The CoII-catalysed synthesis and crystal structure is reported for the title compound, which features a symmetric N⋯H+⋯N unit.
Keywords: 1,3,4-oxadiazole; cyclo-desulfurization; Hirshfeld surface analysis; crystal structure
Abstract
The title compound, C14H12N4O2·0.5HCl·H2O or H(C14H12N4O2)2 +·Cl−·2H2O, arose from the unexpected cyclization of isonicotinoyl-N-phenyl hydrazine carbothioamide catalysed by cobalt(II) acetate. The organic molecule is almost planar and a symmetric N⋯H+⋯N hydrogen bond links two of them together, with the H atom lying on a crystallographic twofold axis. The extended structure features N—H⋯O and O—H⋯Cl hydrogen bonds, which generate [001] chains. Weak C—H⋯Cl interactions cross-link the chains. The chloride ion has site symmetry 2. The major contributions to the Hirshfeld surface are from H⋯H (47.1%), Cl⋯H/H⋯Cl (total 10.8%), O⋯H/H⋯O (7.4%) and N⋯H/H⋯N (6.7%) interactions.
1. Chemical context
1,3,4-Oxadiazole derivatives have been studied in recent years for their diverse biological activities (Gond et al., 2023 ▸; Abd-Ellah et al., 2017 ▸; Bitla et al., 2020 ▸). As a result of their electron-accepting properties, high quantum yield, and good thermal and chemical stabilities, they have also been used in electroluminescent, optical and electron-transporting materials and chelating agents (Najare et al., 2020 ▸; Wu et al., 2012 ▸). Several methods for the synthesis of 1,3,4-oxadiazoles from acyclic precursors are available, which include oxidative cyclization of acylhydrazones (Jedlovská & Leško, 1994 ▸) and acylthiosemicarbazides (Omar et al., 1996 ▸, Paswan et al., 2015 ▸). In the presence of a strong acid, an N-acylhydrazine carbodithioate is converted into a thiadiazole whereas in the presence of a weak acid or base or on complexation they can be cyclized into oxadiazole (Reid & Heindel, 1976 ▸; Jasinski et al., 2011 ▸).
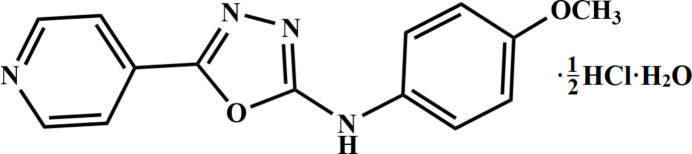
We have previously reported the cyclo-desulfurization of several N-acylhydrazine carbodithioates into the corresponding 1,3,4-oxadiazole in the presence of manganese(II) acetate via the loss of H2S where the MnII ion presumably behaves as a weak Lewis acid (Paswan et al., 2015 ▸, 2016 ▸; Gond et al., 2022 ▸). In the present work, a similar reaction is reported in presence of CoII chloride. Similar CoII-assisted cyclization reactions are also reported in the literature (Li et al., 2021 ▸, 2023 ▸; Bharty et al., 2012 ▸).
2. Structural commentary
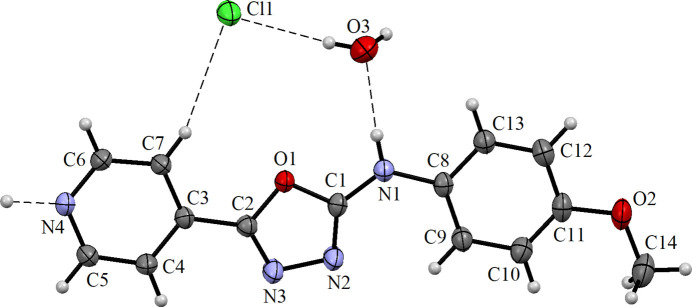
The compound crystallizes in the monoclinic crystal system in space group C2/c. The asymmetric unit consists of one organic molecule, half an equivalent of HCl and one water molecule (Fig. 1 ▸). The C3–C7/N4 pyridyl, C1/C2/N2/N3/O1 oxadiazole and C8–C13 phenyl rings are close to co-planar with the dihedral angles between pyridyl and oxadiazole rings being 4.88 (9)°, oxadiazole and phenyl rings 4.27 (10)° and pyridyl and phenyl rings 2.27 (9)°. The bond distances and angles of the 1,3,4-oxadiazole ring [C1—N2 = 1.298 (2); C2—N3 = 1.277 (2) Å] are in good agreement with values reported previously (Jasinski et al., 2011 ▸; Paswan et al., 2015 ▸, 2016 ▸; Singh et al., 2007 ▸). The C—N bond distance in the pyridine ring, C5—N4 = 1.336 (2) Å, is slightly longer than the corresponding bond in a similar compound (1.326 (2) Å; Singh et al., 2006 ▸), probably due to the N⋯H interaction (Fig. 2 ▸).
Figure 1.
