Abstract
By binding to the transactivation response element (TAR) RNA, the transcriptional transactivator (Tat) from the human immunodeficiency virus increases rates of elongation rather than initiation of viral transcription. Two cyclin-dependent serine/threonine kinases, CDK7 and CDK9, which phosphorylate the C-terminal domain of RNA polymerase II, have been implicated in Tat transactivation in vivo and in vitro. In this report, we demonstrate that CDK9, which is the kinase component of the positive transcription elongation factor b (P-TEFb) complex, can activate viral transcription when tethered to the heterologous Rev response element RNA via the regulator of expression of virion proteins (Rev). The kinase activity of CDK9 and cyclin T1 is essential for these effects. Moreover, P-TEFb binds to TAR only in the presence of Tat. We conclude that Tat–P-TEFb complexes bind to TAR, where CDK9 modifies RNA polymerase II for the efficient copying of the viral genome.
The transcriptional transactivator Tat from human immunodeficiency viruses (HIV-1 and HIV-2) and simian immunodeficiency virus is a highly conserved and unique transcriptional activator in that it binds to RNA rather than to DNA to increase rates of elongation rather than initiation of transcription (17). Its RNA target, the transactivation response element (TAR), forms a stable stem-loop at the 5′ end of all viral transcripts (17). Tat is absolutely essential for viral replication and has been a major focus of research on new anti-HIV therapeutic strategies (10). For example, dominant negative Tat proteins (5), TAR decoys (19), new phenothiazines (16), and kinase inhibitors (22) have all been demonstrated to decrease the ability of Tat to function on the viral promoter, the 5′ long terminal repeat (HIV-1 LTR).
Tat binds to TAR via its RNA-binding domain in the C-terminal half of the protein (17). Additionally, Tat exists in multiprotein complexes in cells. One of these comprises the RNA polymerase II (RNAPII) holoenzyme (7, 11), which contains up to 100 polypeptides and can initiate transcription from promoters in the presence of general transcription factors TFIID and TFIIB (7). In the holoenzyme complex, Tat interacts with TFIIH and activates CDK7, a cyclin H-dependent serine/threonine kinase that is known to phosphorylate the C-terminal domain (CTD) of RNAPII (7, 12, 27, 35). The conversion from a nonphosphorylated CTD (RNAPIIa) to a phosphorylated CTD (RNAPIIo) is a hallmark of the transition from initiation to elongation of transcription (26). Tat also exists in a second complex, which contains CDK9, which, together with cyclin T1, hyperphosphorylates the CTD (22, 34, 38). Transcriptionally active CDK9 is a component of the positive transcription elongation factor b (P-TEFb), which was defined first in in vitro transcription systems from Drosophila melanogaster (23). Cyclin T1 also binds to Tat, increases the affinity of Tat for TAR in vitro, and rescues the block to Tat transactivation in rodent cells (33). Tat-TFIIH and Tat–P-TEFb complexes might function independently and sequentially, such that the activation of CDK7 leads to promoter clearance and the recruitment of CDK9 for efficient elongation, or dependently, such that CDK7 directly or indirectly modifies the activity of CDK9 or vice versa.
Given these data, several critical questions remained. Which of these Tat complexes can interact with TAR, and which kinase can function via RNA to transactivate the HIV-1 LTR? In this study, we demonstrate that CDK9 can function via RNA but only when its kinase domain is intact. Cyclin T1 is also required for the function of CDK9. Finally, we demonstrate that the Tat–P-TEFb complex interacts specifically with TAR in a manner that depends on the 5′ bulge and central loop of TAR. We conclude that Tat–P-TEFb complexes are functional at TAR and suggest that Tat-TFIIH complexes act at an earlier step, such as promoter clearance.
MATERIALS AND METHODS
Plasmid constructions.
