Abstract
Brome mosaic virus (BMV), a positive-strand RNA virus, encodes two replication proteins: the 2a protein, which contains polymerase-like sequences, and the 1a protein, with N-terminal putative capping and C-terminal helicase-like sequences. These two proteins are part of a multisubunit complex which is necessary for viral RNA replication. We have previously shown that the yeast two-hybrid assay consistently duplicated results obtained from in vivo RNA replication assays and biochemical assays of protein-protein interaction, thus permitting the identification of additional interacting domains. We now map an interaction found to take place between two 1a proteins. Using previously characterized 1a mutants, a perfect correlation was found between the in vivo phenotypes of these mutants and their abilities to interact with wild-type 1a (wt1a) and each other. Western blot analysis revealed that the stabilities of many of the noninteracting mutant proteins were similar to that of wt1a. Deletion analysis of 1a revealed that the N-terminal 515 residues of the 1a protein are required and sufficient for 1a-1a interaction. This intermolecular interaction between the putative capping domain and itself was detected in another tripartite RNA virus, cucumber mosaic virus (CMV), suggesting that the 1a-1a interaction is a feature necessary for the replication of tripartite RNA viruses. The boundaries for various activities are placed in the context of the predicted secondary structures of several 1a-like proteins of members of the alphavirus-like superfamily. Additionally, we found a novel interaction between the putative capping and helicase-like portions of the BMV and CMV 1a proteins. Our cumulative data suggest a working model for the assembly of the BMV RNA replicase.
While the sequences of many viral replication proteins have been identified, we have only a minimal understanding of the higher-order interactions between them. The interactions of the replicase subunits of the negative-strand influenza virus have been elucidated at the biochemical level (12, 31). However, the interactions of the subunits of the replicases of positive-strand RNA viruses, including those of the well-characterized coliphage Qβ, are poorly understood (5). We have focused on dissecting the interactions between the replication proteins of the monocot-infecting brome mosaic virus (BMV). These studies are essential for the eventual comparison and understanding of the three-dimensional structure and function of viral RNA replicases.
The BMV genome is composed of three genomic positive-strand RNAs designated RNAs 1, 2, and 3. The genomic RNAs serve dual functions as mRNAs for translation and as templates for the synthesis of the complementary negative-strand RNAs. To complete the replication cycle, the negative-strand RNAs then serve as templates for positive-strand RNA synthesis. RNAs 1 and 2 encode the replication proteins 1a (109 kDa) and 2a (96 kDa), which are sufficient for BMV RNA replication in protoplasts (16). The two proteins have evolved to work specifically with each other, because heterologous combinations of 1a and 2a from the closely related BMV and cowpea chlorotic mottle virus (CCMV) exhibit RNA synthesis defects (7).
Several domains of the 1a and 2a proteins have been identified based on sequence similarities. The 2a protein contains two nonconserved regions flanking a centrally conserved domain which shares sequence motifs with many polymerases, including the presence of the Mg2+-binding GDD motif (3). The N terminus of 1a resembles the nsP1 protein of Sindbis virus, indicating its possible involvement in RNA capping functions (10, 20, 27). The 1a C terminus has sequence homology to many viral and cellular helicases (8). Mutations in 1a and 2a have been shown to abolish or greatly reduce RNA replication levels (17, 32).
We have previously shown that BMV 1a and 2a interact both in vitro and in the yeast two-hybrid system (15, 21). Furthermore, we used a set of well-characterized 1a mutants (PK mutants [17]) to show an absolute correlation between the in vivo phenotypes of these mutants and their ability to interact with 2a in the two-hybrid system (22). In this study, we use the PK mutants to demonstrate a perfect correlation between their in vivo phenotypes and their ability to interact with 1a in the two-hybrid system. We also map the 1a-1a interaction to the N-terminal putative capping domain (previously referred to as the methyltransferase-like domain) of both BMV and cucumber mosaic virus (CMV) and show the interaction to be species specific. These results, along with those from previous mapping data, are placed in the context of the predicted secondary structure of the 1a protein. Finally, we describe a novel interaction between the putative capping and helicase-like domains of the 1a protein.
MATERIALS AND METHODS
Strains, reagents, and two-hybrid procedures.
Saccharomyces cerevisiae strains, plasmids, growth conditions, and reporter protein assays have been described previously (22). In our previous work characterizing the 1a-2a interaction, DNA coding for each PK mutant was individually fused to the LexA DNA binding domain in plasmid pBTM116 (22). For our current analysis, we created fusions of the PK mutants to the GAL4 transcription activation domain in plasmid pGAD424 by using the previously described primers and cloning procedures (Table 1) (22). In general, viral DNA fragments flanked by appropriate restriction sites were produced by PCR and then cloned into the above-mentioned two-hybrid plasmids (kind gifts of Stan Fields). pGBT9 (Clonetech Laboratories, Inc.), which contains a GAL4 DNA binding domain, was used in some experiments. The fusions were then tested for interaction with the appropriate wild-type 1a (wt1a) fusion construct by their ability to activate the β-galactosidase and/or HIS3 reporter genes.
TABLE 1.