The molecular structure of the title compound showing 30% probability displacement ellipsoids with hydrogen bonds indicated by dashed lines.
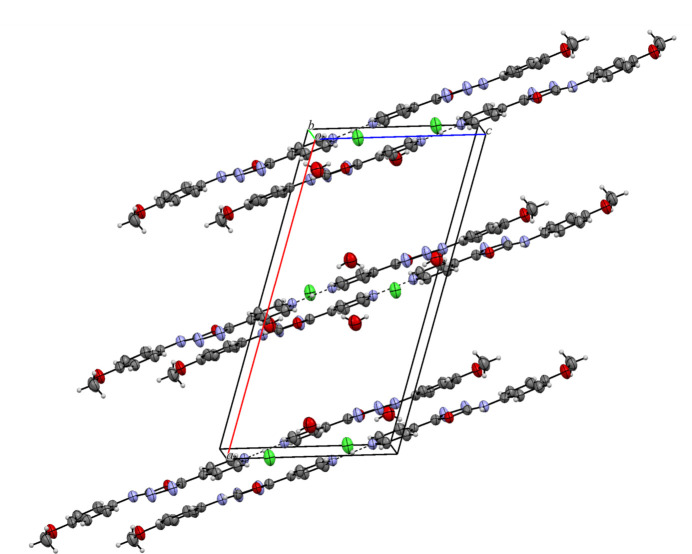
Figure 2.
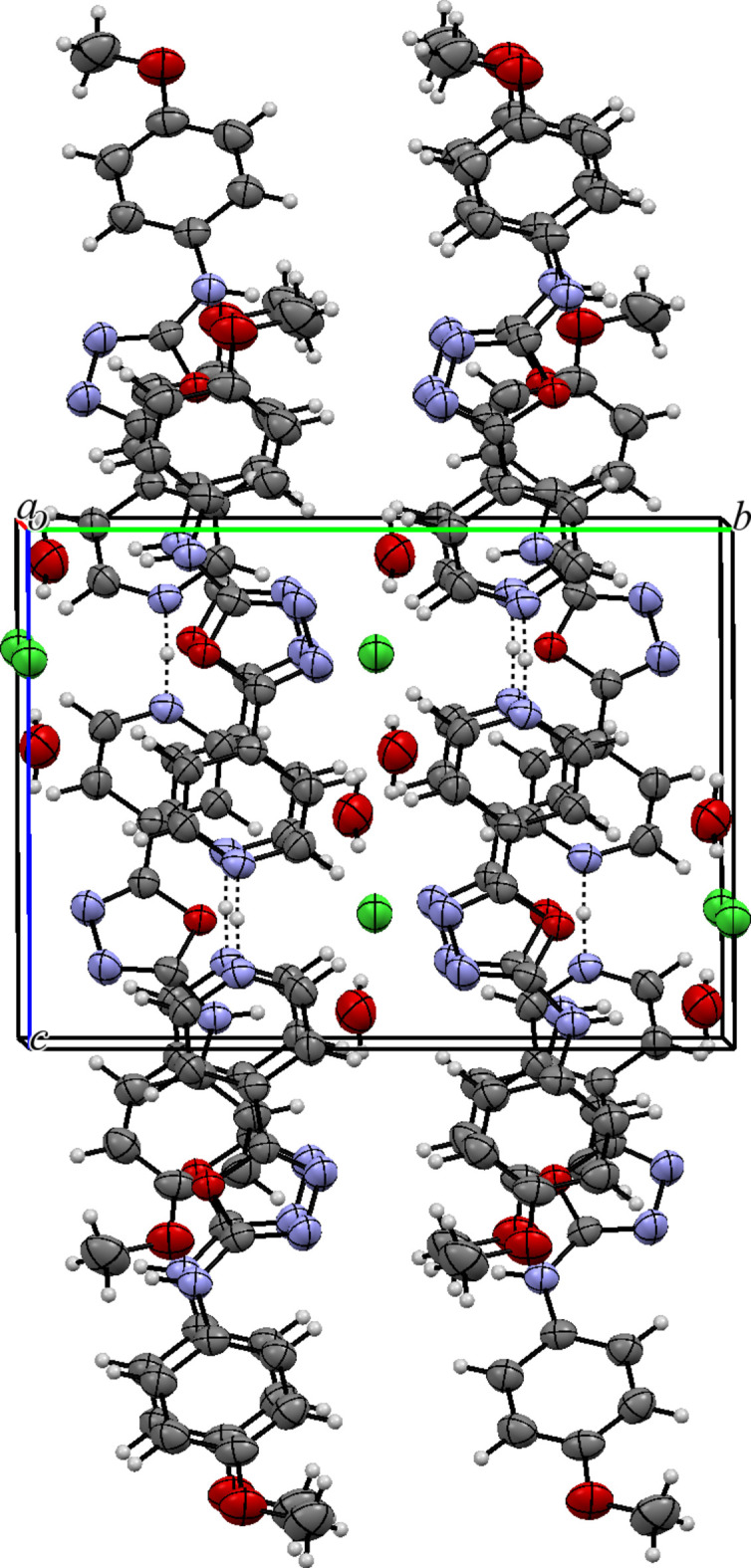
The packing of the title compound viewed along the a-axis direction.
