Abstract
The species Monoserius pennarius (Linnaeus, 1758), is particularly abundant in the tropical Indo-West Pacific east of Sri Lanka, yet very limited genetic information exists for this species. Here, we report the assembled-linear mitochondrial genome of M. pennarius collected from the East China Sea. The 15,197 bp mitogenome contains 13 protein-coding genes (PCGs), two tRNA genes, and two rRNA genes. Notably, the gene order in this mitogenome differs from that of other hydrozoans within the same taxonomic order. Phylogenetic analysis, based on 13 concatenated mitochondrial PCGs, recovered M. pennarius as a sister of Nemalecium lighti (Hargitt, 1924), outside the other Leptothecata hydrozoans, suggesting paraphyly of Leptothecata. The mitogenome of M. pennarius, serving as the first publicly available for the family Aglaopheniidae, holds foreseeable value for investigating Leptothecata evolution.
Keywords: Mitochondrial genome, hydrozoa, Leptothecata, gene rearrangement
1. Introduction
Within the class Hydrozoa, sister group relationships remain highly contentious, especially at the order level (Kayal et al. 2015). Therefore, additional molecular studies are imperative to elucidate the evolutionary relationships among hydrozoan taxa. The species Monoserius pennarius (Hydrozoa: Leptothecata: Aglaopheniidae), a macrobenthic hydroid, is widely distributed in the tropical Indo-West Pacific east of Sri Lanka (Tang 1991). Its elongated body and dense growth on the seafloor create a unique ecological habitat known as the ‘seafloor prairie’ (Zhao 1998). This environment attracts various epizoans and climbing animals, forming a vertically layered assemblage (Tang 1991). M. pennarius, as a keystone species, contributes to the stability of this assemblage. However, the genetic information for M. pennarius or its close relatives is rather lacking in public databases. Specifically, only 10 mitogenomes are available for the order Leptothecata and none for the family Aglaopheniidae in the GeneBank database. In this study, we report the mitochondrial genome of M. pennarius, which will enhance our knowledge of this species and provide molecular data for further taxonomic and phylogenetic studies of hydrozoans.
2. Materials and methods
A specimen of M. pennarius (Figure 1) was collected by bottom trawl from the East China Sea (124.50°E, 29.75°N) in October 2018 aboard the cruise ‘Xiangyanghong 18’, and was identified by morphological features. The sample (Accession No.: FIO-201810-PN5; Contact person: Qinzeng Xu, xuqinzeng@fio.org.cn) was preserved in 95% ethanol and stored at −20 °C in First Institute of Oceanography, Ministry of Natural Resources.
Figure 1.
The specimen of Monoserius pennarius from East China Sea (124.50°E, 29.75°N), photographed by Wenge Shi. (a) Gonotheca; (b) detail of hydrothecal rim showing cusps; (c) hydrocaulus; (d) hydrotheca.
Total DNA was extracted using a DNeasy Blood & Tissue DNA kit (QIAGEN, Germany). The DNA library was then constructed and sequenced on the Illumina HiSeq 2500 Platform (Illumina, Hayward, CA). Adapter sequences and low-quality reads were trimmed from the raw data using Trimmomatic v0.38 (Bolger et al. 2014). The mitochondrial genome was assembled by combining the Getorganelle v1.7.5 (Jin et al. 2020) and Mitofinder v1.4.1 (Allio et al. 2020) and was annotated using the MITOS2 (Bernt et al. 2013). Annotated results were manually verified and corrected using MEGA7 (Kumar et al. 2016) and tRNAscan-SE (Chan et al. 2021). Finally, the mitochondrial genome of M. pennarius was submitted to GenBank. The samtools v1.7 (Li et al. 2009) was used to calculate the depth per base and the OGDRAW (Greiner et al. 2019) was used to make a mitogenome map. Telomeric structures and inverted repeat sequences (IRSs) were identified utilizing TelFinder (Sun et al. 2023) and Inverted Repeats Finder (https://tandem.bu.edu/irf/home) software tools, respectively.
Nineteen mitogenomes of Cnidaria species were downloaded from NCBI Genbank, namely 17 Hydrozoa species and 2 Scyphozoa species as outgroups. The phylogenetic analysis and gene order analysis were conducted using PhyloSuite v1.2.3 (Zhang et al. 2020). The 13 protein-coding genes (PCGs) sequences were aligned with MAFFT v7.505 (Katoh and Standley 2013) and then trimmed using Gblocks prior to concatenation. ModelFinder v2.2.0 (Kalyaanamoorthy et al. 2017) was used to pick the optimal nucleotide partitions and substitution model. Finally, the maximum likelihood (ML) phylogenetic tree was constructed using IQ-TREE v2.2.0 (Trifinopoulos et al. 2016) under the Edge-linked partition model with 5000 ultrafast bootstrap replicates. The references for mitogenomes used in the phylogenetic tree are provided in Supplementary Table S1.