The plasmids pRevTat, pHIVSCAT, pRRESCAT, and pCyclin T1 have been described previously (18, 21, 28, 32). To construct pSPORTRev, Rev coding sequences (350 bp) were amplified by PCR from pcRev (32), restricted with AflII and SacI, and cloned into the PstI and SacI sites of pSV · SPORT 1 (Gibco BRL, Gaithersburg, Md.), which contains modified polylinker sequences (sequences available upon request). Functional expression of Rev was confirmed as described elsewhere (20). Full-length wild-type CDK9 and its kinase-deficient counterpart (D167N) were amplified by PCR from pRc/CMVCDK9-HA (pCDK9) and pRc/CMVCDK9D-NHA [pCDK9(D167N)] (13), respectively, with the oligonucleotides 5′ GCGATCcagctgGAGGCGGCCATGGCAAAGCAGTACGACTCGGT 3′ (lowercase letters indicate the PvuII site) and 5′ CAGACGGAGTTTGAGCGCGTCTTCCTCGAGAGGGGCCGGCG 3′. Amplified products (1.1 kbp) were restricted with PvuII and subcloned into pSPORTRev (PvuII). The identities of the resulting plasmids [pRevCDK9 and pRevCDK9(D167N)] were confirmed by dideoxynucleotide DNA sequencing (Amersham, Arlington Heights, Ill.). The expression of fusion proteins in COS cells was evaluated by Western blotting with anti-Rev and anti-CDK9 antibodies. Wild-type (+1 to +80) or mutant TAR sequences derived from HIV-1SF2 were cloned into pGEM7fZ(+) (SphI-HindIII; Promega, Madison, Wis.) with oligonucleotides containing SphI (5′) and HindIII (3′) linkers. Sequences of the resulting plasmids pGEM7WT (wild-type TAR), pGEM7ΔB (bulge deletion), and pGEM7ΔL (loop deletion) were confirmed by DNA sequencing (see Fig. 4A).
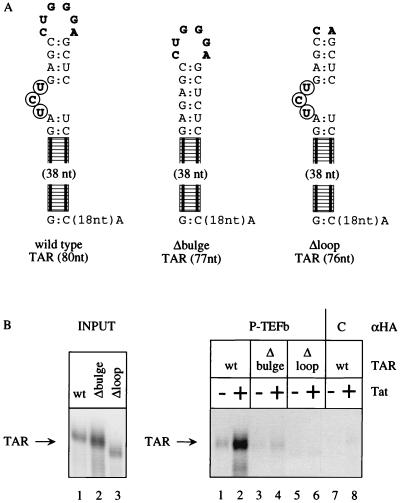
FIG. 4.
CDK9 as part of P-TEFb binds to TAR RNA in the presence of Tat. (A) Different TAR sequences were used in our RNA binding studies. The wild-type TAR contained the entire TAR (60 nucleotides) and an additional 20 nucleotides to the HindIII site at its 3′ end. Δbulge lacked the 5′ bulge (circled boldface letters). Δloop lacked 4 nucleotides of the central loop (boldface letters). Only relevant nucleotides from the upper stem are presented; the rest of the loop is schematized by the railroad track. (B) All three TAR sequences were labeled equivalently, and Δloop migrated faster on these 10% polyacrylamide gels. Immunoprecipitated P-TEFb complex (from HeLa cells expressing HA-tagged CDK9) bound to the wild-type but not mutant TAR sequences only in the presence of Tat (compare lanes 1 and 2 to lanes 3, 4, 5, and 6). In the absence of HA-tagged CDK9 (wild-type HeLa cells), no binding to TAR was observed even in the presence of Tat (lanes 5 and 6). wt, wild type; nt, nucleotide(s).
Transfection and CAT assays.
HeLa cells were transfected with plasmid reporters (0.1 μg) and plasmid effector (0.1 μg) in the presence or absence of pTat (18) with Lipofectamine as recommended by the manufacturer (Gibco BRL). For Fig. 3 and 4, increasing amounts of competitor plasmids were also used (see legends to Fig. 3 and 4). Forty-eight hours after the transfection, cells were harvested and lysed (0.25 M Tris-HCl at pH 7.5, 0.1% Triton X-100). Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (25). As an internal control, 0.2 μg of pCMVβGal was cotransfected with CAT plasmid reporters. β-Galactosidase assays were performed as follows. Cell lysates were mixed with the same amount of 2× β-galactosidase reaction mixture (200 mM NaPO4 [pH 7.3], galactopyranoside) and incubated at 37°C for 2 h. β-Galactosidase activities were quantified by measuring the absorbance at 450 nm by the enzyme-linked immunosorbent assay plate reader. All CAT data were normalized to this internal control.
