Abstract
Helper-dependent herpes simplex virus (HSV) vectors (amplicons) show considerable promise to provide for long-term transduced-gene expression in most cell types. The current packaging system of choice for these vectors involves cotransfection with a set of five overlapping cosmids that encode the full HSV type 1 (HSV-1) helper virus genome from which the packaging (pac) elements have been deleted. Although both the helper virus and the HSV amplicon can replicate, only the latter is packaged into infectious viral particles. Since the titers obtained are too low for practical application, an enhanced second-generation packaging system was developed by modifying both the helper virus and the HSV amplicon vector. The helper virus was reverse engineered by using the original five cosmids to generate a single HSV-bacterial artificial chromosome (BAC) clone in Escherichia coli from which the pac elements were deleted to generate a replication-proficient but packaging-defective HSV-1 genome. The HSV amplicon was modified to contain the simian virus 40 origin of replication, which acts as an HSV-independent replicon to provide for the replicative expansion of the vector. The HSV amplicon is packaged into infectious particles by cotransfection with the HSV-BAC helper virus into the 293T cell line, and the resulting cell lysate is free of detectable helper virus contamination. The combination of both modifications to the original packaging system affords an eightfold increase in the packaged-vector yield.
Herpes simplex virus (HSV) is a neurotropic herpesvirus that can establish lifelong infections in its human host through latent maintenance in the ganglia of sensory neurons. The virus is relatively well characterized, with a genome of ∼155 kb that is maintained as a concatemerized circular or linear episome in infected cells (27). Because of their wide host range, efficient infection, long-term persistence, capacity to accommodate large amounts of foreign DNA, and ability to deliver genes to postmitotic cell types, considerable effort has been expended to develop gene transfer vectors that can exploit the natural biology of HSV (1, 10, 19). Both helper-dependent and helper-independent vectors are currently in wide use. Helper-independent HSV vectors contain the complete viral genome but have deletions in one or more essential viral genes and can be used to express three or more cDNAs by replacing other, nonessential viral genes (17). Helper-independent HSV vectors thus have a replication-defective viral phenotype and can establish a productive infection only on complementing cell lines that express the corresponding deleted viral gene (5). The major problem associated with this class of vectors, in general, is that there is leaky or inappropriate expression of some of the immediate-early and early HSV genes in noncomplementing cell types, which ultimately results in death of the infected cell even though no viral replication has occurred (14–16, 42). Such cytotoxicity can be completely eliminated when all of the immediate-early HSV genes are deleted, but this results in very low levels of transduced reporter gene expression (28, 29). Thus, entry into the latent cycle may be mandatory for gene expression and maintenance of helper-independent HSV vectors, effectively limiting their use to neuronal cell types. Even so, the factors controlling the complete latency-associated shutdown of viral gene expression have not been clearly defined for HSV, and limited progress has been made in bypassing this shutdown to facilitate long-term transduced-gene expression (18, 35, 36).
Helper-dependent HSV vectors (commonly known as HSV amplicons) contain only the cis elements required for HSV replication and packaging, the oriS and pac elements, respectively (8, 34) (Fig. 1B). Because the size of the vector backbone is only a small fraction of that of the HSV genome, usually <10%, HSV amplicons have the potential to express a large number of genes or cDNAs. Moreover, since they contain no virus-encoded genes, these vectors have the potential to provide long-term gene expression in transduced cells by obviating the latency-associated shutdown of HSV gene expression and are thus well positioned to take full advantage of the ability of HSV to infect virtually every type of cell (2, 8, 11). The first practical packaging system for HSV amplicons utilized a replication-defective with HSV ICP4 deleted as a helper virus to provide the necessary viral replication and packaging functions (9). In this system, the amplicon vector is transfected into an ICP4-complementing cell line, which is then superinfected with the helper virus, and the resulting infectious particles are harvested following growth of the virus. Although this approach can yield recombinant-virus titers approaching 109 PFU/ml, unless there is a mechanism in place to provide for the selective replication and packaging of the amplicon vector over that of the helper virus, the transducing lysates produced are heavily contaminated with the helper virus (9, 20). Even when selective replication and packaging of the amplicon vector is provided for, the helper virus represents 10 to 25% of the total virus yield produced (25, 44). Similar to the situation with helper-independent vectors, this helper virus contamination leads to significant delivery-associated cytotoxicity in transduced cells. Although this problem can potentially be eliminated through the use of less cytotoxic forms of HSV to package amplicon vectors, such mutants typically grow much more slowly and the packaged-amplicon yield is correspondingly lower (38).
FIG. 1.
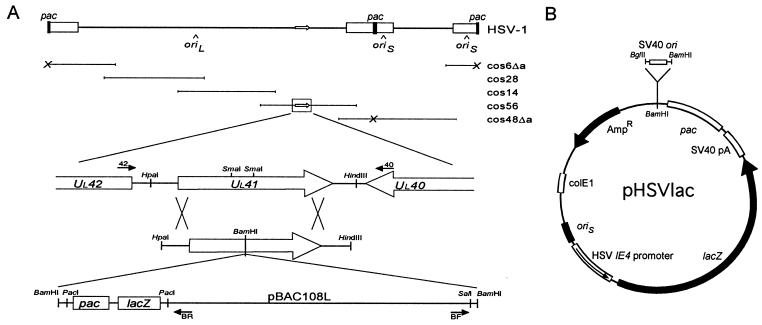
Recombinant HSV-1 constructs. (A) Structure of the UL41-BAC targeting vector. At the top is a schematic representation of the HSV-1 genome and the corresponding Δa cosmid set, comprised of cos6Δa, cos48Δa, cos14, cos28, and cos56. The UL41 gene is located within cos56, as shown enlarged in the middle. Open reading frames are indicated by open arrows. The bottom shows the structure and orientation of the UL41-BAC targeting construct, including the pac cassette containing the HSV-1 pac element and the α fragment of the bacterial lacZ gene. The large crosses indicate the regions of homology between the targeting vector and cos56 that will facilitate homologous recombination to generate a packaging-proficient recombinant virus. The relative location and orientation of the oligonucleotide primers used for PCR are as indicated. Primers 40 and 42 are located outside of the region of homology with the UL41-BAC targeting construct. (B) Structure of the pHSVlac amplicon vector. The HSV-1 oriS and pac elements provide for helper-dependent packaging of the vector. Expression of the bacterial lacZ reporter gene is controlled by the HSV-1 IE4 promoter and SV40 polyadenylation signals, as indicated. The SV40 ori element was cloned into the unique BamHI site adjacent to the HSV pac element.