3. Supramolecular features
In the extended structure, two organic molecules are linked through their pyridine nitrogen atoms via the proton of the hydrochloric acid, which lies on a crystallographic twofold axis. This strong, symmetrical, almost linear N4⋯H4N⋯N4 hydrogen bond (Table 1 ▸) leads to a rod-like dimeric structure. These units form a layer-like structure when viewed along b axis of the unit cell (Fig. 3 ▸). The water molecules and chloride ions (site symmetry 2) are embedded in the space between the chains and are connected to them via N—H⋯O and O—H⋯Cl hydrogen bonds, thereby generating [001] chains. Weak C—H⋯Cl interactions are also observed (Table 1 ▸; Fig. 2 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H4N⋯N4i | 1.34 (1) | 1.34 (1) | 2.675 (3) | 178 (3) |
| N1—H1⋯O3 | 0.86 | 2.02 | 2.875 (2) | 172 |
| O3—H22⋯Cl1 | 0.82 (4) | 2.43 (4) | 3.233 (2) | 166 (3) |
| O3—H21⋯Cl1ii | 0.81 (4) | 2.55 (4) | 3.359 (3) | 172 (4) |
| C7—H7⋯Cl1 | 0.93 | 2.93 | 3.7940 (18) | 155 |
| C6—H6⋯Cl1iii | 0.93 | 2.82 | 3.7137 (18) | 163 |
| C5—H5⋯N3iv | 0.93 | 2.56 | 3.329 (2) | 140 |
| C9—H9⋯N2 | 0.93 | 2.34 | 2.969 (2) | 125 |
Symmetry codes: (i)
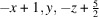 ; (ii)
; (ii)
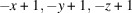 ; (iii)
; (iii)
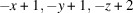 ; (iv)
; (iv)
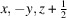 .
.
Figure 3.
The packing of title compound viewed along the b-axis direction.
4. Hirshfeld Surface Analysis
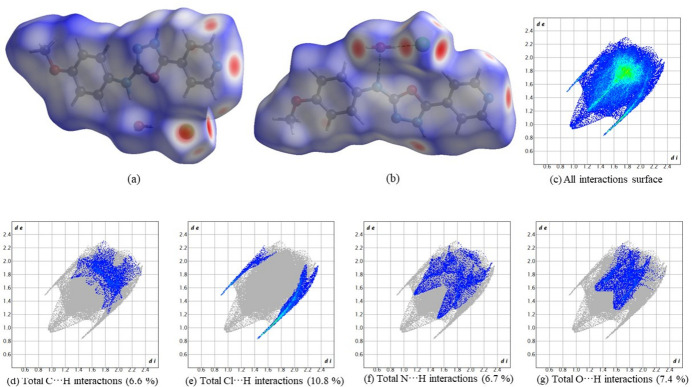
To gain further insight into the intermolecular interactions, a Hirshfeld surface analysis was performed using Crystal Explorer 17.5 (Spackman et al., 2021 ▸). Fig. 4 ▸ a,b shows the Hirshfeld surface mapped over d norm. The red spots show the various hydrogen bonds noted above.
Figure 4.
(a), (b) Two views of the Hirshfeld surface of the title compound mapped over dnorm , (c) fingerprint plot showing the total contribution of individual interactions and those delineated into (d) C⋯H/H⋯C interactions (6.6%), (e) Cl⋯H/H⋯Cl interactions (10.8%), (f) N⋯H/H⋯N interactions (6.7%) and (g) O⋯H/H⋯O interactions (7.4%).
The two-dimensional fingerprint plots are presented in Fig. 4 ▸ c–g. The H⋯H (van der Waals) contacts dominate at 47.1%. Among the directional interactions present in the structure, the Cl⋯H/H⋯Cl (total 10.8%) contact is the most significant. Other contacts include 6.6% for C⋯H/H⋯C, 6.7% N⋯H/H⋯N and 7.4% for O⋯H/H⋯O interactions.
