Abstract
Previous studies by our group showed that infection of human and rodent cells by human adenovirus type 5 (Ad5) results in the induction of p53-independent apoptosis and cell death that are dependent upon transactivation of early region 4 (E4). To identify which E4 products are involved, studies were conducted with p53-deficient human SAOS-2 cells infected with various Ad5 E4 mutants. An E4orf6-deficient mutant was defective in cell killing, whereas another that expressed only E4orf6 and E4orf4 killed like wild-type virus, suggesting that E4orf6 may be responsible for cytotoxicity; however, a mutant expressing only E4orf4 induced high levels of cell death, indicating that this E4 product may also be able to induce cytotoxicity. To define the E4 cell death-inducing functions more precisely, cDNAs encoding individual E4 products were introduced into cells by DNA transfection in the absence of other Ad5 proteins. In cotransfections with a cDNA encoding firefly luciferase, enzymatic activity was high in all cases except with E4orf4, where luciferase levels were less than 20% of those in controls. In addition, drug selection of several cell types following transfection with retroviral vector DNA encoding individual E4 products as well as puromycin resistance yielded a large number of cell colonies except when E4orf4 was expressed. These data demonstrated that E4orf4 is the only E4 product capable of independent cell killing. Cell death induced by E4orf4 was due to apoptosis, as evidenced by 4′,6-diamidino-2-phenylindole (DAPI) staining of cell nuclei in E4orf4-expressing cells. Thus, although E4orf6 may play some role, these results suggested that E4orf4 may be the major E4 product responsible for induction of p53-independent apoptosis.
Many viruses induce apoptosis as part of their natural life cycle (67). Killing of infected cells by this process is advantageous to the virus because it reduces both the inflammatory response and exposure to the host immune system and degradative enzymes. Human adenoviruses (Ads) have developed complex processes that regulate both the induction and suppression of apoptosis. Expression of early region 1A (E1A) products causes a rise in p53 levels, due at least in part to the stabilization of p53, and induces p53-dependent apoptosis (5, 13, 23–25, 40, 58). Recently we (55) and others (11) mapped such effects in rodent cells to the region of E1A products involved in binding the p300 family of cellular transcriptional modulators. We have also found that in normal human cells, binding of the retinoblastoma tumor suppressor (RB) family alone can elicit this effect (55), suggesting that p53 stabilization may be an unavoidable by-product of the induction of unscheduled DNA synthesis caused by complex formation with p300 or RB family members (30, 81). The occurrence of p53-dependent apoptosis early during infection would severely reduce virus production and thus limit the spread of virus in the infected host. E1A is oncogenic in rodent cells, but its ability to yield stable transformants is greatly limited by this p53-dependent cell death. Ads have evolved at least three individual viral products that suppress p53-dependent pathways and thus promote viral replication and cell transformation. Early region 1B (E1B) encodes two such proteins. The E1B 55-kDa protein binds to p53 (62) and blocks both p53-mediated activation of gene expression (68, 69, 79–81) and apoptosis (41, 68). The E1B 19-kDa protein appears to suppress apoptosis by a mechanism functionally analogous to that of the cellular proto-oncogene product Bcl-2 (4, 49, 56, 78). Cells infected with Ad mutants that fail to express the 19-kDa protein display enhanced cytotoxicity and extensive degradation of both cellular and viral DNA into nucleosome-sized fragments, a characteristic of apoptosis (17, 49, 52, 65, 77, 78). Recently, the orf6 protein encoded by early region 4 (E4orf6) has also been found to bind to and inactivate p53 (15, 54). Such interactions block p53-dependent transactivation activity (15, 48) and induce the rapid degradation of p53 protein (48, 54).
In addition to p53-dependent apoptosis, both our group (70) and Subramanian et al. (66) showed that in the absence of E1B, E1A products also cause p53-independent cell death. We found (71) that such cell death exhibited all of the hallmarks of apoptosis, including degradation of DNA to nucleosome-sized fragments, extensive chromosomal condensation, and formation of cytoplasmic vacuoles (36, 65). This effect was found to rely on the ability of the large E1A protein to transactivate E4 (42), suggesting that one or more E4 products may be cytotoxic. Furthermore, in the presence of E1B, cell killing at the final stages of the infectious cycle was prevented or greatly delayed in the absence of E4 products (42), suggesting that this E4-dependent, p53-independent apoptosis may represent a major mechanism for the ultimate death of the infected cell and spread of progeny virions.
Figure 1 shows that E4 encodes seven known proteins as determined by analysis of cloned cDNAs deriving from several open reading frames (orfs) (18, 29, 72). E4orf6 is a 34-kDa protein that is involved in host shutoff and transport of viral late mRNAs following complex formation with the E1B 55-kDa polypeptide (7, 9, 53, 59, 60). As noted above, E4orf6 also binds to and inactivates p53 (15, 54) and can cooperate with E1A to enhance cell transformation (48). E4orf3 is an 11-kDa species that also interacts with the E1B 55-kDa protein (38) and is associated with the nuclear matrix (38, 61). It is involved in accumulation of late viral mRNA (16, 62) and, along with E4orf4 and E4orf6, appears to affect viral DNA synthesis (18). E4orf6/7 is a 17-kDa species that binds as a homodimer to transcription factor E2F to promote E2 promoter activity by ensuring correct spacing and orientation (12, 31, 43, 47, 50, 57). E4orf4 is a 14-kDa species that has been reported to bind to and activate protein phosphatase 2A (35). It may play a role in the regulation of DNA synthesis (8) and AP-1 activity (46). Mutants defective for E4orf4 produce an enhanced cytopathic effect (46). E4orf1 of Ad9 appears to be capable of cell transformation (32, 33, 73–75), possibly through the formation of complexes with several unidentified cellular proteins (75). Little is known about the E4orf2 or E4orf3/4 products. In the present studies, we found that E4orf4, even when expressed in the absence of other viral proteins, is highly cytotoxic, although E4orf6 may also play some role in Ad-induced cell death.
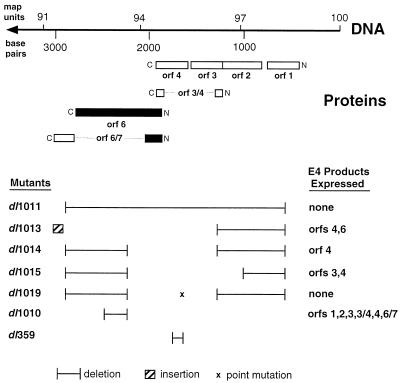
FIG. 1.
Ad5 E4 and E4 mutants. At the top is right end of the Ad5 genome including positions in base pairs and map units. The locations of two SmaI restriction enzyme sites used in the preparation of E4 mutants are also presented. Below are the positions of several open reading frames for E4 proteins that are encoded from right (amino terminus) to left (carboxy terminus). At the bottom are the structures of several E4 deletion mutants. The E4 proteins expressed by these mutants are summarized at the right.