A second-generation packaging system was recently developed for helper-dependent HSV vectors that yields transducing lysates free of helper virus contamination, making it the packaging system of choice for HSV amplicons (7). The system is centered around a set of five overlapping HSV-1 cosmid clones that together encode the complete wild-type viral genome (4) (Fig. 1A). By specifically deleting the pac element in the repetitive a sequences of two of the cosmids, the cosmid set is rendered packaging defective, and when it is cotransfected into mammalian cells along with an HSV amplicon vector, only the latter is packaged into mature viral particles. The deletion of the pac elements addresses the requirement for selective packaging of the vector over the helper virus. However, it does not provide for any selective replication of the amplicon vector, and this may be why the overall yield of packaged vector particles is quite low, usually <106/ml.
In this report, we describe the development of an enhanced second-generation packaging system for helper-dependent HSV vectors, which we achieved by modifying both the packaging-defective helper virus and HSV amplicon vector components of the current system. We modified the helper virus by cloning the entire packaging-defective HSV-1 genome as a single infectious plasmid in bacteria. We modified the amplicon vector by incorporating the simian virus 40 (SV40) origin of replication, which acts as an HSV-independent replicon to provide for the replicative expansion of the vector prior to packaging into infectious HSV-1 particles. The combination of both modifications affords an eightfold increase in the packaged-vector yield and provides sufficient material in benchtop-scale production to enable in vivo studies for most applications.
MATERIALS AND METHODS
Cell lines and plasmids.
The BHK(TK−) cell line (hamster kidney) was provided by Paul Johnson and Theodore Friedman (15). The SV40 T-antigen-positive 293T-17 cell line (human embryonic kidney) was provided by Doug Bell and David Baltimore and was cultured with 400 μg of Geneticin/ml (24). The Vero cell line (African green monkey kidney) was provided by Dora Ho. Each cell line was cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and was split 1:4 every 3 to 4 days by standard methods. The PC12 cell line (rat adrenal pheochromocytoma) was cultured in Dulbecco modified Eagle medium supplemented with 5% fetal calf serum and 5% horse serum and was differentiated by adding murine nerve growth factor to a final concentration of 50 ng/ml (Harlan Bioproducts for Science Inc., Madison, Wis.) (39).
The pUL41 plasmid, encoding the HSV-1 vhs gene, was provided by James Smiley (31). The pα4βgal/ori vector, which contains the HSV-1 pac element, was provided by Dora Ho (12). Bacterial artificial chromosome (BAC) cloning vector pBAC108L and BAC host strain Escherichia coli HS996 were provided by Melvin Simon and Hiroaki Shizuya (30). HSV-1 cosmid set C was provided by Andrew Davison, and the modified cos6Δa and cos48Δa constructs and the pHSVlac vector were provided by Cornel Fraefel and Alfred Geller (4, 7). The latter was modified to include the minimal SV40 ori element by ligating the BglII-HindIII fragment of the pGL3-Promoter vector (Promega Inc., Madison, Wis.) into the unique BamHI site to generate pHSVlacOri (Fig. 1B).
Construction of the targeting vector.
The BAC vector was constructed by subcloning a cassette containing the HSV-1 pac element and the α fragment of the bacterial lacZ gene into the SalI site of pBAC108L as a SalI-XhoI fragment (Fig. 1A). The HSV pac cassette was constructed by using the pcDNAII expression vector (Invitrogen Inc., La Jolla, Calif.), in which the two NsiI sites within the multiple cloning site were fused, the NaeI site was converted to SalI, and the TfiI site was converted to PacI-XhoI by using oligonucleotide linkers. The HSV-1 pac element from pα4βgal/ori was ligated into the SalI site as a SalI-XhoI fragment by using oligonucleotide linkers, after which the SalI site was converted to SalI-BamHI-PacI. The completed BAC construct was subcloned between the two SmaI sites of pUL41 as a linearized BamHI fragment. To accommodate the BAC, an oligonucleotide linker was used to first convert the SmaI sites of pUL41 to BamHI sites, such that the BAC can be excised as an intact fragment. The targeting vector was linearized by digestion with HindIII prior to cotransfection into BHK cells with the HSV Δa cosmid set at a ratio of 1:5.
Packaging of HSV amplicons.
All plasmid, cosmid, and BAC DNA was prepared by using a modified alkaline lysis protocol, followed by purification over a proprietary ion-exchange column in accordance with the manufacturer’s (Qiagen Inc., Valencia, Calif.) protocols. The pBAC-HSV constructs were further treated to remove contaminating bacterial endotoxin by using a proprietary reagent (Qiagen Inc.) and stored in small aliquots at −20°C to minimize shearing of the DNA due to repeated freeze-thawing. The HSV-1 cosmids were digested with PacI and repurified by phenol-chloroform extraction prior to transfection. The pHSVlac vectors were packaged into infectious particles by cotransfection with either the HSV Δa cosmid set or pBAC-V2 at a ratio of 1:4 by using a proprietary cationic liposome formulation (LipofectAMINE) in accordance with the manufacturer’s (Life Technologies Inc., Bethesda, Md.) protocols. Cells were transfected with 2 μg of DNA plus 15 μl of LipofectAMINE in one well of a six-well culture dish when they reached 80 to 90% confluence. Following transfection, N,N′-hexamethylene-bis-acetamide (Sigma Chemical Co., St. Louis, Mo.) was added to a final concentration of 2 mM to stimulate immediate-early viral gene expression (23). The medium was changed and fresh N,N′-hexamethylene-bis-acetamide was added 24 h following transfection, and the cultures were grown until evidence of the viral cytopathic effect was noted (usually a further 1 to 3 days). Viral particles were harvested by scraping the cells and supernatant into a sterile tube and lysing the cells in two rapid freeze-thaw cycles. Cellular debris was removed by centrifugation at 1,000 × g for 10 min, and the viral particles in the supernatant were aliquoted for storage at −80°C (11). The infectious-vector and/or infectious-virus yield was determined by assaying serial dilutions for β-galactosidase activity by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) histochemistry (21) or for plaque-forming activity (11), respectively, on Vero cells.