Summary of plasmids and encoded polypeptides
| Plasmida | Amino acids inserted or deletedb | Encoded fusion polypeptide | Source |
|---|---|---|---|
| DNA binding domain fusions | |||
| pBTM116 | NA | LexA (aa 1–211) | Stan Fields |
| pGBT9 | NA | GAL4 binding domain | Clonetech, Inc. |
| pLex-Δ516-961 (pLex-BMT) | Δ516-961 | LexA-Δ516-961 | This work |
| pGB-Δ516-961 | Δ516-961 | GAL4-Δ516-961 | This work |
| pGB-B1a | None | GAL4-B1a | This work |
| pLex-CMT | Δ534-993 | LexA-Δ534-993 | This work |
| pLex-CHel | Δ1-560 | LexA-Δ1-560 | This work |
| pEOΔ500 | Δ1-500 | LexA-Δ1-500 | 21 |
| Transcription activation domain fusions | |||
| pGAD424 | NA | GAL4 activation domain | Stan Fields |
| pGAL-CMT | Δ534-993 | GAL4-Δ534-993 | This work |
| pGAL-BHel | Δ1-500 | GAL4-Δ1-500 | This work |
| BMV 1a insertional mutations | |||
| pG-PK1 | G-S (267) | GAL4-PK1 | This work |
| pG-PK2 | D-P (507) | GAL4-PK2 | This work |
| pG-PK3 | G-S (5) | GAL4-PK3 | This work |
| pG-PK4 | G-S (492) | GAL4-P4K | This work |
| pG-PK6 | G-S (95) | GAL4-PK6 | This work |
| pG-PK7 | G-S (239) | GAL4-PK7 | This work |
| pG-PK9 | G-S (154) | GAL4-PK9 | This work |
| pG-PK10 | G-P (198) | GAL4-PK10 | This work |
| pG-PK11 | G-P (203) | GAL4-PK11 | This work |
| pG-PK13 | W-A-H (403) | GAL4-PK13 | This work |
| pG-PK14 | G-P (556) | GAL4-PK14 | This work |
| pG-PK15 | W-A-H (651) | GAL4-PK15 | This work |
| pG-PK16 | G-P-T (464) | GAL4-PK16 | This work |
| pG-PK17 | D-P (869) | GAL4-PK17 | This work |
| pG-PK18 | G-S (912) | GAL4-PK18 | This work |
| pG-PK19 | G-S (670) | GAL4-PK19 | This work |
| pG-PK20 | D-P (905) | GAL4-PK20 | This work |
| pG-PK21 | D-P (542) | GAL4-PK21 | This work |
| BMV 1a deletion mutations | |||
| pG-Δ738-961 | Δ738-961 | GAL4-Δ738-961 | This work |
| pG-Δ622-961 | Δ622-961 | GAL4-Δ622-961 | This work |
| pG-Δ564-961 | Δ564-961 | GAL4-Δ564-961 | This work |
| pG-Δ516-961 (pG-BMT) | Δ516-961 | GAL4-Δ516-961 | This work |
| pG-Δ480-961 | Δ480-961 | GAL4-Δ480-961 | This work |
| pG-Δ445-961 | Δ445-961 | GAL4-Δ445-961 | This work |
| pG-Δ1-45 | Δ1-45 | GAL4-Δ1-45 | This work |
| pG-Δ1-296 | Δ1-296 | GAL4-Δ1-296 | This work |
Insertional mutations were first described by Kroner et al. (17). “pLex-” indicates fusions created in pBTM116; “pG-” indicates fusions created in pGAD424; “pGB-” indicates fusions created in pGBT9. Alternative names are in parentheses.
Amino acids inserted or deleted. For 1a insertion mutations, numbers in parentheses indicate the amino acid which occurs immediately prior to the insertion. NA, not applicable.
Western blot analysis.
Yeast extracts were prepared by scraping three-day-old yeast colonies (100 to 200 mg) from plates and suspending them in 3.0 ml of YPD broth. After growth for 2 h at 30°C, the protein synthesis inhibitor cycloheximide (50 μg/ml) was added followed by another hour of incubation. The cells were then washed with ice-cold water and suspended in 200 μl of cracking buffer (40 mM Tris-HCl [pH 6.8], 0.1 mM EDTA, 5% [wt/vol] sodium dodecyl sulfate (SDS), 8 M urea, 0.05 M β-mercaptoethanol, 0.4 mg bromophenol blue/ml, and the protease inhibitors pepstatin A [1 μg/ml], leupeptin [3 μM], benzamidine [14.5 mM], aprotinin [37 μg/ml], and phenylmethylsulfonyl fluoride [33.4 mg/ml]). The cell suspension was heated at 70°C for 10 min, and then the cells were lysed by vortexing in the presence of glass beads as previously described (22). The extracts were then separated on a SDS–8% polyacrylamide gel, which was then blotted onto polyvinylidene difluoride membrane (Millipore) at 31 V for 5 h in Western transfer buffer (39 mM glycine, 48 mM Tris-HCl [pH 8.3], 0.0037% SDS, 20% methanol). After the transfer, exhausted gels were stained with Coomassie blue to ensure even transfer. The blots were then probed with a polyclonal LexA-specific antibody (kindly provided by Barak Cohen) and a secondary antibody linked to horseradish peroxidase. Bands were visualized by chemiluminescence (U.S. Biochemical Corp.). Quantitation was performed with a densitometer (ImageQuant; Molecular Dynamics). The abundance of each mutant protein was determined relative to that of wt1a. A cross-reactive cellular band of approximately 40 kDa was used as an internal control to normalize for the amount of protein loaded. Each number shown is an average of two independent experiments.
Construction of deletion series.
The deletion series of 1a was made by Wang and French, using pGAD424 and pGBT9 (Clonetech Laboratories, Inc.). A DNA segment encoding the entire BMV 1a reading frame was generated by PCR with primers to introduce BamHI restriction sites (upstream primer B1a, 5′-CAACAGGATCCCAAGTTCTA-3′ [native BMV sequences are underlined]; downstream primer B1aRev, 5′-AGACAGGATCCTCACTTAAC-3′). After amplification, the PCR fragments were liberated with BamHI and cloned in frame with the GAL4 transcriptional activation domain in pGAD424 to generate pGAD1a. The BamHI fragment from pGAD1a was then cloned into the BamHI site of pGBT9 to generate pGB1a. The orientation of the insert was determined by restriction analysis, and the junctions of the fusions were sequenced to ensure that the coding frame was correct.
C-terminal truncations of 1a were constructed from pGAD1a by using a series of restriction sites unique to the 1a coding sequence and the pGAD polylinker. The resulting plasmids were then religated, sometimes by filling in the cut sites with T4 DNA polymerase and deoxynucleoside triphosphates. Details of this cloning procedure will be made available upon request.
An N-terminal deletion of 1a, Δ1-45, was generated in pGAD424 with primers B1aRev and b1.1 (5′-CCGGGGATCCTCAACGTTCGCAATAAG-3′) to generate pG-Δ1-45. A second N-terminal deletion removing residues 1 to 296 was created by digesting pGAD1a with SmaI, and an EcoRI linker (5′-GGAATTCC-3′) was then added, followed by digestion with EcoRI. The resulting plasmid, pG-Δ1-296, was religated with BMV 1a sequences fused in frame to the GAL4 transcriptional activation domain between the EcoRI and BamHI sites of pGAD424.
Secondary structure analysis.
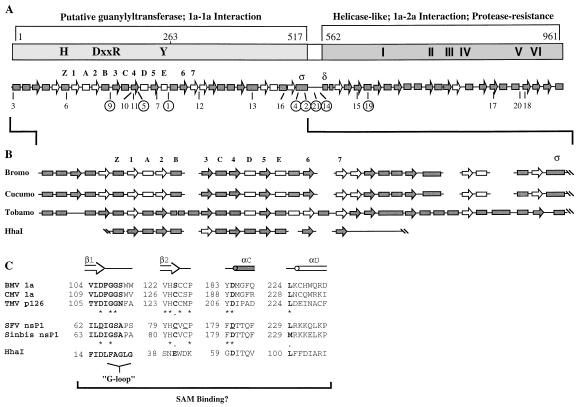
Predictions of secondary structure were generated by the method of Rost and Sander (24, 25), which is more than 70% accurate. The 1a-like proteins from three related plant virus families were analyzed: bromoviruses, cucumoviruses, and tobamoviruses. The specific sequences evaluated can be found in the GenBank database. Bromovirus sequences used were BMV (strain Japanese), CCMV, and broad bean mottle virus (strain BA). Cucumovirus sequences used were CMV (strain fny), peanut stunt virus (strain J), and tomato aspermy virus. Tobamovirus sequences used were tobacco mosaic virus (TMV) (strain Korean), pepper mild mottle virus (strain Spain), tobacco mild green virus (strain U2), and cucumber green mottle virus (strain watermelon).