MATERIALS AND METHODS
Cells and viruses.
Human SAOS-2 cells (ATCC HTB 85) that are deficient for p53 (and pRB) expression were cultured on 60-mm-diameter dishes (Corning Glass Works, Corning, N.Y.) in alpha-modified minimal essential medium supplemented with 10% fetal calf serum, as were LR73 Chinese hamster ovary (CHO) cells, human HeLa cells, 293 cells, and Ad5 E1A-expressing 1A.A3 and 1A.A6 mouse embryo fibroblast cells that were derived from p53-null mice and grown under selective conditions in the presence of 100 μg of hygromycin per ml (39). Normally, cells were infected with mutant or wild-type Ad5 at a multiplicity of infection of 35 PFU per cell (if subjected to titer determination on 293 cells) or 10 PFU per cell (if subjected to titer determination on W162 cells), as described previously (69). The virus used as wild type has been described by Harrison et al. (28). Ad5 E4 mutants are illustrated in Fig. 1 and include dl1019 and dl1011, which express no E4 products; dl1013, which expresses only E4orf4 and E4orf6; dl1014, which expresses only E4orf4; dl1015, which expresses only E4orf3 and E4orf4; dl1010, which expresses all E4 products except E4orf6; and dl359 (26), which is defective for E4orf4 alone. The profiles of these mutants in terms of expression of E4orf4, E4orf6, and E4orf6/7 were confirmed by Western blotting analysis with antisera that recognize these viral products. All these mutants were propagated on W162 monkey cells, as described previously (6), with the exception of dl359, which was grown on 293 cells (21).
Ad vectors expressing E4 products.
cDNAs expressing E4orf6 (or other E4 products) were subcloned into pCA14, a cytomegalovirus (CMV) promoter-containing plasmid that was then used to produce Ad vectors expressing E4 products (AdE4orf6, AdE4orf1, etc.), as previously described (1, 41, 54).
Cell viability assays.
Cells were infected with wild-type or mutant Ad5 in 24-well plates containing cells at about 80% confluence. At various times following infection, adherent and nonadherent cells were pooled and viability was assessed by trypan blue exclusion. At least 300 cells were counted at each time point.
Plasmids.
cDNAs expressing individual E4 products were obtained from Goran Akusjärvi, who showed that they all yielded appropriate mRNA products (51). The E4 cDNAs were subcloned into the multicloning site of pcDNA3, a CMV-driven mammalian expression vector that also expresses the neo drug resistance gene (Invitrogen). From pcDNA3, the E4-specific cDNAs were subcloned into pBABEpuro (45), a retrovirus expression vector that contains the puromycin drug resistance gene. In addition, a hemagglutinin (HA) epitope tag was inserted at the 5′ end of the E4orf4 coding sequence by standard PCR techniques (10). The HA tag was found to have no effect on the ability of E4orf4 to induce cell killing (37). The Rous sarcoma virus LTR-driven luciferase plasmid used in the firefly luciferase assay has been described previously (20).
Luciferase assay.
The luciferase killing assay was carried out with 1A.A3 or 1A.A6 cells plated the day before at a density of 1.5 × 105 cells per well in six-well plates. The DNA mixture was introduced into cells by calcium phosphate transfection (22) and consisted of 0.5 μg of Rous sarcoma virus LTR-driven luciferase plasmid DNA and 3 μg of pcDNA3 plasmid DNA (or pcDNA3 containing an E4 orf), with 2.5 μg of sonicated salmon sperm DNA as a carrier. The cells were harvested 48 h posttransfection, and luciferase activity was determined after freeze-thaw disruption of the cells, as described previously (36).
Cell colony-forming assays.
Mouse 1A.A3 and 1A.A6 cells and human SAOS-2 and HeLa cells were plated at a density of 10% in six-well dishes, and each well was transfected with 1.5 μg of DNA from pBABEpuro (or an E4-containing derivative) and 1.5 μg of sonicated salmon sperm DNA carrier. The cells were replated 48 h posttransfection on 100-mm dishes in the presence of puromycin (1 μg/ml). Two weeks after the onset of drug selection, the cells were fixed in methanol-acetic acid (3:1, vol/vol) and stained with Giemsa (0.15 mg/ml in phosphate-buffered saline [PBS]) and colonies were counted.
Analysis of DNA content by DAPI staining.
CHO and human 293 cells were transfected with 12 μg of DNA from pcDNA3 expressing HA-tagged E4orf4 together with 3 μg of pHOOK vector DNA (Invitrogen), and 24 h after transfection the cells were magnetically sorted by using Capture-Tec beads (Invitrogen) to isolate pHOOK-expressing cells. The cells were then plated on glass coverslips, and 24 h later they were washed in PBS containing 1 mM MgCl2 and then fixed for 20 min in 3.7% formaldehyde in PBS. The cells were permeabilized in 0.02% Triton X-100–PBS for 5 min, after which mouse anti-HA HA.11 (Babco) was added followed by Texas Red-conjugated anti-mouse immunoglobulin G (Molecular Probes) to detect HA-orf4. The nuclear morphology of the cells was analyzed by DNA staining with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes).
Antisera and immunoblotting.
HeLa cells growing in 60-mm-diameter dishes were infected with wild-type or E4 mutant Ad5, and at 16 h postinfection (p.i.) the cells were harvested by scraping into PBS, washed, and lysed in 10 mM HEPES–KOH (pH 7.4), containing 142 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 0.2% Nonidet P-40. Clarified cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 50 μg of protein per lane. Separated proteins were transferred to nitrocellulose, and the filters were immunoblotted with appropriate antibodies. For E1A proteins, mouse monoclonal antibody M73 (27) was used at a 1/2,000 dilution. For E4orf4, a rabbit polyclonal antibody raised against the carboxy terminal 28 residues fused to glutathione S-transferase was prepared for this study and used at a 1/200 dilution. For E4orf6, a rabbit polyclonal serum raised against the amino-terminal 46 residues of E4orf6 fused to glutathione S-transferase was used at a 1/500 dilution.
RESULTS
Analysis of cell killing by Ad5 E4 mutants.
