Abstract
Objective:
The aim of this study was to develop a deep neural network (DNN)-based parallel imaging reconstruction for highly accelerated 2D turbo spin echo (TSE) data in prostate MRI without quality degradation compared to conventional scans.
Methods:
155 participant data were acquired for training and testing. Two DNN models were generated according to the number of acquisitions (NAQ) of input images: DNN-N1 for NAQ = 1 and DNN-N2 for NAQ = 2. In the test data, DNN and TSE images were compared by quantitative error metrics. The visual appropriateness of DNN reconstructions on accelerated scans (DNN-N1 and DNN-N2) and conventional scans (TSE-Conv) was assessed for nine parameters by two radiologists. The lesion detection was evaluated at DNNs and TES-Conv by prostate imaging-reporting and data system.
Results:
The scan time was reduced by 71% at NAQ = 1, and 42% at NAQ = 2. Quantitative evaluation demonstrated the better error metrics of DNN images (29–43% lower NRMSE, 4–13% higher structure similarity index, and 2.8–4.8 dB higher peak signal-to-noise ratio; p < 0.001) than TSE images. In the assessment of the visual appropriateness, both radiologists evaluated that DNN-N2 showed better or comparable performance in all parameters compared to TSE-Conv. In the lesion detection, DNN images showed almost perfect agreement (κ > 0.9) scores with TSE-Conv.
Conclusions:
DNN-based reconstruction in highly accelerated prostate TSE imaging showed comparable quality to conventional TSE.
Advances in knowledge:
Our framework reduces the scan time by 42% of conventional prostate TSE imaging without sequence modification, revealing great potential for clinical application.
Introduction
Prostate cancer is one of the most common diseases and requires screening at an early stage. 1 Conventional approaches to prostate cancer screening have involved prostate-specific antigen (PSA) testing, but PSA-based screening remains controversial due to high overdiagnosis rates. 2,3 Recently, studies that utilized MRI for prostate cancer detection before biopsy have been increasingly performed. 4–9 In particular, multiparametric MRI (mp-MRI) has been regarded as a standard clinical protocol for prostate MR imaging, which includes 2D T 2 weighted turbo spin echo (TSE) sequences, diffusion-weighted imaging, and dynamic contrast-enhanced 3D T 1 weighted imaging. 4,7,10 However, mp-MRI requires scanning for several imaging sequences, leading to long scan times. Additionally, prostate MR imaging requires high-resolution data to detect the small size of lesions, which causes extension of scan times. 8,9 Such long scan times for mp-MRI lead to sensitivity to motion and patient discomfort.
As an effort to reduce the scan time of MR acquisition, a few acceleration techniques have been proposed. 11–16 The most common approach for MR acceleration is parallel imaging reconstruction. The approach accelerates the scan times by undersampling k-space along the phase encoding direction and reconstructs the images by utilizing the coil sensitivity profiles (referred to as sensitivity encoding; SENSE) 13 or assuming the linear property of k-space data (referred to as generalized autocalibrating partially parallel acquisitions; GRAPPA). 14 However, those methods showed performance degradation in highly accelerated data, revealing the limits of scan time reduction. 17,18 Unlike conventional parallel imaging reconstruction, a few methods have changed the sampling scheme of MR acquisition to reduce aliasing artifacts such as controlled aliasing in volumetric parallel imaging (CAIPIRINHA) 11 and compressed sensing (CS). 16 Although those methods show improved performance over conventional parallel imaging methods, they require sequence modification or a huge computational cost and have limited performance on highly accelerated MR data. 19,20
To improve the reconstruction performance of parallel imaging, recent studies have proposed deep neural network (DNN)-based approaches that leverage image features from large amounts of data. 21–27 Studies have shown that DNNs outperform conventional approaches in highly accelerated scan data, demonstrating the potential usage of DNNs in parallel imaging reconstruction. This is because the image features from large amounts of data may have better representations for reconstruction than conventional mathematical priors. 28 Therefore, DNN approaches can be a promising tool for reducing substantial scan times of MR acquisition without image degradation.
In this study, we developed a DNN-based parallel imaging reconstruction for highly accelerated 2D TSE data in prostate MR imaging. We demonstrate the performance of our DNN model for both quantitative evaluation in retrospectively undersampled data and qualitative evaluation in prospectively accelerated data.
Methods and materials
Participants
This study was approved by the institutional review board of The Catholic University of Korea, Eunpyeong St. Mary’s Hospital and informed consent was waived. We collected 2D T2 weighted prostate MR data from 155 patients (mean age = 71 years; range = 38–92 years) from January 2020 to February 2021. The patients were suspected to have prostate disease, and prostate MRI was performed as prebiopsy MRI. Demographic information was collected through a review of the medical records.
Data acquisition
A total of 155 subjects were scanned using a 3 T scanner (MAGNETOM Vida, Siemens Healthineers, Erlangen, Germany) with a 30-channel body coil and a 32- or 72-channel spine coil. The scanned data were stored in raw k-space data format by the Yarra framework (http://yarraframework.com/), which is public software for transfer of the MRI raw k-space data to external storage. For the training and evaluation of the DNN model, conventional reference scans only (from 115 subjects) or both reference scans and accelerated scans (from 40 subjects) were acquired. The reference scan and accelerated scan were acquired by a 2D TSE sequence in the axial plane with different acceleration factors (R = 2 for the reference scan; R = 4 for the accelerated scan). Other parameters for both scans of the 2D TSE sequence were as follows: TR ranging from 2510 to 2890 ms, TE = 103 ms, FOV = 180 x 180 mm2, matrix size = 640 x 640 using zero-filling interpolation (ZIP), slice thickness = 3 mm with a total of 26 to 30 slices, autocalibrating signal (ACS) lines = 32, and the number of acquisitions (NAQ) = 2. Note that the conventional scan time was 196 s while the accelerated scan times were 57 s for NAQ = 1 and 114 s for NAQ = 2.