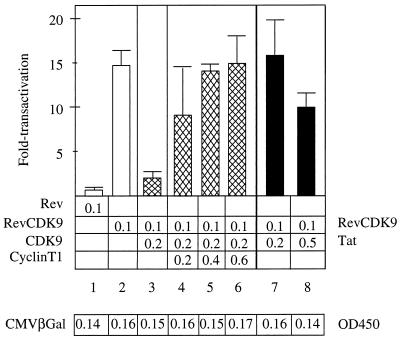
FIG. 3.
Squelching assays revealed functional interactions between CDK9 and cyclin T1 in cells. The expression of free CDK9 reduced effects of the hybrid RevCDK9 protein on SLIIB (15- to 3-fold transactivation [lanes 2 and 3, white and cross-hatched bars]). However, the coexpression of increasing amounts of cyclin T1 together with CDK9 and the chimera restored levels of transactivation from 8- to 15-fold (lanes 4 to 6, cross-hatched bars). The coexpression of Tat had limited effects on the hybrid RevCDK9 protein (16-fold to 10-fold transactivation [lanes 7 and 8, black bars]). A total of 0.1 μg of the plasmid effector was used. Amounts of plasmid effectors are given below the bar graph. The introduction of various proteins had minimal effects on the expression of LacZ from the cotransfected pCMVβGal, which served as our internal control plasmid (table; expressed as absorbance units). Experiments are representative of two independent transfections performed in duplicate where the standard errors of the means are given. OD450, optical density at 450 nm.
Cell culture.
Human 293 cells stably expressing CDK9-HA (PITALRE-HA) have been described earlier (13). Briefly, 293 cells were transfected with pRc/CMVCDK9-HA and selected in G418-containing culture medium (13).
RNA capture assays.
Wild-type and mutant TAR RNA species were prepared from the pGEM7 plasmids by transcribing linearized templates (HindIII) with T7 polymerase in the presence of [α-32P]UTP with the MAXIscript kit (Ambion, Austin, Tex.). Labeled RNA was purified from a 5% denaturing polyacrylamide gel and eluted (1.95 M NH4OAc, 1% sodium dodecyl sulfate, 60 μg of tRNA per ml) for 6 h at 60°C. After phenol and chloroform extractions, the RNA was precipitated with 2.5 volumes of EtOH and resuspended in RNA capture buffer (50 mM HEPES-KOH at pH 7.6, 50 mM KCl, 0.1 mM EDTA, 5% glycerol, 7 mM Mg2Cl, 10 mM dithiothreitol). Nuclear extracts (9) were prepared from 293 cells stably expressing hemagglutinin (HA)-tagged wild-type CDK9 or the parental vector. CDK9 complexes were immunoprecipitated (50 μg of nuclear extract/treatment) onto protein A-Sepharose beads (Pharmacia, Piscataway, N.J.). These beads were washed at least three times in the wash buffer (50 mM Tris-HCl at pH 7.4, 0.25 M NaCl, 0.1% Triton X-100, 5 mM EDTA, 10 mM dithiothreitol, 1 mM sodium metabisulfite, 0.2 mM phenylmethylsulfonyl fluoride) and two times in RNA capture buffer before being resuspended in 250 μl of binding buffer. RNA capture assays were performed in the presence or absence of recombinant Tat protein (500 ng) (7), wild-type or mutant [α-32P]UTP-RNA (106 to 108 cpm/μg, respectively), and 10 ng of tRNA per ml. Binding reaction mixtures were incubated at 25°C for 1 h with gentle rotation and then washed extensively with RNA capture buffer. Complexes were extracted with phenol (pH 5.0), concentrated by precipitation (0.3 M sodium acetate, 20 μg of tRNA, 2.5 volumes of EtOH), and resolved on 10% denaturing polyacrylamide gels. Captured RNA was visualized following exposure of the dried gel to X-ray film.