RESULTS AND DISCUSSION
Effects of two- to three-amino-acid (aa) insertions on BMV 1a-1a interaction.
We have previously established that the two-hybrid system is a convenient and suitable tool for the dissection of the BMV replicase structure. Using this tool, we discovered a novel interaction between two 1a proteins of BMV and those of CMV and CCMV (22). Furthermore, the interaction of the 1a proteins of BMV, CCMV, and CMV occurred in a species-specific manner (22).
To assess the biological relevance of the 1a-1a interaction (22), we utilized a series of well-characterized 2- or 3-aa insertions in 1a previously generated by Kroner and colleagues (17) (Fig. 1A). The ability of these PK mutants to replicate in protoplasts and to interact with 2a both in vitro and in the two-hybrid system has been well documented (14, 17, 22). Briefly, all of the mutants which are replication competent in protoplasts (Fig. 1A) also allow for interaction with 2a in vitro and in the two-hybrid system (14, 22).
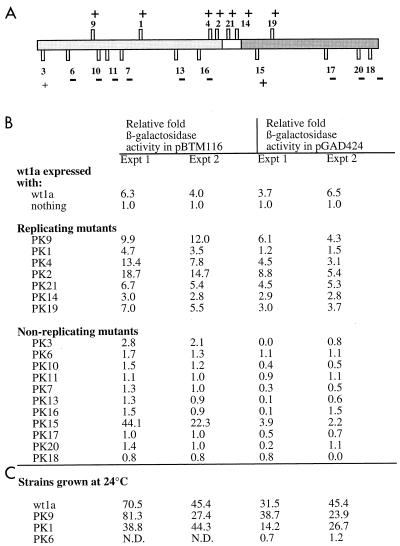
FIG. 1.
(A) Locations of insertion mutations and their replication phenotypes in protoplasts. The box represents the 1a open reading frame. The lightly shaded N-terminal portion contains sequences putatively involved in capping, while the darkly shaded C-terminal portion contains helicase-like sequences. The vertical bars represent the positions of the 2- to 3-aa insertions made and characterized by Kroner and colleagues (17). Bars pointing up represent viable mutants, while bars pointing down represent mutants unable to replicate in barley protoplasts. PK1, -4, and -19 were found to be temperature sensitive for replication at 35°C. The ability (+) or inability (−) of each PK mutant to interact with wt1a in the two-hybrid system is indicated. (B) β-Galactosidase activity detected when each of the PK mutants is coexpressed with wt1a in yeast strain Y835. The results from two independent experiments (Expt) are shown. These activities are presented as fold activity over that of wt1a in the DNA binding domain plasmid. β-Galactosidase activity is shown as micromoles of O-nitrophenyl galactoside hydrolyzed per minute per milligram of protein. (C) β-Galactosidase activity detected for strains grown at 24°C. PK1, a replication-competent mutant in protoplasts which did not interact with wt1a as a LexA fusion at 30°C, did interact at 24°C. PK9, a replicating mutant, and PK6, a nonreplicating mutant, serve as positive and negative controls, respectively. N.D., not determined.
PK mutants 9, 1, 4, 2, 21, 14, and 19, listed according to their positions from the N to C termini in 1a, were competent for replication in protoplasts (17). All seven of these mutants interacted with wt1a when fused to the LexA DNA binding domain, as determined by relative activities ranging from 2.8- to 12.0-fold over background in quantitative β-galactosidase assays (Fig. 1B) and by induction of HIS3, as detected by growth of the strains on plates lacking histidine (data not shown). When fused with the GAL4 transcription activation domain, PK9, -4, -2, -21, -14, and -19 readily interacted with wt1a, with relative activities ranging from 2.8- to 8.8-fold over background (Fig. 1B). However, GAL4-PK1 did not interact with wt1a at 30°C. We noted that PK1 was originally found to be replication competent at 24 but not 35°C (17). Therefore, we tested this mutant along with wt1a at 24 and 30°C. PK9, which was not temperature sensitive, and PK6, a noninteracting mutant, were included as a positive and negative control, respectively. We were not able to assess interactions in yeast cells grown at the nonpermissive temperature tested by Kroner et al. (17) in plant protoplasts because at 35°C, the interaction between the wt1a and wt2a proteins was nearly undetectable. At 24°C, GAL4-PK1 was able to interact with wt1a with high relative activities of 14.2- and 26.7-fold over background (Fig. 1B). These increased levels of activity were paralleled by those of wt1a interacting with itself and with GAL4-PK9 (Fig. 1B). Relative specific activities for the 1a-1a interaction ranged from 31.5- to 70.5-fold over background at 24°C compared with 3.7- to 6.5-fold at 30°C (Fig. 1B). The negative control, GAL4-PK6, still yielded relative activities at or below background levels at either 24 or 30°C (Fig. 1B). Two additional temperature-sensitive mutants, PK4 and PK19, had no relative increases in specific activity when compared to wt1a at 24 versus 30°C (data not shown). Thus, temperature sensitivity is only observed for GAL4-PK1, suggesting that PK1 is more stable as a LexA fusion.
Of the nonreplicating mutants, only PK3 and PK15 allowed for interaction with wt1a. For PK3 this interaction was seen only with fusions to LexA and at levels of approximately 2.5-fold over background. PK15 allowed for interaction with wt1a when fused in either of the two-hybrid plasmids. LexA-PK15 gave high levels of relative activity, at 44.1- and 22.3-fold over background, while GAL4-PK15 gave activities of 3.9- and 2.2-fold over background (Fig. 1B). The observation that PK15 retained interaction with 1a is consistent with our previous observation that this mutation does not grossly affect 1a structure as determined by partial proteolysis assays (21).
All of the replicating mutants can interact with themselves and each other.
The replication-competent PK mutants were tested for interaction with themselves and with each other. Self-interactions were expected, since the original replication results of Kroner et al. (17) were obtained in protoplast transformations which had no source of wt1a. All of the replicating mutants retained interaction with themselves (i.e., PK9-PK9), as detected by the ability of the double transformants to turn blue in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and to grow on plates lacking histidine (data not shown). This is consistent with the hypothesis that 1a-1a interaction is necessary for replication. The PK1-PK1 interaction was observed at 24 but not at 30°C, consistent with our previous results showing GAL4-PK1 to be temperature sensitive (Fig. 1B and data not shown). Additionally, all replicating mutants retained the ability to interact with each other (i.e., PK9-PK21), while PK11 (a nonreplicating mutant) could not interact with itself or any other PK mutant (data not shown). Therefore, all of our results are consistent and the PK mutants which allowed for replication in plant cells also allowed for the 1a-1a interaction between themselves.
