Abstract
We used the yeast interaction trap system to identify a novel human 70-kDa protein, termed NS1-binding protein (NS1-BP), which interacts with the nonstructural NS1 protein of the influenza A virus. The genetic interaction was confirmed by the specific coprecipitation of the NS1 protein from solution by a glutathione S-transferase–NS1-BP fusion protein and glutathione-Sepharose. NS1-BP contains an N-terminal BTB/POZ domain and five kelch-like tandem repeat elements of ∼50 amino acids. In noninfected cells, affinity-purified antibodies localized NS1-BP in nuclear regions enriched with the spliceosome assembly factor SC35, suggesting an association of NS1-BP with the cellular splicing apparatus. In influenza A virus-infected cells, NS1-BP relocalized throughout the nucleoplasm and appeared distinct from the SC35 domains, which suggests that NS1-BP function may be disturbed or altered. The addition of a truncated NS1-BP mutant protein to a HeLa cell nuclear extract efficiently inhibited pre-mRNA splicing but not spliceosome assembly. This result could be explained by a possible dominant-negative effect of the NS1-BP mutant protein and suggests a role of the wild-type NS1-BP in promoting pre-mRNA splicing. These data suggest that the inhibition of splicing by the NS1 protein may be mediated by binding to NS1-BP.
Replication of viruses can induce drastic changes in the metabolism of the infected host cell. The analysis of the replication cycle of viruses by molecular biological techniques has facilitated the identification and study of viral gene products which modulate and affect cellular functions (31). A well-studied example is the NS1 protein of influenza A viruses. The NS1 protein is the only nonstructural protein of the virus and is abundantly expressed in infected cells (35). Several regulatory functions of the NS1 protein have been identified. The NS1 protein influences multiple steps of gene expression, including pre-mRNA splicing (19, 36), nucleocytoplasmic transport of poly(A) RNA (19, 48), and translation (9, 15). In addition, it was recently shown that NS1 can block the activation of the double-stranded RNA-activated protein kinase (PKR), presumably due to its double-stranded RNA binding activity (37). The activation of PKR results in a downregulation of translation and is part of the cellular antiviral defense mechanism. The NS1 protein may counteract this cellular response in order to synthesize high levels of viral proteins in the infected cell (37). These pleiotropic effects may, singly or combined, provide the molecular basis for the role which the NS1 protein plays in determining the host range and virulence of influenza virus strains (54, 61).
Despite the plethora of activities identified in NS1, little is known about the cellular factors that are recognized by the NS1 protein and that may therefore be central to NS1 functions. We have used the yeast interaction trap (26, 74) to screen for cellular proteins that interact with the NS1 protein. Here we report on the identification and characterization of a novel human 70-kDa protein, termed NS1-binding protein (NS1-BP). In noninfected cells, NS1-BP was found to accumulate in discrete nuclear domains which are enriched with pre-mRNA splicing factors. However, in influenza A virus-infected cells, NS1-BP relocalized throughout the nucleoplasm. The addition of a truncated NS1-BP inhibited pre-mRNA splicing in HeLa cell nuclear extracts in vitro, possibly as the result of a dominant-negative effect on the endogenous protein. These results suggest a role for NS1-BP in pre-mRNA splicing and support a model in which the NS1–NS1-BP interaction has a role in mediating the splicing-inhibitory effect of the NS1 protein.
MATERIALS AND METHODS
Viruses, cells, and extracts.
Influenza A/WSN/33 virus was grown in the allantoic cavities of 10-day-old embryonated chicken eggs. HeLa, HEp-2, and 293 cells were passaged in Dulbecco’s modified Eagle’s tissue culture medium (DMEM) containing 10% fetal calf serum. For immunoblot analysis, confluent cell monolayers grown in 35-mm dishes were lysed in radioimmunoprecipitation assay buffer containing 150 mM NaCl, 1.0% Nonidet P-40 (NP-40), 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and 50 mM Tris-HCl (pH 8.0). Lysates were clarified by centrifugation for 10 min at 13,000× g, and supernatants were used for immunoblot analysis.
Yeast strains, E. coli strains, and plasmids.
Escherichia coli strains used for cloning and expression were MH3 (trpC araD lacX hsdR galU galK), DH5α [F− Φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) supE44 λ− thi-1 gyrA96 relA1], BL26 [F− ompT hsdSB (rB− mB−) gal dcm], and XL1-Blue recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ pro AB lacIqZΔM15 Tn10 (Tetr)].
Saccharomyces cerevisiae EGY48 (MATa trp1 ura3 his3 LEU2::pLEX-Aop6-LEU2), plasmids pSH18-34 and pRFHM1, and the HeLa cell cDNA expression library constructed in pJG4-5 were kindly provided by R. Brent (Harvard Medical School) and have been described previously (26, 74). The constructions of plasmids pLexA-NS1, pcDNA3-NS1, and pGEX-NS1 have been described elsewhere (69). SP6-MINX (75) was used as a template for transcription of synthetic pre-mRNA. Construction of plasmids followed standard cloning procedures (2). Plasmid pGEX-NS1-BP was made by subcloning the HeLa cDNA from the library plasmid (see below) into pGEX-5X-1 (Pharmacia). The bacterial expression plasmids pGEX-NS1-BP-REP and pMAL-NS1-BP-REP were generated by inserting NS1-BP cDNA corresponding to amino acids 1 to 368 (nucleotide positions 1 to 1104) between the EcoRI/XhoI sites of pGEX-5X-1 and the EcoRI/SalI sites of pMAL-c2 (New England Biolabs), respectively.
Identification and isolation of NS1-interacting cDNA clones using the yeast interaction trap.
The yeast interaction trap was used to identify and to isolate HeLa cell cDNAs encoding NS1 binding factors as was previously described (69). In brief, EGY48 was transformed with the bait plasmid pLexA-NS1 and the lacZ reporter plasmid pSH18-34. Subsequently, this strain was transformed with a plasmid library in which HeLa cell cDNAs were conditionally expressed as fusions with an acidic activation domain from a GAL1 promoter. A total of 3.3 × 105 primary transformants were screened for interaction, as determined by their ability to grow on minimal synthetic medium in the absence of leucine and to activate the lacZ reporter gene specifically on plates containing galactose but not glucose. The library plasmid p59-1 was isolated from one selected clone by transformation in E. coli MH3 as described elsewhere (44). The specificity of the interaction was examined by retransformation of p59-1 into EGY48 harboring pSH18-34 together with pLexA-NS1 or with pRFHM1, which expresses an unrelated fusion of LexA with the bicoid protein of Drosophila melanogaster. p59-1 activated the lacZ reporter gene specifically in the presence of galactose in combination with pLexA-NS1 but not in combination with pRFHM1. p59-1 was sequenced by using a standard chain termination protocol. p59-1 originated from the same screen that led to the isolation of cDNA corresponding to another NS1-interacting protein, NS1-I, as described elsewhere (69).
Cloning of NS1-BP 5′ end cDNA.
cDNA corresponding to the 5′ end of NS1-BP mRNA was obtained by a 5′-RACE (rapid amplification of cDNA ends) procedure using a 5′-RACE kit (Gibco-BRL). The 59-1-specific DNA oligonucleotide 59GSP1 (dCATTCCTCTCTGTTATAGCC, corresponding to positions 1123 to 1142 of NS1-BP cDNA; 2.5 pmol) was hybridized to 1 μg of HeLa poly(A)+ RNA to prime first-strand cDNA synthesis by Moloney murine leukemia virus reverse transcriptase. The cDNA was tailed with dC by using terminal transferase. The product was used as a template to amplify double-stranded cDNA by PCR with the nested primer 59GSP2 (dCCACCTGCAGCTATGAG, positions 1108 to 1124) and the 5′-RACE anchor primer. The resulting product was subcloned into pGEM-T (Promega) to generate pGEM-NS1-BP-5′RACE plasmids. The NS1-BP cDNA was sequenced by the standard dideoxy method.