Data preprocessing
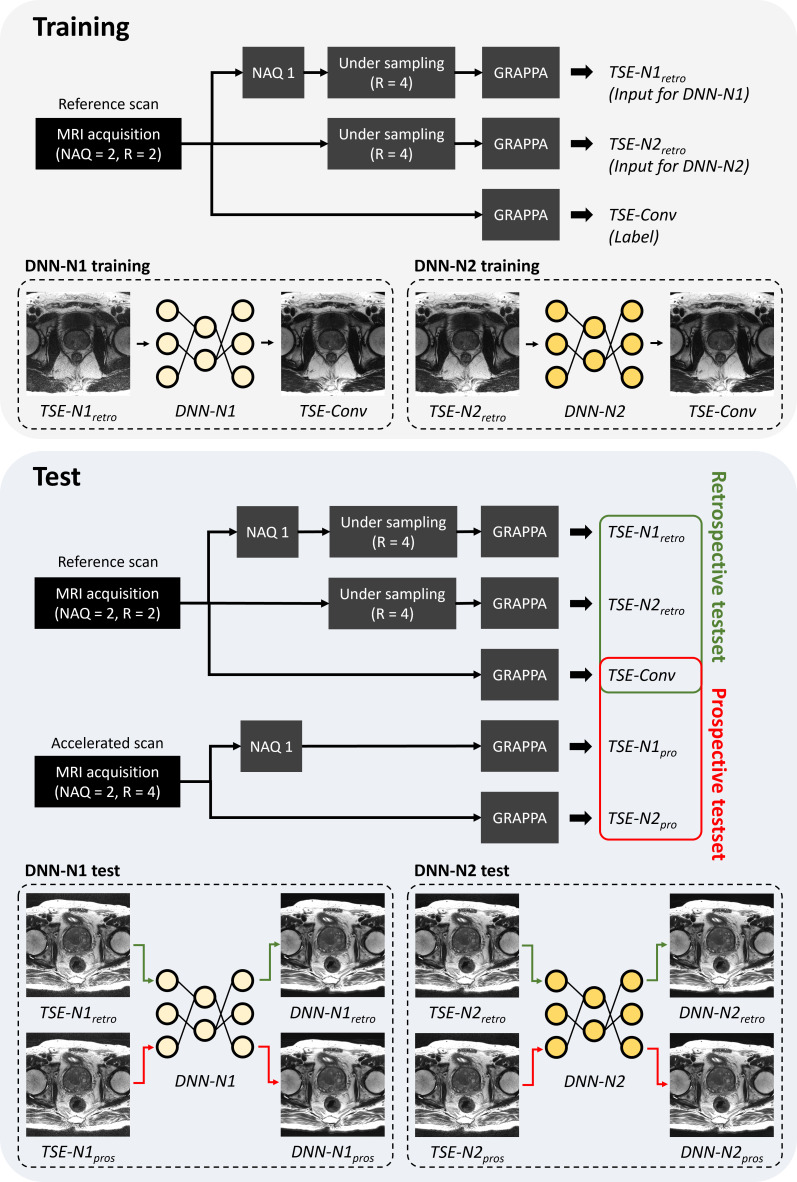
For parallel imaging reconstruction on a high acceleration factor (R = 4), a DNN framework was proposed. Figure 1 summarizes all procedures for training and testing of the DNN. To evaluate the feasibility of DNN-based parallel imaging reconstruction, two different DNN models, referred to as DNN-N1 and DNN-N2, were generated with different NAQs (NAQ = 1 and 2). In the NAQ = 1 experiment, the raw k-space data of the reference scan were sampled with NAQ = 1. Then, retrospective undersampling was performed with R = 4, followed by GRAPPA reconstruction and root-sum-of squares (RSS) channel combining. The resulting coil-combined magnitude images were named TSE-N1retro and used for training inputs of DNN-N1. In the NAQ = 2 experiment, retrospective undersampling (R = 4) was performed in two separate k-spaces. In GRAPPA reconstruction of NAQ = 2, the two separate k-spaces were averaged followed by RSS channel combining, named TSE-N2retro. For the labels of both DNN-N1 and DNN-N2, conventional TSE (TSE-Conv) images were generated by GRAPPA reconstruction on the reference scan followed by RSS channel combining. All details of the data preprocessing are described in Supplementary Material 1 (Supplementary Figure S1).
Figure 1.
Schematic diagram of the experimental setup. DNN, deep neural network; NAQ, number of acquisitions; TSE, turbo spin echo.
To evaluate DNN-N1 and DNN-N2, data for 40 subjects were acquired by both reference scans and accelerated scans and utilized. The retrospective test data were generated using the same data preprocessing of TSE-N1retro and TSE-N2retro, while the prospective test data were generated without retrospective undersampling on the accelerated scan (TSE-N1pros and TSE-N2pros). TSE-Conv images from the reference scan were utilized as a reference for both retro- and prospective test sets.
Deep neural network
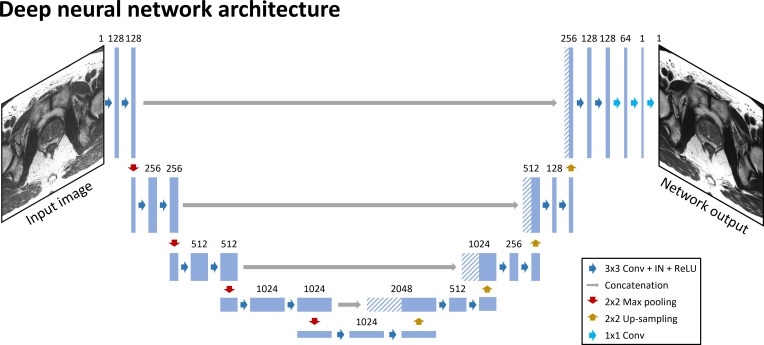
The proposed deep learning framework utilized 2D U-net 29 as a foundational architecture of DNN (Figure 2). Note that U-net has been commonly utilized for DNN-based parallel imaging reconstruction. 30 The DNN architecture is designed with 18 convolutional blocks, 4 max-pooling layers (pool size = 2 x 2), 4 upsampling layers (kernel size = 2 x 2), 4 feature concatenations, and 3 convolutional layers (kernel size = 1 x 1). The convolutional block consists of a convolutional layer with kernel size = 3 x 3, instance normalization (IN), and rectified linear unit (ReLU) activation function. In the first four parts of the network, each part consists of two convolutional blocks followed by a max-pooling layer. The number of channels was 128 in the first part, and it doubled for each subsequent part. The middle part of the network consists of two convolutional blocks while maintaining the number of channels. Then, the feature concatenation was applied to an upsampling layer followed by two convolutional blocks in the next four parts. Finally, three 1 × 1 convolutional layers were applied to reconstruct the output images.
Figure 2.
Deep neural network architecture for prostate imaging in accelerated scans. IN, instance normalization; ReLU, rectified linear unit.
In the DNN training process, a total of 2856 slices and 260 slices were used for the training and validation, respectively. The loss function of DNN was defined as the structure similarity index (SSIM). The learning rate was exponentially decayed from 10−3 at a step size of 20 and decaying ratio = 0.1. Other hyperparameters for training were set as follows: batch size = 4, training epochs = 30, and Adam for optimization. 31 PyTorch 32 and four NVIDIA Titan Xp GPUs with 12 GB memory (NVIDIA Corporation, Santa Clara, CA) were utilized for DNN training. DNN processing time for the test was measured at about 0.3 s for a single slice.
Image quality evaluation
To evaluate the DNN model, quantitative and qualitative analyses were performed. In the quantitative evaluation, four different types of images, DNN-N1retro, TSE-N1retro, DNN-N2retro, and TSE-N2retro, were reconstructed in the retrospective test set. Quantitative error metrics such as normalized root mean squared error (NRMSE), SSIM, and peak signal-to-noise ratio (PSNR) were calculated with TSE-Conv images as a reference.