5. Synthesis and crystallization
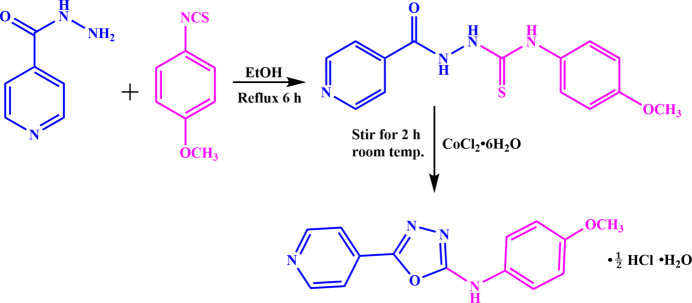
2-Isonicotinoyl-N-(4-methoxyphenyl)hydrazine-1-carbothioamide was prepared by adding 1.652 g (10.00 mmol) of 4-methoxy phenyl isothiocyanate in ethanol solution to 1.370 g (10.00 mmol) of isonicotinohydrazide and the reaction mixture was refluxed for 6 h at 333 K. Upon cooling, a white precipitate of 2-isonicotinoyl-N-(4-methoxyphenyl)hydrazine-1-carbothioamide was obtained (Fig. 5 ▸), which was filtered off and washed with a 50:50 v/v mixture of water and ether. Then, 1.00 mmol of 2-isonicotinoyl-N-(4-methoxyphenyl)hydrazine-1-carbothioamide was dissolved in a 50:50 v/v mixture of methanol and chloroform, and a methanolic solution of 0.5 mmol of CoCl2·6H2O was added and stirred for 2 h, during which time the smell of H2S was noted. The clear solution obtained was kept for crystallization and after 15 days, pale-pink blocks of the title compound were grown. Yield: 60.6%; m.p. 495–498 K. Analysis calculated for C14H12N4O2.0.5 HCl·H2O: C, 55.21; H, 4.79; N, 18.39%; found: C, 55.25; H, 4.50; N, 18.55%.
Figure 5.
Synthesis scheme for the title compound.
The IR spectrum (KBr disc) shows an absorption band at 3280 cm−1 due to the NH group. The C=O band is absent and a new band is observed at 1623 cm−1 corresponding to the C=N bond. In addition, a blue shift is observed for the N=N band at 1179 cm−1 compared to the single bond in the thiosemicarbazide intermediate (Fig. 1 in the supporting information). All these data indicate that the carbothioamide moiety has been transformed into the corresponding oxadiazole (Chandra et al., 2022 ▸; Jaiswal et al., 2023a ▸ b ▸).
The 1H NMR spectrum of the title compound in DMSO-d6 displays peaks at δ 10.69 ppm due to the NH proton, at δ 8.91 and 7.90 ppm due to the pyridyl ring protons and at δ 7.55 and 6.98 ppm due to phenyl ring protons. The methoxy protons appear at δ 3.74 ppm. (Fig. 2 in the supporting information). In the 13C NMR spectrum, peaks at δ 156.9 and 155.2 ppm arise from oxadiazole ring carbon atoms, the methoxy C atom appears at 55.7 ppm and the phenyl and pyridyl carbon atoms are observed in the range δ 114.8–132.4 ppm (Fig. 3 in the supporting information). An absorption at 338 nm in the electronic spectrum of the title compound can be attributed to its π–π* transition (Fig. 4 in the supporting information). It displays fluorescence at 418 nm upon excitation at 338 nm (Fig. 5 in the supporting information) when dissolved in 10−5 M DMSO solution.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Atom H4N was freely refined. Other H atoms were placed in idealized locations (N—H = 0.86 Å, C—H = 0.93–0. 96 Å) and refined using a riding model with U iso(H) =1.2U eq(C,N) or 1.5U eq(C-methyl). Asymmetric N—H⋯N/N⋯H—N refinements with the H atom displaced towards one of the N atoms were inconclusive and atom H4N was placed on the twofold axis.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C28H25N8O4 +·Cl−·2H2O |
| M r | 609.04 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 20.5406 (9), 13.7457 (5), 10.5650 (3) |
| β (°) | 107.006 (4) |
| V (Å3) | 2852.54 (19) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.19 |
| Crystal size (mm) | 0.32 × 0.26 × 0.24 |
| Data collection | |
| Diffractometer | Bruker multiwire proportional |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 16772, 3121, 2026 |
| R int | 0.029 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.041, 0.119, 0.98 |
| No. of reflections | 3009 |
| No. of parameters | 205 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.18 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024002044/hb8088sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024002044/hb8088Isup2.hkl
Spectra. DOI: 10.1107/S2056989024002044/hb8088sup3.docx
Supporting information file. DOI: 10.1107/S2056989024002044/hb8088Isup4.cml
CCDC reference: 2238764
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
MK expresses gratitude to BHU for the financial support provided through the IoE Research Grant for Faculty Development Scheme No. 6031.