Northern blot analysis.
One microgram of HeLa cell poly(A)+ RNA was electrophoresed on a 1% agarose-formaldehyde gel, transferred onto a Nytran (Amersham) nylon membrane, and immobilized by UV cross-linking. A 32P-labeled NS1-BP-specific probe comprising positions 1038 to 2215 was used to detect NS1-BP mRNA by hybridization as described elsewhere (2).
Coprecipitation of NS1 protein with GST–NS1-BP by glutathione-Sepharose.
NS1-BP (amino acids 347 to 619) was expressed from pGEX-NS1-BP as a glutathione S-transferase (GST) fusion protein in E. coli BL26. Synthesis of GST–NS1-BP was induced by the addition of 1 mM isopropyl-β-d-galactopyranoside (IPTG). Bacterial cell lysate containing the GST–NS1-BP fusion protein was adsorbed to glutathione-Sepharose (Pharmacia) according to the protocol supplied by the manufacturer. Contaminating proteins were removed by three washes with phosphate-buffered saline (PBS). NS1 protein was synthesized and labeled with [35S]methionine in coupled 50-μl transcription-translation reactions (TNT; Promega), programmed with pcDNA3-NS1. The translation reaction was mixed with 10 μl of coated glutathione-Sepharose beads in 750 μl of HN100 buffer (20 mM HEPES [pH 8.0], 100 mM NaCl, 0.01% NP-40) for 2 h at 4°C. The beads were washed three times with PBS–0.01% NP-40, and the precipitated proteins were separated by SDS gel electrophoresis and visualized by autoradiography.
Anti-NS1-BP-serum and immunoblot analyses.
The GST–NS1-BP–REP fusion protein, carrying amino acids 1 to 368 of NS1-BP, was expressed in E. coli BL26 transformed with pGEX-NS1-BP-REP and affinity purified on glutathione-Sepharose resin (Pharmacia) as recommended by the manufacturer. A 6-month-old female rabbit was immunized with 400 μg of purified GST–NS1-BP–REP fusion protein in complete Freund’s adjuvant followed by booster injections of 250 μg of fusion protein in incomplete adjuvant at 4-week intervals. NS1-BP-specific antibodies were purified from serum by affinity chromatography using an antigen resin. For the construction of this matrix, a MAL–NS1-BP–REP fusion protein, in which the maltose-binding protein of E. coli was fused to amino acids 1 to 368 of NS1-BP, was expressed in E. coli XL1-Blue cells and affinity purified on an amylose affinity column (New England Biolabs). The MAL–NS1-BP–REP fusion protein was immobilized on CNBr-activated Sepharose (Pharmacia), and the resulting resin was used for the affinity purification of NS1-BP-specific antibodies as described elsewhere (28). The purified antibodies were diluted 1:200 for immunoblot experiments using standard procedures (28).
Indirect immunofluorescence microscopy.
HeLa cells were grown to 50% confluency on glass coverslips in D-MEM containing 10% fetal calf serum. Where indicated, cells were infected at a multiplicity of 10 with influenza A/WSN/33 virus diluted in PBS for 1 h at 37°C. Infection was continued in tissue culture medium at 37°C. Cells were processed for immunofluorescence analysis by fixation in 2.5% methanol-free formaldehyde (Polysciences Inc.) diluted in PBS, and cells were permeabilized in 0.1% Triton X-100 as described elsewhere (69). Cells were stained with primary antibodies diluted in PBS–3% bovine serum albumin. Affinity-purified anti-NS1-BP antibodies and the NS1-specific monoclonal antibody IA7 (a kind gift of Jonathan Yewdell, National Institutes of Health) were used at 1:100 dilutions. The anti-SC35 antibody (21) was purchased from Pharmingen Inc. and used at a dilution of 1:1,000. The cells were washed and incubated with fluorescein isothiocyanate (FITC)-conjugated sheep anti-rabbit immunoglobulin G (IgG) and/or Texas red-conjugated goat anti-mouse IgG. Subsequently, the coverslips were washed and mounted in Mowiol 4-88 (Calbiochem) as described elsewhere (28). For conventional immunofluorescence analysis, cells were viewed on a Zeiss Axiovert 100 fluorescence microscope with a 63× objective, and photographs were captured by a CF8/10× video camera (Kappa GmbH). A Zeiss LSM 410 Invert microscope equipped with a 100× objective lens was used for confocal laser scanning microscopy. Digitized images were pseudocolored by using Photoshop software (Adobe Systems Inc.).
Spliceosome assembly and splicing of a 32P-labeled pre-mRNA in the presence of GST proteins.
GST, GST-NS1, and GST–NS1-BP fusion proteins were expressed in E. coli BL26 and affinity purified on glutathione-Sepharose (Pharmacia) columns as recommended by the manufacturer. GST proteins were eluted with 20 mM glutathione in 50 mM Tris-HCl (pH 8.0), dialyzed against buffer D (20 mM HEPES [pH 8.0], 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol), and stored at −80°C. The purity of the prepared proteins was tested by SDS gel electrophoresis and staining with Coomassie blue. HeLa cell nuclear extract was prepared as described elsewhere (12). 32P-labeled MINX pre-mRNA (an artificial mini-exon construct derived from the major late transcription unit of adenovirus 2) was synthesized as described elsewhere (68). In a typical splicing reaction, 4 ng of pre-mRNA was incubated in a 100-μl volume containing 40% HeLa cell nuclear extract, 3.2 mM MgCl2, 0.5 mM ATP, 20 mM phosphocreatine, and 60 mM KCl. Eight micrograms of GST or equimolar amounts of GST fusion proteins were added where indicated, and the reactions were incubated at 30°C. The formation of splicing complexes was analyzed after treatment with heparin (1 mg/ml) by electrophoresis on native acrylamide-agarose gels (43). For RNA analysis, splicing products were purified and analyzed by electrophoresis on denaturing 13% acrylamide-urea gels.
Sequence comparisons.
The NS1-BP cDNA and its derived amino acid sequence were compared to the GenBank and EMBL databases by using FASTA and TFASTA software (10). The PILEUP and PRETTY programs of the Genetics Computer Group (GCG) (University of Wisconsin, Madison) were used to align the repeat elements of NS1-BP and to create a consensus sequence.
RESULTS
Isolation of NS1 binding factors.
The yeast interaction trap system (26, 74) was used to identify cellular proteins that bind to the NS1 protein of the influenza A virus (69). We used a constitutively expressed LexA-NS1 fusion protein to screen a HeLa cell cDNA plasmid library, in which cDNA-encoded proteins were conditionally expressed as translational fusions with an acidic activation domain from a GAL1 promoter. Expression of the acidic domain fusion proteins is induced in the presence of galactose and repressed by glucose. A total of 3.3 × 105 primary yeast transformants were screened for the galactose-dependent activation of LEU2 and lacZ reporter genes, which are regulated by LexA-specific operator sites. Three library plasmids were isolated from selected transformants that reproduced the interacting phenotype upon retransformation with pLexA-NS1, but not with the control plasmid pRFHM1. Here we report on the analysis of the human cDNA isolated through one of these library plasmids, p59-1, which encodes a novel human protein, NS1-BP.