Stability of the mutant proteins in vivo.
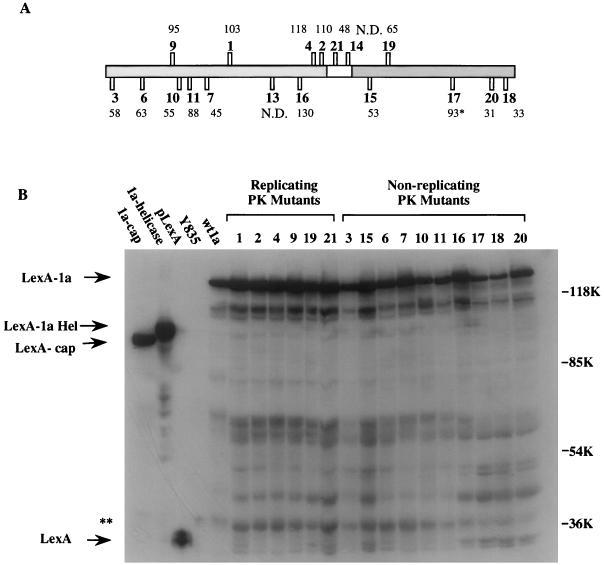
A lack of interaction in the two-hybrid system can simply be due to an unstable fusion protein. To determine if some of the PK mutants were unstable, we checked the abundance of each LexA fusion protein by Western blot analysis. All of the replicating mutants had abundances from 48 to 118% of that of wt1a (Fig. 2). The two nonreplicating mutants which retained the ability to interact with wt1a, PK3 and PK15, were 58 and 53% as abundant as wt1a (Fig. 2). Of the nonreplicating, noninteracting mutants, only PK18 and -20 were somewhat reduced in abundance, present at only 33 and 31% of the abundance of wt1a, respectively (Fig. 2). The observation that many of the noninteracting mutants were as abundant as wt1a indicates that a lack of interaction was not due to a gross reduction in overall protein stability. Thus, the presence of a protein alone is not sufficient for protein-protein interaction.
FIG. 2.
Relative abundance of wt and mutant 1a proteins as observed in Western blots. (A) The basic diagram is the same as that in Fig. 1A. The numbers above or below each PK mutant indicate the percent abundance of each protein relative to that of wt1a. The results shown are an average of the quantitations from two independent Western blots. Quantitation was performed by laser densitometry (Molecular Dynamics). The abundance of PK17 (∗) was somewhat inconsistent between trials, being 150 and 35% that of wt1a. N.D., not determined. (B) Scan of a Western blot showing the abundance of the ca. 140-kDa LexA-PK mutant fusions. The presumed full-length protein is indicated along with the presumed helicase and capping fusions. For unknown reasons the migration of the capping domain was reproducibly faster than expected based on its predicted mass. Lower-molecular-mass bands are presumed to be degradation products. A ca. 40-kDa cellular protein (∗∗) cross-reacted with the antibodies and was used to normalize the amount of protein loaded.
Mapping of the 1a-1a binding domain with deletion analysis.
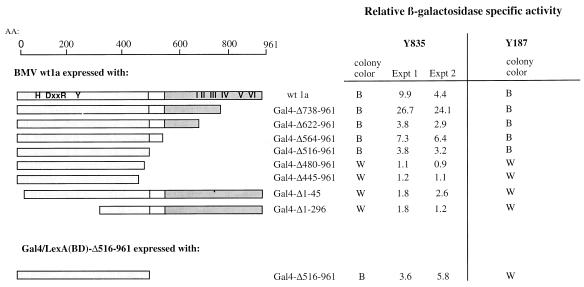
To further identify the region(s) needed for 1a-1a interaction, we tested the abilities of 1a deletions fused to the GAL4 activation domain to interact with wt1a fused to either the LexA or the GAL4 DNA binding domain. Deletions of C-terminal residues 738 to 961, 622 to 961, 564 to 961, and 516 to 961, removing the entire helicase-like domain, all retained interaction with wt1a when tested in yeast strain Y835 (Fig. 3). An additional deletion of 37 residues, producing 1a-Δ480-961, resulted in a loss of interaction with wt1a, indicating that the C-terminal boundary lies between aa 480 and 515 (Fig. 3). At the N terminus of 1a, removal of only 45 residues abolished interaction with full-length 1a. Thus, the region containing putative capping sequences is required for 1a-1a interaction. The same results were obtained when the deletion series was tested by the filter lift assay for the ability to interact with wt1a fused to the GAL4 DNA binding domain (in pGBT9) in strain Y187 (Fig. 3). Negative controls for these assays included the DNA binding domain plasmid in the absence of a partner plasmid and each fusion in combination with a partner plasmid lacking 1a sequences. All of the negative controls resulted in relative specific activities at or below background levels (data not shown). Due to a lack of GAL4-specific antibody, we did not examine the stability of these N- and C-terminal deletions of 1a.
FIG. 3.
Mapping the 1a-1a binding domain by deletion analysis. The domains of the 1a protein are depicted along with various deletions in either the N or C terminus. Fold β-galactosidase levels detected when each of the deletions was expressed with wt1a from two independent experiments are shown relative to those of wt1a alone. For strain Y187 (Clonetech Laboratories, Inc.), only the results of the qualitative filter assays are shown. Δ516-966 retains the ability to interact with both itself and wt1a. B, blue colonies; W, normal yeast colony color. AA, amino acid.
Additionally, we observed that the putative capping domain of 1a (1a-Δ516-961) could not interact with wt2a or any deletion version of the 2a protein (i.e., 2aΔC, 2aΔN/C, and 2aΔN [22]) (data not shown). This observation supports our previous finding that the helicase-like region of 1a (1aΔ1-555) is necessary and sufficient for interaction with 2a (21).
Although the majority of the assays were performed in strain Y835, the 1a truncations were also tested in strain Y187 (Clonetech Laboratories, Inc.) for their ability to interact with the same deletions fused to the GAL4 DNA binding domain in pGBT9. GAL4AD-Δ480-961, which was unable to interact with wt1a, could not interact with itself (GAL4BD-Δ480-961). However, Δ516-961 retained interaction with wt1a and with the same deletion fused to the appropriate partner (Fig. 3). This was also true of the other less severe deletions (data not shown). After generating Δ516-961 fused to LexA, the interaction between LexA-Δ516-961 and GAL4-Δ516-961 was confirmed in strain Y835 and yielded relative activities approximately fourfold higher than background (Fig. 3). Residues 1 to 515 thus contain the sequences necessary and sufficient for 1a-1a interaction in the two-hybrid assay. Additional contact sites are not ruled out by the negative two-hybrid results.