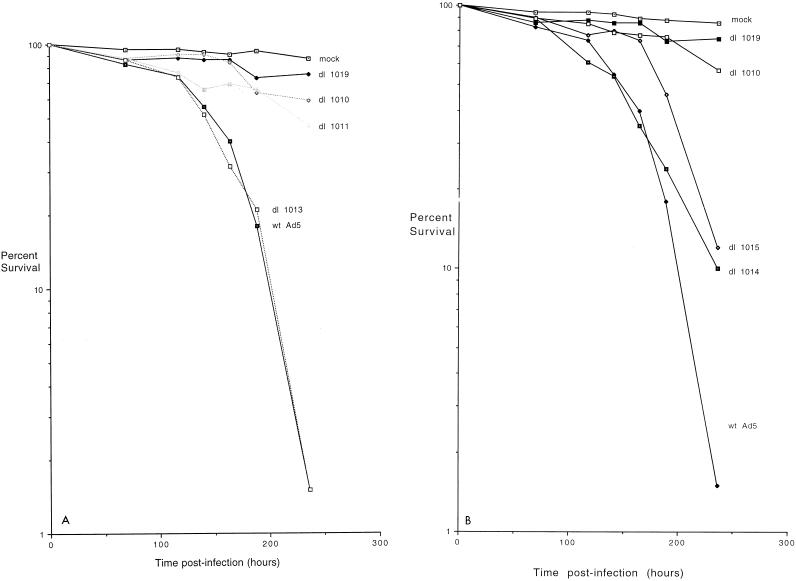
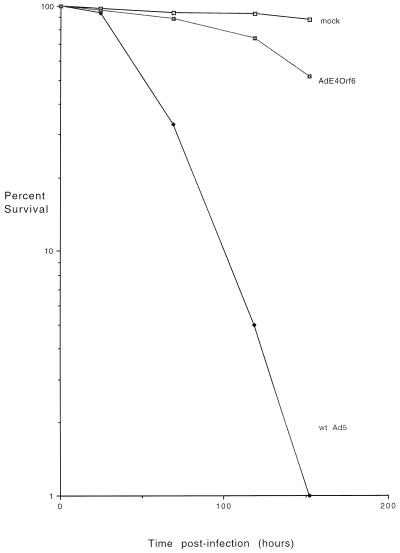
Previous studies indicated that in the presence of E1A proteins, one or more E4 products were required for Ad5 E1A-induced p53-independent apoptosis (42). To identify which E4 products are involved in this p53-independent cell killing, experiments were carried out with various E4 mutants. It was not possible to examine rapid cell killing that occurs in the absence of E1B because E1B-E4 double mutants were not available and were difficult to construct. Therefore, human p53-deficient SAOS-2 cells were infected with wild-type Ad5 or with mutants with defects in the E4 coding region and cell viability was measured by exclusion of trypan blue stain. These viruses all express wild-type E1B products, and thus cell death did not occur until late during infection. Figure 2A shows, as found previously (42), that wild-type virus began to kill cells at about 125 h p.i. and that by 240 h p.i. virtually all the cells were dead, whereas with mutants dl1019 and dl1011, which produce no E4 products, the cells remained almost as viable as in mock-infected cultures. A similar effect was also noted with mutant dl1010 that expresses all E4 products except E4orf6. Cell killing by mutant dl1013, which expresses only E4orf6 and E4orf4, was similar to that by wild-type virus. These results suggested that E4orf6 may be involved in Ad5-induced cell death but left open the possibility that E4orf4 also plays some role.
FIG. 2.
Viability of SAOS-2 cells infected with mutant and wild-type (wt) Ad5. p53-deficient human SAOS-2 cells were mock infected or infected with wild-type Ad5 or a series of mutants which contain defects in E4, and at various times following infection they were tested for viability by a trypan blue exclusion assay, as described in Materials and Methods. The results are presented as the logarithm of the percentage of viable cells. Panels A and B show two separate representative studies involving two sets of E4 mutants.
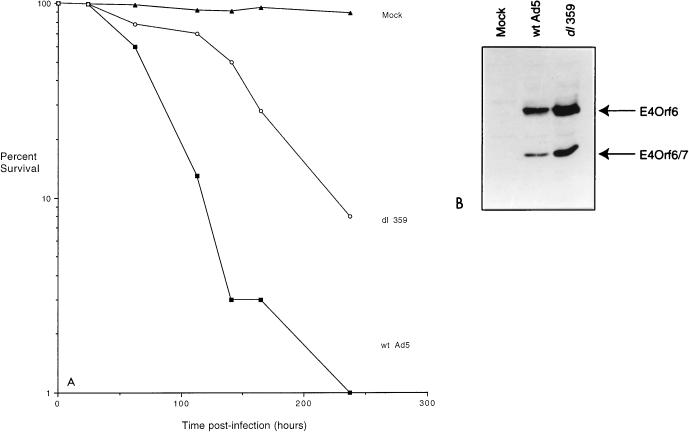
Figure 2B shows the results of a similar experiment with additional E4 mutants. Again, mutant dl1019 failed to kill infected cells as efficiently as did mutant dl1010, which produces all E4 products except E4orf6; however, mutant dl1014, which produces only E4orf4 and none of the other E4 products, killed quite efficiently. Infection with mutant dl1015, which produces only E4orf4 and E4orf3, also resulted in significant cell killing, although over several experiments (data not shown), this killing was always found to be somewhat delayed relative to that by wild-type virus or dl1014. These results indicated that in virus-infected cells E4orf4 alone is able to support efficient cell death and that the presence of E4orf3 might limit this effect somewhat. Thus the data in Fig. 2 suggested that in virus-infected cells E4orf4 and E4orf6 may synergize to produce efficient cell killing. E4orf4 appears to harbor the major killing activity; however, this effect seems to be reduced by another E4 product in the absence of E4orf6. Although the toxic effect of E4orf6 was not tested directly in these experiments, studies that address this issue have been performed (see below). In addition, we were aware of a previous report (46) suggesting that mutant dl359, which expresses all E4 products except E4orf4, produces a greater cytopathic effect than wild-type virus (see below and Fig. 8).
FIG. 8.
Analysis of cell killing and E4 expression by wild-type (wt) Ad5 and mutant dl359. SAOS-2 cells were infected with wild-type Ad5 or mutant dl359 that is defective for E4orf4 expression, and at various times after infection cell viability was assessed by trypan blue exclusion as in Fig. 2. (A) Cell viability by trypan blue staining. Data are expressed as percent cell viability. (B) Expression of E4orf6 and E4orf6/7. Portions of the cultures were harvested at 16 h p.i. and analyzed by immunoblotting as in Fig. 7 with an antiserum that recognizes the amino terminus of E4orf6 and E4orf6/7.
Analysis of cell killing by individual E4 products.