Cloning and analysis of NS1-BP cDNA.
p59-1 had a 1.2-kb cDNA insert containing one long open reading frame of 819 bp followed by 338 bp of an untranslated region that terminated in a run of 20 adenosines (Fig. 1). Northern blot analysis of HeLa cell poly(A) RNA was used to determine if the size of the isolated HeLa cDNA corresponded to a complete copy of NS1-BP mRNA. A 32P-labeled NS1-BP-specific probe hybridized mainly to an RNA species of approximately 3.1 kb (Fig. 2). This result suggested that p59-1 carried an incomplete copy of NS1-BP mRNA. We used a 5′-RACE procedure to generate cDNA derived from the 5′ end of NS1-BP mRNA. The RACE products were subcloned, and six resulting plasmid clones were isolated and sequenced. The longest 5′-RACE clone extended the NS1-BP cDNA to a total of 2,752 bp (Fig. 1). Sequence analysis revealed the presence of one long open reading frame of 1,857 nucleotides that encodes a 619-amino-acid protein with a predicted molecular mass of 69.7 kDa. The initiator ATG codon of the open reading frame is in a sequence context which is compatible with being a translational start site (34). Analysis of the sequence of NS1-BP revealed the presence of five imperfect repeat elements of 47 to 49 amino acids at the C-terminal region between amino acids 368 and 607 (Fig. 3). These tandem repeats are 18 to 41% identical to each other, and five positions are invariant between domains.
FIG. 1.
Nucleotide sequence of NS1-BP cDNA and derived amino acid sequence. The sequence of 2,752 nucleotides was determined by sequencing of two overlapping clones. Nucleotides at positions +1038 to +2215 are derived from the HeLa cDNA insert of the library plasmid p59-1. The 5′ end of the library cDNA is indicated by an arrow. Nucleotides −537 to +1037 were determined by sequencing cloned HeLa cDNA that was generated by 5′-RACE. The open reading frame of 619 amino acids spans positions +1 to +1857. The stop codon is marked by a star.
FIG. 2.
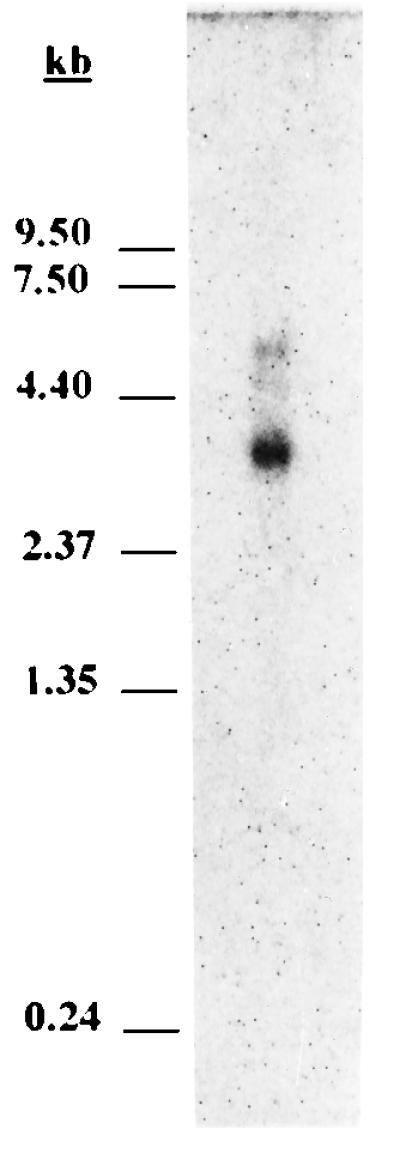
Northern blot analysis of poly(A)-selected HeLa cell RNA with an NS1-BP-specific probe. One microgram of HeLa cell poly(A) RNA was separated by formaldehyde-agarose gel electrophoresis and immobilized on a nylon membrane. A 32P-labeled probe derived from p59-1 was used to detect NS1-BP mRNA by hybridization. RNA size markers are indicated to the left.
FIG. 3.
Alignment of the five tandem repeat elements of NS1-BP. The PILEUP program of the GCG was used to align repetitive sequences between amino acids 368 and 607 of NS1-BP. The PRETTY program of the GCG was used to determine a consensus sequence (Consens.). Capital letters, conserved amino acids; boldface letters, invariant residues.
The cDNA and the deduced amino acid sequence of NS1-BP were compared to sequences in the GenBank and EMBL databases by using the FASTA and TFASTA algorithms (10). Two regions of NS1-BP which had homology to other proteins were identified. First, the N-terminal ∼120 amino acids of NS1-BP are homologous to the BTB (bric-a-brac, tramtrack, broad complex)/POZ (poxviruses and zinc fingers) domain that has been identified in several zinc finger proteins known to act as transcriptional regulators (3, 76). Second, the five tandem repeats located between NS1-BP residues 368 and 607 are homologous to the 50-amino-acid kelch motif that was originally found in the Drosophila Kelch protein (5, 71). The Kelch protein is a component of the intercellular ring canals in the Drosophila egg chamber. Its function is required for the development of fertile oocytes, since mutations in the kelch gene can cause a sterile phenotype in females (71). Interestingly, the Kelch protein also contains a predicted BTB/POZ domain. In total, the amino acid sequence of NS1-BP is 31% identical to that of Kelch.
Several other proteins which have both kelch and BTB/POZ domains were identified. These include the murine ENC-1 protein, which is specifically expressed in the nervous system (29), human and bovine calicin, which are components of the mammalian sperm head (65), the predicted product of the human KIAA0132 gene (42), and the proteins encoded by genes of vaccinia virus (A55R, C2L, and F3L) (22), the Shope fibroma virus (T6, T8, and T9) (62), variola major virus (D16L, C7L, J6R, and B20R) (40), and swinepox virus (C4L and C13L) (39). The functions of the viral gene products are not known. Several cellular kelch repeat proteins containing no BTB/POZ domains were found, including the α- and β-scruin proteins, which are expressed in the sperm of the horseshoe crab Limulus polyphemus (66, 67), and the products of the mouse intracisternal A particle-promoted placenta (MIPP) gene (7) and the spe-26 gene of Caenorhabditis elegans (64).
The NS1 protein binds to NS1-BP in vitro.
To confirm the interaction of NS1 and NS1-BP, in vitro binding assays were performed. NS1-BP cDNA isolated through the library plasmid in the interaction trap screen (corresponding to NS1-BP amino acids 347 to 619) was fused to the GST gene in a bacterial expression vector. GST–NS1-BP fusion protein was expressed in E. coli and adsorbed to glutathione-Sepharose beads. As a control, we prepared glutathione-Sepharose beads complexed with GST protein alone. The NS1 protein was synthesized in vitro and labeled with [35S]methionine through coupled transcription-translation reactions in reticulocyte lysates. The coated glutathione-Sepharose beads were incubated with the radiolabeled NS1 protein. The NS1 protein was efficiently precipitated by the GST–NS1-BP fusion protein but not by GST (Fig. 4, lanes GST and GST–NS1-BP). This result confirms the yeast two-hybrid data and shows that the viral NS1 protein can also physically interact with the cellular NS1-BP.
FIG. 4.