Mapping the 1a-1a interaction in CMV.
We have previously shown that the 1a proteins of both CCMV and CMV interact with themselves in a species-specific manner (22). To determine if the 1a-1a interaction occurred in the putative capping regions of other viruses, we tested for such an interaction in CMV, which is more distantly related to BMV than CCMV, thus providing a more rigorous test of the biological relevance of this interaction. Sequence alignments generated by CLUSTAL W (30) indicated that CMV residues 1 to 533 were homologous to BMV residues 1 to 515. Therefore, PCR was used to generate the CMV DNA fragment flanked by the appropriate restriction enzymes. The fragment containing the putative capping sequences of CMV 1a was then cloned into pBTM116 and pGAD424 to create pLex-CMT and pG-CMT.
The CMV putative capping region (CMT) interacted with the CMV full-length 1a protein (C1a) with relative specific activities of 2.9- to 8.2-fold over background (Table 2). This interaction was observed with CMT fused to either LexA or GAL4. Additionally, CMT could reproducibly interact with itself with a relative specific activity of twofold over background. As a further indication of the validity of these interactions, the transformants all turned blue in the presence of X-Gal and grew on defined media lacking histidine while cells containing either plasmid alone did not (Table 2, Fig. 4, and data not shown).
TABLE 2.
Summary of CMV 1a interaction occurring through the putative capping domain
| Plasmid(s) in strain Y835a | Appearance of colonies in filter assayb | Relative β-galactosidase sp actc
|
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| pB-C1a only | W | ND | ND |
| pB-CMT only | W | ND | ND |
| pB-C1a and pGAD424 | W | 1.00 | 1.00 |
| pB-CMT and pGAD424 | W | 1.00 | 1.00 |
| pBTM116 and pG-C1a | W | 1.4 | 1.4 |
| pBTM116 and pG-CMT | W | 0.95 | 1.4 |
| pB-C1a and pG-C1a | B | 5.15 | 9.59 |
| pB-C1a and pG-CMT | B | 2.87 | 3.49 |
| pB-CMT and pG-C1a | B | 5.49 | 8.17 |
| pB-CMT and pG-CMT | B | 2.11 | 2.09 |
Descriptions of plasmids are in Table 1.
W, normal yeast colony color; B, blue.
Specific activity is shown as micromoles of O-nitrophenyl galactoside hydrolyzed per minute per milligram of protein. Data are shown as fold activity relative to the background activity of pB-C1a or pB-CMT with pGAD424. Results of each experiment are the average of two independent assays. ND, not determined.
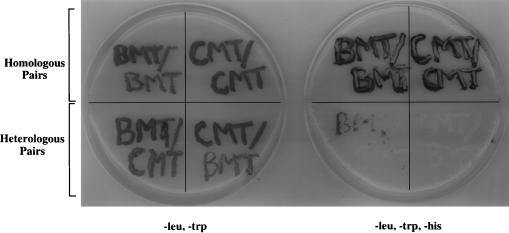
FIG. 4.
Assay for induction of the HIS3 gene in homologous or heterologous pairings of BMV and/or CMV 1a methyltransferase fusions. The plate on the left is supplemented with histidine, while the one on the right is not. Each doubly transformed strain was written onto plates to indicate the identity of the 1a fusion proteins. The LexA fusion is indicated above the slash, and the GAL4 fusion is indicated below the slash (i.e., LexA/GAL4).
When combinations of BMV (BMT) and CMV (CMT) capping fusions were tested, only yeast strains containing homologous pairings of proteins grew on plates lacking histidine while those containing heterologous pairings either did not grow or grew poorly (Fig. 4). Thus, the 1a-1a interaction occurs through the same domain in the more distantly related CMV and is specific to each virus species.
Secondary structure analysis of BMV 1a and related proteins.
Currently, the only structural analysis of BMV 1a comes from protease digestion studies, which indicate that a globular domain exists from aa 556 to 961 in the helicase-like portion of the protein which is needed for interaction with the N terminus of 2a both in vitro and in the two-hybrid system (21) (Fig. 5A). Further analysis would be greatly enhanced by secondary structure predictions which are at least 70% accurate (24, 25). This analysis is also necessitated by the discrepancy in the boundaries encoding the various activities of the putative capping and helicase-like domains (compare the figures in references 1 and 22 with those in references 4, 26, and 29). The discrepancy is based on the interpretations of sequence comparisons of the 1a-like proteins from a number of plant-infecting members of the alphavirus-like superfamily. When compared to Semliki Forest virus (SFV) and Sindbis virus, the highly conserved sequences (H, DxxR, and Y, where x is any amino acid) required for capping in BMV 1a are located from residues 1 to 263 (26) (Fig. 5A). Mutation of the conserved histidine in SFV specifically abolished guanylyltransferase activities, while the homologous mutation in Sindbis virus affected both methyltransferase and guanylyltransferase functions (2, 33). Mutation of the other three conserved residues affected both guanylyltransferase and methyltransferase functions in both SFV and Sindbis virus (2, 33). Comparisons of these and other conserved residues in alfalfa mosaic virus, TMV, BMV, CMV, and tobacco rattle virus revealed a large relatively nonconserved region of >400 aa residing between the putative capping and helicase-like sequences (4, 29).
FIG. 5.
Summary of the predicted secondary structures of BMV and related 1a proteins. Boxes, α-helices; arrows, β-sheets; solid shapes, >80% accuracy; open shapes, >50% accuracy. (A) Diagram of the BMV 1a protein and its predicted secondary structures. The previously identified sequences involved in capping and helicase function are indicated. Consistent with Fig. 1A, the putative capping domain required for 1a-1a interaction is lightly shaded while the protease-resistant helicase-like domain required for 1a-2a interaction is darkly shaded. The locations of the PK mutants in the predicted secondary structures are indicated, with the replication-competent mutants circled. The secondary structures which correspond to those found in DNA methyltransferases are numbered according to the system of Schluckebier et al. (28). We also labeled helices ς and δ to facilitate the flow of the text. (B) Expanded picture of the predicted secondary structure of the putative capping domain of the bromo-, cucumo-, and tobamovirus families compared to that of the DNA methyltransferase HhaI. β3 of HhaI, shown as an unshaded arrow, was not present in the originally solved crystal structure and was not predicted by PHD analysis (6). This strand was later added upon comparison of the structure HhaI to other methyltransferase crystal structures (19, 28). Also, there is a 4-aa β-sheet (β4′) following β4 which we have not included in our diagram for simplicity (6). (C) Functional residues involved in SAM binding in HhaI and their putative functional homologs in the SAM binding capping proteins of RNA viruses. Residues conserved between the DNA and RNA SAM binding proteins, including the G-loop, are shown in bold letters. The underlined residues of SFV nsP1 have been mutated to alanine and all, except D180A, have effects on methyltransferase and guanylyltransferase functions. Asterisks indicate identical amino acids, and periods indicate similar amino acids.