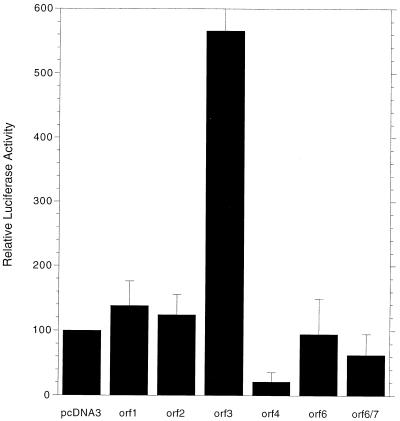
To examine the specific potential of E4 products to induce cell death, cDNAs expressing individual E4 products were subcloned into plasmid pcDNA3, which expresses inserts under the CMV promoter. Previous studies (51) indicated that these cDNAs express appropriate E4 mRNAs. We have confirmed directly by Western blotting analysis that high levels of E4orf4, E4orf6, and E4orf6/7 proteins are synthesized from the corresponding constructs (data not shown). DNA from these plasmids was used to cotransfect p53-null 1A.A3 and 1A.A6 cells expressing Ad5 E1A products, along with DNA encoding firefly luciferase, and after 48 h the cell extracts were assayed for luciferase activity. It was assumed that high luciferase activity was consistent with a low degree of cell killing, whereas if extensive cell death occurred, luciferase activity would be reduced. Figure 3 shows the combined results of five separate experiments. Cells cotransfected with plasmids expressing E4orf1 or E4orf2 exhibited luciferase activities equal to or slightly greater than did cells cotransfected with pcDNA3 alone, suggesting that these E4 products were not highly toxic. Coexpression of E4orf3 resulted in considerably increased levels of luciferase activity. E4orf3 is known to induce effects on mRNA metabolism and splicing (6, 7), and thus this increase may have resulted from an enhancement of expression of luciferase mRNA from the CMV promoter (see below). Coexpression of E4orf6/7 reduced luciferase activity by a modest amount. Interestingly, coexpression of E4orf6 generally had little effect on luciferase activity, although in some experiments a modest reduction was observed. For E4orf4, luciferase activity was reduced by 80 to 90%, suggesting that this E4 product might induce cell death. Coexpression of both E4orf4 and E4orf6 resulted in even slightly lower levels (data not shown). Thus, these data were consistent with the conclusion that E4orf4 is the only E4 product capable of inducing p53-independent cell death when expressed alone. Interpretation of these assays assumed that the E4 products had no direct effect on luciferase expression from the CMV promoter, but this may not be the case. As already described above, increased luciferase activity found with E4orf3 may have resulted from an increase in luciferase expression. Effects of this nature with other E4 products might also affect our ability to interpret cell killing accurately. For example, E4orf4 has been shown to reduce transcription from certain promoters (2, 35), although there is no evidence that the CMV promoter is affected. In addition, E4orf6, in association with the E1B 55-kDa protein, plays a role in enhancing viral mRNA stability and transport (6, 7, 14, 19), and thus cell killing by this protein might be balanced by a concomitant increase in luciferase expression. Clearly, an assay which more directly assesses cell death was warranted.
FIG. 3.
Analysis of cell killing by measurement of luciferase activity. 1A.A6 cells were cotransfected with pcDNA3 plasmids expressing firefly luciferase and individual Ad5 E4 proteins. After 48 h the cells were harvested and the extracts were analyzed for luciferase activity as described in Materials and Methods. The data represent the average of five separate experiments, each involving duplicate samples (standard errors are indicated by lines). The luciferase activity obtained with the pcDNA3 alone control was set at 100%.
One approach was to construct Ad vectors that express each of the E4 products. Such vectors were easily constructed for E4orf1, E4orf2, E4orf3, and E4orf6/7. Only a single plaque was obtained for E4orf6 during five separate attempts; however, as shown previously, such a construct, AdE4orf6, was produced and shown to express wild-type E4orf6 at levels about two- to threefold higher than in Ad5-infected cells (reference 54 and data not shown). We were unable to generate a vector expressing E4orf4 even after many attempts, as might be expected if E4orf4 is highly toxic. The Ad vectors were used to infect SAOS-2 cells; however, none caused any cytopathic effect even after many days (data not shown). Because the data presented above indicated that E4orf6 might play a role in inducing cell death, the AdE4orf6 vector was studied in more detail. SAOS-2 cells were infected with AdE4orf6 or with wild-type virus, and cell killing was measured by the trypan blue exclusion assay as in Fig. 2. Figure 4 shows that cell viability was maintained in AdE4orf6-infected cells to a level similar to that in mock-infected cells, whereas with wild-type virus, cell death occurred as before. These results confirmed that E4orf6, which was expressed at high levels in these cells, possesses little cytotoxic activity when expressed alone.
FIG. 4.
Analysis of cell killing by an Ad vector expressing E4orf6. SAOS-2 cells were infected either with wild-type (wt) Ad5 or with Ad vector AdE4orf6 that expresses only E4orf6. Cell killing was measured by trypan blue exclusion as in Fig. 2.
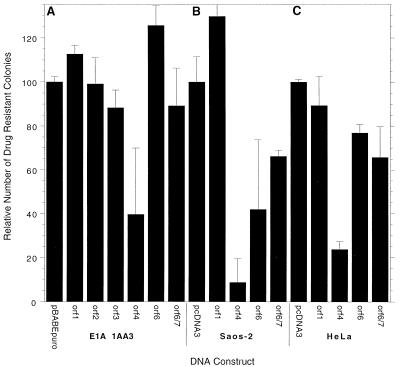
The next approach was to measure the effects of E4 products on colony formation following drug selection. For this assay, cDNAs encoding E4 products were inserted into the pBABE retroviral vector, which also contains the gene for puromycin resistance, and colony formation was measured following selection by puromycin of transfected 1A.A6 mouse cells and human SAOS-2 or HeLa cells. Figure 5A shows that colony formation with mouse 1A.A6 cells following puromycin selection was similar to that of controls with plasmids expressing all of the E4 products except for E4orf4, for which colony formation was inhibited by over 60%. Similar results were also obtained in a more limited study with human SAOS-2 and HeLa cells (Fig. 5B and C). In these cases, colony formation was inhibited by 80 to over 90% by E4orf4, although some reduction was also seen with E4orf6 and E4orf6/7 with SAOS-2 cells. These data indicated that E4orf4 exhibits the greatest capacity to block colony formation and that the other E4 products either had no effect or produced only a modest reduction. Reduction in colony formation could result either from growth arrest or from cell killing. We have not made a detailed study of the properties of the colonies produced with constructs containing E4orf4; however, most were much smaller than those produced by controls. Thus, it was possible that these colonies were produced because of low levels of expression of E4orf4.
FIG. 5.
Analysis of E4 killing by colony inhibition. E1A-expressing, p53-minus 1A.A6 cells (A), SAOS-2 cells (B), or HeLa cells (C) were transfected with the puromycin resistance-containing pBABE retroviral plasmid expressing individual E4 products. The cells were plated and then grown in the presence of puromycin for 14 days, as described in Materials and Methods, after which time the number of colonies was tabulated. The results are the mean of four (A) or two (B and C) experiments involving two plates of each E4orf per experiment (standard error indicated by bars). Colony formation in the presence of pcDNA3, which varied between about 80 and 120 per plate, was set at 100%.