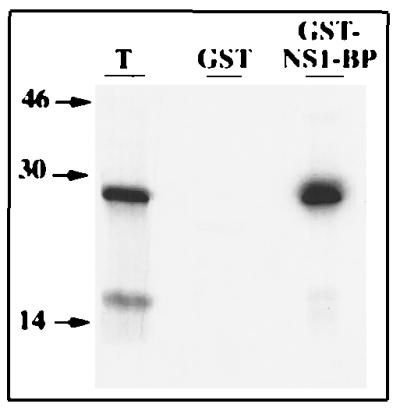
Precipitation of 35S-labeled NS1 protein by GST–NS1-BP fusion protein. Radiolabeled NS1 protein was synthesized in coupled transcription-translation reactions in the presence of [35S]methionine by using pcDNA3-NS1 as a template. The NS1 protein was precipitated by glutathione-Sepharose beads coated with GST (lane GST) or GST–NS1-BP, which carries amino acids 347 to 619 of NS1-BP (lane GST–NS1-BP). The precipitates were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. A 10% aliquot of the total reaction was separated in parallel (lane T). The positions of molecular weight markers (in thousands) are indicated to the left.
NS1-BP is concentrated in intranuclear domains enriched with splicing factors.
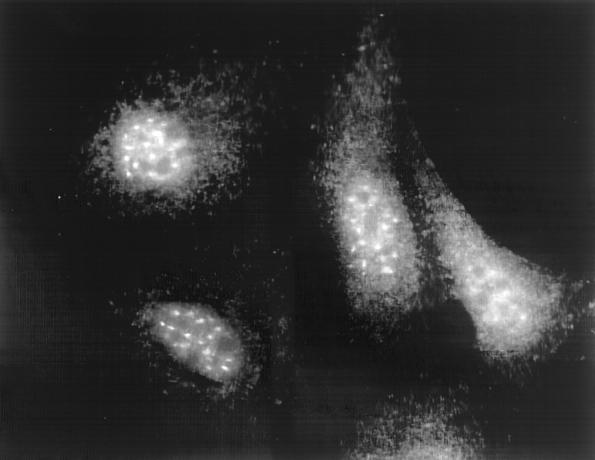
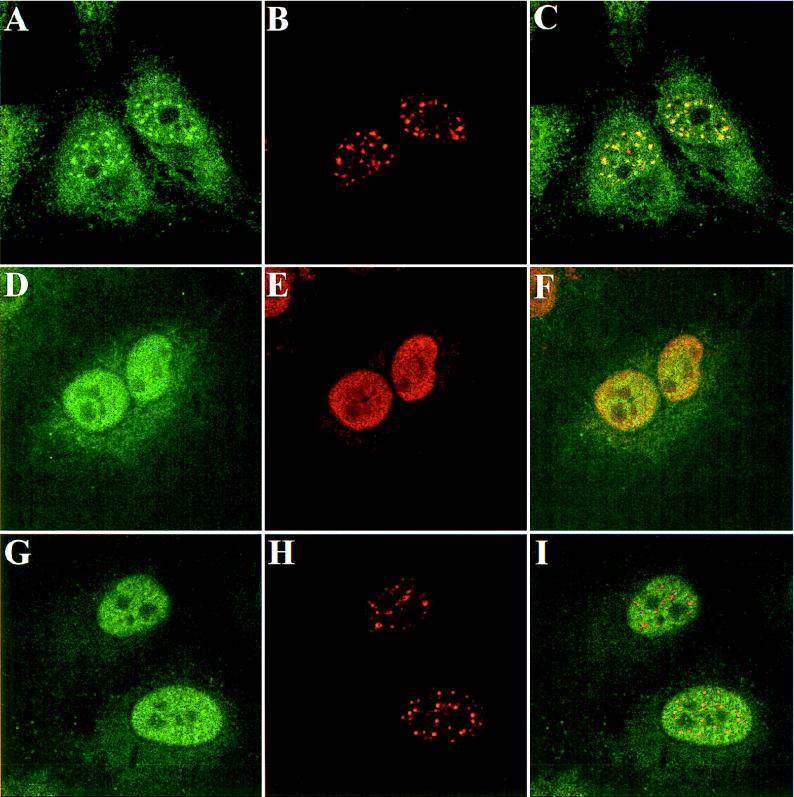
Polyclonal rabbit antibodies were raised against recombinant NS1-BP and used to analyze the concentration and intracellular localization of NS1-BP in mammalian cells. Immunoblot analyses of the human epithelium-derived HEp-2, 293, and HeLa cell lines by NS1-BP-specific antibodies detected a protein doublet band with a molecular mass of about 70 kDa (Fig. 5). This is the predicted size for a protein derived from the NS1-BP open reading frame. Two minor protein bands migrating at 65 and 50 kDa were stained at variable intensities and may correspond to NS1-BP breakdown products. NS1-BP-specific antibodies were affinity purified from immune serum and used for immunofluorescence analysis. In HeLa cells, a punctate nuclear staining pattern that excluded the nucleoli was observed (Fig. 6). In addition, a weak, diffuse staining of the cytoplasm was reproducibly seen. The nuclear staining of NS1-BP was similar in appearance to the “speckled” pattern that was obtained by immunofluorescence staining of cells with antibodies directed against factors involved in pre-mRNA splicing (59). The speckle domains correspond ultrastructurally to interchromatin granules and perichromatin fibrils and are enriched with splicing snRNPs and non-snRNP splicing factors like SC35 and other serine-arginine-rich (SR) proteins (reviewed in reference 58). To determine how NS1-BP localization relates to speckle domains, we double-immunostained HeLa cells with NS1-BP-specific antibodies and a monoclonal antibody raised against the spliceosome assembly factor SC35, which is a known component of speckle domains (21). Confocal laser scanning microscopy demonstrated that the dot-like nuclear NS1-BP signal colocalized with the SC35 signal (Fig. 7A through C). The concentration of multiple proteins involved in pre-mRNA processing in these nuclear regions suggests an important role of this compartment for cellular RNA biogenesis (55). The accumulation of NS1-BP in the same compartment suggests that NS1-BP may be a component of the cellular splicing machinery.
FIG. 5.
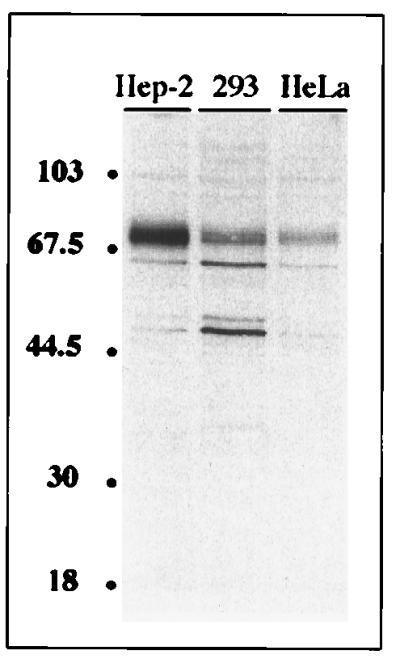
Immunoblot analysis of NS1-BP. Confluent monolayers of HEp-2, 293, and HeLa cells were lysed in radioimmunoprecipitation assay buffer. Soluble proteins from equivalent volumes of extract corresponding to 5 × 104 cells were separated by SDS gel electrophoresis, transferred to a nitrocellulose membrane, and probed with affinity-purified NS1-BP-specific antibodies. The positions of marker proteins (in kilodaltons) are indicated to the left.
FIG. 6.