Analysis of viral DNA.
Infected cells from eight P150 culture dishes were harvested by scraping and centrifugation at 1,000 × g for 10 min and then resuspended in 8 ml of TE buffer (10 mM Tris-HCl [pH 7.8], 100 mM EDTA) and lysed after adding a further 8 ml of TE buffer supplemented with 2% Triton X-100 by using a Dounce homogenizer. Cellular debris was removed by centrifugation at 2,000 × g for 10 min, and capsids were pelleted by centrifugation at 17,000 × g for 90 min. The capsids were resuspended in 0.5 ml of TEN buffer (10 mM Tris-HCl [pH 7.8], 10 mM EDTA, 150 mM NaCl), and the viral capsids were removed by digestion with 2-mg/ml proteinase K and 0.1% (final concentration) sodium dodecyl sulfate at 60°C for 1 h, followed by phenol extraction and dialysis against three changes of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). For ligation reactions, 100 ng of DNA was utilized in a 20-μl reaction volume to promote recircularization. After incubation overnight at 14°C, 5 μl of the reaction mixture was electroporated into competent HS996 bacterial cells by using standard protocols. For field inversion gel electrophoresis, 100 ng of DNA was loaded onto a 1.0% agarose gel in 0.5× Tris-borate-EDTA (TBE), and the gel was run for 20 h at 200 V and 14°C. Forward pulse times were ramped from 0.9 to 6.3 s with a forward-to-reverse pulse time ratio of 3:1.
PCR amplifications.
HSV-specific primers were targeted to the UL40 gene (primer 40 [ACCATAGCCAATCCATGACC]) and the UL42 gene (primer 42 [GTCGTGAGGAAGAACTTGAGG]) and were designed to amplify across the entire UL41 gene. BAC-specific primers were targeted to the forward region (primer BF [TATTGACATGTCGTCGTAACC]) and the reverse region (primer BR [ATGTCGGCAGAATGCTTAATG]) flanking the PacI cassette and, in conjunction with primers 40 and 42, respectively, were designed to amplify across the sites of BAC integration into the HSV-1 UL41 gene. A high-processivity Taq polymerase cocktail (Expand Long Template PCR System; Boehringer Mannheim GmbH, Mannheim, Germany) was used for all reactions. The annealing temperature for all primer pairs was 60°C, with 100 ng of BAC template DNA or 30 ng of cosmid template DNA in each reaction mixture. Dimethyl sulfoxide was included to a final concentration of 10% for those reactions that involved extension across the GC-rich HSV-1 pac element, but otherwise, the thermal profile and all other parameters for amplification were in accordance with the manufacturer’s recommendations. PCR products were analyzed by agarose gel electrophoresis by standard methods.
RESULTS
Cloning strategy.
The development of a packaging system for helper-dependent HSV vectors that could provide lysates that are free of contaminating helper virus was a significant breakthrough in terms of facilitating the efficient, nontoxic delivery of infectious vector particles (7). However, there are a number of limitations inherent to this system that make it technically demanding to use and that limit the ultimate packaged amplicon vector yield. (i) Packaging requires the transfection of five overlapping HSV-1 cosmid clones, which must recombine to form a functional HSV-1 genome prior to replication. (ii) Some of the cosmid clones are unstable when propagated in bacteria, and the preparation of high-quality DNA for transfections is therefore somewhat laborious. (iii) Transfection efficiencies approaching 100% are required to maximize the packaged amplicon vector yield. We hypothesized that it should be possible to surmount these limitations by cloning the entire packaging-defective HSV-1 genome as a single plasmid DNA by using a BAC cloning vector (30). This would provide a technically simple and much more efficient method of reconstituting the helper virus in the packaging cell. The BAC vector was developed in response to the need to stably propagate large genomic DNA fragments in bacteria, and thus, it is likely that the cloned HSV-1 genome will be more stable as a BAC than in the presently used cosmids.
The approach we pursued was to reverse engineer a single HSV-BAC clone from the five original HSV-1 cosmid clones with the pac elements already deleted and is based on the ability of the HSV-1 cosmid set to generate a functional viral genome through recombination in mammalian cells (4). Since it has been established that only one pac element is required for packaging of the HSV-1 genome (7) and that pac elements retain their function when placed at an alternative locus (3, 32), we inferred that it should be possible to reconstitute a packaging-proficient HSV-1 genome from the Δa cosmid set by providing a suitable pac element. Our strategy was to generate a functional virus from the HSV Δa cosmid set by targeting a BAC vector containing a single pac element to a nonessential HSV-1 gene through homologous recombination. The BAC vector facilitates the subsequent cloning of the resulting recombinant viral genome in bacteria as a single construct. The pac element can then be removed from this construct in vitro to generate a packaging-defective HSV-1 clone that can be used as a helper virus for the packaging of HSV amplicon vectors.
Generation of an infectious HSV-1 clone in a BAC vector.