supplementary crystallographic information
Crystal data
| C28H25N8O4+·Cl−·2H2O | F(000) = 1276 |
| Mr = 609.04 | Dx = 1.418 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 20.5406 (9) Å | Cell parameters from 8519 reflections |
| b = 13.7457 (5) Å | θ = 2.5–26.2° |
| c = 10.5650 (3) Å | µ = 0.19 mm−1 |
| β = 107.006 (4)° | T = 293 K |
| V = 2852.54 (19) Å3 | Block, pinkish |
| Z = 4 | 0.32 × 0.26 × 0.24 mm |
Data collection
| Bruker multiwire proportional diffractometer | Rint = 0.029 |
| Radiation source: sealed tube | θmax = 27.0°, θmin = 2.5° |
| phi and ω scans | h = −25→25 |
| 16772 measured reflections | k = −17→13 |
| 3121 independent reflections | l = −13→11 |
| 2026 reflections with I > 2σ(I) |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.041 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.119 | w = 1/[σ2(Fo2) + (0.0602P)2 + 1.1461P] where P = (Fo2 + 2Fc2)/3 |
| S = 0.98 | (Δ/σ)max < 0.001 |
| 3009 reflections | Δρmax = 0.20 e Å−3 |
| 205 parameters | Δρmin = −0.18 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.500000 | 0.49848 (5) | 0.750000 | 0.0696 (3) | |
| O1 | 0.60240 (6) | 0.24882 (8) | 0.73971 (10) | 0.0438 (3) | |
| O2 | 0.73500 (8) | 0.20297 (12) | 0.12535 (14) | 0.0738 (4) | |
| N4 | 0.52376 (7) | 0.20300 (10) | 1.14485 (13) | 0.0447 (4) | |
| N1 | 0.63480 (7) | 0.26868 (11) | 0.55203 (13) | 0.0470 (4) | |
| H1 | 0.627028 | 0.329212 | 0.562248 | 0.056* | |
| N2 | 0.62683 (8) | 0.11322 (11) | 0.64893 (14) | 0.0567 (4) | |
| O3 | 0.59808 (11) | 0.46965 (13) | 0.5629 (3) | 0.0817 (5) | |
| N3 | 0.60728 (9) | 0.08972 (11) | 0.76220 (15) | 0.0579 (4) | |
| C3 | 0.57017 (8) | 0.18290 (12) | 0.92666 (15) | 0.0395 (4) | |
| C1 | 0.62287 (8) | 0.20733 (13) | 0.64050 (15) | 0.0423 (4) | |
| C2 | 0.59381 (8) | 0.16960 (12) | 0.81088 (16) | 0.0421 (4) | |
| C7 | 0.55180 (9) | 0.27308 (13) | 0.96298 (16) | 0.0465 (4) | |
| H7 | 0.554704 | 0.328207 | 0.913884 | 0.056* | |
| C8 | 0.65878 (8) | 0.24517 (13) | 0.44342 (16) | 0.0438 (4) | |
| C6 | 0.52914 (9) | 0.27987 (13) | 1.07303 (16) | 0.0481 (4) | |
| H6 | 0.517179 | 0.340647 | 1.097866 | 0.058* | |
| C4 | 0.56536 (9) | 0.10281 (13) | 1.00308 (17) | 0.0489 (4) | |
| H4 | 0.577659 | 0.041269 | 0.981505 | 0.059* | |
| C5 | 0.54218 (9) | 0.11591 (13) | 1.11088 (17) | 0.0504 (5) | |
| H5 | 0.539154 | 0.062199 | 1.162390 | 0.060* | |
| C13 | 0.66769 (10) | 0.32122 (14) | 0.36423 (17) | 0.0526 (5) | |
| H13 | 0.656561 | 0.384241 | 0.382362 | 0.063* | |
| C11 | 0.70949 (9) | 0.21120 (15) | 0.23119 (17) | 0.0537 (5) | |
| C9 | 0.67449 (9) | 0.15188 (14) | 0.41413 (17) | 0.0514 (5) | |
| H9 | 0.668165 | 0.100040 | 0.465852 | 0.062* | |
| C12 | 0.69280 (10) | 0.30451 (15) | 0.25909 (19) | 0.0578 (5) | |
| H12 | 0.698590 | 0.356168 | 0.206558 | 0.069* | |
| C10 | 0.69972 (10) | 0.13515 (15) | 0.30768 (18) | 0.0560 (5) | |
| H10 | 0.710046 | 0.072074 | 0.288146 | 0.067* | |
| C14 | 0.75778 (13) | 0.11035 (19) | 0.0984 (2) | 0.0827 (7) | |
| H14A | 0.774283 | 0.114498 | 0.022456 | 0.124* | |
