Abstract
The proteasome degrades proteins, which is essential for cellular homeostasis. Ubiquitin independent proteolysis degrades highly disordered and misfolded proteins. A decline of proteasomal activity has been associated with multiple neurodegenerative diseases due to the accumulation of misfolded proteins. In this work, cyclic peptide proteasome stimulators (CyPPSs) that enhance the clearance of misfolded proteins were discovered. In the initial screen of predicted natural products (pNPs), several cyclic peptides were found to stimulate the 20S core particle (20S CP). Development of a robust structural activity relationship led to the identification of potent, cell permeable CyPPSs. In-vitro assays revealed that CyPPSs stimulate degradation of highly disordered and misfolded proteins without affecting ordered proteins. Furthermore, using a novel flow-based assay for proteasome activity, several CyPPSs were found to stimulate the 20S CP in-cellulo. Overall, this work describes the development of CyPPSs as chemical tools capable of stimulating the proteasome and provides strong support for proteasome stimulation as a therapeutic strategy for neurodegenerative diseases.
Keywords: macrocycles, peptides, proteasome, stimulation
Graphical Abstract

Cyclic peptide proteasome stimulators: Cyclic peptides inspired by natural products activate ubiquitin independent degradation of disordered proteins. Development and utilization of a flow-based proteasome assay enabled confirmation of the activity of cyclic peptides in cells, making them valuable chemical tools for studying proteasome activation.
Introduction
The proteasome is essential for the degradation of cellular proteins1–3 and maintenance of normal cellular functions3–11. Protein degradation via the proteasome is done by either a ubiquitin-proteasome system (UPS)12–14 or a ubiquitin-independent proteasome system (UIPS)15–18. The 26S and 30S isoforms1,19,20 of the proteasome, which consist of the 19S regulator particle (19S RP) and the 20S core particle (20S CP), are responsible for UPS12,21,22 (Figure 1A). The 20S CP can also degrade proteins alone via UIPS. However, the proteins must be at least partially disordered to pass through the gate formed by the α-subunits15,23–27. Homeostasis between the UPS and UIPS is essential for the maintenance of healthy cells28. During the aging process the production of the 19S RP diminishes29,30, leaving UIPS as a major pathway for protein degradation. This decline in UPS activity, along with other mechanisms of UPS inactivation, has been associated with many neurodegenerative diseases including Huntington’s Disease31, Alzheimer’s Disease32,33, and Parkinson’s Disease34,35. These proteopathies are hypothesized to be caused by the accumulation of misfolded proteins36–38. Literature suggests that the associated proteins (e.g. α-synuclein and tau) can be degraded by the proteasome23. However, the UIPS is not strongly activated, limiting its ability to clear these proteins. For this reason, there is an effort to identify UIPS activators that can increase the rate of clearance of misfolded proteins. Multiple small molecules and a handful of peptides (Figure 1B) have been observed to stimulate the proteasome8,39–43. While these small molecules and peptides have been useful for answering foundational questions about the mechanism of proteasome stimulation39,40,44,45, the majority of proteasome stimulators reported to date either have low potency (e.g. AM-404 and ursolic acid), are not selective (e.g. cyclosporine), or have limited activity in cell-based assays (e.g. betulinic acid, chloropromazine, and MK-866). Target-engagement studies with these molecules have also proven challenging, with little-to-no evidence of their direct interaction with the proteasome. A cyclic peptide stimulator could provide regions to easily append cross-linking moieties or other pull-down handles for these future important studies.
Figure 1.
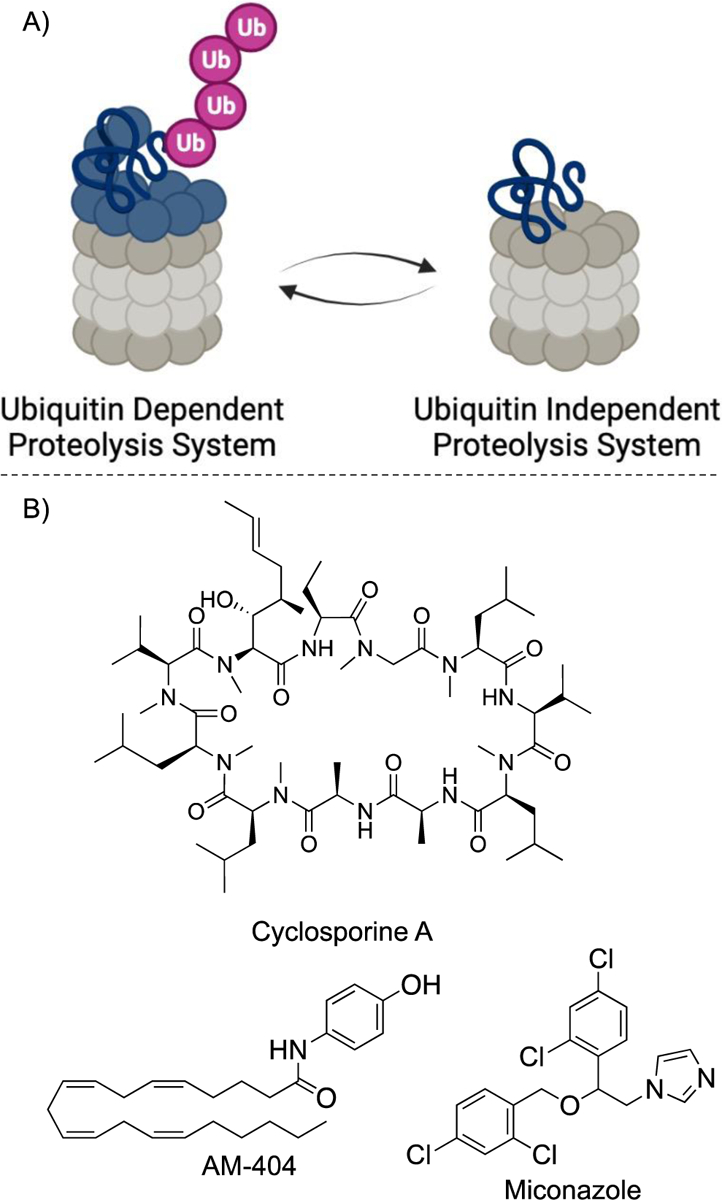
A) The 26S proteasome (left) consists of the 19S regulatory particle (blue) and the 20S core particle (gray). Ubiquitin independent proteolysis (right) only consists of the 20S core particle. B) Known 20S core particle proteasome stimulators.