Intracellular localization of NS1-BP as determined by indirect immunofluorescence analysis of HeLa cells. Subconfluent HeLa cells were fixed and stained with affinity-purified NS1-BP-specific rabbit antibodies, followed by visualization using FITC-conjugated secondary antibodies.
FIG. 7.
Intracellular distribution of cellular NS1-BP, the SC35 protein, and the viral NS1 protein in noninfected or influenza A virus-infected HeLa cells. Confocal micrographs show noninfected (A through C) or influenza A/WSN/33 virus-infected HeLa cells at 10 h postinfection (D through I). The intranuclear localization of NS1-BP was visualized by staining with NS1-BP-specific primary rabbit antibodies and FITC-conjugated secondary antibodies (A, D, and G). The cellular SC35 protein (B and H) and the viral NS1 protein (E) were labeled by monoclonal mouse antibodies and visualized by Texas red-conjugated anti-mouse IgG. Micrographs in the right column (C, F, and I) show confocal overlays of the FITC and Texas red signals from the left and middle columns.
NS1-BP relocalizes to the entire nucleoplasm in influenza A virus-infected cells.
The viral NS1 protein accumulates in the nuclei of cells infected with influenza A virus (24, 72). We examined whether the speckled nuclear localization of cellular NS1-BP would be affected in virus-infected cells expressing the NS1 protein. Influenza A virus-infected cells were double-immunostained with antibodies directed against NS1-BP and either the viral NS1 or the cellular SC35 protein (Fig. 7). As expected, the NS1 protein localized predominantly to the nucleoplasm, with some additional nucleolar signal (Fig. 7E). For the NS1-BP staining, a remarkable change was observed after infection by influenza virus. The cellular NS1-BP was no longer found concentrated in the nuclear speckles but was instead distributed throughout the nucleoplasm. Its distribution pattern was similar to that of the viral NS1 protein except that there was no nucleolar signal (Fig. 7D and F). This redistribution of NS1-BP was observed in a few cells as early as 4 h postinfection. With ongoing infection, most of the cells expressing the viral NS1 protein had an NS1-BP staining pattern similar to that shown in Fig. 7D. The intensity of the nuclear NS1-BP signal appeared to increase slightly in infected cells. However, we did not detect an increase in the amount of NS1-BP in virus-infected cells by immunoblotting (data not shown). This suggests that NS1-BP epitopes are more easily accessible to antibodies in the nuclei of infected cells.
The intranuclear relocalization of NS1-BP in infected cells raised the question of whether the distribution of other proteins which normally localize to speckles would also change. Gross redistribution of proteins might occur if speckles break down during influenza virus infection. However, the staining of virus-infected cells with anti-SC35 antibody at 10 h postinfection (Fig. 7H) showed only a small change from the normal pattern. The average size of the speckles appeared to be slightly decreased, with a concomitant increase in the number of these domains. Essentially the same observation was made in a previous study that examined the distribution of splicing factors in influenza A virus-infected cells (20). These findings suggest that the redistribution of NS1-BP to the nucleoplasm in infected cells is not merely a consequence of speckle disintegration. It is tempting to speculate that cellular NS1-BP is relocalized via the binding to the viral NS1 protein. The intracellular relocalization is likely to have an impact on NS1-BP function in virus-infected cells.
A truncated NS1-BP protein inhibits pre-mRNA splicing at a step after spliceosome assembly.
It has been demonstrated that the NS1 protein can inhibit pre-mRNA splicing in vivo and in vitro (19, 36). The block in splicing was assigned to a step after the assembly of spliceosomes, but before the first catalytic event (36). We hypothesized that the binding of the NS1 protein to a cellular protein(s) whose function is essential for splicing leads to the observed block in splicing. Given the intranuclear colocalization of NS1-BP with well-known factors of the mRNA splicing apparatus, we wondered if NS1-BP could play a role in pre-mRNA splicing, as analyzed by in vitro splicing assays using HeLa cell nuclear extracts. We decided to use a truncated NS1-BP (amino acid positions 347 to 619) in the form of a GST–NS1-BP fusion protein as a potential dominant-negative inhibitor of the endogenous NS1-BP. Such a strategy has been used previously to examine a functional role of a protein in RNA splicing (73).
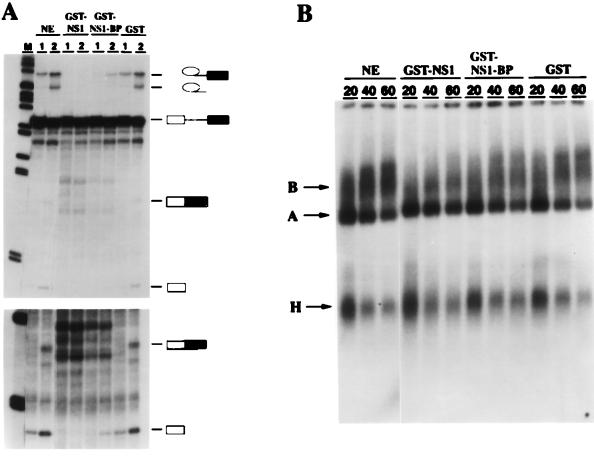
The splicing of a 32P-labeled pre-mRNA in HeLa cell nuclear extracts was analyzed in the presence of GST–NS1-BP, GST-NS1, or control GST protein (Fig. 8). We also examined the formation of heparin-resistent splicing complexes in the same reactions by native gel electrophoresis. In the control reaction, the intron lariat-exon 2 splicing intermediate was easily detected after a 1-h incubation. At the 2-h time point, the accumulation of spliced mRNA and the intron lariat was observed (Fig. 8A, lanes NE). Native gel electrophoresis showed that both A- and B-type spliceosomes were formed normally at 20, 40, and 60 min, with a higher proportion of radiolabeled mRNAs shifting into B-complex bands at later time points (Fig. 8B, lanes NE). The A complex contains U2 snRNP, and the B complex, which represents the fully assembled spliceosome, contains the U2, U4/U6, and U5 snRNPs (32). H represents a heterogenous class of RNA-protein complexes, some of which may be precursors of A- and B-type spliceosomes (Fig. 8B) (32). The addition of GST protein to nuclear extracts did not change the splicing of the pre-mRNA, nor did it interfere with the assembly of spliceosomes (Fig. 8, lanes GST). However, no splicing intermediates or products accumulated in the presence of equimolar amounts of affinity-purified GST-NS1 protein (Fig. 8A, lanes GST-NS1). This effect was not due to a defect in spliceosome assembly, because both A- and B-type complexes assembled, although B bands formed at a slightly reduced rate (Fig. 8B, lanes GST-NS1). Interestingly, an almost identical result was obtained in splicing reactions complemented with the same concentration of purified GST–NS1-BP. There were no splicing products detectable after 1 h of incubation, and only trace amounts of the exon 1 and intron lariat-exon 2 splicing intermediates were detected at the 2-h time point (Fig. 8, lanes GST–NS1-BP). The RNP gel analysis showed that the formation of B-type splicing complexes occurred in the presence of GST–NS1-BP (Fig. 8B, lanes GST–NS1-BP and GST-NS1). This result demonstrates that truncated NS1-BP and the viral NS1 proteins block a cellular activity required at the same step of pre-mRNA splicing. We suggest that the observed splicing inhibition by truncated NS1-BP is the result of a dominant-negative effect on the splicing function of the endogenous wild-type protein.
FIG. 8.