For qualitative evaluation, prospectively accelerated scan data were utilized to generate DNN-N1pros, TSE-N1pros, DNN-N2pros, and TSE-N2pros images. Note that TSE-Conv images from the reference scan data (NAQ = 2 and R = 2) were utilized as references. To investigate the effects of NAQ on image quality, the prospective test set was divided into two groups: (DNN-N1pros, TSE-N1pros, TSE-Conv) and (DNN-N2pros, TSE-N2pros, TSE-Conv). In each group, two experienced radiologists with more than 9 years of experience in prostate MRI blindly assessed image quality for the randomly presented image set of each patient. They reviewed the data set in the first group and then reviewed the data set in the second group. Eight image quality parameters, including overall image quality, perceived signal-to-noise ratio (SNR), artifacts, and sharpness of prostate, seminal vesicle, rectal wall, urinary bladder, and obturator muscle, were scored by a 5-point Likert scale. The scoring criteria of each image quality parameter were defined as shown in Table 1. Note that higher scores indicate better image quality in all parameters.
Table 1.
Scoring criteria for qualitative evaluation
| Score | Overall image quality | Perceived SNR | Artifacts | Image sharpness |
|---|---|---|---|---|
| 1 | Non-diagnostic | Poor | Severe | Very severe blurring, nondiagnostic |
| 2 | Poor, but still interpretable | Fair | Moderate | Severe blurring, affecting diagnosis |
| 3 | Fair | Average | Mild | Moderate blurring |
| 4 | Good | Good | Minimal | Minimal blurring |
| 5 | Excellent | Excellent | None | Sharp boundary |
SNR, signal-to-noise ratio.
To demonstrate the impact of DNN reconstruction on the images in patients with lesions, the presence of clinically significant cancer was evaluated in TSE-Conv, DNN-N1pros, and DNN-N2pros images, based on prostate imaging-reporting and data system (PI-RADS) v. 2.1 for T 2 weighted imaging (T 2WI) 33 by the two radiologists (Table 2).
Table 2.
Presence of clinically significant cancer scored by PI-RADS v. 2.1 in T 2 weighted imaging
| Score | PI-RADS v 2.1 |
|---|---|
| 1 | Very low (clinically significant cancer is highly unlikely to be present) |
| 2 | Low (clinically significant cancer is unlikely to be present) |
| 3 | Intermediate (the presence of clinically significant cancer is equivocal) |
| 4 | High (clinically significant cancer is likely to be present) |
| 5 | Very high (clinically significant cancer is highly likely to be present) |
PI-RADS, prostate imaging-reporting and data system.
Statistical analysis
To evaluate the performance of DNN, statistical analyses were performed in both quantitative and qualitative evaluations. In the quantitative evaluation, a paired t-test was performed between the quantitative error metrics of DNN-N1retro and TSE-N1retro (or DNN-N2retro and TSE-N2retro) to demonstrate significant differences. In the qualitative evaluation, image quality parameters of DNN-N1pros (or N2pros), TSE-N1pros (or N2pros), and TSE-Conv were compared by a Wilcoxon signed-rank test. For multiple statistical analyses, Bonferroni correction was applied. The inter- and intraobserver agreement were demonstrated by intraclass correlation coefficients (ICCs) defined as poor (0.00–0.20), fair (0.21–0.40), moderate (0.41–0.60), substantial (0.61–0.80), and almost perfect (0.81–1.00). Intraobserver agreement was performed by one radiologist with a 3-month period between the first and second evaluations. In the lesion evaluation by T 2WI PI-RADS, intermethod and interobserver agreements were estimated by weighted Cohen’s κ (κ) statistics. All statistical analyses were performed by SPSS, v. 28.0 (IBM Corporation, Armonk, NY), and statistically significant differences were determined based on a threshold of 0.05.
Results
Demographic characteristics
From the data set, the images from 115 subjects were used for deep neural network training and validation (mean age±standard deviation = 71.4±8.8 years), and those from 40 subjects were used for quantitative and qualitative evaluation (mean age±standard deviation = 70.3±9.7 years).
Quantitative evaluation
Table 3 reports the quantitative error metrics of DNN-N1retro, TSE-N1retro, DNN-N2retro, and TSE-N2retro. The mean and standard deviation of NRMSE, SSIM, and PSNR across the subjects were compared. Note that the comparison was performed independently according to the NAQ. Compared to TSE, DNN showed lower NRMSE, higher SSIM, and higher PSNR with significant differences in both NAQ = 1 and NAQ = 2 (all, p < 0.001), revealing improved performance in DNN-based reconstruction.
Table 3.
Comparison of quantitative error metrics in the retrospective test set between DNN and TSE
| DNN-N1retro | TSE-N1retro | p-value | DNN-N2retro | TSE-N2retro | p-value | |
|---|---|---|---|---|---|---|
| (DNN-N1retro
vs TSE-N1retro) |
(DNN-N2retro
vs TSE-N2retro) |
|||||
| NRMSE (%) | 7.7 ± 1.0 | 13.4 ± 2.5 | <0.001 | 5.7 ± 0.9 | 8.0 ± 1.9 | <0.001 |
| SSIM (0–1) | 0.92 ± 0.03 | 0.81 ± 0.06 | <0.001 | 0.95 ± 0.02 | 0.91 ± 0.04 | <0.001 |
| PSNR (dB) | 34.9 ± 1.5 | 30.1 ± 1.9 | <0.001 | 37.4 ± 1.7 | 34.6 ± 2.2 | <0.001 |
N represents the number of acquisitions; DNN, a deep neural network; NRMSE, normalized root mean squared error; PSNR, peak signal-to-noise-ratio; SSIM, structure similarity index; TSE, turbo-spin echo images;retro, the retrospective undersampling was applied to the data.
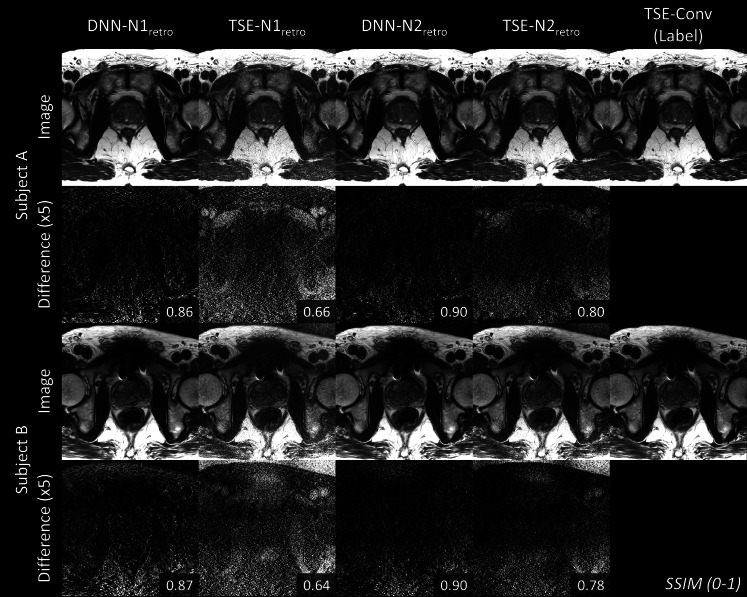
Figure 3 displays DNN-N1retro, TSE-N1retro, DNN-N2retro, TSE-N2retro, and TSE-Conv images from the two representative subjects. In the second and fourth rows, the error maps were calculated with the TSE-Conv images as references. SSIM error metrics are shown in the right corner of the error maps. The highest SSIM values are shown in DNN-N2retro, confirming the best performance of DNN-N2 reconstruction among the methods.