Recently, we reported synthetic natural product inspired cyclic peptides (SNaPP), a useful tool for predicting novel natural product-like molecules with biological activity46. Given that linear peptides have previously been identified as proteasome stimulators, we hypothesized that our cyclic peptide predicted natural products (pNPs) might be capable of stimulating the 20S CP. Cyclic peptides have several advantages over linear peptides, including their generally greater stability to proteases47–49, their improved cell permeability47–51, and their rigidity47–49,52,53, which allows for increased affinity for their targets. Described herein, we utilized pNPs in our previously developed in vitro proteasome stimulation assay41 and found several cyclic pNPs that efficiently stimulate the 20S CP. Derivatives were synthesized to evaluate any structure activity relationship and revealed more potent cyclic pNPs. In vitro protein degradation assays revealed that these molecules stimulate proteasomal degradation of disordered proteins (e.g. α-synuclein) while having little to no effect on ordered proteins (e.g. lysozyme and GAPDH). Finally, cell-based assays revealed cyclic pNPs stimulate the 20S CP.
Results and Discussion
Development of Cyclic Peptide Proteasome Stimulators
The TAS-1 biochemical assay54 was utilized to analyze the 20S CP proteasome stimulation. This assay relies on a fluorescent-based probe, which incorporates a rhodium fluorescent probe conjugated to a proteasome peptide substrate on one side and a peptoid for solubility on the other. Active proteasome cleaves the peptide, resulting in fluorescence of the rhodium probe. This enables monitoring of the real-time activity of the proteasome, with greater intensity of fluorescence corresponding to higher proteasome activity.
Utilizing the TAS-1 biochemical assay, we screened a library of 45 cyclic peptides that were generated by SNaPP46 (Figure S1). In our library, we had nine peptides that stimulated the proteasome 20% more than the DMSO control, giving us a hit rate of 20%. This is in stark contrast to many other proteasome stimulator screens, which typically have hit rates of less than 1%40,55. Of the hits identified in the initial screen, six cyclic peptides stimulated the proteasome greater than 140%. These top compounds all contained an arginine, a polar uncharged amino acid, and an aromatic amino acid. Furthermore, four of the six with the highest percent stimulation contained six amino acids (Figure 2). Analogous to the peptides in Figure 2, many peptide proteasome stimulators have an arginine56. Another common motif throughout peptide proteasome stimulators is a tyrosine, which has shown to open or stabilize the gate of the 20S CP45,57. Although, none of the top pNP stimulators have tyrosine, they do have a conserved phenylalanine, which may be acting similarly.
Figure 2.
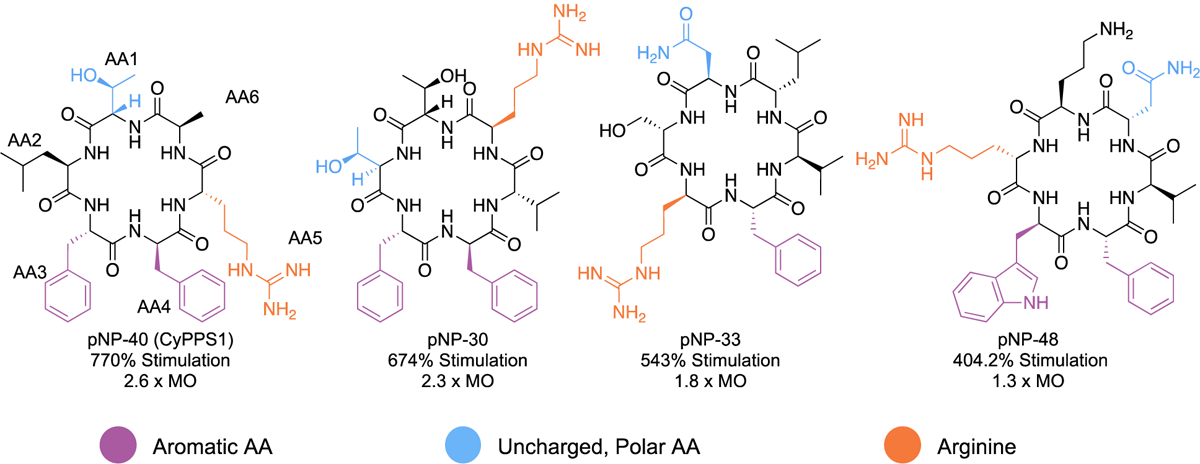
Structural activity relationship of the initial screen of the SNaPP library. Each peptide is a hexamer that consists of at least one aromatic amino acid (purple), uncharged polar amino acid (blue), and arginine (orange).
Because pNP-40 (renamed Cyclic Peptide Proteasome Stimulator 1, CyPPS1) had the highest percent stimulation of the identified hits, we decided to pursue derivatives of CyPPS1 to develop a structural activity relationship (SAR). An alanine scan was first performed to determine the residues that were necessary for activity (CyPPS2–6, Figure 3A). Each amino acid was substituted with an alanine, except for arginine. Unfortunately, the derivative where arginine is replaced with alanine is insoluble and thus could not be evaluated. For this reason, we utilized a derivative that switched the position of arginine with alanine (CyPPS6). The stimulatory activity for CyPPS6 was preserved, suggesting that the position of the arginine is not essential. When each amino acid was substituted with an alanine, the activity decreased, suggesting that all the amino acids are essential for the stimulatory affect (Figure 3A–B). Specifically, replacing either the D-leucine (CyPPS3) or the D-phenylalanine (CyPPS5) with D-alanine resulted in the greatest loss of stimulatory activity, suggesting that these amino acids are particularly important for activity. Additionally, a linear version of CyPPS1 (PPS1) was explored and found to have little-to-no stimulatory effect. This provides strong evidence for the necessity of cyclizing these peptides for activity and suggests that cyclization likely holds them in an active conformation.
Figure 3.