3. Results and discussion
3.1. Mitogenome organization
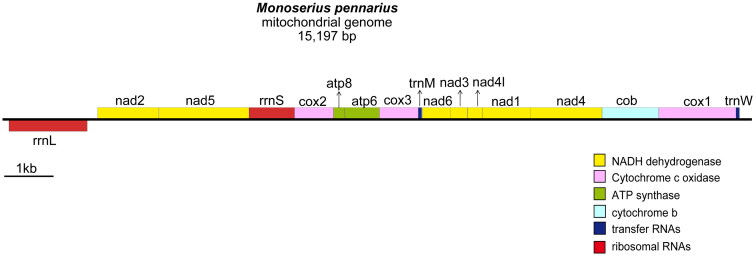
The coverage-depth map (Supplementary Figure S1) showed that the average depth per base is 2,167×. The mitochondrial genome of M. pennarius (GenBank accession no.: OR501924) encodes 13 PCGs, two tRNA genes (tRNATrp and tRNAMet), and two rRNA genes (Figure 2), which is consistent with the feature of ‘the loss of all but one or two tRNA genes’ in medusozoans mitogenome (Kayal et al. 2012). All gene was encoded compactly on the heavy strand but the rrnL. This mitogenome is 15,197 bp in length, with a base composition of 31.5% A, 42.6% T, 11.9% C, and 14.1% G. ATG is the main start codon except for cox3 (ATT), nad3/nad4 (TTG), and atp6 (ATC). TAA is the stop codon for all PCGs but cox1, cox3, and nad3 (TAG). Based on the high sequencing depth (2,167×) and multiple assembly strategies (includes the use of various assembly software, sequence extension with samtools, and gap filled with cap3), the mitogenome could not be circularized, indicating that it may also be linearly structured as reported in most hydrozoan species (Kayal et al. 2012, Macher et al. 2021, Seo et al. 2021a, Seo et al. 2021b). Five repeat sequences were identified at termini, including [TTTTA]15, [TTTATA]8, and [GAATCA]2 (Supplementary Table S2), which might serve as telomeric motifs that facilitate replication and maintenance of the termini linear mtDNA. Notably, the mitochondrial genes of M. pennarius exhibited a unique arrangement different from other three gene orders within Leptothecata (Supplementary Figure S2). Moreover, several rearranged genes – ATP8, cox2, nad2, and rrnS have altered lengths compared with homologues of other Leptothecata species, and IRS were identified around/in these genes, likely representing vestiges of rearrangement (Supplementary Table S3).
Figure 2.
Gene map of the mitochondrial genome of Monoserius pennarius. Genes encoded on the heavy and the light strands were shown above and below the chain, respectively.
3.2. Phylogenetic analysis
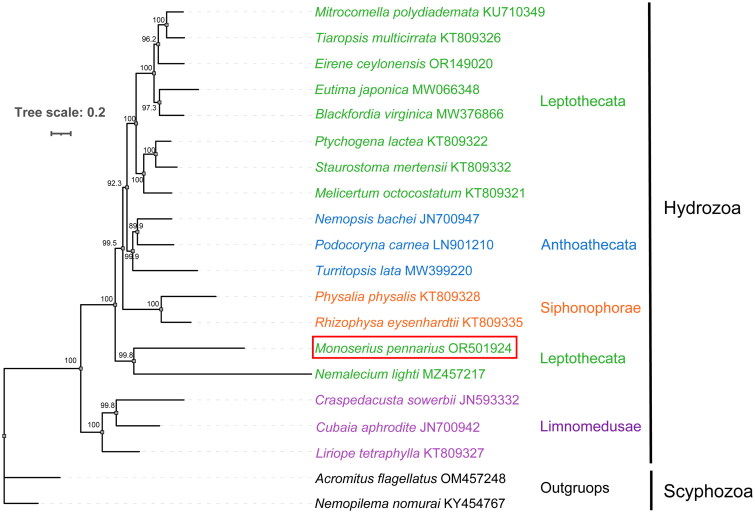
The order Leptothecata has always been considered monophyletic (Collins et al. 2008, Maronna et al. 2016). However, our phylogenetic analysis (Figure 3) recovered M. pennarius as a sister of Nemalecium lighti (Hargitt, 1924), outside of other Leptothecata hydrozoans, suggesting paraphyly of Leptothecata, as proposed by Macher et al. (2021). This result indicates that the phylogenetic position of M. pennarius and N. lighti may need to be reconsidered and underscores the necessity for comprehensive phylogenetic investigations of Leptothecata hydrozoans. The mitochondrial genome of M. pennarius in our study, serving as the first publicly available for the family Aglaopheniidae, provides a valuable resource for investigating Leptothecata evolution.