We selected the HSV-1 UL41 gene as the site of integration of the BAC vector. The UL41 gene product is a tegument protein that is incorporated into mature viral particles and is involved in the degradation of host cell mRNA to provide the viral host shutoff (vhs) function (31). Since the ultimate use of the helper virus will be to provide efficient delivery of the packaged HSV amplicon, we expect that the elimination of the potentially toxic vhs function from the infectious particle should have a positive effect on survival of the transduced cells. We designed a UL41 targeting vector that contained an HSV pac cassette within a standard BAC cloning vector and then transfected it into BHK cells along with the HSV Δa cosmid set (Fig. 1A). The resulting recombinant HSV-BAC viral progeny were expanded on Vero cells to obtain sufficient quantities for purification of the viral genomic DNA. We elected not to clone out individual viral recombinants at this stage but rather to select for the fittest recombinants in the population by growing them as a single pool. This strategy ensures that the fittest viral progeny during growth in vitro will thus be overrepresented when cloned out in bacteria. Since the recombinant HSV-BAC viral progeny have the BAC cloning vector integrated into the UL41 gene, these were cloned simply by recircularizing the linear viral genome and electroporating the DNA into BAC host strain HS996.
We recovered three independent clones and characterized them by restriction endonuclease fingerprinting with BamHI to determine which contained a complete and intact HSV-1 genome. Since the three had very similar patterns (data not shown), we first determined if any were still infectious and able to generate plaques when transfected back into BHK cells. One of the clones was able to produce a functional virus in this assay and was designated pBAC-V1. We assessed the relative efficiency of plaque formation by the pBAC-V1 clone by comparing it to that of the original, packaging-proficient HSV-1 cosmid set. The DNA constructs were transfected into BHK cells in a single well of a six-well culture dish, and after 4 days, the viral particles were harvested and the total virus yield was determined by assaying serial dilutions for plaque formation on Vero cell monolayers. Under these conditions, the pBAC-V1 and HSV cosmid set yielded 5.0 × 103 and 6.0 × 102 PFU/ml, respectively. Thus, despite the fact that it has a deletion at the UL41 locus and contains only a single pac element, the pBAC-V1 clone produced eightfold more virus particles when transfected into BHK cells than did the HSV cosmid set, which contains an intact viral genome that includes two pac elements. This result substantiates our initial expectation that an intact viral genomic fragment should be intrinsically more efficient at generating a functional virus than five overlapping genomic fragments.
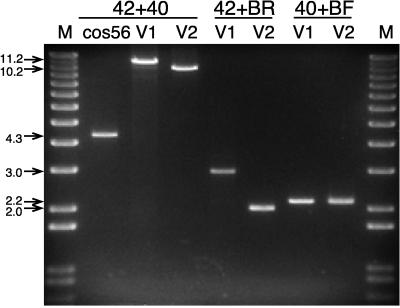
We used three independent analyses to determine if the pBAC-V1 clone contains the full HSV-1 genome. First, we characterized the site of integration of the BAC vector into the HSV-1 genome by using PCR amplification (Fig. 2). Amplification across the entire UL41 locus by using the 42-40 primer set yielded a 4.3-kb product in cos56 which corresponds to the intact UL41 gene and an 11.2-kb product in pBAC-V1 which corresponds to integration of the complete BAC targeting construct within the UL41 gene. Furthermore, amplification across the integration junctions of the BAC targeting construct using the 40-BF and 42-BR primer sets yielded products of 2.2 and 3.0 kb, respectively, confirming that the targeting construct correctly localized the BAC in the intended location within the UL41 gene. Second, we analyzed the size of the PacI-digested pBAC-V1 clone by using field inversion gel electrophoresis and found it to be identical to that of the genomic DNA of the original pooled HSV-BAC recombinant virus at ∼155 kb (Fig. 3A). Third, we compared the BamHI restriction endonuclease fingerprint of the clone to that of the genomic DNA of the original pooled HSV-BAC recombinant virus (Fig. 3B). Although a few minor differences in the patterns were noted, these are likely attributable to heterogeneity in the pooled virus sample. For example, note that the fragments at 11.0, 10.5, 9.0, and 8.5 kb in the pooled HSV-BAC viral DNA are present at half of the expected intensity, whereas in the pBAC-V1 sample, only the 10.5- and 9.0-kb fragments are present but, in this case, at full intensity. These differences are due to recombination in the repeat regions of the HSV-1 genome that generate four alternative isomers of the viral DNA sample (3, 26). The BamHI fingerprinting analysis confirms that there is equal representation of all four isomers in the pooled viral DNA sample and that only one of the isomers is represented in the pBAC-V1 clone. The results of all three analyses are consistent with the conclusion that the pBAC-V1 clone contains an otherwise intact HSV-1 genome in which the UL41 gene has been inactivated. We have also done similar analyses by using pBAC-V1 clones that have been serially subcultured in bacteria and have never recovered any rearrangements (data not shown), suggesting that the HSV-1 genome is completely stabilized in the BAC vector. Thus, we proceeded to the next step in developing a helper virus-free packaging system by rendering pBAC-V1 packaging defective through deletion of the single pac element present.
FIG. 2.
Integration of the BAC vector into the HSV-1 UL41 gene. PCR products were generated from HSV cos56 and pBAC-V1 and pBAC-V2 with the indicated primer pairs and separated through a 0.8% agarose gel. The loss of 1.0 kb from the PCR products derived from pBAC-V2 is due to excision of the pac cassette from this clone. The marker used in lane M is the 1-kb Plus Ladder (Life Technologies Inc.). Molecular sizes are shown on the left in kilobases.
FIG. 3.