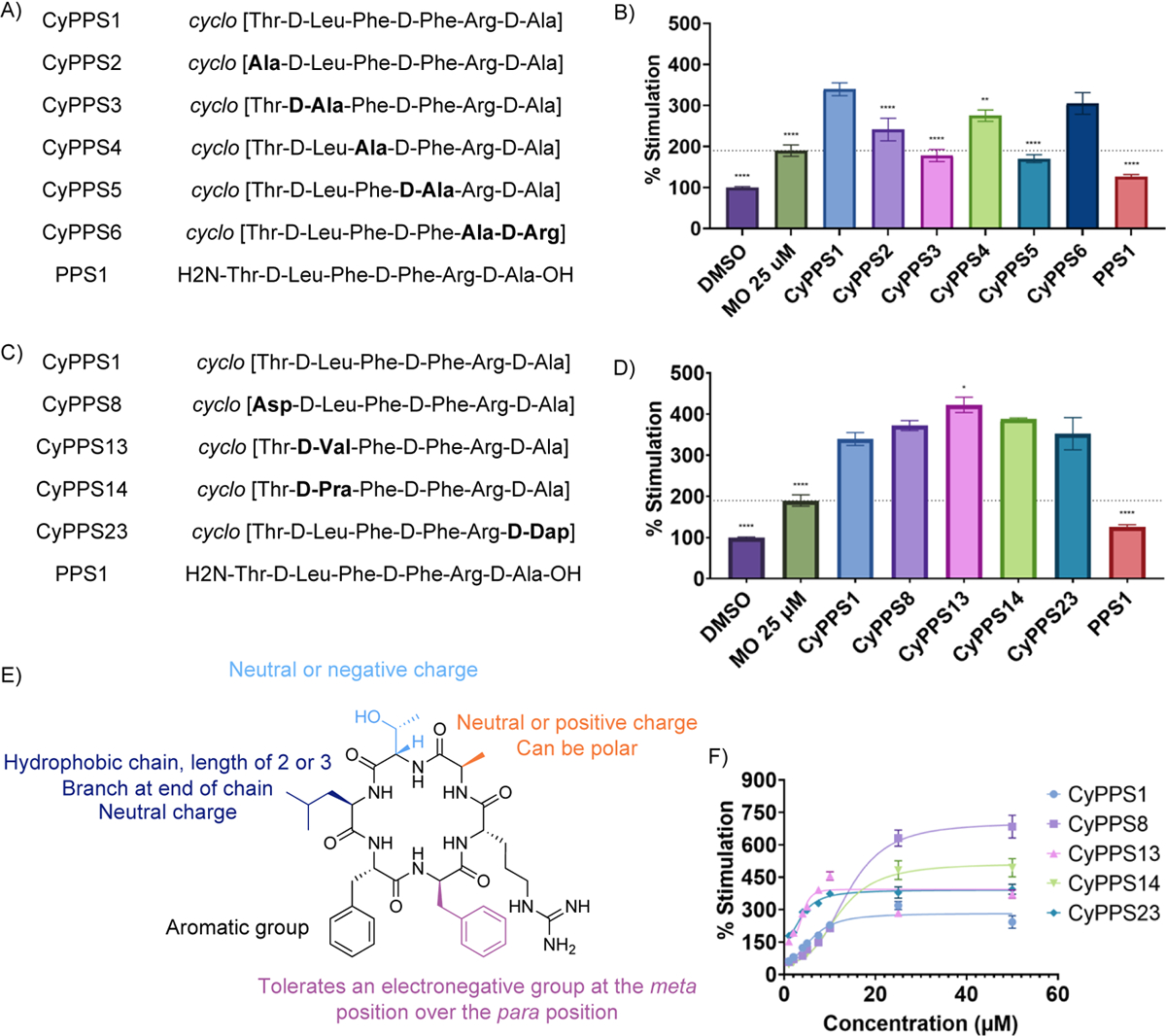
A) Structures of the peptides in Figure 3B. B) TAS-1 20S CP stimulation of the alanine scan of CyPPS1. All compounds except miconazole (MO) were tested at 10 μM. 2-way ANOVA analysis performed comparing to CyPPS1 (n=3 with S.E.M. indicated) **** P<0.0001, *** P<0.001, ** P<0.01, * P<0.05, and ns P > 0.05. Average of 3 independent replicates. Abbreviations: MO = miconazole. C) Structures of the peptides in Figure 3D. D) TAS-1 20S CP stimulation of the top five 20S CP stimulators and the linear of CyPPS1. All compounds except MO were tested at 10 μM. 2-way ANOVA analysis performed comparing to CyPPS1 (n=3 with S.E.M. indicated) **** P<0.0001, *** P<0.001, ** P<0.01, * P<0.05, and ns P > 0.05. Average of 3 independent replicates. Abbreviations: MO = miconazole. E) Structural activity relationship of CyPPS1. F) Dose response curve of the top 5 20S CP stimulators.
The SAR was further explored by substituting other amino acids at each position (Figure 3C–E, Table S1–2). The threonine at position 1 was substituted with lysine (CyPPS7), aspartic acid (CyPPS8), and valine (CyPPS9). The lysine substitution decreased activity, suggesting basic amino acids are not tolerated at position 1. Both CyPPS8 and CyPPS9 maintained stimulatory activity, suggesting that position 1 tolerates both acidic and aliphatic amino acids, in addition to the polar threonine. The D-leucine at position 2 was substituted with D-aspartic acid (CyPPS10), D-lysine (CyPPS11), D-isoleucine (CyPPS12), and D-valine (CyPPS13). Only CyPPS13 retained activity, with the other derivatives having decreased activity. While the lack of activity with CyPPS12 is surprising, these results overall suggest that an aliphatic amino acid with branching is preferred. Position 2 was also substituted with D-propargylglycine (CyPPS14), which also retained stimulatory activity, supporting the necessity for an aliphatic side chain. All substitutions at position 3 decreased activity except for 4-fluorophenylalanine (CyPPS15). Tyrosine (CyPPS16/CyPPS17) was also substituted for position 3 and 4. Interestingly, a decline in stimulatory activity was observed, indicating CyPPSs are likely not acting at the same location as the previously studied tyrosine-containing peptides. Substitutions of D-4-fluorophenylalanine (CyPPS18) at position 4 decreased activity. Other substitutions such as D-3-flurophenylalanine (CyPPS19) and D-3,4-difluorophenylalanine (CyPPS20) retained activity, suggesting that electron-withdrawing groups are tolerated. We also substituted this position for benzophenone (CyPPS21) and found that the 20S CP stimulatory activity was maintained. Substitution of the D-alanine at position 6 with D-serine (CyPPS22) and D-diaminopropionic acid (CyPPS23) maintained stimulatory activity, suggesting that small polar substitutions are tolerated. We were unable to substitute position 5 with any amino acids other than basic amino acids due to solubility issues. The basic amino acid diaminopropionic acid (CyPPS24) maintained stimulatory activity. Finally, a cyclic pentapeptide (CyPPS25) was explored but had decreased activity, demonstrating the importance of ring size for activity. Of the 37 derivatives tested, 4 derivatives were found to have similar or increased stimulatory activity compared to the parent CyPPS1 (Figure 3C–D). These were selected to undergo further testing.
Dose response and activity with purified proteins
Initially, the dose-response relationship was determined for the top five hits (Figure 3F). CyPPS13 and CyPPS23 had the lowest EC50 values (4.0 μM and 4.1 μM, respectively), suggesting they are the most potent compounds. However, their maximal response was lower compared to CyPPS8 and CyPPS14 (Table S3 for Emax and EC50 values). While the reasoning for the differences in EC50 and Emax for these molecules is currently unclear, the structural similarity of the molecules suggests they likely have similar binding sites.