Pre-mRNA splicing, but not spliceosome assembly, is inhibited by truncated NS1-BP. 32P-labeled MINX pre-mRNA, which is an artificial mini-exon construct derived from the major late transcription unit of adenovirus 2 (75), was incubated in HeLa cell nuclear extracts under splicing-compatible conditions in the absence (lanes NE) or presence of 80 ng of affinity-purified GST/μl (lanes GST) or equimolar amounts of affinity-purified GST-NS1 (lanes GST-NS1) or GST–NS1-BP fusion protein, which carries amino acids 347 to 619 of NS1-BP (lanes GST-NS1-BP). (A) RNA analysis. RNAs were purified from aliquots of the reactions after a 1-h (lanes 1) or 2-h (lanes 2) incubation period and were analyzed by electrophoresis on denaturing 13% polyacrylamide-urea gels. The positions of the pre-mRNA, the intron-exon 2 and exon 1 intermediates, and the spliced mRNA and lariat products are indicated to the right. The lower panel shows a longer exposure of the gel. M, 32P-labeled size marker DNA fragments. (B) Splicing complex analysis. Aliquots of the splicing reactions were taken after 20, 40, and 60 min, and heparin was added to a final concentration of 1 mg/ml. The samples were analyzed by electrophoresis on a native agarose-polyacrylamide gel. The positions of the H-, A-, and B-type splicing complexes (32) are indicated on the left.
DISCUSSION
NS1 is the only nonstructural protein of influenza A viruses which is expressed in infected cells. Although the NS1 protein has been implicated in several different processes, including pre-mRNA splicing and mRNA transport and translation, little is known about specific cellular factors that are recognized by NS1. Since NS1 has pleiotropic effects, it may interact with a variety of proteins in infected cells, thereby affecting different steps of cell metabolism. In order to identify such cellular proteins, we have used the yeast interaction trap to screen a human cDNA expression library, using a LexA-NS1 fusion protein as bait. We have previously characterized NS1-I (NS1-interactor), which is a cytoplasmic 55-kDa protein that binds to the divergent NS1 proteins expressed by influenza A and B viruses (69). NS1-I is derived from the precursor protein of an estradiol 17β-dehydrogenase, and its binding to NS1 may have a function in modulating steroid hormone levels in virus-infected cells (69). Here we report on the identification of NS1-BP through its specific interaction with the NS1 protein. This interaction was confirmed by the use of an in vitro binding assay. The NS1 protein coprecipitated with a GST–NS1-BP fusion protein but not with GST alone, demonstrating that NS1 also physically binds to NS1-BP. We did not detect coimmunoprecipitation of radiolabeled NS1 protein from extracts of virus-infected cells with NS1-BP specific antibodies (data not shown). This result may indicate that NS1–NS1-BP complexes are disrupted by the antibody preparation used or that the interaction of the two proteins is labile in the immunoprecipitation buffer.
The analysis of the primary structure of NS1-BP identified two regions with considerable homology to known proteins. The amino-terminal 120 amino acids of NS1-BP are homologous to the BTB/POZ domain originally identified in a group of proteins that primarily regulate transcription (3, 76). This includes the human proto-oncogenes LAZ3/BCL6 and PLZF and the Drosophila Tramtrack, GAGA, and Broad Complex proteins (for a review, see reference 1). It has been shown that the isolated BTB/POZ domains of bric-a-brac (bab), ZID, LAZ3/BCL6, and Kelch can mediate homo- and/or heterodimerization, suggesting that BTB/POZ domains are a conserved protein-protein interaction motif (3, 8, 11, 13, 50). In the bab protein, the first 51 amino acids of the BTB/POZ domain were found to be sufficient for dimerization (8). Another function of this module may be protein targeting to specific nuclear domains, since the appearance of ZID, LAZ3/BCL6, and hZF5 proteins in “nuclear dots” depended on the integrity of their BTB/POZ domains (3, 8, 11, 13, 60). The nature of the nuclear regions in which these BTB/POZ domain-containing proteins accumulate has not been analyzed so far. We are currently examining how the speckled regions in which the NS1-BP is localized relate to the nuclear dots recognized by antibodies directed against other BTB/POZ proteins.
The human cDNA isolated through the interaction trap encoded amino acids 347 to 619 of NS1-BP, which suggests that this region contains the binding site for the NS1 protein. This part of the NS1-BP consists almost entirely of five imperfect repeats of 47 to 49 amino acids that are homologous to the kelch repeat motif (5). Based on phylogenetic sequence comparisons, it was suggested that kelch repeats take on a conserved three-dimensional fold that was previously identified in procaryotic and eucaryotic enzymes (5). A high-resolution X-ray diffraction analysis for one of these enzymes, galactose oxidase of Dactylium dendroides, revealed that each repeat element folds into a blade-like domain of four-stranded antiparallel β sheets. The blade-like domains are circularly arranged, resulting in a β propeller structure (30). The sequence homology suggests that the five repeats of NS1-BP may also adopt a similar three-dimensional fold.
In spite of the conservation on the sequence level, kelch repeats appear to have divergent functions in the homologous proteins. In galactose oxidase of D. dendroides, the kelch repeat fold contains the catalytic center of the enzyme (30). On the other hand, the kelch elements of the β-scruin protein of L. polyphemus have been shown to bind to actin, which leads to the proposal that kelch repeats may constitute an actin-binding domain (52, 67). However, other proteins that contain kelch repeats, such as the α-scruin or the calicin proteins, are localized to intracellular regions that appear to be devoid of actin (65, 66).
The proteins encoded by different poxviruses that are homologous to NS1-BP have not been studied. The genes of the vaccinia virus A55R, C2L, and F3L open reading frame products could be deleted without affecting viral replication in tissue culture and are therefore considered nonessential (33, 45). However, the presence of homologous proteins in different poxviruses argues for important roles of these proteins. For example, these gene products may increase virus virulence or otherwise play a role in infected animals.
By immunolocalization studies, we discovered that NS1-BP is concentrated in discrete regions in the nuclei of noninfected cells. This intracellular distribution is compatible with a function of NS1-BP in gene regulation. Confocal double-immunostaining analyses of cells demonstrated that NS1-BP colocalized in a speckled pattern with the spliceosome assembly factor SC35 (21). Several immunolocalization studies have shown that a number of other factors involved in pre-mRNA splicing, among them the spliceosomal snRNPs, also accumulate in the 20 to 50 irregularly shaped SC35 domains termed speckles (reviewed in reference 58). As shown by electron-microscopic analysis, the speckle domains correspond to interchromatin granules and perichromatin fibrils (17, 59). Different conclusions have been drawn about the functional significance of the accumulation of splicing factors in specific subnuclear compartments. Since speckle domains localize near genes that are transcribed and spliced, it was suggested that speckles constitute a compartment in which pre-mRNA is actively spliced (70). However, nascent RNA polymerase II transcripts were detected by Br-UTP labeling in a random distribution throughout the nucleoplasm (18). Since splicing is thought to occur cotranscriptionally, it was concluded by this group that pre-mRNA is processed throughout the nucleoplasm. For the speckle domains, a role as a storage or recycling compartment that supplies splicing factors to transcription sites was also proposed (reviewed in reference 55). In any case, the important role of speckle domains for cellular RNA biogenesis is emphasized by their dynamic appearance in response to alterations of cellular gene expression. Stress conditions such as heat shock that result in inhibition of RNA splicing also induce apparent changes in the distribution of splicing factors (4, 59). The localization of NS1-BP in nuclear regions that contain high concentrations of pre-mRNA splicing factors suggests a role for NS1-BP in mRNA splicing.