RESULTS
The hybrid RevCDK9 protein activates transcription from the HIV-1 LTR where TAR was replaced by SLIIB from RRE.
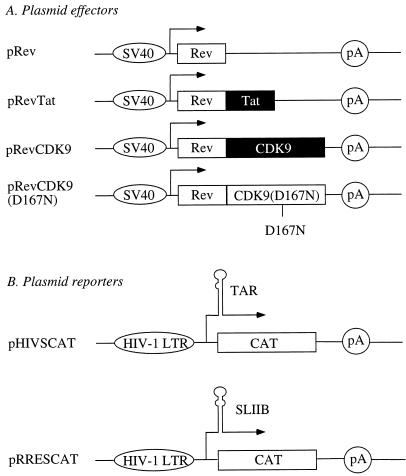
Recently, P-TEFb and especially cyclin T1 were demonstrated to interact with Tat (22, 34, 38). Furthermore, the inhibition of the kinase activity of CDK9 abolished Tat transactivation (22). Since P-TEFb phosphorylates the CTD, we wanted to test whether CDK9 could activate transcription when introduced to the transcription complex via RNA. To this end, the following plasmids were used (Fig. 1). They directed the synthesis of Rev (pRev), hybrid RevTat (pRevTat), RevCDK9 (pRevCDK9), and mutant RevCDK9(D167N) [pRevCDK9(D167N)] proteins from the simian virus 40 early promoter (Fig. 1A). The mutation of aspartic acid at position 167 to asparagine abolishes the ability of CDK9 to phosphorylate the retinoblastoma protein (13) and the CTD (data not presented). However, this same mutation does not affect the ability of the mutant CDK9 to form higher-order complexes (data not presented). Additionally, we used pTat and pCDK9. These plasmids are described elsewhere (13, 18). Two major plasmid effectors were used. They contained the wild-type HIV-1 LTR (pHIVSCAT) or the HIV-1 LTR where TAR was replaced by the high-affinity Rev-binding stem-loop IIB (SLIIB) from the Rev response element (RRE) (pRRESCAT) (32) linked to the CAT reporter gene (Fig. 1B).
FIG. 1.
Diagrammatic representation of plasmid effectors and reporters used in this study. (A) Plasmid effectors consisted of pRev, pRevTat, pRevCDK9, and pRevCDK9(D167N), which directed the synthesis of Rev, hybrid RevTat, hybrid RevCDK9, and hybrid Rev-kinase-negative-CDK9 proteins from the simian virus 40 early promoter, respectively. (B) Plasmid reporters consisted of pHIVSCAT, and pRRESCAT, which contained the wild-type HIV-1 LTR from HIV-1SF2 and substituted HIV-1 LTRs, where TAR was replaced by SLIIB from the RRE linked to the CAT reporter gene, respectively. Ovals before arrows denote types of promoters, followed by start sites of transcription (lines with arrows), TAR and SLIIB structures, and reporters (CAT), followed by the polyadenylation site (pA). D167N stands for a mutation of aspartic acid to asparagine at position 167 of CDK9, which destroys the kinase activity of CDK9.