Analysis of recombinant BAC clones. (A) Analysis of recombinant and cloned HSV-1 constructs by field inversion gel electrophoresis. Purified viral genomic DNA from the pooled HSV-BAC recombinants was analyzed without further processing, whereas the pBAC-V1 and pBAC-V2 clones were linearized by digestion with PacI prior to loading onto the gel. The arrow indicates the position of the linearized HSV-1 genome at ∼155 kb. The 1-kb PacI fragment corresponding to the pac cassette excised from pBAC-V1 is not resolved in this analysis. The marker used in lane M is Midrange PFG Marker I (New England BioLabs, Beverly, Mass.), and the corresponding sizes in kilobases are indicated on the right. (B) Restriction endonuclease fingerprinting of recombinant and cloned HSV-1 constructs. DNA from the various constructs was digested with BamHI and resolved by electrophoresis through a 0.6% agarose gel. In order to provide adequate contrast for the different fragment sizes down the entire gel, the image shown is a composite of two different photographic exposures. The closed arrows indicate the 11.0-, 10.5-, 9.0-, and 8.5-kb fragments corresponding to the different isomers in the pooled recombinant HSV-BAC genomes, and the larger closed arrows indicate the 10.5- and 9.0-kb fragments corresponding to the isomer cloned in the pBAC-V1 and -V2 constructs. The open arrow indicates the 7.5-kb fragment corresponding to the excised BAC vector in the pBAC-V1 sample, and the white lines indicate changes in the size of this fragment in the pooled recombinant HSV-BAC and pBAC-V2 samples. In pBAC-V2, the BAC vector is reduced in size by 1 kb due to removal of the pac cassette, and in the HSV-BAC sample, the BAC vector is reduced in size by 0.2 kb due to cleavage of the recombinant viral genome within the pac element. The marker used in lane M is the 1-kb Plus Ladder (Life Technologies Inc.), and the sizes of the corresponding bands are indicated in kilobases on the left.
Generation of a packaging-defective HSV-1 clone.
The BAC cloning strategy we developed was designed to facilitate the efficient removal of the single pac element in the pBAC-V1 clone. The HSV pac cassette is flanked by PacI restriction endonuclease sites, and since PacI does not cut anywhere else in the BAC vector or in the HSV-1 genome, the cassette can be excised by digestion with PacI, followed by recircularization of the construct. To facilitate the identification of clones with pac deletions, we incorporated the α fragment of the bacterial lacZ gene into the original PacI cassette. The pBAC-V1 clones that contain the cassette are blue on X-Gal-containing plates, whereas clones that have lost the cassette are white. We recovered a number of such clones by using this simple screen and characterized two of them with respect to their BamHI restriction endonuclease fingerprints to determine if the HSV-1 genome was intact. Except for a 1-kb reduction of the 7.5-kb BAC-derived fragment corresponding to the loss of the HSV pac cassette, both clones were identical to the parental pBAC-V1 construct. The BamHI fingerprint of one of these, designated pBAC-V2, is shown in Fig. 3B. Analysis of the PacI-linearized clone by field inversion gel electrophoresis confirms that its size is the same as that of pBAC-V1 (Fig. 3A).
We further characterized pBAC-V2 by using the same methodology described for the parental pBAC-V1 clone. PCR amplification across the entire UL41 locus by using the 42-40 primer set yielded a 10.2-kb product, and amplification across the integration junctions of the BAC targeting construct by using the 42-BR primer set yielded a 2.0-kb product for pBAC-V2 (Fig. 2). The loss of 1.0 kb from the PCR products derived from pBAC-V2 in comparison to pBAC-V1 is consistent with the loss of the HSV pac cassette from this clone. In addition, we transfected pBAC-V2 into BHK cells to determine if it was still infectious and able to generate plaques, as described previously for pBAC-V1 and the HSV cosmid set. As expected from the combined data indicating that the pac element was deleted from this clone, no infectious HSV-1 particles were produced. These results are consistent with the conclusion that pBAC-V2 contains a replication-competent but packaging-defective HSV-1 genome and that this construct can be used to package amplicon vectors that will be free of helper virus contamination.
Packaging of an HSV amplicon vector into infectious particles.
Since the packaging-defective HSV Δa cosmid set can be used as a helper virus to package HSV amplicons (7) and the resulting cellular lysates remain free of helper virus contamination, we evaluated the ability of the pBAC-V2 clone to function in a similar role. When the prototypical pHSVlac amplicon vector was used as a substrate, pBAC-V2 was able to provide appropriate helper functions and package the vector to a titer of 8.0 × 104 blue-forming units (BFU)/ml and with no detectable helper virus contamination. In comparison with the HSV Δa cosmid set, which yielded a titer of 2.0 × 104 BFU/ml in a parallel transfection, pBAC-V2 was fourfold more proficient at packaging the pHSVlac vector into infectious particles. We have repeated this experiment many times with a variety of cell lines and with a number of different HSV amplicon vectors, and although there is considerable variation in the titer between experiments, pBAC-V2 consistently yields two- to eightfold more packaged vector than does the HSV Δa cosmid set.
BHK cells were used in these experiments because they routinely provide transfection efficiencies surpassing 90% in our studies. However, as described for the cosmid packaging set, any cell line can be used for amplicon packaging (7). We have found that similar high transfection efficiencies can be achieved by using 293T cells (24), whereas those in other laboratories utilize the Vero-based 2-2 cell line (7, 33). Regardless of the cell line used, we have never observed any difference in transfection efficiency for either pBAC-V2 or the HSV Δa cosmid set when it is used as a helper virus (data not shown). It is unlikely that the increased amplicon packaging efficiency of the former can be attributed to differences in transfectability, although given the greater technical difficulties in preparing DNA from the HSV Δa cosmid set, this remains a possibility. A more likely interpretation of this finding is that the packaged amplicon vector yield of either system is directly related to the infectivity of the transfected helper virus DNA. The pBAC-V2 construct is therefore more efficient than the HSV Δa cosmid set because it does not require any recombination events to generate the replication-competent viral genome that is required to package amplicon vectors.