Figure 3.
Reconstructed images of two representative subjects in the retrospective test data set are displayed. DNN-N1retro, TSE-N1retro, DNN-N2retro and TSE-N2retro images are shown with TSE-Conv images as references (last column). The second and fourth rows display the difference map with the label. For visualization, the difference maps were multiplied by 5. SSIM is reported in the right corner of each difference map. DNN shows higher SSIM than TSE on the retrospective undersampled data. DNN, deep neural network; SSIM, structure similarity index; TSE, turbo spin echo.
Qualitative evaluation
Tables 4 and 5 show the results of qualitative evaluation of the two radiologists in NAQ = 1 and NAQ = 2 prospective test sets. In both NAQ = 1 and NAQ = 2 data evaluations, the images from conventional scans (NAQ = 2 and R = 2) were used as the gold-standard. In NAQ = 1 results, two radiologists evaluated that DNN-N1pros showed improved overall image quality, perceived SNR, and sharpness of the structures compared to TSE-N1pros with statistically significant differences (all, p < 0.001; Table 4). As assessed by radiologist 1, the artifact score of DNN-N1pros was significantly higher than that of TSE-N1pros. In the comparison between DNN-N1pros and TSE-Conv, DNN-N1pros showed an improved perceived SNR according to both radiologists (p < 0.003). Although the overall image quality of DNN-N1pros showed incompatible results between radiologists 1 (significantly higher than TSE-Conv) and 2 (significantly lower than TSE-Conv), the median score of overall image quality in both image sets was 4 or higher.
Table 4.
Image quality comparison in the prospective test set with number of acquisitions = 1 (Median [Q1–Q3])
| Accelerated Scan | p-value | |||||
|---|---|---|---|---|---|---|
| Qualitative metrics | Reader | DNN-N1pros | TSE-N1pros | TSE-Conv | DNN-N1pros vs TSE-N1pros | DNN-N1pros vs TSE-Conv |
| Overall image quality | ||||||
| 1 | 5 [4–5] | 3 [3–3] | 4 [4–4] | <0.001 | <0.001 | |
| 2 | 4 [4–5] | 3 [2–3] | 5 [5–5] | <0.001 | <0.001a | |
| Perceived SNR | ||||||
| 1 | 5 [5–5] | 3 [3–3] | 4 [4–4] | <0.001 | <0.001 | |
| 2 | 5 [5–5] | 3 [3–3] | 5 [4–5] | <0.001 | <0.004 | |
| Artifacts | ||||||
| 1 | 3 [2–4] | 3 [2–4] | 3 [3–4] | 0.002b | 0.825 | |
| 2 | 4 [4–4] | 4 [3–4] | 4 [4–5] | 0.102 | <0.001a | |
| Sharpness | ||||||
| Prostate | 1 | 5 [4–5] | 4 [3–4] | 4 [4–5] | <0.001 | 0.052 |
| 2 | 5 [4–5] | 3 [3–4] | 5 [5–5] | <0.001 | 0.013 | |
| Seminal vesicle | 1 | 5 [4–5] | 4 [3–4] | 5 [3–5] | <0.001 | 0.272 |
| 2 | 5 [4–5] | 3 [3–3] | 5 [4–5] | <0.001 | 0.366 | |
| Rectal wall | 1 | 5 [4–5] | 4 [3–4] | 4 [4–5] | <0.001 | 0.134 |
| 2 | 5 [4–5] | 4 [3–4] | 5 [5–5] | <0.001 | 0.132 | |
| Urinary bladder | 1 | 4 [4–5] | 4 [4–4] | 4 [4–4] | <0.001 | 0.007 |
| 2 | 4 [4–5] | 3 [3–4] | 4 [4–5] | <0.001 | 0.285 | |
| Obturator muscle | 1 | 5 [5–5] | 5 [4–5] | 5 [5–5] | <0.001 | >0.999 |
| 2 | 5 [5–5] | 4 [4–4] | 5 [5–5] | <0.001 | 0.025 | |
N represents the number of acquisitions; DNN, a deep neural network; SNR, Signal-to-noise ratio; TSE, turbo spin echo images; TSE-Conv, turbo spin echo images from conventional scans;pros, the prospective test set was acquired with accelerated scans.
Bold numbers indicate statistically significant p-value with Bonferroni correction (p < 0.05/2).
indicates that DNN shows significantly lower values than TSE.
DNN-N1pros shows significantly higher artifact scores than TSE-N1pros (Averaged score: DNN-N1pros = 3.3 and TSE-N1pros = 2.8).
Table 5.
Image quality comparison in the prospective test set with number of acquisitions = 2 (Median [Q1–Q3])
| Accelerated scan | p-value | |||||
|---|---|---|---|---|---|---|
| Qualitative metrics | Reader | DNN-N2pros | TSE-N2pros | TSE-Conv | DNN-N2pros vs TSE-N2pros | DNN-N2pros vs TSE-Conv |
| Overall image quality | ||||||
| 1 | 5 [5–5] | 3 [3–3] | 4 [4–4] | <0.001 | <0.001 | |
| 2 | 5 [5–5] | 4 [4–4] | 5 [5–5] | <0.001 | 0.083 | |
| Perceived SNR | ||||||
| 1 | 5 [5–5] | 3 [3–3] | 4 [4–4] | <0.001 | <0.001 | |
| 2 | 5 [5–5] | 4 [4–4] | 5 [5–5] | <0.001 | 0.157 | |
| Artifacts | ||||||
| 1 | 4 [3–4] | 3 [3–4] | 3 [3–4] | 0.041 | 0.005 | |
| 2 | 4 [4–5] | 4 [4–4] | 4 [4–4.25] | <0.001 | 0.102 | |
| Sharpness | ||||||
| Prostate | 1 | 5 [4–5] | 4 [4–4] | 4 [4–5] | <0.001 | 0.011 |
| 2 | 5 [5–5] | 4 [4–4] | 5 [5–5] | <0.001 | 0.317 | |
| Seminal vesicle | 1 | 5 [4–5] | 4 [3–5] | 5 [4–5] | 0.001 | 0.132 |
| 2 | 5 [4–5] | 4 [4–4] | 5 [4–5] | <0.001 | 0.655 | |
| Rectal wall | 1 | 5 [4–5] | 4 [4–4] | 4 [4–5] | <0.001 | 0.029 |
| 2 | 5 [4–5] | 4 [4–4] | 5 [4–5] | <0.001 | 0.739 | |
| Urinary bladder | 1 | 4 [4–5] | 4 [4–4] | 4 [4–4.25] | <0.001 | 0.046 |
| 2 | 5 [4–5] | 4 [4–4] | 4 [4–5] | <0.001 | 0.034 | |
| Obturator muscle | 1 | 5 [5–5] | 5 [4–5] | 5 [5–5] | 0.005 | 0.257 |
| 2 | 5 [5–5] | 4 [4–5] | 5 [5–5] | <0.001 | >0.999 | |
N represents the number of acquisitions; DNN, a deep neural network; SNR, signal-to-noise ratio; TSE, turbo spin echo images; TSE-Conv, turbo spin echo images from conventional scans;pros, the prospective test set was acquired with accelerated scans.