Several lines of evidence suggest that the BMV capping domain is more extensive than the conserved residues in aa 1 to 263. First, Sindbis virus and SFV require ca. 515 aa for capping functions (2, 33). Secondly, our present analysis indicates that aa 1 to 515 of BMV 1a exist as a domain which is necessary for the 1a-1a interaction. These observations led us to use the PHD method of Rost and Sander (24, 25) to predict the secondary structures of 1a, especially those in the putative capping domains of several bromo-, cucumo-, and tobamoviruses (Fig. 5). All α-helices and β-sheets shown are predicted with greater than 50% reliability, and those predicted with more than 80% reliability are indicated.
The predicted secondary structure of the 1a protein grossly resembles earlier drawings containing two large domains separated by a small hinge region (14, 17, 21). However, the lengths and positions of these domains are different from those in previous drawings based solely on primary sequence. Previously, the putative capping domain of BMV was predicted to span from aa 1 to 425 (1). Secondary structure analysis indicates that the putative capping domain actually spans from aa 1 to 517. Although BMV does not have much primary sequence similarity to related viruses from aa 426 to 516, the same general order of structural elements is conserved (Fig. 5B). The bromo- and cucumoviral 1a proteins, including BMV, CCMV, broad bean mottle virus, CMV, peanut stunt virus, and tomato aspermy virus, have identical structural elements despite being up to 61% dissimilar at the amino acid level (Fig. 5B and data not shown). The p126-like proteins, including those from TMV, pepper mild mottle virus, cucumber green mosaic virus, and tomato mild green mosaic virus, were identical to one another but varied slightly from the bromo- and cucumovirus 1a proteins. Compared to the bromo- and cucumoviruses, the longer tobamovirus proteins contain several additional α-helices. Further, the tobamovirus proteins had an α-helix in place of β4 and was missing a predicted β-sheet, which was found at the N terminus of the bromo- and cucumovirus 1a proteins. The remaining structures found in the bromo- and cucumovirus 1a proteins were present in regions of tobamovirus sequence that were up to 84% dissimilar (Fig. 5B and data not shown).
Correlation of predicted domain borders with function.
The predicted secondary structures of the putative capping domain agree remarkably well with our functional analysis of protein-protein interaction. Our deletion analysis places the end of the putative capping domain at aa 515 (Fig. 3). Only the truncations which retain all of the predicted capping domain intact can interact with 1a. The minimally functional truncation, aa 1 to 515, which removes 2 aa of ας, does not grossly disturb its structure (Fig. 5A). Removal of an additional 34 aa completely removes ας and is not functional for 1a-1a interaction. An N-terminal truncation of 45 residues in GAL4-Δ1-45 removes the first two α-helices and is unable to interact with 1a. Our mapping and structural prediction data suggest that BMV residues 1 through 517 exist as a functional domain.
Based on primary sequence comparisons with Sindbis virus, the predicted helicase-like domain of BMV 1a was reported to span from residues 510 to 961 (1). However, secondary structure analysis places the helicase-like domain within residues 562 to 961 (Fig. 5A). Results from previous protease digestion studies and two-hybrid analysis correlate with this prediction. We found that aa 556 to 961, which include αδ of the helicase-like domain, could interact with 2a in vitro and in the two-hybrid system (21). We found that aa 567 to 961, which shortens αδ from 19 to 12 residues, could not interact with 2a in vitro (21). However, in the two-hybrid system we found that aa 567 to 961 could still interact with 2a, and we postulated that this was due to the fusion of LexA to aa 567 (21). In fact, secondary structure prediction indicates that αδ (Fig. 5A) is fortuitously lengthened from 12 to 16 residues when it is fused to LexA (data not shown). αδ is completely removed by a further truncation, aa 580 to 961, which cannot interact in vitro or in the two-hybrid system (21). Our mapping and prediction data suggest that the helicase-like domain spans at least residues 562 to 961.
A putative hinge was previously placed between residues 426 and 509 (1). In our analysis, a confidently predicted loop region exists between BMV 1a residues 517 and 562 (Fig. 5A). This predicted loop is consistent with our previous observation that an unusually high number of prolines are present from aa 514 to 560 (21). This region of 39 aa is likely to be a flexible hinge that separates the capping and helicase-like domains.
Effect of 2- to 3-aa insertions on predicted secondary structures.
We noted that two of the replicating mutants, PK21 and -14, are present in the newly positioned hinge region and, as expected, have no affect on the predicted secondary structure (data not shown). We wondered if similar correlations could be drawn from other PK mutants. Four of the eight replicating mutants, PK9, -1, -21, and -14, are located in predicted loops and have no affect on predicted α-helices or β-sheets (Fig. 5A). The other four replicating mutants, PK5, -4, -2, and -19, are located within predicted α-helices or β-sheets, but their insertions do not disrupt the predicted structures. All of the nonreplicating mutants had predicted adverse effects on the elements they occupied and/or nearby elements (Fig. 5A and data not shown). Thus, a perfect correlation exists between the maintenance of the predicted secondary structure and the ability to replicate in protoplasts. Since we have shown that all PK mutants are somewhat stable (Fig. 2), it is likely that the perturbations of secondary structure are affecting function.
PK15, which cannot replicate in protoplasts (17), retains the protease-resistant domain and the ability to interact with both 1a and 2a (14, 21). The PK15 insertion is located near helicase motif I, which is thought to be a nucleoside triphosphate binding site. Our analysis of secondary structure indicates that PK15 completely disrupts the β-sheet in which it resides. These results support our previous suggestion that PK15 affects a specific activity within 1a (21).
Comparisons of predicted elements to those found in animal virus and DNA methyltransferases.
Portions of the predicted secondary structures in the putative capping domain of the plant-infecting viruses appear to resemble structures important for S-adenosyl-l-methionine (SAM) binding found in DNA methyltransferases (Fig. 5). The resemblance between RNA and DNA methyltransferases was first observed when the structure of the VP39 protein of vaccinia virus was solved (11). The bromo-, cucumo-, and tobamovirus 1a-like proteins all contain the alternating α-β-α-β structures universally found in SAM-dependent methyltransferases (19, 28) (Fig. 5B). We used a group γ methyltransferase, HhaI, to elaborate this comparison because its crystal structure complexed with SAM has been solved (6, 19). All of HhaI’s known elements were predicted accurately by the PHD program, including the absence of β3, which was not present in the originally reported crystal structure (6) (Fig. 5B). However, this strand was later added based on comparisons to other methyltransferase crystal structures (6a, 19, 28). The bromo- and cucumovirus 1a proteins retain the secondary structure elements in the same order as HhaI with a single insertion of an α-helix between αE and β6 (Fig. 5B). The TMV-like proteins have several additional α-helices between αB and β3, αE and β6, and β6 and β7 (Fig. 5B). It is interesting to speculate that these and the other extra α-helical TMV elements shown in Fig. 5B may be related to the effects of this domain on viral pathogenesis proposed for TMV p126 (4, 29).