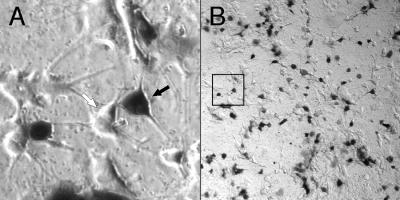
We utilized PC12 cells to examine the ability of infectious particles generated by the pBAC-V2 helper virus genome to deliver the packaged pHSVlac vector to postmitotic cells. When grown in media containing nerve growth factor, PC12 cells exit the cell cycle and differentiate to resemble sympathetic neurons in many aspects of cellular physiology (39). We compared the plating efficiencies of the packaged pHSVlac vector on differentiated PC12 cells and Vero cells. Since the β-galactosidase transducing titers were identical, we concluded that the pBAC-V2 helper virus genome provides all of the necessary HSV-1 proteins for efficient infection of neuronal cell types. Moreover, the vector-transduced cells show no detectable signs of toxicity at 3 days postinfection (Fig. 4). In this experiment, the lowest dilution of the packaged pHSVlac vector that was plated resulted in a multiplicity of infection of 0.8 particle/cell, such that the majority of transduced PC12 cells were targeted by a single infectious particle. Because the HSV-1 vhs function is deleted from the pBAC-V2 helper virus genome, we expect that the infectious HSV-1 particles generated will be tolerated equally well at much higher multiplicities of infection, for example, as can occur in vivo during direct injection of packaged vector material into the mammalian central nervous system. We now have data from rat and hamster models using a variety of different amplicon vectors that support this premise (unpublished data).
FIG. 4.
Transduced reporter gene expression in postmitotic cells. Differentiated PC12 cells were infected at a multiplicity of ∼0.8 in a single well of a P24 culture dish, and 2 days later, the cells were fixed and stained for pHSVlac-derived β-galactosidase reporter gene activity by using X-Gal histochemistry. The black and white arrows indicate positive and negative staining of cells, respectively. The boxed area in panel B indicates the location of the field shown at a higher magnification in panel A.
An alternative viral replicon enhances HSV amplicon vector packaging.
The inefficiency of first-generation HSV packaging systems is due to their inability to provide for the selective replication and packaging of the amplicon vector. The HSV cosmid set and HSV-BAC packaging systems work in part because they specifically address the latter deficiency through the use of a packaging-defective helper virus. Since they do not select for replication of the amplicon vector, we postulated that the packaged amplicon vector yield may be limited by ineffective competition with the helper virus for HSV-specific replication factors. If this is the case, then a simple solution is to specifically increase the copy number of the amplicon vector relative to that of the helper virus, in essence ensuring that subsequent replication is skewed to favor the vector rather than the helper virus. Our approach was to use an alternative, HSV-independent replicon to afford such a competitive advantage to the amplicon and thus increase the overall yield of packaged vector particles. It is well established that the SV40 origin of replication can facilitate the selective amplification of virtually any plasmid DNA in cells that express the corresponding SV40 T antigen (40). Accordingly, we utilized the SV40 system to determine if this would have a similar effect on amplicon packaging.
The pHSVlac vector was modified to include the minimal elements of the well-characterized SV40 ori element, which includes the basal SV40 early promoter but not the enhancer (Fig. 1B). We compared the efficiency of packaging of pHSVlac and that of modified pHSVlacOri in BHK and 293T cells (Table 1). In the T-antigen-positive 293T cell line (24), the packaged amplicon vector yield is increased almost twofold when the vector contains the SV40 ori element. This increase is not seen in the T-antigen-negative BHK cell line, where the pHSVlac vector is packaged more efficiently than modified pHSVlacOri. In independent transient transfection experiments, we determined that the copy number of pHSVlacOri is increased fourfold over that of pHSVlac by 48 h posttransfection in 293T cells but is unchanged in BHK cells (data not shown). Therefore, we concluded that the enhanced packaging of pHSVlacOri in 293T cells is due to T-antigen-dependent prereplication of the vector and that this relatively small increase in vector copy number effectively overcomes the slight packaging disadvantage of pHSVlacOri observed in BHK cells. Since the vector copy number can be increased in other expression systems by several orders of magnitude by using the SV40 ori element (40), this suggests that further increases in packaging efficiency may be attainable by increasing the replicative efficiency of the added replicon, for example, by increasing the activity of the SV40 ori element by incorporating the full SV40 enhancer or by incorporating a more efficient viral replicon into the vector.
TABLE 1.
Enhanced packaging of an HSV amplicon vector
| Vector | Cell linea | Vector titerb (104 BFU/ml of lysate) | Virus titerc |
|---|---|---|---|
| pHSVlac | 293T | 1.5 | <10 |
| pHSVlacOri | 293T | 2.7 | <10 |
| pHSVlac | BHK | 2.4 | <10 |
| pHSVlacOri | BHK | 1.8 | <10 |
The pBAC-V2 clone was used as the HSV-1 helper construct in all transfections.
Serial 10-fold dilutions of the lysates were plated on a confluent lawn of Vero cells, and the cells were assayed for β-galactosidase activity after 2 days by using X-Gal histochemistry.
Serial 10-fold dilutions of the lysates were plated on a confluent lawn of Vero cells, and plaques were counted after 3 days. A titer of <10 reflects the fact that no helper virus was detected in a 10−1 dilution of the lysate, which was the lowest dilution tested in this experiment.
DISCUSSION
The development of gene-based therapies is currently being pursued in fields as diverse as infectious disease, autoimmune disease, cardiovascular disease, and neurodegenerative disease. The availability of an efficient gene delivery and expression system is an essential enabling technology that unites all of these applications, and as a result, this has been an area of intense research in recent years. HSV-based vectors show considerable promise in this regard because of their wide host rang, efficient infection, long-term persistence, capacity to accommodate large amounts of foreign DNA, and ability to deliver genes to postmitotic cell types (5, 10, 13). After further optimization of culture and transfection conditions and only a modest upscaling of our methods, we have succeeded in reproducibly obtaining titers of 107 transducing particles/ml of lysate for a variety of different HSV amplicon vectors by using the pBAC-V2 helper construct. Thus, the HSV packaging system is now efficient and economical enough to provide sufficient material for most in vivo studies, and we anticipate that it will be possible to extend these to include expression of a therapeutic gene in various animal models of disease. In this regard, future developments will likely lie in refinements to the vector backbone itself in order to stabilize the HSV amplicon and thus afford extended transduced-gene expression. One approach that we and others have taken is to incorporate an alternative viral replicon, for example, the episomal replicon of the Epstein-Barr virus (37, 41) or the integrating replicon from the adeno-associated virus (6), and it is likely that additional hybrid HSV amplicon vectors are already in development in other laboratories. Given the recent improvements in helper-independent HSV vectors that eliminate all delivery-associated cytotoxicity (28, 29), it will be interesting to compare the relative efficiency of long-term transduced-gene expression from this platform with that obtained by using the newer hybrid amplicon vector systems.