| H14B | 0.793818 | 0.088726 | 0.173472 | 0.124* | |
| H14C | 0.720706 | 0.064931 | 0.081080 | 0.124* | |
| H22 | 0.5797 (18) | 0.477 (2) | 0.621 (3) | 0.129 (14)* | |
| H21 | 0.578 (2) | 0.478 (3) | 0.485 (4) | 0.159 (18)* | |
| H4N | 0.500000 | 0.205 (2) | 1.250000 | 0.092 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.1079 (7) | 0.0494 (4) | 0.0619 (4) | 0.000 | 0.0413 (4) | 0.000 |
| O1 | 0.0531 (7) | 0.0434 (7) | 0.0391 (6) | 0.0010 (5) | 0.0201 (5) | −0.0004 (5) |
| O2 | 0.0879 (11) | 0.0901 (11) | 0.0591 (8) | 0.0004 (9) | 0.0458 (8) | −0.0021 (8) |
| N4 | 0.0507 (9) | 0.0484 (9) | 0.0375 (7) | 0.0013 (7) | 0.0168 (6) | −0.0010 (6) |
| N1 | 0.0547 (9) | 0.0476 (8) | 0.0434 (8) | −0.0006 (7) | 0.0218 (7) | 0.0003 (7) |
| N2 | 0.0838 (12) | 0.0460 (10) | 0.0518 (9) | −0.0039 (8) | 0.0380 (8) | −0.0053 (7) |
| O3 | 0.1057 (15) | 0.0670 (11) | 0.0814 (13) | 0.0140 (9) | 0.0413 (13) | −0.0026 (9) |
| N3 | 0.0875 (12) | 0.0457 (9) | 0.0520 (9) | −0.0028 (8) | 0.0386 (9) | −0.0040 (7) |
| C3 | 0.0413 (9) | 0.0428 (9) | 0.0346 (8) | −0.0022 (7) | 0.0115 (7) | −0.0011 (7) |
| C1 | 0.0421 (9) | 0.0488 (10) | 0.0371 (9) | −0.0036 (8) | 0.0133 (7) | −0.0054 (7) |
| C2 | 0.0471 (10) | 0.0406 (9) | 0.0395 (9) | −0.0022 (7) | 0.0141 (8) | −0.0009 (7) |
| C7 | 0.0604 (11) | 0.0395 (9) | 0.0421 (9) | 0.0029 (8) | 0.0189 (8) | 0.0033 (7) |
| C8 | 0.0408 (9) | 0.0546 (11) | 0.0361 (8) | −0.0040 (8) | 0.0116 (7) | −0.0016 (8) |
| C6 | 0.0597 (11) | 0.0424 (10) | 0.0457 (10) | 0.0045 (8) | 0.0209 (9) | −0.0018 (8) |
| C4 | 0.0628 (12) | 0.0398 (10) | 0.0493 (10) | 0.0033 (8) | 0.0245 (9) | 0.0018 (8) |
| C5 | 0.0633 (12) | 0.0445 (10) | 0.0476 (10) | 0.0016 (9) | 0.0228 (9) | 0.0079 (8) |
| C13 | 0.0592 (12) | 0.0552 (11) | 0.0469 (10) | −0.0007 (9) | 0.0208 (9) | 0.0019 (8) |
| C11 | 0.0504 (11) | 0.0751 (14) | 0.0397 (9) | −0.0067 (9) | 0.0195 (8) | −0.0036 (9) |
| C9 | 0.0597 (12) | 0.0548 (12) | 0.0446 (10) | −0.0049 (9) | 0.0231 (9) | 0.0002 (8) |
| C12 | 0.0639 (13) | 0.0655 (13) | 0.0484 (10) | −0.0049 (10) | 0.0235 (9) | 0.0070 (9) |
| C10 | 0.0595 (12) | 0.0595 (12) | 0.0534 (11) | −0.0021 (9) | 0.0236 (9) | −0.0085 (9) |
| C14 | 0.0935 (18) | 0.0996 (19) | 0.0706 (14) | −0.0045 (15) | 0.0485 (13) | −0.0172 (13) |
Geometric parameters (Å, º)
| O1—C1 | 1.3634 (19) | C7—H7 | 0.9300 |
| O1—C2 | 1.3637 (19) | C8—C9 | 1.379 (3) |
| O2—C11 | 1.371 (2) | C8—C13 | 1.384 (2) |
| O2—C14 | 1.414 (3) | C6—H6 | 0.9300 |
| N4—C6 | 1.324 (2) | C4—C5 | 1.369 (2) |
| N4—C5 | 1.336 (2) | C4—H4 | 0.9300 |
| N4—H4N | 1.3379 (15) | C5—H5 | 0.9300 |
| N1—C1 | 1.334 (2) | C13—C12 | 1.374 (3) |
| N1—C8 | 1.412 (2) | C13—H13 | 0.9300 |
| N1—H1 | 0.8600 | C11—C10 | 1.371 (3) |
| N2—C1 | 1.298 (2) | C11—C12 | 1.382 (3) |
| N2—N3 | 1.407 (2) | C9—C10 | 1.388 (2) |
| O3—H22 | 0.82 (4) | C9—H9 | 0.9300 |
| O3—H21 | 0.81 (4) | C12—H12 | 0.9300 |
| N3—C2 | 1.277 (2) | C10—H10 | 0.9300 |
| C3—C7 | 1.382 (2) | C14—H14A | 0.9600 |
| C3—C4 | 1.386 (2) | C14—H14B | 0.9600 |
| C3—C2 | 1.454 (2) | C14—H14C | 0.9600 |