Figure 3.
Maximum-likelihood phylogenetic tree based on 13 concatenated mitochondrial genes from 20 hydrozoa species. The scyphozoan Acromitus flagellatus and Nemopilema nomurai were used as outgroups. The Monoserius pennarius is highlighted in red box. Numbers at nodes indicate bootstrap values. The tree was constructed using IQ-TREE with 5000 ultrafast bootstraps replicates and visualized using iTOL (https://itol.embl.de/).
Supplementary Material
Acknowledgements
We are grateful to Dr. Xikun Song from the Institute of Deep-Sea Science and Engineering, Chinese Academy of Sciences for his assistance in specimen classification.
Funding Statement
This work was supported by National Natural Science Foundation of China [42176135].
Authors’ contributions
Specimen collection and DNA extraction were carried out by Xuetao Wang and Wenge Shi. Jing Mo, Min Lu and Anning Mou were involved in the conception and design, analysis and interpretation of the data. Jing Mo drafted the paper. Qinzeng Xu provided the funds, participated in the formal analysis and data curation of the project. Xuelei Zhang reviewed this paper and gave final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethical approval
All animal experiments in this study were approved by the Ethical Committee of Ministry of Natural Resources of the People’s Republic of China.
Data availability statement
The genome sequence data are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OR501924. The associated ‘BioProject,’ ‘BioSample’ and ‘SRA’ numbers are PRJNA1009986, SAMN37176710, and SRR25774581 respectively.
References
- Allio R, Schomaker BA, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F.. 2020. MitoFinder: efficient automated large‐scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20(4):892–905. doi: 10.1111/1755-0998.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Bolger A, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lin BY, Mak AJ, Lowe TM.. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49(16):9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AG, Bentlage B, Lindner A, Lindsay D, Haddock SH, Jarms G, Norenburg JL, Jankowski T, Cartwright P.. 2008. Phylogenetics of Trachylina (Cnidaria: hydrozoa) with new insights on the evolution of some problematical taxa. J Mar Biol Ass. 88(8):1673–1685. doi: 10.1017/S0025315408001732. [DOI] [Google Scholar]
- Greiner S, Lehwark P, Bock R.. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z.. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG.. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403. doi: 10.7717/peerj.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV.. 2012. Evolution of linear mitochondrial genomes in Medusozoan cnidarians. Genome Biol Evol. 4(1):1–12. doi: 10.1093/gbe/evr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macher J-N, Kayal E, Duijm E, van der Hoorn B, Montano S, Speksnijder A.. 2021. The mitochondrial genome of Nemalecium lighti (Hydrozoa, Leptothecata). Mitochon DNA B Resour. 6(11):3196–3198. doi: 10.1080/23802359.2021.1989335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maronna MM, Miranda TP, Peña Cantero ÁL, Barbeitos MS, Marques A.. 2016. Towards a phylogenetic classification of Leptothecata (Cnidaria, Hydrozoa). Sci Rep. 6(1):18075. doi: 10.1038/srep18075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Eom HJ, Cho JK, Kang HS, Rhee JS.. 2021a. The linear mitochondrial genome of commensal hydroid Eutima japonica (Cnidaria, Hydrozoa, Eirenidae). Mitocho DNA B Resour. 6(3):1082–1084. doi: 10.1080/23802359.2021.1899869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y, Chae J, Ki JS.. 2021b. Complete mitochondrial genome of the hydrozoan jellyfish Blackfordia virginica Mayer, 1910 (Cnidaria; Hydrozoa; Leptothecata) with phylogenetic analysis. Mitocho DNA B Resour. 6(3):1202–1203. doi: 10.1080/23802359.2021.1903363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wang H, Tao S, Xi X.. 2023. Large-scale detection of telomeric motif sequences in genomic data using TelFinder. Microbiol Spectr. 11(2):e03928-22. doi: 10.1128/spectrum.03928-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZC. 1991. Monoserius pennarius assemblage and its ecological and geographical studies on the continental shelf of waters around the Nansha islands, South China Sea. Res Marine Biology Nansha Islands Neighbo Waters Collected Papers (II). 2:255–251. [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ.. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT.. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhao H. 1998. Type and evolution of landscapes of Nansha Islands. Chin GeographSc. 8(2):144–151. doi: 10.1007/s11769-997-0028-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence data are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OR501924. The associated ‘BioProject,’ ‘BioSample’ and ‘SRA’ numbers are PRJNA1009986, SAMN37176710, and SRR25774581 respectively.