Bold numbers indicate statistically significant p-value with Bonferroni correction (p < 0.05/2).
In the NAQ = 2 results, DNN-N2pros showed improved overall image quality, perceived SNR, and sharpness of the structures compared to TSE-N2pros with statistically significant differences for both radiologists (Table 5). As assessed by radiologist 2, the artifact score of DNN-N2pros was significantly higher than that of TSE-N2pros. When DNN-N2pros were compared with TSE-Conv, all qualitative parameters were better than or comparable to those of TSE-Conv.
Interobserver agreements were fair to almost perfect for all image quality parameters in both NAQ = 1 and NAQ = 2 results (ICC = 0.434–0.926, p < 0.002), except for sharpness of the obturator muscle in NAQ = 2 (ICC = 0.224, p > 0.05). Intraobserver agreements were fair to almost perfect for all image quality parameters (ICC = 0.497–0.961, p < 0.001), supporting the reliability of qualitative evaluation. Each ICC value and corresponding 95% confidence interval and p-value are reported in Supplementary Material 1 (Supplementary Tables 1 to 4).
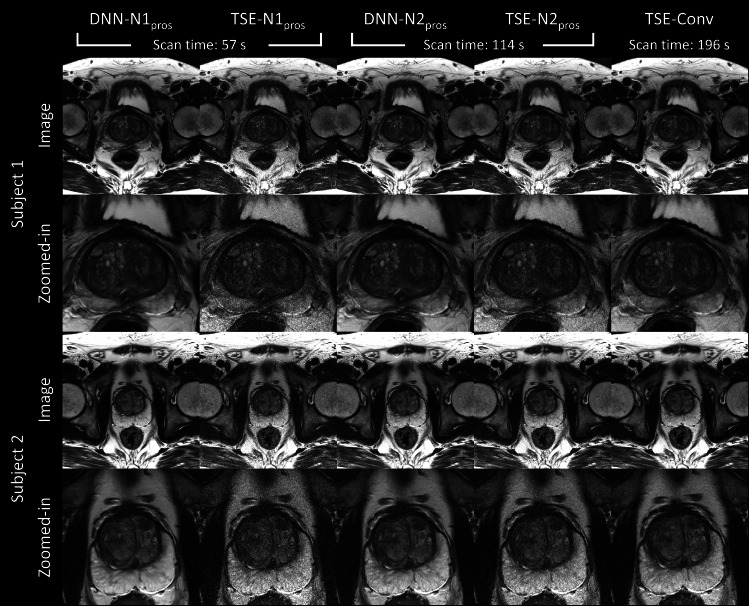
Figure 4 displayed the images of the two representative subjects in the prospective test set. The images from DNN and TSE reconstruction on accelerated scans (NAQ = 1 for first and second columns; NAQ = 2 for third and fourth columns), and TSE-Conv (last column) were shown. Overall, DNN-reconstructed images showed comparable contrasts and quality to those from TSE-Conv images. When zoomed-in for details (second and fourth rows), DNN-N1pros and DNN-N2pros showed better delineation of the structure than TSE-N1pros and TSE-N2pros, supporting the blind-test results from the radiologists in Tables 4 and 5.
Figure 4.
Reconstructed images of two representative subjects (DNN-N1pros, TSE-N1pros, DNN-N2pros, TSE-N2pros, and TSE-Conv) in the prospective test data set are displayed. Scan times were reduced by 71 and 42% in NAQ = 1 and 2, respectively. DNN images showed improved quality in TSE images on the accelerated scans, which is more comparable to TSE-Conv. DNN, deep neural network; TSE, turbo spin echo.
Presence of cancer
Table 6 shows the scores of the presence of clinically significant cancer based on T 2WI PI-RADS v. 2.1 from the two radiologists. The results revealed that intermethod agreement between DNN and TSE-Conv was almost perfect (all, κ > 0.9). In addition, the interobserver agreement between the two radiologists was moderate or substantial (κ, 0.58–0.61), supporting the reliability of the evaluation (Supplementary Material 1- Table 5).
Table 6.
Intermethod agreement of the presence of clinically significant cancer scoring by T2 weighted imaging PI-RADS v. 2.1
| Reader | Method | PI-RADS score | Weighted κ value with TSE-Conv | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||||
| 1 |
First set | DNN-N1 | 32 | 0 | 0 | 5 | 3 | 0.947 |
| TSE-N1 | 32 | 0 | 0 | 5 | 3 | 0.947 | ||
| TSE-Conv | 31 | 0 | 0 | 6 | 3 | - | ||
| 2 |
DNN-N1 | 33 | 2 | 2 | 2 | 1 | 0.901 | |
| TSE-N1 | 33 | 2 | 2 | 2 | 1 | 0.901 | ||
| TSE-Conv | 33 | 1 | 2 | 2 | 2 | - | ||
| 1 |
Second set | DNN-N2 | 29 | 0 | 0 | 8 | 3 | 1.000 |
| TSE-N2 | 29 | 0 | 0 | 8 | 3 | 1.000 | ||
| TSE-Conv | 29 | 0 | 0 | 8 | 3 | - | ||
| 2 |
DNN-N2 | 32 | 2 | 1 | 3 | 2 | 1.000 | |
| TSE-N2 | 32 | 2 | 2 | 2 | 2 | 0.971 | ||
| TSE-Conv | 32 | 2 | 1 | 3 | 2 | - | ||
N represents the number of acquisitions; DNN, a deep neural network; PI-RADS, prostate imaging-reporting and data system;TSE-Conv, turbo spin echo images from conventional scans.
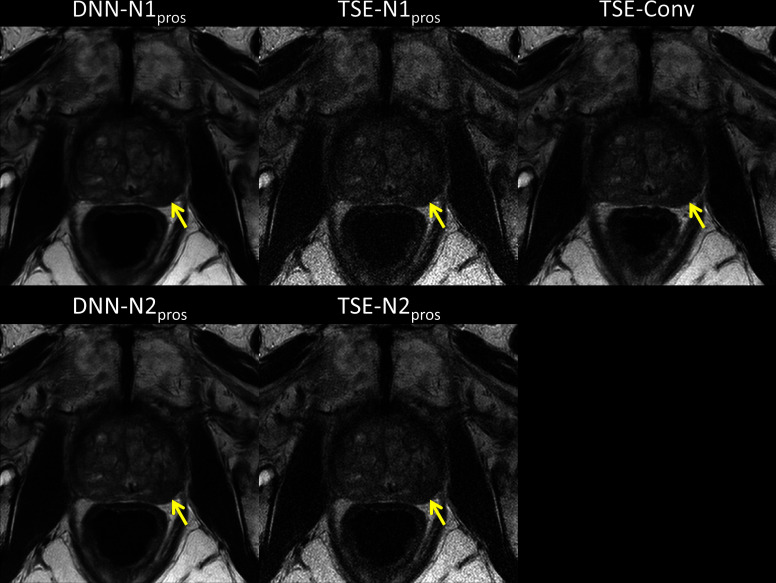
In Figure 5, the images of one representative patient, who had clinically significant lesions in the prostate, in the prospective test data set are displayed. Each image was reconstructed DNN-N1, TSE-N1, DNN-N2, TSE-N2, and TSE-Conv pipelines. When compared to TSE-N1pros or TSE-N2pros, DNN-N1pros and DNN-N2pros showed better contrast and image quality. Additionally, both DNN images showed good agreement with TSE-Conv in lesion delineation as indicated by yellow arrows, supporting the T2WI PI-RADS scoring results in Table 6.