The ability of the SV40 ori element to increase the packaged-vector yield in our packaging system stands in contrast to data obtained by using a fully functional HSV helper virus, where the incorporation of the SV40 ori element has no effect on amplicon packaging (43). Thus, the ability of prereplication to increase the packaged-vector yield may be context specific, for example, dependent upon the use of a packaging-deficient helper virus and/or a specific HSV amplicon. For example, the prereplication by the SV40 ori element may be effective only under those conditions in which replication of the HSV helper virus (and thus the HSV amplicon) is suboptimal, as is the case when a replication-defective or packaging-deficient HSV helper virus is used. In support of this view, we have incorporated the minimal SV40 ori element into other HSV amplicon vectors that our laboratory has engineered and have found that it is more effective in those vectors that are packaged relatively inefficiently in comparison to pHSVlac and can increase packaged-vector yields by as much as 10-fold in such cases (unpublished data).
The strategy we developed for cloning of the HSV-1 genome as a single infectious BAC should be applicable to other DNA viruses. Indeed, during the course of our experiments, a remarkably similar approach was described for the murine cytomegalovirus (mCMV), a herpesvirus related to HSV (22). In this case, the experimental goal was to develop a system in which the mCMV genome could be manipulated in bacteria such that the phenotype of viral mutants could be defined without the requirement for growth in a mammalian cell that is a prerequisite in conventional mutagenesis protocols. In such a strategy, it is imperative that the cloned viral genome be stable in bacteria, such that the incidence of unwanted or cryptic mutations in other viral genes is minimized, since these would confound interpretation of the resulting viral phenotype. In this regard, the growth kinetics of the HSV-BAC virus and the virus derived from the infectious pBAC-V1 clone are identical. Moreover, we have never detected any rearrangements in the pBAC-V2 clone following serial passage in bacteria, suggesting that the cloned HSV genome appears to satisfy this criterion. Thus, we expect that a mutagenesis scheme similar to that described for the cloned mCMV genome can be applied to HSV-1. The fact that we were able to generate a packaging-deficient but otherwise completely functional viral genome illustrates the power of such an approach, since it is otherwise impossible to propagate such a recombinant virus in mammalian cells. Note, however, that the pBAC-V1 and pBAC-V2 clones described in this study have the a and UL41 sequences deleted and are thus not well suited for most other studies.
ACKNOWLEDGMENTS
We thank Marilyn McLeod for excellent technical assistance throughout this project, Aly Cassam for providing the differentiated PC12 cells, Jim Smiley for forwarding his protocol for the purification of HSV genomic DNA, and Greg Dekaban, Mick Bhatia, and Grant McFadden for helpful discussions and comments on the manuscript. We are indebted to Jim Smiley, Dora Ho, Cornel Fraefel, Alfred Geller, Andrew Davison, Melvin Simon, Hiroaki Shizuya, Paul Johnson, Ted Friedman, Doug Bell, and David Baltimore for their generosity in providing the various cell lines and plasmid constructs.
This work was supported by an operating grant to C.A.S. from the Medical Research Council of Canada. C.A.S. is supported by a Cancer Research Society/Medical Research Council scholarship, and T.A.S. is supported by an Ontario Graduate Scholarship.
REFERENCES
- 1.Breakefield X O, DeLuca N A. Herpes simplex virus for gene delivery to neurons. New Biol. 1991;3:203–218. [PubMed] [Google Scholar]
- 2.Chiocca E A, Choi B B, Cai W Z, DeLuca N A, Schaffer P A, DiFiglia M, Breakefield X O, Martuza R L. Transfer and expression of the lacZ gene in rat brain neurons mediated by herpes simplex virus mutants. New Biol. 1990;2:739–746. [PubMed] [Google Scholar]
- 3.Chou J, Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985;41:803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 5.Fink D J, DeLuca N A, Goins W F, Glorioso J C. Gene transfer to neurons using herpes simplex virus-based vectors. Annu Rev Neurosci. 1996;19:265–287. doi: 10.1146/annurev.ne.19.030196.001405. [DOI] [PubMed] [Google Scholar]
- 6.Fraefel C, Jacoby D R, Lage C, Hilderbrand H, Chou J Y, Alt F W, Breakefield X O, Majzoub J A. Gene transfer into hepatocytes mediated by helper virus-free HSV/AAV hybrid vectors. Mol Med. 1997;3:813–825. [PMC free article] [PubMed] [Google Scholar]
- 7.Fraefel C, Song S, Lim F, Lang P, Yu L, Wang Y, Wild P, Geller A I. Helper virus-free transfer of herpes simplex virus type 1 plasmid vectors into neural cells. J Virol. 1996;70:7190–7197. doi: 10.1128/jvi.70.10.7190-7197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geller A I, Breakefield X O. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science. 1988;241:1667–1669. doi: 10.1126/science.241.4873.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geller A I, Keyomarsi K, Bryan J, Pardee A B. An efficient deletion mutant packaging system for defective herpes simplex virus vectors: potential applications to human gene therapy and neuronal physiology. Proc Natl Acad Sci USA. 1990;87:8950–8954. doi: 10.1073/pnas.87.22.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glorioso J C, Goins W F, Fink D J, DeLuca N A. Herpes simplex virus vectors and gene transfer to brain. Dev Biol Stand. 1994;82:79–87. [PubMed] [Google Scholar]
- 11.Ho D. Amplicon-based herpes simplex virus vectors. Methods Cell Biol. 1994;43:191–210. doi: 10.1016/s0091-679x(08)60604-4. [DOI] [PubMed] [Google Scholar]
- 12.Ho D Y, Saydam T C, Fink S L, Lawrence M S, Sapolsky R M. Defective herpes simplex virus vectors expressing the rat brain glucose transporter protect cultured neurons from necrotic insults. J Neurochem. 1995;65:842–850. doi: 10.1046/j.1471-4159.1995.65020842.x. [DOI] [PubMed] [Google Scholar]