In addition to the maintenance of overall secondary structure, many important functional residues found in DNA methyltransferases are conserved in the putative capping enzymes of RNA viruses (Fig. 5C). While the primary sequences of different DNA methyltransferases are quite divergent, their tertiary structures are remarkably similar, especially with regard to the positions of several conserved catalytic residues (19, 29). The proteins with putative capping functions in RNA viruses share many of these functional residues involved in SAM binding (Fig. 5C). Following β1 in the DNA methyltransferases is the loosely conserved “G-loop,” which is crucial for the proper positioning of the adenine ring of SAM (6). The putative capping enzymes of RNA viruses have similar G loops following β1 (Fig. 5C). G loop residue F18 and residue L100 in HhaI form Van der Waals interactions with the adenine ring in SAM. F18, or another suitable hydrophobic residue, and L100 appear to be present in the RNA virus capping enzymes (Fig. 5C). The terminal aspartate of β1 is maintained in the putative capping enzymes of RNA viruses (Fig. 5C). Changing this aspartate to alanine in SFV had adverse affects on both methyltransferase and guanylyltransferase activities (2).
In HhaI, the acidic residues at the end of β2 and beginning of αC form specific hydrogen bonds with SAM. In place of the glutamate at the end of β2, the RNA capping enzymes have a serine or cysteine which should also be capable of acting as a hydrogen acceptor. This substitution may affect the Km for SAM binding in the RNA capping enzymes. Changing the cysteine at the β2 terminus of SFV had detrimental effects on both methyltransferase and guanylyltransferase activities. The aspartate at the beginning of αC is conserved between HhaI and the examined RNA viruses; however, it remains to be determined whether this residue is the actual homolog of HhaI D60, since a D180A mutation in SFV had no effect on methyltransferase activity (2). Thus, all of the functionally important residues involved in SAM binding are maintained by these putative RNA capping enzymes in regions of similar structure, suggesting that they may function in a similar fashion.
An interaction between the helicase- and methyltransferase-like portions of the 1a protein.
In our previous work we found that none of the nonreplicating mutants could interact with 2a (22). This is surprising for PK mutants 6, 7, 10, and 11 because their insertions are in the putative capping domain while the helicase-like domain has been demonstrated to be sufficient for interacting with 2a. Further, Western blotting results show that these mutants are as stable as wt1a (Fig. 3), indicating that there may be an additional requirement for protein-protein interaction. One hypothesis consistent with these observations is that the N- and C-terminal halves of the 1a protein may interact. Thus, mutations in the 1a N terminus affect interactions that occur in the 1a C terminus. To test this possibility, we used pEOΔ500, a LexA fusion retaining the entire hinge and helicase-like domain (21), for interaction with pG-BMT, which retains the putative capping domain fused to GAL4. The product of pEOΔ500, LexA-BHel, was able to interact with GAL4-BMT with a relative activity of greater than 25-fold over background (Table 3). We subcloned sequence coding for 1a helicase into pGAD424, generating pG-BHel. When tested, GAL4-BHel interacted with LexA-BMT with a relative activity of threefold over background. Although many helicases function as multimers (18), we found no evidence for a helicase-helicase interaction (Table 3). Negative controls for these experiments all gave relative activities at or below background (Table 3).
TABLE 3.
Summary of 1a intramolecular interaction occurring between methyltransferase- and helicase-like domains in BMV and CMV
| Expressed fusion proteina | Appearance of colonies in filter assayb | Growth on plates lacking histidine | Relative β-galactosidase sp actc
|
|
|---|---|---|---|---|
| Expt 1 | Expt 2 | |||
| LexA-BMT only | W | No | 1.0 | 1.0 |
| LexA-BHel only | W | No | 1.3 | 0.8 |
| LexA-BHel and GAL4-BHel | W | No | ND | ND |
| LexA-BMT and GAL4-BMT | B | Yes | 2.7 | 3.1 |
| LexA-BHel and GAL4-BMT | B | Yes | 29.6 | 24.8 |
| LexA-BMT and GAL4-BHel | B | Yes | 3.6 | 3.6 |
| LexA-wt1a and GAL4-Δ1-45 | W | No | ND | ND |
| LexA-BMT and GAL4-Δ1-45 | B | Yes | ND | ND |
| LexA-CMT only | W | No | ND | ND |
| LexA-CMT and GAL4-CMT | B | Yes | ND | ND |
| LexA-CHel and GAL4-CMT | B | Yes | ND | ND |
Descriptions of fusion proteins are in Table 1.
W, normal yeast colony color; B, blue.
Specific activity is shown as micromoles of O-nitrophenyl galactoside hydrolyzed per minute per milligram of protein. Data are shown as fold activity relative to the background activity of LexA-BMT alone. Results of each experiment are the average of two independent assays. ND, not determined.
The ability of the putative capping and helicase-like domains to interact with one another is conserved in other tricornaviruses. Yeast strains expressing the putative capping and helicase-like domains of CMV 1a also turned blue in the presence of X-Gal and grew in the absence of histidine (Table 3).
This interaction between the putative capping and helicase-like domains could occur within the same molecule of 1a or between two different 1a molecules. The nature of the two-hybrid system does not allow for a definitive assessment of these two possibilities. However, the results of our deletion analysis indirectly suggest that the interaction is intramolecular in nature. Our deletion analysis has shown that removal of the N-terminal 45 aa of wt1a in GAL4-Δ1-45 caused the loss of interaction with LexA-wt1a (Fig. 4 and Table 3). Therefore, the wt putative capping domain of the LexA-wt1a fusion is not capable of interacting in trans with the helicase-like domain of GAL4-Δ1-45. To test this apparent cis preference, we evaluated the ability of LexA-BMT alone to interact with GAL4-Δ1-45 and found that they could interact (Table 3). Thus, intermolecular interactions between the putative capping and helicase-like domains were only observed when an intramolecular, or cis, interaction was not possible.
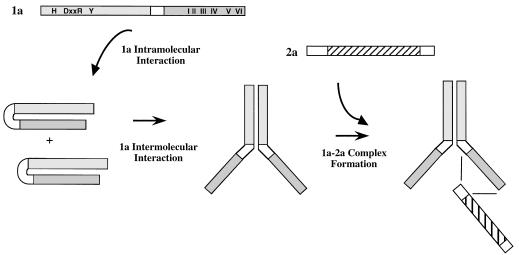
Model for replicase assembly.
Our observations to date suggest a working model for the assembly of the 1a-2a complex (Fig. 6). The presence of the putative intramolecular interaction in 1a may serve to prevent the formation of the 1a-2a complex until the appropriate 1a-1a complex has formed. It is also possible that each step is influenced by host factors and/or RNA interactions.