Figure 5.
Reconstructed images of one representative patient in the prospective test data set are displayed. Note that TSE-Conv images were used as a reference. Although scan times were reduced in DNN-N1pros and DNN-N2pros, both images showed good agreement with TSE-Conv in lesion delineation (yellow arrows) with better image quality than TSE-N1pros and TSE-N2pros. DNN, deep neural network; TSE, turbo spin echo.
Discussion
In this study, we proposed DNN-based parallel imaging reconstruction in highly accelerated 2D TSE prostate imaging. The proposed DNN model is based on the U-net architecture, which aims to reconstruct the images from accelerated scans (R = 4, NAQ = 1 or 2) into the images from conventional scans (R = 2, NAQ = 2). Note that the conventional scan time was 196 s while the accelerated scan times were 57 s for NAQ = 1 and 114 s for NAQ = 2. The high-quality performance of DNN was demonstrated in the accelerated scan by the two analyses: quantitative evaluation in the retrospective test data set and qualitative evaluation in the prospective test data set. Both evaluations showed that DNN reconstruction on NAQ = 2 showed improved or comparable image quality to conventional scans despite a 42% reduction of scan time.
In the results of the quantitative evaluation, DNN images showed better quantitative error metrics, lower NRMSE, higher SSIM, and high PSNR, than TSE images with statistically significant differences in both NAQ = 1 and NAQ = 2 scans; such error metrics have been used to calculate image differences in various aspects. For example, NRMSE represents the averaged voxelwise errors. SSIM reflects the ability of the human visual system to reveal perceptual differences such as distortion and blurring. 34,35 PSNR is highly sensitive for assessing the noise in images. 36 In previous DNN-based MR acceleration studies, DNN showed improved performance compared to conventional parallel imaging reconstruction with 27–65% lower NRMSE, 23,26 2–22% higher SSIM, 23,26,37,38 and 2.8–6.1 dB higher PSNR, 37,38 which is similar to our results (29–43% lower NRMSE, 4–13% higher SSIM, and 2.8–4.8 dB higher PSNR). Thus, the results of quantitative evaluation revealed that the reconstruction of highly accelerated 2D TSE prostate data can be improved by the DNN-based model.
In the qualitative evaluation using the prospective data, both radiologists rated DNN images above TSE images in all parameters except for the artifact observed by reader 2 in DNN-N1pros and by reader 1 in DNN-N2pros. The results proved that DNN reconstruction could significantly improve image quality. When comparing DNN images and TSE-Conv, both DNN-N1pros and DNN-N2pros demonstrated better or comparable performance in most parameters. Higher scores were noted in overall image quality and perceived SNR in both DNN-N1pros and DNN-N2pros than TSE-Conv by reader 1. Reader 2 evaluated the overall image quality of DNN-N2 as comparable to TSE-Conv and that of DNN-N1pros as lower than TSE-Conv, even though the median image quality score of DNN-N1pros was 4, which is a good score. Such different results for the two readers may be explained by the fact that each reader has a different preference for MRI images. The results showed that there was no item in DNN-N2pros with worse results than TSE-Conv, suggesting that DNN reconstruction on NAQ = 2 scans is more clinically feasible than NAQ = 1.
The slightly worse performance of DNN-N1pros may be attributed to two reasons. First, compared to the reference scan, the accelerated scan with NAQ = 1 was acquired with 71% reduction in scan time, leading to a substantially low SNR in the input images. Low SNR images indicate that the amounts of prior information to be used by DNNs have decreased, 39,40 resulting in performance degradation of DNN reconstruction. Second, the motion between the scan interval may affect the quality of DNN-N1pros. Note that DNN-N1 was trained by using NAQ = 1 data as inputs and NAQ = 2 data as labels. Since involuntary anatomic motion from peristalsis and rectal distention occurs during prostate MR scans, 41,42 misregistration between NAQ = 1 and 2 data can be generated. Thus, training by the misregistration data may generate blurring artifacts on the output images, 43,44 leading to low scores in DNN-N1pros. On the other hand, the data preprocessing step of DNN-N2pros is less affected by SNR degradation and misregistration. Therefore, all quantitative parameters showed improved or comparable quality to TSE-Conv according to both readers.
DNN-based image reconstruction, which is advantageous in terms of reducing the acquisition time, should not impair the diagnostic performance of MRI. In this study, the cancer detection analysis by T 2WI PI-RADS score demonstrated that DNN-N2pros and DNN-N1pros showed almost perfect interobserver and intermethod agreement with TSE-Conv. Intermethod and interobserver agreement are important in prebiopsy prostate MRI because performing subsequent biopsies depends on the MRI results. The reason why there is no difference in diagnostic performance despite the difference in image quality evaluation among DNN images and TSE-Conv may be that image quality in all images was secured above a certain level and did not impact diagnosis.
Compared to our study, a few studies have proposed DNN-based acceleration in prostate MRI, revealing similar results in terms of image quality evaluation and diagnostic performance. 45–47 However, none of the previous studies performed a statistical quantitative evaluation with various error metrics. In addition, the DNN architecture of the previous studies used the k-space data as inputs while our model used image data as inputs, which is easily obtainable as the digital imaging and communications in medicine format. Considering the difficulty of accessing k-space data in a clinical setting, our model may be more feasible for clinical application.
There are a few limitations to our study. First, while the proposed framework demonstrated the improved performance of the U-net architecture, we did not compare our results to the state-of-the-art DNN models. 21,22,25–27 To optimize the reconstruction performance, a comparison with various DNN models needs to be conducted. Second, the generalization of DNN on scan parameters or sequences was not demonstrated in this study. It is worth demonstrating the proposed DNN framework on different sequences since the mp-MRI scan protocol including diffusion-weighted imaging and dynamic contrast-enhanced imaging is recommended as a routine sequence. For example, DNN reconstruction on highly accelerated prostate diffusion-weighted imaging has been demonstrated. 44,47 One caveat is that the input image quality affects DNN-based reconstruction. 48 Thus, the accelerated scan parameters need to be set up differently across the sequence protocols. Reducing the scan times of all mp-MRI sequences by DNN reconstruction may have the potential to reduce patient discomfort originating from the long scan times involved in prostate imaging. Third, the undersampling factor on the accelerated scan was limited to R = 4 because the reference data was acquired with R = 2. If the full-sampled k-space data set is collected, DNN reconstruction on various acceleration factors can be tested. Fourth, the data preprocessing for training DNN requires the k-space data. Thus, the accessibility to the k-space data, which depends on vendor and institution, may limit the DNN development using the proposed framework. Lastly, pathology results were unavailable in this study as we included patients who underwent prostate MRI for suspected prostate cancer regardless of whether the following biopsy was performed. However, all MRI examinations were performed without prior biopsy and diagnostic performance was evaluated by two radiologists. We thought that the results showed the usefulness of DNN-based reconstruction in the clinical diagnostic process using prebiopsy MRI.