- 13.Huard J, Goins W F, Glorioso J C. Herpes simplex virus type 1 vector mediated gene transfer to muscle. Gene Ther. 1995;2:385–392. [PubMed] [Google Scholar]
- 14.Johnson P A, Miyanohara A, Levine F, Cahill T, Friedmann T. Cytotoxicity of a replication-defective mutant of herpes simplex virus type 1. J Virol. 1992;66:2952–2965. doi: 10.1128/jvi.66.5.2952-2965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson P A, Wang M J, Friedmann T. Improved cell survival by the reduction of immediate-early gene expression in replication-defective mutants of herpes simplex virus type 1 but not by mutation of the virion host shutoff function. J Virol. 1994;68:6347–6362. doi: 10.1128/jvi.68.10.6347-6362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson P A, Yoshida K, Gage F H, Friedmann T. Effects of gene transfer into cultured CNS neurons with a replication-defective herpes simplex virus type 1 vector. Brain Res Mol Brain Res. 1992;12:95–102. doi: 10.1016/0169-328x(92)90072-j. [DOI] [PubMed] [Google Scholar]
- 17.Krisky D M, Marconi P C, Oligino T, Rouse R J, Fink D J, Glorioso J C. Rapid method for construction of recombinant HSV gene transfer vectors. Gene Ther. 1997;4:1120–1125. doi: 10.1038/sj.gt.3300497. [DOI] [PubMed] [Google Scholar]
- 18.Lachmann R H, Efstathiou S. Utilization of the herpes simplex virus type 1 latency-associated regulatory region to drive stable reporter gene expression in the nervous system. J Virol. 1997;71:3197–3207. doi: 10.1128/jvi.71.4.3197-3207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leib D A, Olivo P D. Gene delivery to neurons: is herpes simplex virus the right tool for the job? Bioessays. 1993;15:547–554. doi: 10.1002/bies.950150808. [DOI] [PubMed] [Google Scholar]
- 20.Lim F, Hartley D, Starr P, Lang P, Song S, Yu L, Wang Y, Geller A I. Generation of high-titer defective HSV-1 vectors using an IE 2 deletion mutant and quantitative study of expression in cultured cortical cells. BioTechniques. 1996;20:460–469. doi: 10.2144/19962003460. [DOI] [PubMed] [Google Scholar]
- 21.MacGregor G R, Mogg A E, Burke J F, Caskey C T. Histochemical staining of clonal mammalian cell lines expressing E. coli beta galactosidase indicates heterogeneous expression of the bacterial gene. Somat Cell Mol Genet. 1987;13:253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- 22.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson T, Everett R D. A prominent serine-rich region in Vmw175, the major transcriptional regulator protein of herpes simplex virus type 1, is not essential for virus growth in tissue culture. J Gen Virol. 1990;71:1775–1783. doi: 10.1099/0022-1317-71-8-1775. [DOI] [PubMed] [Google Scholar]
- 24.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pechan P A, Fotaki M, Thompson R L, Dunn R, Chase M, Chiocca E A, Breakefield X O. A novel ’piggyback’ packaging system for herpes simplex virus amplicon vectors. Hum Gene Ther. 1996;7:2003–2013. doi: 10.1089/hum.1996.7.16-2003. [DOI] [PubMed] [Google Scholar]
- 26.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16:481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 27.Roizman B, Sears A B, Whitley R J. Fields virology. 3rd ed. New York, N.Y: Lippincott-Raven; 1996. [Google Scholar]
- 28.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 32.Smiley J R, Lavery C, Howes M. The herpes simplex virus type 1 (HSV-1) a sequence serves as a cleavage/packaging signal but does not drive recombinational genome isomerization when it is inserted into the HSV-2 genome. J Virol. 1992;66:7505–7510. doi: 10.1128/jvi.66.12.7505-7510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 34.Spaete R R, Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci USA. 1985;82:694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr P A, Lim F, Grant F D, Trask L, Lang P, Yu L, Geller A I. Long-term persistence of defective HSV-1 vectors in the rat brain is demonstrated by reactivation of vector gene expression. Gene Ther. 1996;3:615–623. [PubMed] [Google Scholar]
- 36.Stevens J G. Human herpesviruses: a consideration of the latent state. Microbiol Rev. 1989;53:318–332. doi: 10.1128/mr.53.3.318-332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strathdee C A. 15th Annual Meeting of the American Society for Virology. 1996. Development of a hybrid herpesvirus vector for gene expression in mammalian tissues, abstr. W31-8. [Google Scholar]
- 38.Strathdee, C. A. Unpublished data.
- 39.Tischler A S, Greene L A. Morphologic and cytochemical properties of a clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Lab Invest. 1978;39:77–89. [PubMed] [Google Scholar]
- 40.Tsui L C, Breitman M L, Siminovitch L, Buchwald M. Persistence of freely replicating SV40 recombinant molecules carrying a selectable marker in permissive simian cells. Cell. 1982;30:499–508. doi: 10.1016/0092-8674(82)90247-1. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Vos J M. A hybrid herpesvirus infectious vector based on Epstein-Barr virus and herpes simplex virus type 1 for gene transfer into human cells in vitro and in vivo. J Virol. 1996;70:8422–8430. doi: 10.1128/jvi.70.12.8422-8430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu N, Watkins S C, Schaffer P A, DeLuca N A. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70:6358–6369. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X, Leduc Y, Cynader M, Tufaro F. Examination of conditions affecting the efficiency of HSV-1 amplicon packaging. J Virol Methods. 1995;52:219–229. doi: 10.1016/0166-0934(94)00166-e. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, O’Shea H, Entwisle C, Boursnell M, Efstathiou S, Inglis S. An efficient selection system for packaging herpes simplex virus amplicons. J Gen Virol. 1998;79:125–131. doi: 10.1099/0022-1317-79-1-125. [DOI] [PubMed] [Google Scholar]