| C7—C6 | 1.376 (2) | ||
| C1—O1—C2 | 102.04 (13) | C7—C6—H6 | 118.8 |
| C11—O2—C14 | 117.79 (17) | C5—C4—C3 | 118.79 (16) |
| C6—N4—C5 | 118.91 (15) | C5—C4—H4 | 120.6 |
| C6—N4—H4N | 124.9 (13) | C3—C4—H4 | 120.6 |
| C5—N4—H4N | 116.2 (13) | N4—C5—C4 | 122.43 (16) |
| C1—N1—C8 | 127.16 (15) | N4—C5—H5 | 118.8 |
| C1—N1—H1 | 116.4 | C4—C5—H5 | 118.8 |
| C8—N1—H1 | 116.4 | C12—C13—C8 | 120.64 (18) |
| C1—N2—N3 | 105.04 (14) | C12—C13—H13 | 119.7 |
| H22—O3—H21 | 122 (4) | C8—C13—H13 | 119.7 |
| C2—N3—N2 | 107.14 (14) | C10—C11—O2 | 125.05 (19) |
| C7—C3—C4 | 118.62 (16) | C10—C11—C12 | 119.59 (17) |
| C7—C3—C2 | 122.13 (15) | O2—C11—C12 | 115.36 (18) |
| C4—C3—C2 | 119.25 (15) | C8—C9—C10 | 120.20 (18) |
| N2—C1—N1 | 131.13 (16) | C8—C9—H9 | 119.9 |
| N2—C1—O1 | 113.05 (14) | C10—C9—H9 | 119.9 |
| N1—C1—O1 | 115.82 (15) | C13—C12—C11 | 120.19 (18) |
| N3—C2—O1 | 112.73 (15) | C13—C12—H12 | 119.9 |
| N3—C2—C3 | 127.75 (15) | C11—C12—H12 | 119.9 |
| O1—C2—C3 | 119.50 (14) | C11—C10—C9 | 120.31 (19) |
| C6—C7—C3 | 118.86 (16) | C11—C10—H10 | 119.8 |
| C6—C7—H7 | 120.6 | C9—C10—H10 | 119.8 |
| C3—C7—H7 | 120.6 | O2—C14—H14A | 109.5 |
| C9—C8—C13 | 119.05 (17) | O2—C14—H14B | 109.5 |
| C9—C8—N1 | 123.71 (16) | H14A—C14—H14B | 109.5 |
| C13—C8—N1 | 117.23 (16) | O2—C14—H14C | 109.5 |
| N4—C6—C7 | 122.38 (16) | H14A—C14—H14C | 109.5 |
| N4—C6—H6 | 118.8 | H14B—C14—H14C | 109.5 |
| C1—N2—N3—C2 | −0.2 (2) | C5—N4—C6—C7 | 1.3 (3) |
| N3—N2—C1—N1 | −179.21 (18) | C3—C7—C6—N4 | −0.6 (3) |
| N3—N2—C1—O1 | 0.22 (19) | C7—C3—C4—C5 | 0.5 (3) |
| C8—N1—C1—N2 | −3.2 (3) | C2—C3—C4—C5 | −179.14 (16) |
| C8—N1—C1—O1 | 177.42 (14) | C6—N4—C5—C4 | −1.2 (3) |
| C2—O1—C1—N2 | −0.20 (18) | C3—C4—C5—N4 | 0.3 (3) |
| C2—O1—C1—N1 | 179.33 (14) | C9—C8—C13—C12 | −1.0 (3) |
| N2—N3—C2—O1 | 0.0 (2) | N1—C8—C13—C12 | 178.02 (16) |
| N2—N3—C2—C3 | 178.69 (16) | C14—O2—C11—C10 | −5.0 (3) |
| C1—O1—C2—N3 | 0.08 (18) | C14—O2—C11—C12 | 175.28 (18) |
| C1—O1—C2—C3 | −178.69 (14) | C13—C8—C9—C10 | 0.9 (3) |
| C7—C3—C2—N3 | −174.38 (18) | N1—C8—C9—C10 | −178.10 (15) |
| C4—C3—C2—N3 | 5.2 (3) | C8—C13—C12—C11 | 0.0 (3) |
| C7—C3—C2—O1 | 4.2 (2) | C10—C11—C12—C13 | 1.1 (3) |
| C4—C3—C2—O1 | −176.23 (15) | O2—C11—C12—C13 | −179.10 (17) |
| C4—C3—C7—C6 | −0.3 (3) | O2—C11—C10—C9 | 178.98 (18) |
| C2—C3—C7—C6 | 179.26 (15) | C12—C11—C10—C9 | −1.3 (3) |
| C1—N1—C8—C9 | −0.8 (3) | C8—C9—C10—C11 | 0.3 (3) |
| C1—N1—C8—C13 | −179.82 (16) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4N···N4i | 1.34 (1) | 1.34 (1) | 2.675 (3) | 178 (3) |
| N1—H1···O3 | 0.86 | 2.02 | 2.875 (2) | 172 |
| O3—H22···Cl1 | 0.82 (4) | 2.43 (4) | 3.233 (2) | 166 (3) |
| O3—H21···Cl1ii | 0.81 (4) | 2.55 (4) | 3.359 (3) | 172 (4) |
| C7—H7···Cl1 | 0.93 | 2.93 | 3.7940 (18) | 155 |
| C6—H6···Cl1iii | 0.93 | 2.82 | 3.7137 (18) | 163 |
| C5—H5···N3iv | 0.93 | 2.56 | 3.329 (2) | 140 |
| C9—H9···N2 | 0.93 | 2.34 | 2.969 (2) | 125 |
Symmetry codes: (i) −x+1, y, −z+5/2; (ii) −x+1, −y+1, −z+1; (iii) −x+1, −y+1, −z+2; (iv) x, −y, z+1/2.