Expression of E4orf4 induces apoptosis.
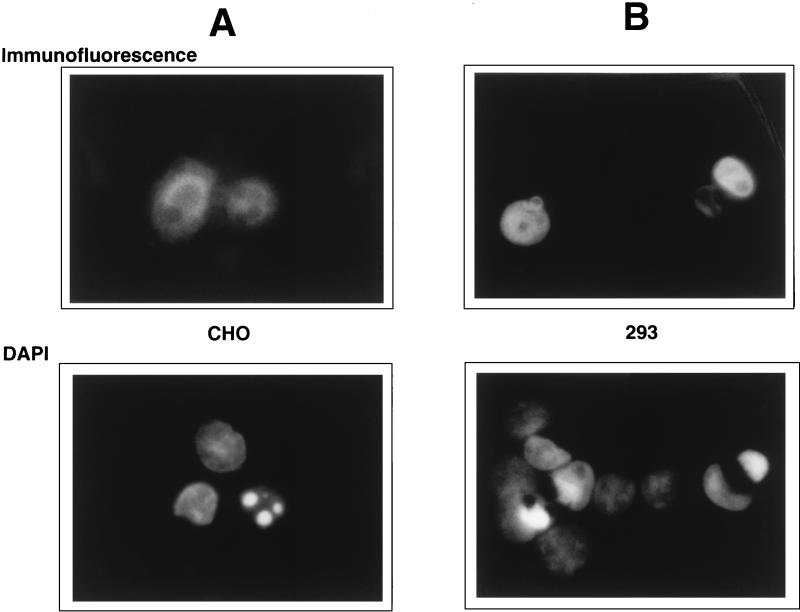
The assays described so far indicated that E4orf4 alone among the E4 products exhibited significant effects on luciferase expression and colony formation that were consistent with cell killing. It was not possible in these types of experiments to measure cell death directly, since only a small percentage of cells in such cultures express E4orf4 introduced exogenously. We believed from previous studies (42, 69) that the effects caused by E4orf4 were due to p53-independent apoptosis. To determine if expression of E4orf4 induces apoptosis, CHO cells and human 293 cells were cotransfected with DNA from a pcDNA3 plasmid that expresses an HA-tagged version of E4orf4 together with that of the pHOOK vector. Cells were magnetically sorted by using Capture-Tec beads to isolate cells expressing pHOOK. Selected cells were examined both by immunofluorescence with an anti-HA antibody and by DAPI staining. As is well known, the DAPI technique allows identification of apoptotic cells because of the presence of irregularly shaped nuclei, in some cases containing foci of bright fluorescence. Figure 6 shows representative fields of cells treated by these two methods. The upper panels in Fig. 6A (CHO cells) and Fig. 6B (293 cells) show the pattern of immunofluorescence obtained with anti-HA antibody. Expression of E4orf4 was largely nuclear, with some diffuse cytoplasmic staining. The lower panels show the same fields visualized by DAPI staining. It is clear that cells that lack detectable levels of E4orf4 exhibited DAPI-stained nuclei of normal morphology; however, cells that expressed E4orf4 contained nuclei that either were irregular in shape or contained brightly stained regions that are characteristic of highly condensed chromatin. This correlation was consistent throughout the entire cell population. These results suggested that expression of E4orf4 induces apoptosis. This contention was confirmed in extensive separate studies with CHO cell lines expressing E4orf4 under an inducible promoter (37). Following induction of E4orf4 expression, cells exhibited a number of changes characteristic of apoptosis, including rapid cell death, DNA degradation, and the appearance of phosphatidylserine on the outer cell surface, as shown by staining with annexin 5. It is clear therefore that E4orf4 is toxic and induces apoptosis.
FIG. 6.
CHO (A) or 293 (B) cells were transfected with plasmid pcDNA3 expressing an HA-tagged version of E4orf4 together with the pHOOK plasmid to facilitate magnetic sorting of transfected cells (see Materials and Methods). After sorting, the cells were analyzed either by immunofluorescence with an antibody directed against HA or by staining for DNA with DAPI. A single representative field for each cell line is presented.
Further analysis of the roles of E4orf4 and E4orf6 in cell killing.
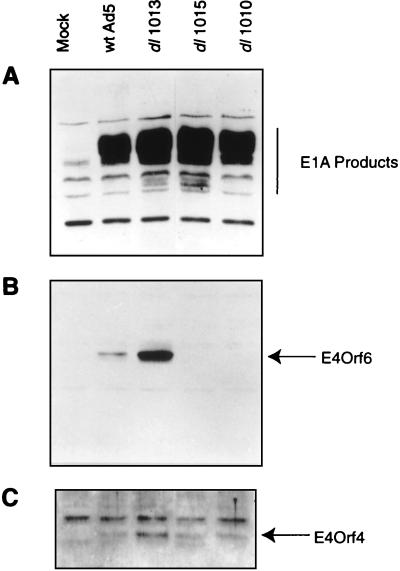
The results described above indicated that E4orf4 appeared to be the major E4 product involved in the induction of p53-independent apoptosis. The question remained of why mutant dl1010, which encodes E4orf4 but not E4orf6, failed to kill cells in the experiments in Fig. 2. One explanation was that in the absence of E4orf6, but in the presence of the other E4 products, expression of E4orf4 could be reduced. To study this possibility, HeLa cells were infected with wild-type Ad5 or with the E4 mutant dl1010, dl1013, or dl1015 and at 16 h p.i. the cells were harvested and extracts were analyzed for the presence of the E1A proteins and E4orf6 and E4orf4 by immunoblotting using appropriate antisera. Figure 7A shows that wild-type and all mutant viruses contained approximately equal amounts of E1A products. The levels of E4orf6 (Fig. 7B) and E4orf4 (Fig. 7C) with dl1013 were somewhat elevated relative to those of the wild-type. Mutants dl1010 and dl1015 produced no E4orf6, as expected; however, the levels of E4orf4 were similar to that seen with the wild type. Similar results were also obtained with SAOS-2 cells, although the overall levels of all Ad5 proteins, including E4orf4, were lower than with HeLa cells (data not shown). Thus, the failure of cell killing by dl1010 must be due to some mechanism other than a reduction in E4orf4 expression (see Discussion).
FIG. 7.
Analysis of viral protein expression with wild-type and E4 mutant Ad5. HeLa cells were infected by wild-type Ad5 or mutants dl1010, dl1013, or dl1015 and harvested at 16 h p.i. Cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted. (A) Anti-E1A M73 monoclonal antibody. (B) Anti-E4orf6 serum. (C) Anti-E4orf4 serum.