Given the potency of the CyPPSs in the TAS-1 assay, we chose to explore their abilities to degrade proteins using an in vitro degradation assay. The 20S CP typically degrades proteins that are highly disordered. To ensure the selectivity of the CyPPSs for disordered proteins, the ability of these molecules to induce degradation of disordered proteins (e.g. α-synuclein) and ordered proteins (e.g. GAPDH and lysosyme) was explored. CyPPS1, CyPPS14, and CyPPS23 greatly enhanced proteasomal degradation of α-synuclein while having little-to-no effect on GAPDH and lysozyme (Figure 4 and Figure S2). The lack of activity of CyPPS8 in this assay is unsurprising given that molecules were tested at 10 μM, a concentration where CyPPS8 has little effect (Figure 3F). Overall, this suggests that CyPPS1, CyPPS14, and CyPPS23 are excellent leads for proteasome stimulation. The top hits were also tested for toxicity in mammalians cells. Based on their selectivity for disordered proteins, we would not expect these molecules to be toxic. Gratifyingly, no toxicity was observed in HEK293 cells (Table S2). Additionally, no hemolysis was observed with human red blood cells, further supporting these as excellent lead molecules (Table S2).
Figure 4.
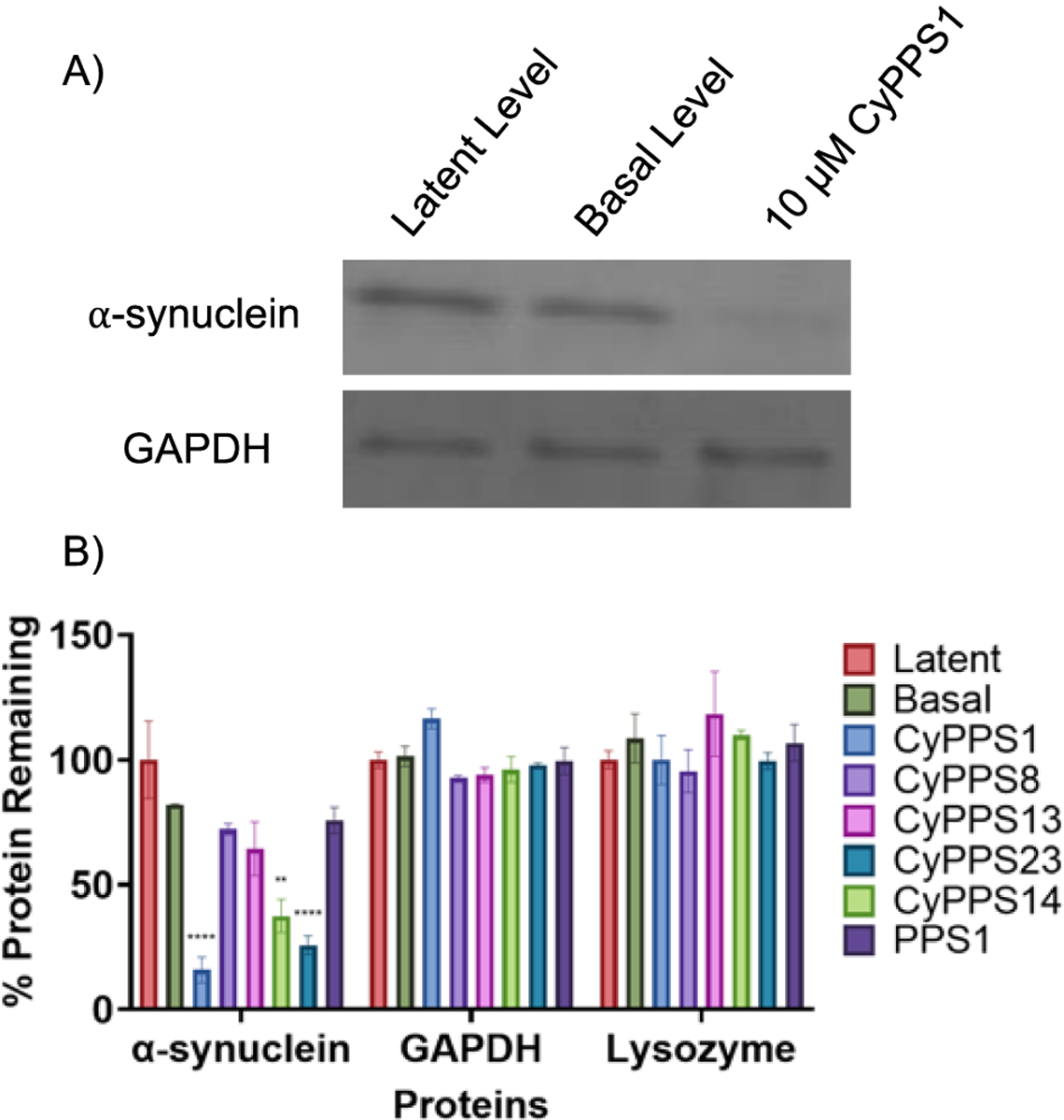
A) Coomassie gel (representative of 3 independent replicates) and B) quantification of the degradation of highly disordered (⍺-synuclein) and low disordered (GAPDH and lysozyme) proteins. All compounds were tested at 10 μM. 2-way ANOVA performed comparing to basal activity (n=3 with S.E.M. indicated) **** P<0.0001, *** P<0.001, ** P<0.01, * P<0.05, and ns P > 0.05. Average of 3 independent replicates.
Cell-Based Activity
While many cyclic peptides are cell permeable, it is challenging to predict a priori the peptides that are capable of entering cells. Because the proteasome is an intracellular target, we decided to investigate the ability of the CyPPSs to enter cells. To do this, a BODIPY-tagged CyPPS (CyPPS26) was synthesized from the alkyne derivative CyPPS14 (SI Scheme 2). A549 cells were then dosed with CyPPS26 and analyzed for cell permeability via confocal microscopy. Furthermore, we investigated the mechanism by which CyPPS26 was entering the cell. Cellular uptake of cyclic peptides usually occurs either via passive diffusion58,59 or endocytosis60–65. The puncta present suggest that CyPPSs may enter the cell via endosomal uptake. This possibility was further supported by the overlap of the BODIPY and LysoTracker signal (Figure 5). However, further experimentation is needed to confirm this. As some cytosolic fluorescence is observed, it is hypothesized that at least some of the CyPPSs are capable of escaping the endosome, and thus likely capable of engaging with the cytosolic proteasome. However, it does appear like the majority of the peptide remains within the endosome, at least at this timepoint. This is an area that will be optimized in the future. Cytosolic protein accumulation is linked to many neurodegenerative diseases, including α-synuclein accumulation in Parkinson’s disease66. Given that CyPPSs can degrade highly disordered proteins and are cell permeable, we chose to investigate their abilities to stimulate the proteasome in cellulo using a newly established flow cytometry assay.
Figure 5.