In conclusion, we developed DNN-based reconstruction on highly accelerated 2D TSE prostate MR imaging without image quality degradation. Compared to the conventional scan, a 42% reduction in scan time was achieved. The proposed framework has great potential to reduce the overall scan times of prostate MR imaging protocols.
Supplementary Material
Footnotes
Acknowledgment: This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: KMDF_PR_20200901_0062, 9991006735).
Conflict of interest: Woojin Jung, Jingyu Ko, and Geunu Jeong are employees of AIRS Medical. Moon Hyung Choi is currently receiving a research grant from Company Siemens Healthineers for the topic not related to the current reasearch. For the remaining authors none were declared.
Contributor Information
Woojin Jung, Email: jung.woojin@airsmed.com, AIRS Medical, Seoul, Republic of Korea .
Eu Hyun Kim, Email: doorihyun6@gmail.com, Department of Radiology, St.Vincent’s Hospital, College of Medicine, The Catholic University of Korea, Suwon, Gyeonggi-do, Republic of Korea .
Jingyu Ko, Email: ko.jingyu@airsmed.com, AIRS Medical, Seoul, Republic of Korea .
Geunu Jeong, Email: jeong.geunu@airsmed.com, AIRS Medical, Seoul, Republic of Korea .
Moon Hyung Choi, Email: cmh@catholic.ac.kr, Department of Radiology, Eunpyeong St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea .
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A . Cancer statistics, 2019 . CA Cancer J Clin 2019. ; 69: 7 – 34 . doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2. Nordström T, Discacciati A, Bergman M, Clements M, Aly M, et al. . Prostate cancer screening using a combination of risk-prediction, mri, and targeted prostate biopsies (sthlm3-mri): a prospective, population-based, randomised, open-label, non-inferiority trial . Lancet Oncol 2021. ; 22: 1240 – 49 . doi: 10.1016/S1470-2045(21)00348-X [DOI] [PubMed] [Google Scholar]
- 3. US Preventive Services Task Force. Grossman DC, Curry SJ, Owens DK, Bibbins-Domingo K, et al. . Screening for prostate cancer: us preventive services task force recommendation statement . JAMA 2018. ; 319: 1901 – 13 . doi: 10.1001/jama.2018.3710 [DOI] [PubMed] [Google Scholar]
- 4. Panebianco V, Barchetti F, Sciarra A, Ciardi A, Indino EL, et al. . Multiparametric magnetic resonance imaging vs. standard care in men being evaluated for prostate cancer: a randomized study . Urol Oncol 2015. ; 33: 17 . doi: 10.1016/j.urolonc.2014.09.013 [DOI] [PubMed] [Google Scholar]
- 5. Sherrer RL, Glaser ZA, Gordetsky JB, Nix JW, Porter KK, et al. . Comparison of biparametric mri to full multiparametric mri for detection of clinically significant prostate cancer . Prostate Cancer Prostatic Dis 2019. ; 22: 331 – 36 . doi: 10.1038/s41391-018-0107-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fütterer JJ, Briganti A, De Visschere P, Emberton M, Giannarini G, et al. . Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? a systematic review of the literature . Eur Urol 2015. ; 68: 1045 – 53 . doi: 10.1016/j.eururo.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 7. Thestrup KCD, Logager V, Baslev I, Møller JM, Hansen RH, et al. . Biparametric versus multiparametric mri in the diagnosis of prostate cancer . Acta Radiol Open 2016. ; 5: 2058460116663046 . doi: 10.1177/2058460116663046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sciarra A, Barentsz J, Bjartell A, Eastham J, Hricak H, et al. . Advances in magnetic resonance imaging: how they are changing the management of prostate cancer . Eur Urol 2011. ; 59: 962 – 77 . doi: 10.1016/j.eururo.2011.02.034 [DOI] [PubMed] [Google Scholar]
- 9. Dickinson L, Ahmed HU, Allen C, Barentsz JO, Carey B, et al. . Magnetic resonance imaging for the detection, localisation, and characterisation of prostate cancer: recommendations from a european consensus meeting . Eur Urol 2011. ; 59: 477 – 94 . doi: 10.1016/j.eururo.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 10. Vidya Shankar R, Roccia E, Cruz G, Neji R, Botnar R, et al. . Accelerated 3d t2 w-imaging of the prostate with 1-millimeter isotropic resolution in less than 3 minutes . Magn Reson Med 2019. ; 82: 721 – 31 . doi: 10.1002/mrm.27764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bilgic B, Gagoski BA, Cauley SF, Fan AP, Polimeni JR, et al. . Wave-caipi for highly accelerated 3d imaging . Magn Reson Med 2015. ; 73: 2152 – 62 . doi: 10.1002/mrm.25347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lustig M, Pauly JM . SPIRiT: iterative self-consistent parallel imaging reconstruction from arbitrary k-space . Magn Reson Med 2010. ; 64: 457 – 71 . doi: 10.1002/mrm.22428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P . SENSE: sensitivity encoding for fast mri . Magn Reson Med 1999. ; 42: 952 – 62 . doi: [DOI] [PubMed] [Google Scholar]
- 14. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, et al. . Generalized autocalibrating partially parallel acquisitions (grappa) . Magn Reson Med 2002. ; 47: 1202 – 10 . doi: 10.1002/mrm.10171 [DOI] [PubMed] [Google Scholar]
- 15. Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, et al. . ESPIRiT--an eigenvalue approach to autocalibrating parallel mri: where sense meets grappa . Magn Reson Med 2014. ; 71: 990 – 1001 . doi: 10.1002/mrm.24751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lustig M, Donoho DL, Santos JM, Pauly JM . Compressed sensing mri . IEEE Signal Process Mag 2008. ; 25: 72 – 82 . doi: 10.1109/MSP.2007.914728 [DOI] [Google Scholar]
- 17. Nana R, Zhao T, Heberlein K, LaConte SM, Hu X . Cross-validation-based kernel support selection for improved grappa reconstruction . Magn Reson Med 2008. ; 59: 819 – 25 . doi: 10.1002/mrm.21535 [DOI] [PubMed] [Google Scholar]
- 18. Chang Y, Liang D, Ying L . Nonlinear grappa: a kernel approach to parallel mri reconstruction . Magn Reson Med 2012. ; 68: 730 – 40 . doi: 10.1002/mrm.23279 [DOI] [PubMed] [Google Scholar]
- 19. Smith DS, Welch EB, Li X, Arlinghaus LR, Loveless ME, et al. . Quantitative effects of using compressed sensing in dynamic contrast enhanced mri . Phys Med Biol 2011. ; 56: 4933 – 46 . doi: 10.1088/0031-9155/56/15/018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma SD, Fong CL, Tzung BS, Law M, Nayak KS . Clinical image quality assessment of accelerated magnetic resonance neuroimaging using compressed sensing . Invest Radiol 2013. ; 48: 638 – 45 . doi: 10.1097/RLI.0b013e31828a012d [DOI] [PubMed] [Google Scholar]