Another question relates to the previously observed phenotype of cells infected with mutant dl359 that is defective for E4orf4 expression but wild type for other E4 proteins. dl359 has been reported to elicit a greater cytopathic effect than wild-type virus, as evidenced by the extensive destruction of cell monolayers (46). How could E4orf4 represent the major cytotoxic E4 protein if, in its absence, dl359 elicits this effect? To examine this question, SAOS-2 cells were infected either with wild-type Ad5 or with dl359 and cell viability assays were conducted as in Fig. 2. It should be noted that the kinetics of cell killing in these experiments cannot be directly compared to those in Fig. 2 because the viruses used for the present experiment were subjected to titer determination on 293 cells whereas the viruses used in the experiment in Fig. 2 had to be subjected to titer determination on monkey W162 cells. For this assay (as in others), both adherent and nonadherent cells were collected and examined. It was readily apparent by microscopic examination that in dl359-infected cultures, cells began to round up and detach much earlier than did those infected with wild-type virus, so that by about 60 h p.i., when most of the cells in wild-type-virus-infected cultures remained attached, a large percentage of mutant-infected cells had already detached (data not shown). Nevertheless, Fig. 8A shows that in terms of cell viability, dl359-infected cells did not die more rapidly than those infected with wild-type virus but, rather, that death was considerably delayed. As found previously (46), Fig. 8B shows that expression of both E4orf6 and E4orf6/7, as detected by immunoblotting with an antiserum that recognizes the amino terminus of both of these proteins, was higher in dl359-infected cells than in those infected with wild-type Ad5. This effect is believed to be due to the role of E4orf4 in reducing expression of E4 transcripts (2, 3, 76). Thus, while it appeared that dl359 did cause the early detachment of cells from the monolayer, the cells that were released were still viable, at least by the criterion of trypan blue exclusion. These results were entirely consistent with a major role for E4orf4 in cell killing. The data also clearly suggested that additional mechanisms for the generation of cytopathic effects that involve other E4 proteins and/or additional viral products must exist.
DISCUSSION
The major goal of the present studies was to establish which Ad5 E4 products are responsible for the induction of p53-independent apoptosis. Analysis of cell death induced by E4 mutant viruses in the presence of E1B yielded a somewhat paradoxical result. Cell death was inhibited by using a mutant that produced all E4 products except E4orf6, suggesting that this product plays a role in cytotoxicity. A mutant expressing only E4orf6 and E4orf4 killed infected cells as efficiently as did wild-type virus; however, a high degree of cell killing was obtained with a virus expressing E4orf4 as the only E4 product. These results clearly indicated that E4orf4 was sufficient among E4 products for induction of cell death. We recognize that analyses involving virus-infected cells may be complicated by the effects of multiple viral products that may affect E4orf4-dependent cell death or that may participate in parallel death pathways. It is unlikely that results obtained with E4 mutant viruses can be explained by differences in the kinetics of the early phase of the infectious cycle, since previous studies indicated that such mutants exhibit only slight or moderate delays relative to wild-type virus (6). In fact, these earlier studies indicated that the largest effects on progression of the early phase were with dl1014, which killed almost as well as did the wild type. In addition, successful entry into the late phase does not appear to be critical for killing, since our previous work showed that such killing could be demonstrated in infected rodent cells that fail to replicate virus and express significant levels of late viral products (42, 70). We are still uncertain about the roles of other E4 products, but the simplest explanation is that in virus-infected cells, E4orf6 may enhance killing by E4orf4, perhaps by counteracting the effects of one or more of the other E4 products that may suppress this effect. Such suppression was seen to some degree with mutant dl1015, which expresses only E4orf4 and E4orf3 and caused cell death slightly more slowly than did dl1014, which produces only E4orf4. The molecular basis for such suppression is unclear. One possibility was that in the absence of E4orf6 the levels of expression of E4orf4 were reduced, thus limiting the toxic effects of the virus; however, this did not appear to be the case, since the level of E4orf4 synthesized in mutant-infected cells was similar to that obtained with wild-type virus (Fig. 7). It would be of interest to identify which of the E4 products suppresses E4orf4-dependent cell death and how E4orf6 blocks this inhibition or otherwise promotes cytotoxicity. E4orf6 is a multifunctional protein and plays a role in the regulation of p53 function and stability (15, 44, 48, 54, 55, 60); it cooperates with the E1B 55-kDa protein in the regulation of late viral mRNA transport and host cell shutoff (14, 19). Generation of the appropriate E4orf6 mutants should permit the mapping of the E4orf6 effect.
E4orf4 was found to induce apoptosis when expressed alone in the absence of all other Ad5 products. The induction of apoptosis was confirmed by the appearance of abnormal DAPI-stained nuclei in E4orf4-expressing cells. In separate studies involving continuous CHO cell lines expressing E4orf4 under an inducible promoter, we found that expression of E4orf4 induces cell death associated with classic morphological changes, degradation of DNA into nucleosome-sized fragments, and an exchange of phosphatidylserine from the inside to the outside of the plasma membrane as visualized by staining with annexin 5 (37). All of these properties are specific characteristics of apoptosis. Additionally, recent work by Shtrichman and Kleinberger has shown that 293, H1299, and transformed NIH 3T3 cells undergo apoptosis in response to E4orf4 expression (64). Previous studies showed that E4-dependent cell death was due to p53-independent apoptosis (42, 70), and it is likely that E4orf4-induced apoptosis is also unrelated to p53 status since it occurred in both CHO cells that contain very low endogenous levels of p53 and in 293 cells that express high levels of p53 due to the presence of E1A and E1B products (reference 37 and this report).
The role of E4orf6 in promoting cell death remains unclear. Like the other E4 products, E4orf6 exhibited little cytotoxicity when expressed in the absence of other viral products. As discussed above, perhaps its role is simply to enhance E4orf4-dependent cell death. It is also possible that, in the context of viral infection, it plays a more direct role in cell killing. Mutant dl359, which is defective in E4orf4, yielded an interesting phenotype. As found previously (46), dl359-infected cells exhibited a greater cytopathic effect than did wild-type-Ad5-infected cells; however, we found that the cells that detached rapidly from the cell monolayer were not dead by the criterion of trypan blue exclusion but, rather, took longer to die than did those from wild-type-infected cultures. These results supported the conclusion that E4orf4 is a major inducer of cell death, but they also indicated that other mechanisms for Ad5-induced cytopathogenicity must exist. One product that has been implicated in cell death is the E3 14.7-kDa product or “adenovirus death protein” (71). Its role in this process remains unclear, but the absence of cell death with many of the E4 mutants described in Fig. 2 suggests that it may function in cooperation with E4 products. It is known that E4orf4 suppresses E4 transcription (2, 3, 35, 76), and thus with dl359, expression of E4orf6 and E46/7 was found to be somewhat elevated (Fig. 8). It is possible that these higher levels of E4orf6, perhaps in combination with one or more additional early or late viral proteins, result in a second form of Ad5-induced cytotoxicity.