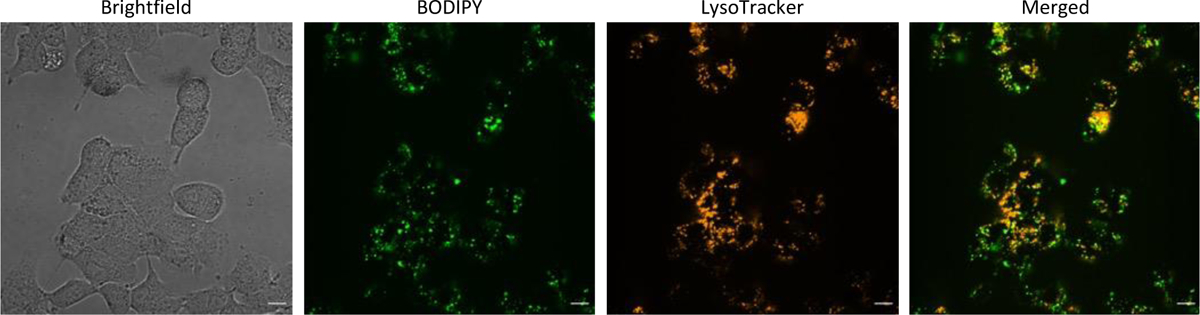
Cell permeability and mechanism of cell uptake of CyPPSs was determined with 15 μM of CyPPS26 in A549 cells after 1.5 hours (representative of 3 independent replicates). Scale bar is 10 μm.
Flow cytometry allows for the study of proteasomal activity in physiologically relevant conditions and presents a high-throughput alternative to traditional gel electrophoresis and western blotting techinques67–69. To test the assay, a covalent fluorescent-based probe70,71 was applied in HEK293T cells to quantify proteasome stimulation of CyPPSs, a known proteasome stimulator (miconazole [MO]) and proteasome inhibitor (MG132) (Figure S3). The covalent fluorescent-based probe is based on the proteasome inhibitor epoxomicin, which interacts with the β5 subunit of the proteasome, with a fluorophore appended to the N-termini. Upon incubation with the proteasome, it forms a covalent bond, thus fluorescently tagging the proteasome72. This probe has previously been shown to act in cells, enabling the current flow-based studies with higher fluorescence indicating higher proteasome activation. The addition of CyPPSs induces a significant shift in the intracellular fluorescence (Figure 6A). This shift validates that CyPPSs are capable of stimulating cytosolic proteasome in cellulo. Interestingly, the same compounds that degrade purified α-synuclein (CyPPS1, CyPPS14, and CyPPS23) show significant cellular stimulation of the proteasome. However, their activity is lower than what would be expected based on their in vitro activity. This may be due to incomplete cellular uptake or escape from the endosomes. Unsurprisingly, the linear PPS1 does not stimulate the proteasome. This is likely due to a combination of effects including the inability to stimulate the proteasome (Figure 3B–D) as well as the generally poor cell permeability and proteolytic stability of linear peptides.
Figure 6.
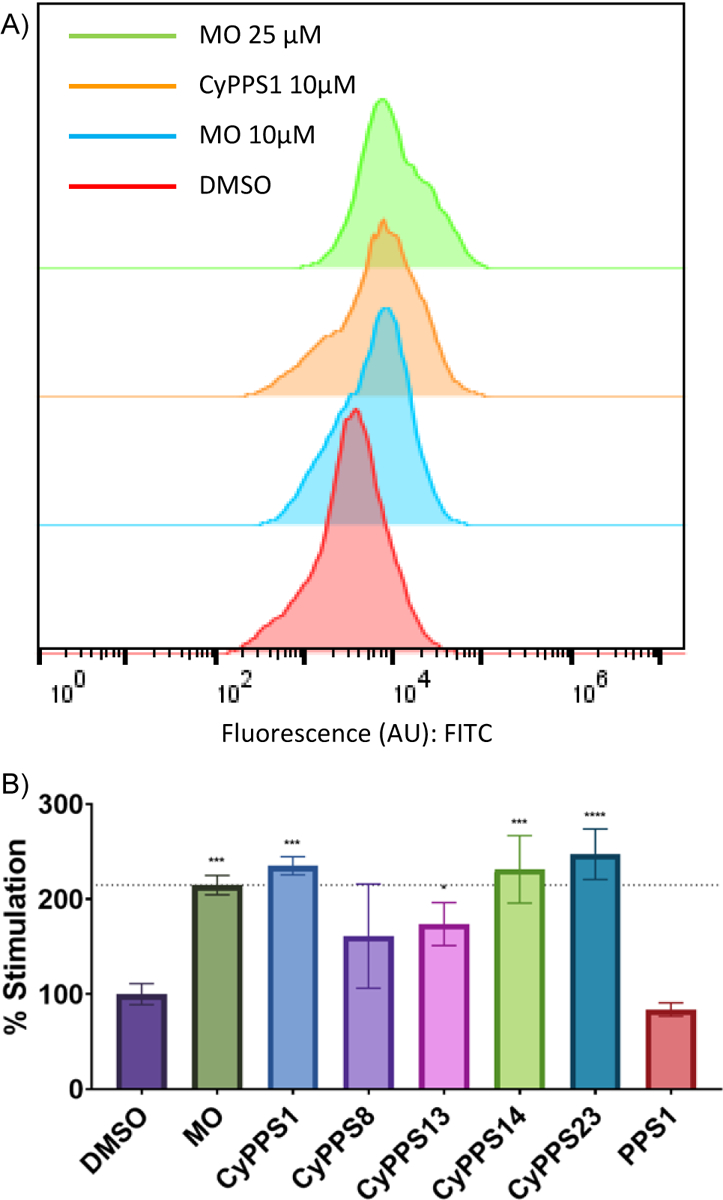
A) Flow cytometry plots showing shifts in fluorescence signal (FITC) in HEK293 T cells (representative of 3 independent replicates). Abbreviations: MO = miconazole. B) Flow cytometry analysis of CyPPSs at 10 μM in HEK293 T cells. 2-way ANOVA performed ((n=3 with S.E.M. indicated) **** P<0.0001, *** P<0.001, ** P<0.01, *<0.05, and ns P > 0.05. Average of 3 independent replicates. P<0.05, and ns P > 0.05. Average of 3 independent replicates. Abbreviations: MO = miconazole.
Conclusions
Described herein, CyPPSs were developed as stimulators of UIPS. Exploration of derivatives enabled development of a robust SAR, resulting in identification of three CyPPSs that selectively degrade highly disordered proteins (i.e. α-synuclein). CyPPSs were found to enter the cell via endocytosis and demonstrated significant endosomal escape into the cytosol. A flow-based proteasome stimulation assay—which allows for high throughput evaluation of proteasome stimulation—was developed and revealed that CyPPSs efficiently stimulate the proteasome in cells. Overall, the CyPPSs are promising leads for potent, cell-active proteasome stimulators.