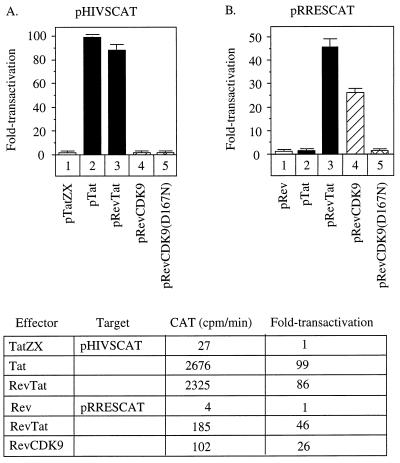
First, we cotransfected our plasmid effectors with the wild-type HIV-1 LTR target (pHIVSCAT). The wild-type Tat increased levels of expression 99-fold over that for the mutant Tat (Fig. 2A, lanes 1 and 2). Similarly, the hybrid RevTat protein transactivated the HIV-1 LTR 86-fold (Fig. 2A, lane 3). Basal levels of expression in this and subsequent experiments were measured by the liquid CAT assay, where the slope was measured by increases in counts per minute (Fig. 2, bottom). As expected, since Rev has no affinity for TAR, neither Rev nor the hybrid RevCDK9 proteins had any effect on the HIV-1 LTR. In sharp contrast, the hybrid RevCDK9 transactivated the substituted HIV-1 LTR (pRRESCAT), where TAR was replaced by SLIIB, 26-fold (Fig. 2B, lane 4). This effect was about one-half of that observed with the hybrid RevTat protein on the same plasmid target (46-fold [Fig. 2B, lane 3]). Importantly, neither wild-type Tat nor the mutant hybrid RevCDK9(D167N) protein had any effect when targeted to the HIV-1 LTR via SLIIB (Fig. 2B, lanes 2 and 5). Together, these data demonstrate that CDK9 can function when tethered to RNA and that the kinase activity of CDK9 is essential for this effect. Interestingly, neither hybrid RevCDK9 nor RevVP16 protein (32) worked as well as the hybrid RevTat protein, suggesting that structural constraints on these fusion proteins might exist.
FIG. 2.
RNA tethering is required for the activity of CDK9 on the HIV-1 LTR. HeLa cells were cotransfected with pHIVSCAT (A) or pRRESCAT (B) and indicated plasmid effectors. (A) Only the wild-type Tat and hybrid RevTat proteins transactivated pHIVSCAT, which contains TAR (99- and 86-fold, respectively [black bars]). Neither the hybrid RevCDK9 nor RevCDK9(D167N) protein transactivated the wild-type HIV-1 LTR (striped bars). (B) Both the hybrid RevTat and RevCDK9 but not Tat and RevCDK9(D167N) proteins increased transcription from pRRESCAT, which contains the binding site for Rev (SLIIB) in place of TAR. The hybrid RevCDK9 protein had about one-half the activity of the hybrid RevTat protein (46-fold versus 26-fold transactivation [black versus striped bars]). A total of 0.1 μg of each plasmid target and effector was used. The table below the bar graphs contains the mean slopes from liquid CAT data and absolute values for the fold transactivation. Experiments are representative of three independent transfections performed in duplicate where the standard errors of the means are given.
Cyclin T1 is also required for the function of CDK9 in cells.
To determine what other components of P-TEFb might also be required for the function of CDK9, we performed the following squelching experiments in cells. Squelching refers to the ability of one domain of a protein to prevent the relevant cofactor(s) from interacting with the target protein. Previously, free Tat could efficiently compete for a limiting coactivator on heterologously tethered hybrid Tat proteins (6, 21). As presented in Fig. 3, a twofold excess of free CDK9 could decrease levels of transactivation of the hybrid RevCDK9 protein by 80% (15- to 3-fold, lanes 2 and 3). Western blots revealed that levels of CDK9 were twofold higher than those of the chimera (data not presented). When increasing amounts of cyclin T1 were coexpressed with both CDK9 proteins, this squelching was eliminated in a dose-dependent fashion (Fig. 3, lanes 4 to 6). Importantly, coexpression of these different proteins had little to no effect on the cotransfected internal control, pCMVβGal (Fig. 3, bottom). We conclude that cyclin T1 is present in limiting amounts in cells and is required for the function of CDK9. Based on the data presented in Fig. 2, this function is the ability of CDK9 to act as a kinase. Additionally, these results suggest strongly that no other component of P-TEFb is limiting or required for the function of CDK9 in cells.
We also asked whether Tat had any effect in this system. One could imagine that Tat could have a positive, a negative, or no effect on the hybrid RevCDK9 protein. However, Tat had virtually no effect on the hybrid RevCDK9 protein, even at a fivefold excess of Tat over the chimera (Fig. 3, lanes 7 and 8). We conclude that Tat does not affect interactions between cyclin T1 and CDK9 and that Tat does not increase the activity of CDK9. This finding is in agreement with previous studies, where no effect of Tat on the activity of CDK9 was reported in vitro (22). In this scenario, Tat would allow for optimal interactions between CDK9, cyclin T1, and TAR RNA and the presentation of P-TEFb to the transcription complex.