The major finding in the present work remains the ability of E4orf4 to induce p53-independent apoptosis independently. The only biochemical function associated with this E4 product is its ability to bind to and activate the trimeric form of PP2A. This activity appears to account for the role of E4orf4 in regulating E4 expression through the dephosphorylation of transcription factors required for E4 mRNA production (2, 3) and/or members of the mitigen-activated protein kinase family which phosphorylate a site on E1A products required for efficient transactivation of the E4 promoter (76). Induction of cell death could relate to induction of protein phosphatase 2A or to some other unidentified function of E4orf4. Studies to identify the biochemical function of E4orf4 involved in induction of apoptosis are under way. It should be noted that because this effect is independent of p53, E4orf4 or derivatives that function in a similar fashion might be amenable to the development of reagents for the treatment of p53-null human cancers.
ACKNOWLEDGMENTS
We are indebted to Scott Lowe for cell lines 1A.A3 and 1A.A6, to Tom Shenk for dl309 and dl359, and to Goran Akusjärvi for the E4 cDNAs. We also thank Denis Paquette, Rachel Charbonneau, and Dennis Takayesu for technical assistance.
This work was supported by grants to P.E.B. and G.C.S. from the National Cancer Institute of Canada and to P.E.B. from the Medical Research Council of Canada. R.C.M. held a Student Research Award from the Glaxo/Burroughs-Wellcome Corp. J.N.L. holds a Medical Research Council of Canada Centenial postdoctoral fellowship, and D.B. is the recipient of a FRSQ postdoctoral fellowship.
REFERENCES
- 1.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondesson M, Öhman K, Manervik M, Fan S, Akusjärvi G. Adenovirus E4 open reading frame 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bondesson M, Svensson C, Linder S, Akusjärvi G. The carboxy-terminal exon of the adenovirus E1A protein is required for the E4F-dependent transcription activation. EMBO J. 1992;11:3347–3354. doi: 10.1002/j.1460-2075.1992.tb05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D’Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 5.Braithwaite A, Nelson C, Skulimowshi A, McGovern J, Pigott D, Jenkins J. Transactivation of the p53 oncogene by E1a gene products. Virology. 1990;177:595–605. doi: 10.1016/0042-6822(90)90525-v. [DOI] [PubMed] [Google Scholar]
- 6.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge E, Ketner G. Interaction of adenoviral E4 and E1b products in late gene expression. Virology. 1990;174:345–353. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- 8.Bridge E, Medghalchi S, Ubol S, Leesong M, Ketner G. Adenovirus early region 4 and viral DNA synthesis. Virology. 1993;193:794–801. doi: 10.1006/viro.1993.1188. [DOI] [PubMed] [Google Scholar]
- 9.Challberg M D, Ketner G. Deletion mutants of adenovirus 2: isolation and initial characterization of virus carrying mutations near the right end of the viral genome. Virology. 1981;114:196–209. doi: 10.1016/0042-6822(81)90265-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Branton P E, Shore G C. Induction of p53-independent apoptosis by hygromycin B: suppression by Bcl-2 and adenovirus E1B 19-kDa protein. Exp Cell Res. 1995;221:55–59. doi: 10.1006/excr.1995.1351. [DOI] [PubMed] [Google Scholar]
- 11.Chiou S-K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis in infected cells. J Virol. 1997;68:6553–6566. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cutt J R, Shenk T, Hearing P. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J Virol. 1987;61:543–552. doi: 10.1128/jvi.61.2.543-552.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 14.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;6:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 16.Downey J F, Rowe D T, Bacchetti S, Graham F L, Bayley S T. Mapping of a 14,000-dalton antigen to early region 4 of the human adenovirus 5 genome. J Virol. 1983;45:514–523. doi: 10.1128/jvi.45.2.514-523.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezoe H, Lai Fatt R B, Mak S. Degradation of intracellular DNA in KB cells infected with cyt mutants of human adenovirus type 12. J Virol. 1981;40:20–27. doi: 10.1128/jvi.40.1.20-27.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fryer G A, Katch Y, Roberts R J. Characterization of the major mRNAs from adenovirus 2 early region 4 by cDNA cloning and sequencing. Nucleic Acids Res. 1984;12:3503–3519. doi: 10.1093/nar/12.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goping I S, Lamontagne S, Shore G C, Nguyen M. A gene-type-specific enhancer regulates the carbamyl phosphate synthetase I promoter by cooperating with the proximal GAG activating element. Nucleic Acids Res. 1995;25:1717–1721. doi: 10.1093/nar/23.10.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 22.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 23.Grand R J, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 24.Grand R J, Lecane P S, Roberts S, Grant M L, Lane D P, Young L S, Dawson C W, Gallimore P H. Overexpression of wild-type p53 and c-Myc in human fetal cells transformed with adenovirus early region 1. Virology. 1993;193:579–591. doi: 10.1006/viro.1993.1166. [DOI] [PubMed] [Google Scholar]
- 25.Grand R J, Owens D, Rookes S M, Gallimore P H. Control of p53 expression by adenovirus 12 early region 1A and early region 1B 54K proteins. Virology. 1996;218:23–34. doi: 10.1006/viro.1996.0162. [DOI] [PubMed] [Google Scholar]
- 26.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow E, Franza B R, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison T, Graham F L, Williams J. Host range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977;77:319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- 29.Hérissé J, Rigolet M, DuPont de Dinechin S, Galibert F. Nucleotide sequence of adenovirus 2 DNA fragment encoding the carboxylic region of the fiber protein and the entire E4 region. Nucleic Acids Res. 1981;9:4023–4042. doi: 10.1093/nar/9.16.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howe J A, Mymryk J S, Egan C, Branton P E, Bayley S T. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc Natl Acad Sci USA. 1990;87:5883–5887. doi: 10.1073/pnas.87.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M M, Herring P. The adenovirus early region 4 open reading frame 6/7 regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- 32.Javier R, Raska K, Jr, Shenk T. Requirement for the adenovirus type 9 E4 region in production of mammary tumors. Science. 1993;257:1267–1271. doi: 10.1126/science.1519063. [DOI] [PubMed] [Google Scholar]
- 33.Javier R T. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J Virol. 1994;68:3917–3924. doi: 10.1128/jvi.68.6.3917-3924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerr J F, Wyllie A H, Currie A R. Apoptosis: a basic biological phenomenon with wide-ranging implication in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinberger T, Shenk T. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol. 1993;67:7556–7560. doi: 10.1128/jvi.67.12.7556-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavoie J N, L’Allemain G, Brunet A, Müller R, Pouysségur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 37.Lavoie J N, Nguyen M, Marcellus R C, Branton P E, Shore G C. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway which is not inhibited by zVAD-fmk. J Cell Biol. 1998;140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leppert, K. Personal communication.