Supplementary Material
Acknowledgments
We would like to acknowledge the Purdue Live Cell Imaging Facility and E. A. David for their assistance with the confocal microscopy experiments, the Purdue Research Instrumentation Center for their assistance with the mass spectrometry, the Purdue Department of Chemistry Cell Culture Facility and M. Alley for their assistance with cell culture, and A. F. Salazar-Chaparro for their assistance with biochemical experiments. This work was supported by a grant from the National Institutes of Health (1R35GM138002-01 to E.I.P. and R01AI150847 to D.J.T. and F31CA247327 to C.S.M.) and the Frederick N. Andrews Fellowship from the Department of Medicinal Chemistry and Molecular Pharmacology at Purdue University (to S.N.). The authors acknowledge the support from the Purdue Center for Cancer Research, NIH grant P30 CA023168.
Footnotes
Conflicts of interest.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Prof. Trader is a shareholder and consultant for Booster Therapeutics, GmbH. Other author declares no conflict of interest.
References
- (1).Coux O; Tanaka K; Goldberg AL Structure and Functions of the 20S and 26S Proteasomes. Ann. Rev. Biochem 1996, 65 (1), 801–847. 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- (2).Rock KL; Gramm C; Rothstein L; Clark K; Stein R; Dick L; Hwang D; Goldberg AL Inhibitors of the Proteasome Block the Degradation of Most Cell Proteins and the Generation of Peptides Presented on MHC Class I Molecules. Cell 1994, 78 (5), 761–771. 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- (3).Ciechanover A The Ubiquitin-Proteasome Proteolytic Pathway. Cell 1994, 79 (1), 13–21. 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- (4).Fenteany G; Schreiber SL Lactacystin, Proteasome Function, and Cell Fate. Journal of Biological Chemistry 1998, 273 (15), 8545–8548. 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- (5).Lee DH; Goldberg AL Proteasome Inhibitors: Valuable New Tools for Cell Biologists. Trends Cell Biol 1998, 8 (10), 397–403. 10.1016/S0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- (6).Naujokat C; Hoffmann S Role and Function of the 26S Proteasome in Proliferation and Apoptosis. Laboratory Investigation 2002, 82 (8), 965–980. 10.1097/01.lab.0000022226.23741.37. [DOI] [PubMed] [Google Scholar]
- (7).Goldberg AL Functions of the Proteasome: From Protein Degradation and Immune Surveillance to Cancer Therapy. Biochem Soc Trans 2007, 35 (1), 12–17. 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- (8).Huang L; Chen CH Proteasome Regulators: Activators and Inhibitors. Curr. Med. Chem 2009, 16 (8), 931–939. 10.2174/092986709787581860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hochstrasser M Ubiquitin, Proteasomes, and the Regulation of Intracellular Protein Degradation. Curr Opin Cell Biol 1995, 7 (2), 7915–223. 10.1016/0955-0674(95)80031-X. [DOI] [PubMed] [Google Scholar]
- (10).Jentsch S The Ubiquitin-Conjugation System. Annu. Rev. Genet 1992, 26 (1), 179–207. https://doi.org/10.1146.annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- (11).Isaksson A; Musti AM; Bohmann D Ubiquitin in Signal Transduction and Cell Transformation. Biochim Biophys Acta 1996, 1288 (1), 21–29. 10.1016/0304-419X(96)00011-X. [DOI] [PubMed] [Google Scholar]
- (12).Nandi D; Tahiliani P; Kumar A; Chandu D The Ubiquitin-Proteasome System. J. Biosci 2006, 31 (1), 137–155. 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- (13).Ciechanover A; Schwartz AL The Ubiquitin-Proteasome Pathway: The Complexity and Myriad Functions of Proteins Death. Proc. natl. Acad. Sci. USA 1998, 95 (6), 2727–2730. 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Finley D; Ciechanover A; Varshavsky A Thermolability of Ubiquitin-Activating Enzyme from the Mammalian Cell Cycle Mutant Ts85. Cell 1984, 37 (1), 43–55. 10.1016/0092-8674(84)90299-X. [DOI] [PubMed] [Google Scholar]
- (15).Ben-Nissan G; Sharon M Regulating the 20S Proteasome Ubiquitin-Independent Degradation Pathway. Biomolecules 2014, 4 (3), 862–884. 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Erales J; Coffino P Ubiquitin-Independent Proteasomal Degradation. Biochimica et Biophysica Acta 2014, 1843 (1), 216–221. 10.1016/J.BBAMCR.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Opoku-Nsiah KA; Gestwicki JE Aim for the Core: Suitability of the Ubiquitin-Independent 20S Proteasome as a Drug Target in Neurodegeneration. Translational Research 2018, 198, 48–57. 10.1016/J.TRSL.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hoyt MA; Coffino P Ubiquitin-Free Routes into the Proteasome. Mol. Life Sci 2004, 61, 1596–1600. 10.1007/s00018-004-4133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Tanaka K The Proteasome: Overview of Structure and Functions. Proc. Jpn. Acad., Ser. B 2009, 85 (1), 12–36. 10.2183/PJAB.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Adams J The Proteasome: Structure, Function, and Role in the Cell. Cancer Treat Rev 2003, 29, 3–9. 10.1016/S0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- (21).Glickman MH; Ciechanover A The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol Rev 2002, 82 (2), 373–428. 10.1152/PHYSREV.00027.2001. [DOI] [PubMed] [Google Scholar]
- (22).Hoffman L; Pratt G; Rechsteiner M Multiple Forms of the 20 S Multicatalytic and the 26 S Ubiquitin/ATP- Dependent Proteases from Rabbit Reticulocyte Lysate. Journal of Biological Chemistry 1992, 267 (31), 22362–22368. 10.1016/s0021-9258(18)41680-8. [DOI] [PubMed] [Google Scholar]
