F, Coen D M. Quantification of herpes simplex virus DNA and transcripts during latent infection in mouse trigeminal ganglia by reverse transcriptase PCR. Doctoral dissertation. Cambridge, Mass: Harvard University; 1995. [Google Scholar]
- 46.Kramer M F, Coen D M. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1995;69:1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 48.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Lagunoff M, Randall G, Roizman B. Phenotypic properties of herpes simplex virus 1 containing a derepressed open reading frame P gene. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mavromara-Nazos P, Roizman B. Activation of herpes simplex virus 1 g2 genes by viral DNA replication. Virology. 1987;161:593–598. doi: 10.1016/0042-6822(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 53.McGeoch D J, Dalrymple M A, Davidson A J, Dolan A J, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 54.McGeoch D J, Dolan A, Donald S, Brauer D H K. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1744. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metzler D W, Wilcox K W. Isolation of herpes simplex virus regulatory protein ICP4 as a homodimeric complex. J Virol. 1985;55:329–337. doi: 10.1128/jvi.55.2.329-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nichol P, Chang J Y, Johnson E M, Jr, Olivo P D. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. J Virol. 1996;70:5476–5486. doi: 10.1128/jvi.70.8.5476-5486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000 and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Hare P, Hayward G S. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J Virol. 1985;56:723–733. doi: 10.1128/jvi.56.3.723-733.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paterson R, Everett R D. Mutational dissection of the HSV-1 immediate-early protein Vmw-175 involved in transcriptional transactivation and repression. Virology. 1988;166:186–196. doi: 10.1016/0042-6822(88)90160-2. [DOI] [PubMed] [Google Scholar]
- 60.Paterson R, Everett R D. The regions of the herpes simplex virus type 1 immediate early protein Vmw175 required for site specific DNA binding closely correspond to those involved in transcriptional regulation. Nucleic Acids Res. 1990;16:11005–11025. doi: 10.1093/nar/16.23.11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pereira L, Wolff M H, Fenwick M, Roizman B. Regulation of herpes virus macromolecular synthesis. V. Properties of a polypeptide made in HSV-1 and HSV-2 infected cells. Virology. 1977;77:733–749. doi: 10.1016/0042-6822(77)90495-0. [DOI] [PubMed] [Google Scholar]
- 62.Rivera-Gonzalez R, Imbalzano A N, Gu B, DeLuca N A. The role of ICP4 repressor activity in temporal expression of the IE-3 and latency-associated transcript promoters during HSV-1 infection. Virology. 1994;202:550–564. doi: 10.1006/viro.1994.1377. [DOI] [PubMed] [Google Scholar]
- 63.Roberts M S, Boundy A, O’Hare P, Pizzorno M C, Ciufo D M, Hayward G S. Direct correlation between a negative response element at the cap site of the herpes simplex virus type IE175 (α4) promoter and a specific binding site for the IE175 (ICP4) protein. J Virol. 1988;62:4307–4320. doi: 10.1128/jvi.62.11.4307-4320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rock S L, Nesburn A B, Ghiasi H, Ong J, Lewis T L, Lokensgard J R, Wechsler S L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits infected with herpes simplex virus type 1. J Virol. 1987;61:3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samaniego L A, Webb A L, DeLuca N A. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol. 1995;69:5705–5715. doi: 10.1128/jvi.69.9.5705-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwyzer M, Vlcek C, Menekse O, Fraefel C, Paces V. Promoter, spliced leader, and coding sequence for BICP4, the largest of the immediate-early proteins of bovine herpesvirus 1. Virology. 1993;197:349–357. doi: 10.1006/viro.1993.1596. [DOI] [PubMed] [Google Scholar]
- 67.Shepard, A. A., and N. A. DeLuca. Unpublished results.
- 68.Shepard A A, Imbalzano A N, DeLuca N A. Separation of primary structural components conferring autoregulation, transactivation and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J Virol. 1989;63:3714–3728. doi: 10.1128/jvi.63.9.3714-3728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith C A, Bates P, Rivera-Gonzalez R, Gu B, DeLuca N A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spivack J G, Fraser N W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987;61:3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stevens J G, Cook M L. Latent herpes simplex virus in spinal ganglia of mice. Science. 1971;173:843–845. doi: 10.1126/science.173.3999.843. [DOI] [PubMed] [Google Scholar]
- 72.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L. RNA complementary to a herpes virus a gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 73.Stevens J G. Overview of herpesvirus latency. Semin Virol. 1994;5:191–196. [Google Scholar]
- 74.Wobbe K K, Digard P, Staknis D, Coen D M. Unusual regulation of expression of the herpes simplex virus DNA polymerase gene. J Virol. 1993;67:5419–5425. doi: 10.1128/jvi.67.9.5419-5425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of the herpes virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia K, DeLuca N, Knipe D K. Analysis of phosphorylation sites of herpes simplex virus type 1 ICP4. J Virol. 1996;70:1061–1071. doi: 10.1128/jvi.70.2.1061-1071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xia K, Knipe D K, DeLuca N. Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J Virol. 1996;70:1050–1060. doi: 10.1128/jvi.70.2.1050-1060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yeh L, Schaffer P A. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J Virol. 1993;67:7373–7382. doi: 10.1128/jvi.67.12.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]