- 39.Lowe S W, Jacks T, Housman D E, Ruley H E. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci USA. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus E1A in mouse thymocytes. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 41.Marcellus R C, Teodoro J G, Charbonneau R, Shore G C, Branton P E. Expression of p53 in Saos-2 osteosarcoma cells induces apoptosis which can be inhibited by Bcl-2 or the adenovirus E1B-55 kDa protein. Cell Growth Differ. 1996;7:1643–1650. [PubMed] [Google Scholar]
- 42.Marcellus R C, Teodoro J G, Wu T, Brough D E, Ketner G, Shore G C, Branton P E. Adenovirus type 5 early region 4 is responsible for E1A-induced p53-independent apoptosis. J Virol. 1996;70:6207–6215. doi: 10.1128/jvi.70.9.6207-6215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marton M J, Baim S B, Ornelles D A, Shenk T. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J Virol. 1990;64:2345–2359. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller U, Kleinberger T, Shenk T. Adenovirus E4orf4 protein reduces phosphorylation of c-Fos and E1a proteins while simultaneously reducing the level of AP-1. J Virol. 1992;66:5867–5878. doi: 10.1128/jvi.66.10.5867-5878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neill S D, Hemstrom C, Virtanen A, Nevins J R. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc Natl Acad Sci USA. 1990;87:2008–2012. doi: 10.1073/pnas.87.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen M, Branton P E, Walton P A, Korsmeyer S J, Shore G C. Role of the membrane anchor domain of Bcl-2 in suppression of apoptosis caused by E1B-defective adenovirus. J Biol Chem. 1994;269:16521–16524. [PubMed] [Google Scholar]
- 50.Obert S, O’Connor R J, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heterodimeric E2F complex. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Öhman K, Nordqvist D, Akusjärvi G. Two adenovirus proteins with redundant activities in growth facilitates tripartite leader mRNA accumulation. Virology. 1993;194:50–58. doi: 10.1006/viro.1993.1234. [DOI] [PubMed] [Google Scholar]
- 52.Pilder S, Logan J, Shenk T. Deletion of the gene encoding the adenovirus 5 early region 1b 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J Virol. 1984;52:664–671. doi: 10.1128/jvi.52.2.664-671.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Querido E, Teodoro J G, Branton P E. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raychaudhuri P, Bagchi S, Neill S D, Nevins J R. Activation of E2F transcription factor in adenovirus infected cells involves E1A-dependent stimulation of DNA binding activity and induction of cooperative binding mediated by an E4 gene product. J Virol. 1990;64:2702–2710. doi: 10.1128/jvi.64.6.2702-2710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Prieto R, Lleonart M, Ramón y Cajal S. Lack of correlation between p53 protein level and sensitivity of DNA-damaging agents in keratinocytes carrying adenovirus E1a mutants. Oncogene. 1995;11:675–682. [PubMed] [Google Scholar]
- 59.Sandler A B, Ketner G. Adenovirus early region 4 is essential for normal stability of late nuclear RNAs. J Virol. 1989;63:624–630. doi: 10.1128/jvi.63.2.624-630.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1b 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarnow P, Hearing P, Anderson C W, Reich N, Levine A J. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J Mol Biol. 1982;162:565–583. doi: 10.1016/0022-2836(82)90389-8. [DOI] [PubMed] [Google Scholar]
- 62.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58Kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54Kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 63.Searle J, Lawson T A, Abbott P J, Harmon B, Kerr J F. An electron microscopic study of the mode of cell death induced by cancer-chemotherapeutic agents in populations of proliferating normal and neoplastic cells. J Pathol. 1975;116:129–138. doi: 10.1002/path.1711160302. [DOI] [PubMed] [Google Scholar]
- 64.Shtrichman R, Kleinberger T. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol. 1998;72:2975–2982. doi: 10.1128/jvi.72.4.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian T, Kuppuswamy M, Gysbergs J, Mak S, Chinnadurai G. 19-kDa tumor antigen coded by early region E1b of adenovirus 2 is required for efficient synthesis and for protection of viral DNA. J Biol Chem. 1984;259:11777–11783. [PubMed] [Google Scholar]
- 66.Subramanian T, Tarodi B, Chinnadurai G. p53-independent apoptotic and necrotic cell deaths induced by adenovirus infection: suppression by E1B 19K and Bcl-2 proteins. Cell Growth Differ. 1995;6:131–137. [PubMed] [Google Scholar]
- 67.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teodoro J G, Branton P E. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J Virol. 1997;71:3620–3627. doi: 10.1128/jvi.71.5.3620-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teodoro J G, Halliday T, Whalen S G, Takayesu D, Graham F L, Branton P E. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J Virol. 1994;68:776–786. doi: 10.1128/jvi.68.2.776-786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teodoro J G, Shore G C, Branton P E. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- 71.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Virtanen A, Gilardi P, Naslund A, LeMoullec J M, Pettersson U, Perricaudet M. mRNAs from human adenovirus 2 early region 4. J Virol. 1984;51:822–831. doi: 10.1128/jvi.51.3.822-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiss R S, Gold M O, Vogel H, Javier R T. Mutant adenovirus type 9 E4 ORF1 genes define three protein regions required for transformation of CREF cells. J Virol. 1997;71:4385–4394. doi: 10.1128/jvi.71.6.4385-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weiss R S, Lee S S, Prasad B V, Javier R T. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J Virol. 1997;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss R S, Javier R T. A carboxy-terminal region required by the adenovirus type 9 E4 Orf1 oncoprotein for transformation mediates direct binding to cellular polypeptides. J Virol. 1997;71:7873–7880. doi: 10.1128/jvi.71.10.7873-7880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whalen S G, Marcellus R C, Ahn N, Whalen A, Ricciardi R P, Branton P E. Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J Virol. 1997;71:3545–3553. doi: 10.1128/jvi.71.5.3545-3553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White E, Cipriani R, Sabbatini P, Denton A. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White E, Sabbatini P, Debbas M, Wold W S M, Kusher D, Gooding L R. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor a. Mol Cell Biol. 1992;12:2570–2580. doi: 10.1128/mcb.12.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early region 1B protein. Nature (London) 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 80.Yew P R, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 81.Yew P R, Liu X, Berk A J. Adenovirus E1B oncogene tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 82.Zerler B, Roberts R J, Mathews M B, Moran E. Different functional domains of the adenovirus E1A gene are involved in regulation of host cell cycle products. Mol Cell Biol. 1987;7:821–829. doi: 10.1128/mcb.7.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]