FIG. 6.
Working model for the assembly of the 1a-2a complex. The putative intramolecular interaction in 1a prevents the formation of the 1a-2a complex until the intermolecular 1a-1a interaction has occurred. The 2a protein interacts with the helicase-like domain of 1a through its N terminus.
Thus far, we have not determined the number of 2a molecules present in the complex. Previous studies suggest that oligomerization of the poliovirus 3D polymerase is required for function (13, 23). Thus, it is possible that each available helicase-like domain interacts with one or more molecules of 2a. The 2a polypeptide has inherent transcription activation activity when fused to the LexA DNA binding domain, and thus, 2a-2a interactions could not be tested in the two-hybrid system (7a). However, the recently published crystal structure of the poliovirus 3D polymerase suggests two possible sites of polymerase oligomerization (9). Using these limited regions as guides, tests for the interaction of the corresponding regions in the BMV 2a protein will be performed.
In an attempt to test this replicase assembly model, we tried to separate intermolecular interactions from intramolecular interactions by creating PK mutants in the context of the putative capping domain alone (i.e., PK9Δhel and PK6Δhel). We expected to find mutants that could support the intramolecular interaction but not the intermolecular interaction. However, none of the truncated mutants were stable, even though the full-length proteins containing the mutations and the wt capping domain alone were stable (Fig. 2 and data not shown). Although indirect, these results also suggest that an intramolecular interaction exists between the capping and helicase-like domains of 1a. This interaction may help to stabilize the insertions in the context of the full-length protein.
Summary.
In this work we have (i) shown a perfect correlation between replication and the ability of the PK mutants to interact with wt1a, (ii) mapped the species-specific intermolecular interaction to the N-terminal putative capping domains of both the BMV and CMV 1a proteins, (iii) demonstrated a high degree of conservation in the predicted secondary structures among members of the plant alphavirus-like superfamily and structures associated with SAM binding in DNA methyltransferases, and (iv) demonstrated a novel interaction between the N- and C-terminal halves of the 1a proteins of both BMV and CMV that is likely to be intramolecular in nature. The predicted secondary structures correlate well with the effects of deletions and insertions on protein-protein interactions and on RNA replication in barley protoplasts. These results predict new borders for the functional domains of the putative capping, helicase-like, and hinge regions of 1a. The figures in this paper reflect these new borders. Additionally, this analysis predicts the positions of functional residues involved in BMV 1a SAM binding. These results contribute further to an understanding of the architecture and assembly of replicase complexes from positive-strand RNA viruses.
ACKNOWLEDGMENTS
This work, including a supplement to E.K.O., was supported by NSF grant MCB 9507344. E.K.O. also acknowledges support from the Plant Sciences Ogg Fellowship from Indiana University.
We thank Paul Ahlquist for use of the PK mutants, Peter Palukaitis for pFny106, Barak Cohen for the LexA antibody, Stan Fields for pBTM116 and pGAD424, Greta Faurote and Jonathan Paul for use of the 2a deletion constructs, Rick Nelson for helpful discussions, and, finally, the IU Cereal Killers, especially Matt Chapman, for helpful discussions and encouragement throughout the course of this work.
REFERENCES
- 1.Ahlquist P, Strauss E G, Rice C M, Strauss J H, Haseloff J, Zimmern D. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J Virol. 1985;53:536–542. doi: 10.1128/jvi.53.2.536-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao Y, Carter S A, Nelson R S. The 126- and 183-kilodalton proteins of tobacco mosaic virus, and not their common nucleotide sequence, control mosaic symptom formation in tobacco. J Virol. 1996;70:6378–6383. doi: 10.1128/jvi.70.9.6378-6383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal T, Carmichael G. RNA replication: function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X, Kumar S, Posfai J, Pflugrath J W, Roberts R J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 6a.Cheng, X. Personal communication.
- 7.Dinant S, Janda M, Kroner P, Ahlquist P. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J Virol. 1993;67:7181–7189. doi: 10.1128/jvi.67.12.7181-7189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Faurote, G. Unpublished data.
- 8.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A novel superfamily of nucleotide triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 10.Haseloff J, Goelet P, Zimmern D, Ahlquist P, Dasgupta R, Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci USA. 1984;81:4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodel A E, Gershon P D, Shi X, Quiocho F A. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85:247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 12.Honda A, Ishihama A. The molecular anatomy of influenza virus RNA polymerase. Biol Chem. 1997;378:483–488. [PubMed] [Google Scholar]
- 13.Hope D A, Diamond S E, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao C C, Quadt R, Hershberger R P, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiberstis P A, Loesch-Fries L S, Hall T C. Viral protein synthesis in barley protoplasts inoculated with native and fractionated brome mosaic virus RNA. Virology. 1981;112:804–808. doi: 10.1016/0042-6822(81)90331-7. [DOI] [PubMed] [Google Scholar]
- 17.Kroner P A, Young B M, Ahlquist P. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J Virol. 1990;64:6110–6120. doi: 10.1128/jvi.64.12.6110-6120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohman T. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 19.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyl-transferases, and suggests a catalytic mechanism. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 20.Mi S, Stollar V. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology. 1991;184:423–427. doi: 10.1016/0042-6822(91)90862-6. [DOI] [PubMed] [Google Scholar]
- 21.O’Reilly E K, Tang N, Ahlquist P, Kao C C. Biochemical and genetic analyses of the interaction between the helicase-like and polymerase-like proteins of brome mosaic virus. Virology. 1995;214:59–71. doi: 10.1006/viro.1995.9954. [DOI] [PubMed] [Google Scholar]
- 22.O’Reilly E K, Paul J D, Kao C C. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol. 1997;71:7526–7532. doi: 10.1128/jvi.71.10.7526-7532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 24.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 25.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–77. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 26.Rozanov M N, Koonin E V, Gorbalenya A E. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 27.Scheidel L M, Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology. 1991;181:490–499. doi: 10.1016/0042-6822(91)90881-b. [DOI] [PubMed] [Google Scholar]
- 28.Schluckebier G, O’Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 29.Shintaku M H, Carter S A, Bao Y, Nelson R S. Mapping nucleotides in the 126-kDa protein gene that control the differential symptoms induced by two strains of tobacco mosaic virus. Virology. 1996;221:218–225. doi: 10.1006/viro.1996.0368. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyoda T, Adyshev D M, Kobayashi M, Iwata A, Ishihama A. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J Gen Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 32.Traynor P, Young B M, Ahlquist P. Deletion analysis of brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J Virol. 1991;65:2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H-L, O’Rear J, Stollar V. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology. 1996;217:527–531. doi: 10.1006/viro.1996.0147. [DOI] [PubMed] [Google Scholar]