The intranuclear localization of NS1-BP was drastically altered in influenza A virus-infected cells that expressed the NS1 protein. The speckled pattern was replaced by a rather homogeneous distribution of NS1-BP throughout the nucleoplasm in a fashion similar to that observed for the viral NS1 protein. In contrast, only subtle changes were detected in the appearance of the SC35 protein in influenza A virus-infected cells. This finding argues that the relocalization of NS1-BP is a specific effect and not the result of a disintegration of the SC35-enriched domains. This notion is further supported by our observations that stress conditions such as heat shock, serum starvation, or the addition of actinomycin D, which inhibits RNA polymerase II transcription, did not disrupt the colocalization of NS1-BP with SC35 (67a). It is conceivable that local changes in NS1-BP concentration in response to an influenza A virus infection also influence its function. We did not detect NS1-BP redistribution in cells that transiently expressed the NS1 protein from an RNA polymerase II-driven plasmid vector (data not shown). This result may indicate that NS1-BP redistribution requires intracellular concentrations of the NS1 protein that are not achievable by the expression system used. Alternatively, another viral gene product or spliced viral mRNA may be needed for the effect to be observed.
The NS1 protein has previously been shown to inhibit pre-mRNA splicing in vitro and in vivo (19, 36). It was speculated that the inhibition of splicing would result in the retention of pre-mRNA in the nuclei of infected cells, thereby increasing the concentration of mRNA cap structures available for cap snatching by the viral RNA polymerase (36). Alternatively, the activity of the NS1 protein may contribute to the observed regulated splicing of the viral mRNAs derived from segments 7 and 8 (56, 63). Because cellular NS1-BP is concentrated in nuclear regions enriched with pre-mRNA splicing factors and because it relocalizes in virus-infected cells, we examined whether NS1-BP has a role in pre-mRNA splicing in vitro. A truncated NS1-BP was used as a potential nonfunctional competitor of the endogenous protein in HeLa cell nuclear extract, and the effects of this probe were compared to the known inhibition of pre-mRNA splicing by the NS1 protein. A similar experimental design has been used before by others to examine the role of the large subunit of RNA polymerase II in pre-mRNA splicing (14, 73). In the present study, we demonstrated that the truncated NS1-BP blocks the splicing of a 32P-labeled pre-mRNA in HeLa cell nuclear extracts at the same step as the NS1 protein does. Splicing complexes formed at only slightly reduced rates in the presence of each of the two proteins. However, the conversion of the pre-mRNA into splicing intermediates or products was greatly reduced. This finding suggests that both proteins act on the same stage of the spliceosome pathway, i.e., they block an activity required for the first catalytic step. The shortened NS1-BP we used lacks the 346 N-terminal amino acids of the wild-type protein and was fused to the 26-kDa GST protein. This mutant NS1-BP protein is therefore unlikely to retain the full activity of the wild-type protein. However, the truncated NS1-BP may still be able to interact with other essential splicing factors, thereby preventing their association with the wild-type NS1-BP. The bacterially expressed full-length NS1-BP was insoluble (data not shown). Therefore, we were unable to assess whether the full-length NS1-BP had an effect on pre-mRNA splicing similar to that of the truncated protein. Nevertheless, our results are compatible with a role of the wild-type NS1-BP in pre-mRNA splicing.
How does the observed inhibition of splicing by the viral NS1 protein relate to these findings? In normal cells, NS1-BP is concentrated in intranuclear domains that are enriched with multiple splicing factors. We have demonstrated that cellular NS1-BP is specifically relocalized in influenza A virus-infected cells that express the NS1 protein. Redistribution of NS1-BP is likely to alter its function or activity. We propose a model in which the influenza A virus inhibits host cell splicing in infected cells by the association of the viral NS1 protein with cellular NS1-BP. The NS1 protein may downregulate NS1-BP activity directly by blocking its normal association with spliceosomes. Alternatively, a mechanism can be envisioned in which the viral NS1 protein removes cellular NS1-BP from centers of active splicing, thereby lowering its availability for participation in cellular mRNA splicing processes. In both models, the relocalization of NS1-BP may reflect its disrupted function. It has previously been suggested that the NS1 protein inhibits splicing by binding to U6 snRNA (36, 49), which is a key component of the catalytic core within the spliceosome (25, 53). Our proposal to explain the inhibition of splicing by the NS1 protein through the binding to NS1-BP does not exclude an NS1-U6 interaction. It is estimated that at least 80 to 100 different factors are involved in pre-mRNA splicing (23, 53). For that reason it has been difficult to dissect their dynamic and complex interactions during the assembly of a spliceosome and the catalysis of the splicing reaction. It is possible that interactions of the NS1 protein with both NS1-BP and U6 snRNA contribute to the inhibition of pre-mRNA splicing.
Like the influenza A virus NS1 protein, the essential ICP27 protein of herpes simplex virus type 1 (HSV-1) has been implicated in impairing cellular pre-mRNA splicing, possibly as part of a host cell shutoff mechanism (27). One could therefore ask if there are parallels between lytic infections by influenza A virus and HSV-1. As has been shown for the NS1 protein, the ICP27 protein has been found to have pleiotropic regulatory effects. Roles for ICP27 in mRNA 3′-end processing (6, 41) and in mRNA export (47, 57) have been suggested in addition to an inhibitory effect on pre-mRNA splicing. Interestingly, the expression of ICP27 induces the redistribution of SC35 and spliceosomal snRNPs from the known speckled pattern to a few condensed intranuclear structures, in which they colocalize with the ICP27 protein (46, 51). This pattern is basically the opposite of the situation observed in influenza A virus-infected cells. In the present study, the number of SC35 domains appears to increase, with a concomitant decrease in size (reference 20 and this study), and in contrast to ICP27, the NS1 protein is localized throughout the nucleus in a diffuse pattern.
We will need further analyses to explore the precise function of NS1-BP in pre-mRNA splicing and the role of the NS1–NS1-BP interaction in virus-infected cells. For example, we will examine if other splicing factors that associate with NS1-BP during the course of splicing can be identified. Influenza A viruses with temperature sensitivity (ts) mutations in the NS1 gene have been isolated (summarized in reference 38). We want to examine if any of the identified ts lesions in NS1 have an effect on the binding to NS1-BP. These experiments will reveal if it is feasible to correlate a weakened interaction between NS1 and NS1-BP with a ts phenotype and if this is paralleled by a lack of NS1-BP relocalization at the nonpermissive temperature. Since the NS1 gene of the influenza A virus is now amenable to genetic manipulation (16), it should also be possible to mutate the NS1-BP binding site within the NS1 protein and to study the consequences for virus infection. These studies will expand our knowledge of how viruses interact with cellular components and will also help us to learn more about the cellular splicing apparatus.
ACKNOWLEDGMENTS
We thank R. Brent (Massachusetts General Hospital) for providing yeast strains and two-hybrid vectors and J. Gyuris for the acidic domain-HeLa cell cDNA library. T.W. thanks H.-D. Klenk (Institute of Virology, Philipps-University Marburg) for his generous support and help in the immunization of rabbits.
This work was supported by a fellowship of the AIDS-Stipendienprogramm of the German Ministry for Education, Science, Research, and Technology (T.W.) and by a grant from the Deutsche Forschungsgemeinschaft (SFB286).