References
- Abd-Ellah, H. S., Abdel-Aziz, M., Shoman, M. E., Beshr, E. A. M., Kaoud, T. S. & Ahmed, A. F. F. (2017). Bioorg. Chem. 74, 15–29. [DOI] [PubMed]
- Bharty, M. K., Bharti, A., Dani, R. K., Dulare, R., Bharati, P. & Singh, N. K. (2012). J. Mol. Struct. 1011, 34–41.
- Bitla, S., Sagurthi, S. R., Dhanavath, R., Puchakayala, M. R., Birudaraju, S., Gayatri, A. A., Bhukya, V. K. & Atcha, K. R. (2020). J. Mol. Struct. 1220, 128705.
- Bruker (2004). FRAMBO. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chandra, S., Gond, M. K., Jaiswal, S., Bharty, M. K., Maiti, B., Kushwaha, D. & Butcher, R. J. (2022). J. Mol. Struct. 1249, 131637.
- Gond, M. K., Rai, N., Chandra, B., Gautam, V., Garai, S., Butcher, R. J. & Bharty, M. K. (2023). Dalton Trans. 52, 10213–10221. [DOI] [PubMed]
- Gond, M. K., Shukla, A., Pandey, S. K., Bharty, M. K., Maiti, B., Acharya, A., Tiwari, N., Katiyar, D. & Butcher, R. J. (2022). J. Mol. Struct. 1249, 131547.
- Jaiswal, S., Pandey, S. K., Minocha, T., Chandra, S., Bharty, M. K., Yadav, S. K., Kushwaha, D. & Butcher, R. J. (2023a). J. Mol. Struct. 1281, 135075.
- Jaiswal, S., Pandey, S. K., Prajapati, J., Chandra, S., Gond, M. K., Bharty, M. K., Tiwari, I. & Butcher, R. J. (2023b). Appl. Organom Chem. 37, e7085.
- Jasinski, J. P., Bharty, M. K., Singh, N. K., Kushwaha, S. K. & Butcher, R. J. (2011). J. Chem. Crystallogr. 41, 6–11.
- Jedlovská, E. & Leško, J. (1994). Synth. Commun. 24, 1879–1885.
- Li, J. L., Li, H. Y., Zhang, S. S., Shen, S., Yang, X. L. & Niu, X. (2023). J. Org. Chem. 88, 14874–14886. [DOI] [PubMed]
- Li, X., Liao, S., Chen, Y., Xia, C. & Wang, G. (2021). J. Chem. Res. 45, 1038–1041.
- Najare, M. S., Patil, M. K., Nadaf, A. A., Mantur, S., Garbhagudi, M., Gaonkar, S., Inamdar, S. R. & Khazi, I. A. M. (2020). J. Mol. Struct. 1199, 127032.
- Omar, F. A., Mahfouz, N. M. & Rahman, M. A. (1996). Eur. J. Med. Chem. 31, 819–825. [DOI] [PubMed]
- Paswan, S., Bharty, M. K., Gupta, S. K., Butcher, R. J. & Jasinski, J. P. (2016). IUCrData, 1, x161724.
- Paswan, S., Bharty, M. K., Kumari, S., Gupta, S. K. & Singh, N. K. (2015). Acta Cryst. E71, o880–o881. [DOI] [PMC free article] [PubMed]
- Reid, J. R. & Heindel, N. D. (1976). J. Heterocycl. Chem. 13, 925–926.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. A71, 3–8.
- Singh, N. K., Butcher, R. J., Bharty, M. K., Srivastava, A. K. & Tripathi, P. (2006). Acta Cryst. E62, o3473–o3474.
- Singh, N. K., Butcher, R. J., Tripathi, P., Srivastava, A. K. & Bharty, M. K. (2007). Acta Cryst. E63, o782–o784.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Wu, C. A., Chou, H. H., Shih, C. H., Wu, F. I., Cheng, C. H., Huang, H. L., Chao, T. C. & Tseng, M. R. (2012). J. Mater. Chem. 22, 17792–17799.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024002044/hb8088sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024002044/hb8088Isup2.hkl
Spectra. DOI: 10.1107/S2056989024002044/hb8088sup3.docx
Supporting information file. DOI: 10.1107/S2056989024002044/hb8088Isup4.cml
CCDC reference: 2238764
Additional supporting information: crystallographic information; 3D view; checkCIF report