Complexes containing Tat and P-TEFb bind to TAR specifically.
Since we could establish a functional link among cyclin T1, CDK9, and the ability of CDK9 to act via RNA, we wondered if P-TEFb could bind to TAR RNA in vitro. Recently, recombinant Tat and cyclin T1 were demonstrated to interact with TAR, and this binding depended on an intact 5′ bulge and central loop in TAR (33). Moreover, the mutant Tat protein lacking its RNA binding arginine-rich domain no longer facilitated these interactions (33). However, the ability of the P-TEFb complex to bind to TAR had not been tested. To this end, we created 293 cells which constitutively express influenza virus HA-tagged CDK9. In these cells, P-TEFb can be immunoprecipitated with anti-HA antibodies. As reported previously, the immunoprecipitation of CDK9 from nuclear extracts purifies P-TEFb, which can hyperphosphorylate the CTD in vitro (38). TAR and mutant TAR RNA species were created (Fig. 4A). Anti-HA immunoprecipitates were tested for their binding to these labeled TAR RNA species in the presence and absence of Tat.
As observed from data presented in Fig. 4B, P-TEFb could bind to the wild-type TAR RNA only in the presence of Tat (lanes 1 and 2). Our Tat was purified by affinity chromatography following its overexpression in Escherichia coli, which yielded a single stained band on gel electrophoresis (with the Coomassie blue dye [7]). This Tat–P-TEFb complex could not bind to a TAR RNA where the 5′ bulge was deleted (Δbulge, Fig. 4B, lanes 3 and 4) or where the central loop was absent (Δloop, Fig. 4B, lanes 5 and 6). Importantly, in the absence of the HA-tagged CDK9, immunoprecipitates from wild-type HeLa cells demonstrated no binding to TAR in the presence or absence of Tat (Fig. 4B, lanes 7 and 8). Additionally, the mutant Tat containing only the activation domain of Tat (N-terminal 48 residues) did not facilitate these interactions (data not presented). We conclude that P-TEFb can bind to the wild-type TAR, which contains an intact 5′ bulge and central loop, and that these interactions depend on the presence of the wild-type Tat protein.
DISCUSSION
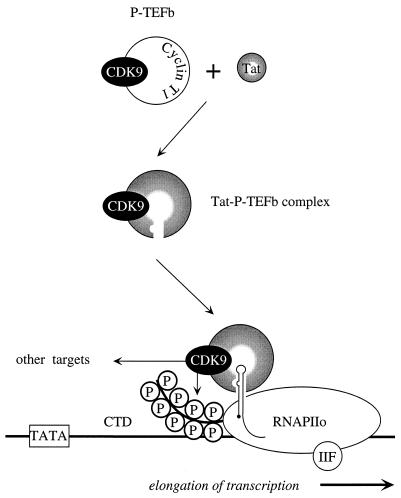
In this study, we demonstrated that CDK9 can function via RNA provided that its kinase domain is intact. However, since no effect of CDK9 or the hybrid RevCDK9 proteins was observed in the absence of SLIIB, RNA tethering is required for these effects. Additionally, cyclin T1 was required for the activity of CDK9 via RNA. Although Tat had no effect on this heterologous tethering system, P-TEFb complexes, which contain CDK9, could bind specifically to TAR RNA only in the presence of Tat. This binding required the 5′ bulge and central loop in TAR. We conclude that Tat–P-TEFb complexes bind to TAR where CDK9 phosphorylates the CTD and possibly other proteins (Fig. 5).
FIG. 5.
A model for interactions among Tat, P-TEFb, and TAR. CDK9 exists in the P-TEFb complex in cells. This complex minimally contains CDK9 and cyclin T1. Tat binds to P-TEFb in the absence of TAR. This complex has also been referred to as the Tat-TAK (Tat-associated kinase) complex. The assembly of the Tat–P-TEFb complex results in the formation of a high-affinity surface for the binding to TAR RNA. Both the 5′ bulge and central loop are required for these protein-RNA interactions. Upon binding to TAR, CDK9 can then phosphorylate the CTD of RNAPIIo and possibly other targets, which results in efficient elongation of viral transcription. Importantly, the kinase activity of CDK9 and the presence of cyclin T1 are required for these effects.