REFERENCES
- 1.Albagli O, Dhordain P, Deweindt C, Lecoq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R E, Kingston D D, Moore J G, Seidman J A, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 3.Bardwell V J, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 4.Bond U. Heat shock but not other stress inducers leads to the disruption of a subset of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Doolittle R F. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 6.Brown C R, Nakamura M S, Mosca J D, Hayward G S, Straus S E, Perera L P. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J Virol. 1995;69:7187–7195. doi: 10.1128/jvi.69.11.7187-7195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang-Yeh A, Mold D E, Huang R C C. Identification of a novel murine IAP-promoted placenta-expressed gene. Nucleic Acids Res. 1991;19:3667–3672. doi: 10.1093/nar/19.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Zollman S, Couderc J-L, Laski F A. The BTB domain of bric à brac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.De La Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deveraux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhordain P, Albagli O, Ansieau S, Koken M H M, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J-P, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 12.Dignam D L, Lebovitz R M, Roeder R D. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong H-J, Wang Z-Y, Licht J, Waxman S, Chomienne C, Chen Z, Zelent A, Chen S-J. Amino-terminal protein-protein interaction motif (POZ domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-α fusion protein. Proc Natl Acad Sci USA. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du L, Warren S L. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enami M, Palese P. High-efficiency formation of influenza virus transfectants. J Virol. 1991;65:2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 18.Fay F S, Taneja K L, Shenoy S, Lifshitz L, Singer R H. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- 19.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortes P, Lamond A I, Ortin J. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J Gen Virol. 1995;76:1001–1007. doi: 10.1099/0022-1317-76-4-1001. [DOI] [PubMed] [Google Scholar]
- 21.Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 22.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 23.Green M R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 24.Greenspan D, Palese P, Krystal M. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 26.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 27.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 29.Hernandez M-C, Andres-Barquin P J, Martinez S, Bulfone A, Rubenstein J L R, Israel M A. ENC-1: a novel mammalian Kelch-related gene specifically expressed in the nervous system encodes an actin-binding protein. J Neurosci. 1997;17:3038–3051. doi: 10.1523/JNEUROSCI.17-09-03038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito N, Phillips S E V, Stevens C, Ogel Z B, McPherson M J, Keen J N, Yadav K D S, Knowles P F. Novel thioether bond revealed by a 1.7 Å crystal structure of galactose oxidase. Nature. 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- 31.Knipe D M. Virus-host cell interactions. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Publishers; 1996. pp. 273–299. [Google Scholar]
- 32.Konarska M M, Sharp P A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 33.Kotwal G, Moss B. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology. 1988;167:524–537. [PubMed] [Google Scholar]
- 34.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarowitz S G, Compans R W, Choppin P W. Influenza virus structural and nonstructural proteins in infected cells and their plasma membranes. Virology. 1971;46:830–843. doi: 10.1016/0042-6822(71)90084-5. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Qian X-Y, Krug R M. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig S, Vogel U, Scholtissek C. Amino acid replacements leading to temperature-sensitive defects of the NS1 protein of influenza A virus. Arch Virol. 1995;140:945–950. doi: 10.1007/BF01314970. [DOI] [PubMed] [Google Scholar]
- 39.Massung R F, Jayarama V, Moyer R W. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 40.Massung R F, Liu L, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 41.McLauchlan J, Phelan A, Loney C, Sandri-Goldin R M, Clements J B. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 43.Nelson K K, Green M R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988;2:319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- 44.O’Neill R E, Palese P. NPI-1, the human homologue of SRP1, interacts with influenza virus nucleoprotein. Virology. 1994;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 45.Perkus M E, Goebel S J, Davis S W, Johnson G P, Norton E K, Paoletti E. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology. 1991;180:406–410. doi: 10.1016/0042-6822(91)90047-f. [DOI] [PubMed] [Google Scholar]
- 46.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A I, Clements J B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phelan A, Clements J B. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 48.Qiu Y, Krug R M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu Y, Nemeroff M, Krug R M. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson D N, Cooley L. Drosophila kelch is an oligomeric ring canal organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid M F, Agris J M, Jakana J, Matsudeira P, Chiu W. Three-dimensional structure of a single filament in the Limulus acrosomal bundle: scruin binds to homologous helix-loop-beta motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp P A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu K, Mullinix M G, Chanock R M, Murphy B R. Temperature-sensitive mutants of influenza A/Udorn/72 (H3N2) virus. III. Genetic analysis of temperature-dependent host range mutants. Virology. 1983;124:35–44. doi: 10.1016/0042-6822(83)90288-x. [DOI] [PubMed] [Google Scholar]
- 55.Singer R H, Green M R. Compartmentalization of eucaryotic gene expression: causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 56.Smith D B, Inglis S C. Regulated production of an influenza virus spliced mRNA mediated by virus-specific products. EMBO J. 1985;4:2313–2319. doi: 10.1002/j.1460-2075.1985.tb03932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and the cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spector D L. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 59.Spector D L, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugiura K, Muro Y, Nagai Y, Kamimoto T, Wakabayashi T, Ohasi M, Hagiwara M. Expression cloning and intracellular localization of a human ZF5 homologue. Biochim Biophys Acta. 1997;1352:23–26. doi: 10.1016/s0167-4781(97)00045-6. [DOI] [PubMed] [Google Scholar]
- 61.Treanor J J, Snyder M H, London W T, Murphy B R. The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology. 1989;171:1–9. doi: 10.1016/0042-6822(89)90504-7. [DOI] [PubMed] [Google Scholar]
- 62.Upton C, Macen J L, Wishart D S, McFadden G. Myxoma virus and malignant rabbit fibroma virus encode a serpin-like protein important for virus virulence. Virology. 1990;179:618–631. doi: 10.1016/0042-6822(90)90129-f. [DOI] [PubMed] [Google Scholar]
- 63.Valcarel J, Portela A, Ortin J. Regulated M1 mRNA splicing in influenza virus-infected cells. J Gen Virol. 1991;72:1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]
- 64.Varkey J P, Muhlrad P J, Minniti A N, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- 65.von Bülow M, Heid H, Hess H, Franke W W. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- 66.Way M, Sanders M, Chafel M, Tu Y-H, Knight A, Matsudaira P. β-Scruin, a homologue of actin crosslinking protein scruin, is localized to the acrosomal vesicle of Limulus sperm. J Cell Sci. 1995;108:3155–3162. doi: 10.1242/jcs.108.10.3155. [DOI] [PubMed] [Google Scholar]
- 67.Way M, Sanders M, Garcia C, Sakai J, Matsudaira P. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J Cell Biol. 1995;128:51–60. doi: 10.1083/jcb.128.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67a.Wolff, T. Unpublished data.
- 68.Wolff T, Bindereif A. Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J. 1992;11:345–359. doi: 10.1002/j.1460-2075.1992.tb05057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolff T, O’Neill R E, Palese P. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. J Virol. 1996;70:5363–5372. doi: 10.1128/jvi.70.8.5363-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xing Y, Johnson C V, Moen P T, McNeil J A. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;6:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 72.Young J F, Desselberger U, Palese P, Ferguson B, Shatzman A R, Rosenberg M. Efficient expression of influenza virus NS1 nonstructural proteins in Escherichia coli. Proc Natl Acad Sci USA. 1983;80:6105. doi: 10.1073/pnas.80.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich protein. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zervos A S, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 75.Zillmann M, Zapp M L, Berget S M. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol. 1988;8:814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zollmann S, Godt D, Prive G G, Couderc J-L, Laski F. The BTB domain, found primarily in zinc finger proteins, defines an evolutionary conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]