- (23).Coleman RA; Mohallem R; Aryal UK; Trader DJ Protein Degradation Profile Reveals Dynamic Nature of 20S Proteasome Small Molecule Stimulation. RCS Chemical Biology 2021, 2 (2), 636–644. 10.1039/d0cb00191k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Pickering AM; Davies KJA Degradation of Damaged Proteins: The Main Function of the 20S Proteasome. Prog Mol Biol Transl Sci 2012, 109, 227–248. 10.1016/B978-0-12-397863-9.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Davies KJ A. Degradation of Oxidized Proteins by the 20S Proteasome. Biochimie 2001, 83 (3–4), 301–310. 10.1016/S0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- (26).Grune T; Catalgol B; Licht A; Ermak G; Pickering AM; Ngo JK; Davies KJA HSP70 Mediates Dissociation and Reassociation of the 26S Proteasome during Adaptation to Oxidative Stress. Free Radic Biol Med 2011, 51 (7), 1355–1364. 10.1016/J.FREERADBIOMED.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Aiken CT; Kaake RM; Wang X; Huang L Oxidative Stress-Mediated Regulation of Proteasome Complexes. Molecular & Cellular Proteomics 2011, 10 (5). 10.1074/MCP.M110.006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Coleman RA; Trader DJ All about the Core: A Therapeutic Strategy to Prevent Protein Accumulation with Proteasome Core Particle Stimulators. ACS Pharmacol Transl Sci 2018, 1 (2), 140–142. 10.1021/ACSPTSCI.8B00042/ASSET/IMAGES/LARGE/PT-2018-00042Y_0002.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Huang Q; Figueiredo-Pereira ME Ubiquitin/Proteasome Pathway Impairment in Neurodegeneration: Therapeutic Implications. Apoptosis 2010, 15, 1292–1311. 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Husom AD; Peters EA; Kolling EA; Fugere NA; Thompson LV; Ferrington DA Altered Proteasome Function and Subunit Composition in Aged Muscle. Arch Biochem Biophys 2004, 421 (1), 67–76. 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- (31).Arrasate M; Finkbeiner S Protein Aggregates in Huntington’s Disease. 2011. 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gadhave K; Bolshette N; Ahire A; Pardeshi R; Thakur K; Trandafir C; Istrate A; Ahmed S; Lahkar M; Muresanu DF; Balea M The Ubiquitin Proteasomal System: A Potential Target for the Management of Alzheimer’s Disease. J Cell Mol Med 2016, 20 (7), 1392–1407. 10.1111/JCMM.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Palmieri G; Cocca E; Gogliettino M; Valentino R; Ruvo M; Cristofano G; Angiolillo A; Balestrieri M; Rossi M; Di Costanzo A Low Erythrocyte Levels of Proteasome and Acyl-Peptide Hydrolase (APEH) Activities in Alzheimer’s Disease: A Sign of Defective Proteostasis? Journal of Alzheimer’s Disease 2017, 60 (3), 1097–1106. 10.3233/JAD-170389. [DOI] [PubMed] [Google Scholar]
- (34).McNaught K. St. P.; Belizaire R; Isacson O; Jenner P; Olanow WC Altered Proteasomal Function in Sporadic Parkinson’s Disease. Exp Neurol 2003, 179 (1), 38–46. 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- (35).McNaught K St. P.; Jenner, P. Proteasomal Function Is Impaired in Substantia Nigra in Parkinson’s Disease. Neurosci Lett 2001, 297, 191–194. 10.1016/S0304-3940(00)01701-8. [DOI] [PubMed] [Google Scholar]
- (36).Holmes BB; Furman JL; Mahan TE; Yamasaki TR; Mirbaha H; Eades WC; Belaygorod L; Cairns NJ; Holtzman DM; Diamond MI Proteopathic Tau Seeding Predicts Tauopathy in Vivo. PNAS 2014, 111 (41), E4376–E4385. 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Abbasbeigi S Misfolded Structures | A Brief Insight into Protein Aggregation Criteria, Which May Lead to Proteopathy Diseases. Journal of Chemical Reviews 2021, 3 (1), 97–108. 10.22034/JCR.2020.256367.1092. [DOI] [Google Scholar]
- (38).Jucker M; Walker LC Pathogenic Protein Seeding in Alzheimer Disease and Other Neurodegenerative Disorders. Ann Neurol 2011, 70 (4), 532–540. 10.1002/ANA.22615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Halder S; Macatangay NJ; Zerfas BL; Salazar-Chaparro AF; Trader DJ Oleic Amide Derivatives as Small Molecule Stimulators of the Human Proteasome’s Core Particle. RCS Medicinal Chemistry 2022, 13 (9), 1077–1081. 10.1039/d2md00133k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jones CL; Njomen E; Sjö B; Dexheimer TS; Tepe JJ Small Molecule Enhancement of 20S Proteasome Activity Targets Intrinsically Disordered Proteins. ACS Chem Biol 2017, 12 (9), 2240–2247. 10.1021/acschembio.7b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Coleman RA; Trader DJ Development and Application of a Sensitive Peptide Reporter to Discover 20S Proteasome Stimulators. ACS Combinational Science 2018, 20 (5), 269–276. 10.1021/acscombsci.7b00193. [DOI] [PubMed] [Google Scholar]
- (42).Leestemaker Y; de Jong A; Witting KF; Penning R; Schuurman K; Rodenko B; Zaal EA; van de Kooij B; Laufer S; Heck AJR; Borst J; Scheper W; Berkers CR; Ovaa H Proteasome Activation by Small Molecules. Cell Chem Biol 2017, 24 (6), 725–736. 10.1016/J.CHEMBIOL.2017.05.010. [DOI] [PubMed] [Google Scholar]
- (43).Njomen E; Tepe JJ Proteasome Activation as a New Therapeutic Approach To Target Proteotoxic Disorders. J Med Chem 2019, 62 (14), 6469–6481. 10.1021/acs.jmedchem.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Dal Vechio FH; Cerqueira F; Augusto O; Lopes R; Demasi M Peptides That Activate the 20S Proteasome by Gate Opening Increased Oxidized Protein Removal and Reduced Protein Aggregation. Free Radic Biol Med 2014, 67, 304–313. 10.1016/J.FREERADBIOMED.2013.11.017. [DOI] [PubMed] [Google Scholar]
- (45).Witkowska J; Giżyńska M; Grudnik P; Golik P; Karpowicz P; Giełdoń A; Dubin G; Jankowska E Crystal Structure of a Low Molecular Weight Activator Blm-Pep with Yeast 20S Proteasome-Insights into the Enzyme Activation Mechanism. Sci Rep 2017, 7 (1), 1–11. 10.1038/s41598-017-05997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hostetler MA; Smith C; Nelson S; Budimir Z; Modi R; Woolsey I; Frerk A; Baker B; Gantt J; Parkinson EI Synthetic Natural Product Inspired Cyclic Peptides. ACS Chem Biol 2021, 16 (11), 2604–2611. 10.1021/acschembio.1c00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Malde AK; Hill TA; Iyer A; Fairlie DP Crystal Structures of Protein-Bound Cyclic Peptides. Chem Rev 2019, 119 (17), 9861–9914. 10.1021/ACS.CHEMREV.8B00807. [DOI] [PubMed] [Google Scholar]