The hybrid RevCDK9 protein functioned efficiently via RNA. In this context, CDK9 was required but was by itself not sufficient for activity, i.e., both an intact kinase domain and cyclin T1 were required for transactivation. This finding implies that RNA tethering is involved in catalysis, e.g., in the phosphorylation of the CTD of RNAPII. Since RNA must pass by the CTD for cotranscriptional processing (31), it is not surprising that RNA presentation would represent an optimal way of bringing kinases to the CTD. On the other hand, most transcriptional activators recruit general transcription factors and RNAPII to the promoter (29) and therefore do not function via RNA. So far, the only exception is VP16, which can also interact with TFIIH and possibly other kinases (35).
Our squelching results suggest that components of P-TEFb, especially cyclin T1, are also limiting in cells. This finding has some relevance to the replication of HIV and the establishment of molecular latency. It is possible that HIV enters cells where levels of P-TEFb are low, e.g., nonactivated cells (34). In this scenario, there might be insufficient P-TEFb to support Tat transactivation, leading to a block in viral transcription. In these cells, optimal Tat transactivation would occur only upon cellular activation. Finally, direct binding studies revealed that P-TEFb and Tat form a complex that binds specifically to TAR, and this cooperative binding depends on the intact 5′ bulge and central loop of TAR. Since CDK9 is encoded on the human chromosome 4 (4), our data also support the finding that cyclin T1 is the protein encoded on the human chromosome 12, which rescues Tat transactivation in rodent cells (33). Moreover, our studies extend this finding by revealing that Tat is required for the binding of P-TEFb to TAR RNA.
Tat has placed the control of transcriptional elongation at center stage. After the initiation of transcription, there exist a series of well-orchestrated steps leading to productive elongation of transcription (2, 3, 14, 15, 30). TFIIH plays the major role in promoter clearance (1, 35), which in HIV is increased by Tat (8, 12, 27). CDK7 partially phosphorylates the CTD, and the activity of CDK7 is increased by Tat (8, 12, 27). In this study, we did not present data on RNA tethering of CDK7. However, the hybrid RevCDK7, RevCyclinH, and RevMat1 proteins were all inactive on pRRESCAT. These data could mean that all these chimeras rendered CDK7 inactive or that the work of CDK7 is completed by the time that SLIIB, which is longer than TAR, is synthesized (14, 37). The latter finding would agree nicely with studies that revealed that TFIIH dissociates from RNAPII early after promoter clearance (37).
In a subsequent step, CDK9 hyperphosphorylates the CTD and possibly other targets. In the case of HIV, the entrance of P-TEFb into the transcription complex depends on Tat and TAR. The conversion of RNAPII from the IIa to the IIo form then results in an exchange of cellular proteins that bind to the CTD (24, 36). Studies of Tat will allow for the dissection of these steps and the characterization of participating proteins. One intriguing possibility is that RNA presentation leads to proper positioning and longer association between cyclin-dependent kinases, CTD, and the transcription complex. How other promoters, which are also regulated by attenuation, such as c-myc, c-myb, and c-fos, recruit their kinase complexes to RNAPII remains to be investigated (3, 30).
Our study is compatible with the following model for interactions among Tat, TAR, and P-TEFb (Fig. 5). However, it does not exclude an important role for interactions between Tat and CDK7 (Tat-CAK or Tat-TFIIH complexes) at an earlier step in transcription from the HIV promoter. Tat and P-TEFb are known to assemble independently of TAR in cells (34, 38). These interactions result in structural changes of the Tat–P-TEFb complex that allow for its binding to TAR. When TAR is made, Tat–P-TEFb complex is recruited to TAR, where CDK9 hyperphosphorylates the CTD of RNAPII and possibly other targets. Efficient elongation of transcription results. To extend the model, other components of P-TEFb must be characterized, targets of CDK9 must be identified, and the elongating RNAPIIo must be analyzed. In the process, new therapeutic strategies against HIV might be revealed.
ACKNOWLEDGMENTS
We thank Michael Armanini for expert secretarial assistance and other members of the laboratory for helpful discussions and comments on the manuscript.
This work was supported in part by grants from the NIH (AI38532-01 to B.M.P. and GM35500 to D.H.P.).
REFERENCES
- 1.Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 2.Aso T, Conaway J W, Conaway R C. The RNA polymerase II elongation complex. FASEB J. 1995;9:1419–1428. doi: 10.1096/fasebj.9.14.7589983. [DOI] [PubMed] [Google Scholar]
- 3.Bentley D L. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 4.Bullrich F, MacLachlan T K, Sang N, Druck T, Veronese M L, Allen S L, Chiorazzi N, Koff A, Heubner K, Croce C M, Giordano A. Chromosomal mapping of members of the cdc2 family of protein kinases, cdk3, cdk6, PISSLRE, and PITALRE, and a cdk inhibitor, p27Kip1, to regions involved in human cancer. Cancer Res. 1995;55:1199–1205. [PubMed] [Google Scholar]
- 5.Caputo A, Grossi M P, Bozzini R, Rossi C, Betti M, Marconi P C, Barbanti-Brodano G, Balboni P G. Inhibition of HIV-1 replication and reactivation from latency by tat transdominant negative mutants in the cysteine rich region. Gene Ther. 1996;3:235–245. [PubMed] [Google Scholar]
- 6.Carroll R, Peterlin B M, Derse D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. J Virol. 1992;66:2000–2007. doi: 10.1128/jvi.66.4.2000-2007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1480. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gait M J, Karn J. Progress in anti-HIV structure-based drug design. Trends Biotechnol. 1995;13:430–438. doi: 10.1016/S0167-7799(00)88998-2. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Martinez L F, Ivanov D, Gaynor R B. Association of Tat with purified HIV-1 and HIV-2 transcription preinitiation complexes. J Biol Chem. 1997;272:6951–6958. doi: 10.1074/jbc.272.11.6951. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garriga J, Mayol X, Grana X. The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem J. 1996;319:293–298. doi: 10.1042/bj3190293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 15.Greenblatt J, Nodwell J R, Mason S W. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 16.Haubrich R H, Flexner C, Lederman M M, Hirsch M, Pettinelli C P, Ginsberg R, Lietman P, Hamzeh F M, Spector S A, Richman D D. A randomized trial of the activity and safety of Ro 24-7429 (Tat antagonist) versus nucleoside for human immunodeficiency virus infection. The AIDS Clinical Trials Group 213 Team. J Infect Dis. 1995;172:1246–1252. doi: 10.1093/infdis/172.5.1246. [DOI] [PubMed] [Google Scholar]
- 17.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 18.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by Tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 19.Lee S W, Gallardo H F, Gaspar O, Smith C, Gilboa E. Inhibition of HIV-1 in CEM cells by a potent TAR decoy. Gene Ther. 1995;2:377–384. [PubMed] [Google Scholar]
- 20.Luo Y, Madore S J, Parslow T G, Cullen B R, Peterlin B M. Functional analysis of interactions between Tat and the trans-activation response element of human immunodeficiency virus type 1 in cells. J Virol. 1993;67:5617–5622. doi: 10.1128/jvi.67.9.5617-5622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madore S J, Cullen B R. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 24.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 25.Neumann J R, Morency C A, Russian K O. A novel rapid assay for chloramphenicol acetyltransferase gene expression. BioTechniques. 1987;5:444–447. [Google Scholar]
- 26.O’Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 27.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 28.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 30.Spencer C A, Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990;5:777–785. [PubMed] [Google Scholar]
- 31.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 32.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 33.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 34.Yang X Z, Gold M O, Tang D N, Lewis D E, Herrmann C H, Rice A P. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promyelocytic cell lines. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yankulov K Y, Pandes M, McCraken S, Bouchard D, Bentley D. TFIIH functions in regulating transcriptional elongation by RNA polymerase II in Xenopus oocytes. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]