- (48).Zhang H; Zhao Q; Bhattacharya S; Waheed AA; Tong X; Hong A; Heck S; Curreli F; Goger M; Cowburn D; Freed EO; Debnath AK A Cell-Penetrating Helical Peptide as a Potential HIV-1 Inhibitor. J Mol Biol 2008, 378 (3), 565–580. 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Tsomaia N Peptide Therapeutics: Targeting the Undruggable Space. Eur J Med Chem 2015, 94, 459–470. 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- (50).Verdine GL; Hilinski GJ All-Hydrocarbon Stapled Peptides as Synthetic Cell-Accessible Mini-Proteins. Drug Discov Today Technol 2012, 9 (1), e41–e47. 10.1016/j.ddtec.2012.01.004. [DOI] [PubMed] [Google Scholar]
- (51).Karatas H; Li Y; Liu L; Ji J; Lee S; Chen Y; Yang J; Huang L; Bernard D; Xu J; Townsend EC; Cao F; Ran X; Li X; Wen B; Sun D; Stuckey JA; Lei M; Dou Y; Wang S Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)–Mixed Lineage Leukemia (MLL) Protein–Protein Interaction. J Med Chem 2017, 60 (12), 4818–4839. 10.1021/acs.jmedchem.6b01796. [DOI] [PubMed] [Google Scholar]
- (52).Hill TA; Shepherd NE; Diness F; Fairlie DP Constraining Cyclic Peptides To Mimic Protein Structure Motifs. Angewandte Chemie International Edition 2014, 53 (48), 13020–13041. 10.1002/anie.201401058. [DOI] [PubMed] [Google Scholar]
- (53).Fairlie DP; Abbenante G; March DR Macrocyclic Peptidomimetics Forcing Peptides into Bioactive Conformations. Curr Med Chem 1995, 2 (2), 654–686. 10.2174/0929867302666220218001506. [DOI] [Google Scholar]
- (54).Zerfas BL; Coleman RA; Salazar-Chaparro AF; Macatangay NJ; Trader DJ Fluorescent Probes with Unnatural Amino Acids to Monitor Proteasome Activity in Real-Time. ACS Chem Biol 2020, 15 (9), 2588–2596. 10.1021/acschembio.0c00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Trader DJ; Simanski S; Dickson P; Kodadek T Establishment of a Suite of Assays That Support the Discovery of Proteasome Stimulators. Biochim Biophys Acta 2017, 1861 (4), 892–899. 10.1016/J.BBAGEN.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Osmulski PA; Karpowicz P; Jankowska E; Bohmann J; Pickering AM; Gaczynska M New Peptide-Based Pharmacophore Activates 20S Proteasome. Molecules 2020, 25 (6), 1439. 10.3390/MOLECULES25061439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Rabl J; Smith DM; Yu Y; Chang SC; Goldberg AL; Cheng Y Mechanism of Gate Opening in the 20S Proteasome by the Proteasomal ATPases. Mol Cell 2008, 30 (3), 360–368. 10.1016/J.MOLCEL.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Smolewski P; Robak T The Discovery and Development of Romidepsin for the Treatment of T-Cell Lymphoma. Expert Opin Drug Discov 2017, 12 (8), 859–873. 10.1080/17460441.2017.1341487. [DOI] [PubMed] [Google Scholar]
- (59).Faulds D; Goa KL; Benfield P Cyclosporin: A Review of Its Pharmacodynamic and Pharmacokinetic Properties, and Therapeutic Use in Immunoregulatory Disorders. Drugs 1993, 45 (6), 953–1040. 10.2165/00003495-199345060-00007. [DOI] [PubMed] [Google Scholar]
- (60).Traboulsi H; Larkin H; Bonin M-A; Volkov L; Lavoie CL; Marsault E Macrocyclic Cell Penetrating Peptides: A Study of Structure-Penetration Properties. Bioconjug Chem 2015, 26 (3), 405–411. 10.1021/acs.bioconjchem.5b00023. [DOI] [PubMed] [Google Scholar]
- (61).Mandal D; Shirazi AN; Parang K Cell-Penetrating Homochiral Cyclic Peptides as Nuclear-Targeting Molecular Transporters. Angewandte Chemie International Edition 2011, 50 (41), 9633–9637. 10.1002/ANIE.201102572. [DOI] [PubMed] [Google Scholar]
- (62).Qian Z; Liu T; Liu Y-Y; Briesewitz R; Barrios AM; Jhiang SM; Pei D Efficient Delivery of Cyclic Peptides into Mammalian Cells with Short Sequence Motifs. ACS Chem Biol 2012, 8 (2), 423–431. 10.1021/cb3005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Qian Z; Martyna A; Hard RL; Wang J; Appiah-Kubi G; Coss C; Phelps MA; Rossman JS; Pei D Discovery and Mechanism of Highly Efficient Cyclic Cell-Penetrating Peptides. Biochemistry 2016, 55 (18), 2601–2612. 10.1021/acs.biochem.6b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Qian Z; Larochelle JR; Jiang B; Lian W; Hard RL; Selner NG; Luechapanichkul R; Barrios AM; Pei D Early Endosomal Escape of a Cyclic Cell-Penetrating Peptide Allows Effective Cytosolic Cargo Delivery. Biochemistry 2014, 53 (24), 4034–4046. 10.1021/bi5004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Liu T; Liu Y; Kao H-Y; Pei D Membrane Permeable Cyclic Peptidyl Inhibitors against Human Peptidylprolyl Isomerase Pin1. J. Med. Chem 2010, 53 (6), 2494–2501. 10.1021/jm901778v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Stefanis L α-Synuclein in Parkinson’s Disease. Cold Spring Harb Perspect Med 2012, 2 (2), a009399. 10.1101/CSHPERSPECT.A009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Berkers CR; Van Leeuwen FWB; Groothuis TA; Peperzak V; Van Tilburg EW; Borst J; Neefjes JJ; Ovaa H Profiling Proteasome Activity in Tissue with Fluorescent Probes. Molecular Pharmaceuticals 2007, 4 (5), 739–748. 10.1021/mp0700256. [DOI] [PubMed] [Google Scholar]
- (68).Gilman-Sachs A Flow Cytometery. Anal Chem 1994, 66 (13), 700–707. 10.1021/ac00085a002. [DOI] [PubMed] [Google Scholar]
- (69).Ambrose WP; Cai H; Goodwin PM; Jett JH; Habbersett RC; Larson EJ; Grace WK; Werner JH; Keller RA Flow Cytometric Sizing of DNA Fragments. Topics in Fluorescence Spectroscipy 2003, 7, 239–268. 10.1007/0-306-47947-8_8. [DOI] [Google Scholar]
- (70).Salazar-Chaparro AF; Halder S; Maresh ME; Trader DJ Solid-Phase Synthesis and Application of a Clickable Version of Epoxomicin for Proteasome Activity Analysis. ChemBioChem 2022, 23 (7), e202100710. 10.1002/CBIC.202100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Salazar-Chaparro AF; Halder S; Trader DJ Synthesis and Application of a Clickable Epoxomicin-Based Probe for Proteasome Activity Analysis. Curr Protoc 2022, 2 (7), e490. 10.1002/cpz1.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Bo Kim K; Fonseca FN; Crews CM Development and Characterization of Proteasome Inhibitors. Methods Enzymol 2005, 399, 585–609. 10.1016/S0076-6879(05)99039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