- 21. Akçakaya M, Moeller S, Weingärtner S, Uğurbil K . Scan-specific robust artificial-neural-networks for k-space interpolation (raki) reconstruction: database-free deep learning for fast imaging . Magn Reson Med 2019. ; 81: 439 – 53 . doi: 10.1002/mrm.27420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muckley MJ, Riemenschneider B, Radmanesh A, Kim S, Jeong G, et al. . Results of the 2020 fastmri challenge for machine learning mr image reconstruction . IEEE Trans Med Imaging 2021. ; 40: 2306 – 17 . doi: 10.1109/TMI.2021.3075856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ji S, Jeong J, Oh S-H, Nam Y, Choi SH, et al. . Quad-contrast imaging: simultaneous acquisition of four contrast-weighted images (pd-weighted, t₂-weighted, pd-flair and t₂-flair images) with synthetic t₁-weighted image, t₁- and t₂-maps . IEEE Trans Med Imaging 2021. ; 40: 3617 – 26 . doi: 10.1109/TMI.2021.3093617 [DOI] [PubMed] [Google Scholar]
- 24. Aggarwal HK, Mani MP, Jacob M . MoDL: model-based deep learning architecture for inverse problems . IEEE Trans Med Imaging 2019. ; 38: 394 – 405 . doi: 10.1109/TMI.2018.2865356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eo T, Jun Y, Kim T, Jang J, Lee H-J, et al. . KIKI-net: cross-domain convolutional neural networks for reconstructing undersampled magnetic resonance images . Magn Reson Med 2018. ; 80: 2188 – 2201 . doi: 10.1002/mrm.27201 [DOI] [PubMed] [Google Scholar]
- 26. Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, et al. . Learning a variational network for reconstruction of accelerated mri data . Magn Reson Med 2018. ; 79: 3055 – 71 . doi: 10.1002/mrm.26977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sriram A, Zbontar J, Murrell T, Defazio A, Zitnick CL, et al. . End-to-end variational networks for accelerated mri reconstruction . ArXiv Prepr 2020. . doi: https://doi.org/ArXiv:200406688 [Google Scholar]
- 28. Ulyanov D, Vedaldi A, Lempitsky V . Deep image prior . Int J Comput Vis 2020; 128: 1867–88. 10.1007/s11263-020-01303-4 [DOI] [Google Scholar]
- 29. Ronneberger O, Fischer P, Brox T . U-net: convolutional networks for biomedical image . Int Conf Med Image Comput Comput Interv 2015. ; 234 – 41 . doi: 10.1007/978-3-319-24574-4 [DOI] [Google Scholar]
- 30. . Zbontar J, Knoll F, Sriram A, Murrell T, Huang Z, Muckley MJ, et al. . fastMRI: An Open Dataset and Benchmarks for Accelerated MRI . Arxiv 2018. . [Google Scholar]
- 31. Kingma DP, Ba J . Adam: a method for stochastic optimization . ArXiv Prepr 2014. . doi: https://doi.org/ArXiv:14126980 [Google Scholar]
- 32. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, et al. . PyTorch: animperative style, high-performance deep learning library . Adv Neural Inf Process Syst 2019. ; 8024 – 35 . [Google Scholar]
- 33. Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, et al. . Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2 . Eur Urol 2019. ; 76: 340 – 51 . doi: 10.1016/j.eururo.2019.02.033 [DOI] [PubMed] [Google Scholar]
- 34. Brooks AC, Zhao X, Pappas TN . Structural similarity quality metrics in a coding context: exploring the space of realistic distortions . IEEE Trans Image Process 2008. ; 17: 1261 – 73 . doi: 10.1109/TIP.2008.926161 [DOI] [PubMed] [Google Scholar]
- 35. Wang Z, Bovik AC, Sheikh HR, Simoncelli EP . Image quality assessment: from error visibility to structural similarity . IEEE Trans Image Process 2004. ; 13: 600 – 612 . doi: 10.1109/tip.2003.819861 [DOI] [PubMed] [Google Scholar]
- 36. Hore A, Ziou D . Image quality metrics: psnr vs ssim . 2010 20th Int Conf Pattern Recognit 2010. ; 2366 – 69 . doi: 10.1109/icpr.2010.579 [DOI] [Google Scholar]
- 37. Pawar K, Egan GF, Chen Z . Domain knowledge augmentation of parallel mr image reconstruction using deep learning . Comput Med Imaging Graph 2021. ; 92: 101968 . doi: 10.1016/j.compmedimag.2021.101968 [DOI] [PubMed] [Google Scholar]
- 38. Han Y, Sunwoo L, Ye JC . K -space deep learning for accelerated mri . IEEE Trans Med Imaging 2020. ; 39: 377 – 86 . doi: 10.1109/TMI.2019.2927101 [DOI] [PubMed] [Google Scholar]
- 39. Koonjoo N, Zhu B, Bagnall GC, Bhutto D, Rosen MS . Boosting the signal-to-noise of low-field mri with deep learning image reconstruction . Sci Rep 2021. ; 11( 1 ): 8248 . doi: 10.1038/s41598-021-87482-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jung W, Bollmann S, Lee J . Overview of quantitative susceptibility mapping using deep learning: current status, challenges and opportunities . NMR Biomed 2020. : e4292 . doi: 10.1002/nbm.4292 [DOI] [PubMed] [Google Scholar]
- 41. Padhani AR, Khoo VS, Suckling J, Husband JE, Leach MO, et al. . Evaluating the effect of rectal distension and rectal movement on prostate gland position using cine mri . Int J Radiat Oncol Biol Phys 1999. ; 44: 525 – 33 . doi: 10.1016/s0360-3016(99)00040-1 [DOI] [PubMed] [Google Scholar]
- 42. Caglic I, Hansen NL, Slough RA, Patterson AJ, Barrett T . Evaluating the effect of rectal distension on prostate multiparametric mri image quality . Eur J Radiol 2017. ; 90: 174 – 80 . doi: 10.1016/j.ejrad.2017.02.029 [DOI] [PubMed] [Google Scholar]
- 43. Wang G, Gong E, Banerjee S, Martin D, Tong E, et al. . Synthesize high-quality multi-contrast magnetic resonance imaging from multi-echo acquisition using multi-task deep generative model . IEEE Trans Med Imaging 2020. ; 39: 3089 – 99 . doi: 10.1109/TMI.2020.2987026 [DOI] [PubMed] [Google Scholar]
- 44. Kaye EA, Aherne EA, Duzgol C, Häggström I, Kobler E, et al. . Accelerating prostate diffusion-weighted mri using a guided denoising convolutional neural network: retrospective feasibility study . Radiol Artif Intell 2020. ; 2: e200007 . doi: 10.1148/ryai.2020200007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gassenmaier S, Afat S, Nickel MD, Mostapha M, Herrmann J, et al. . Accelerated t2-weighted tse imaging of the prostate using deep learning image reconstruction: a prospective comparison with standard t2-weighted tse imaging . Cancers (Basel) 2021. ; 13( 14 ): 3593 . doi: 10.3390/cancers13143593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gassenmaier S, Afat S, Nickel D, Mostapha M, Herrmann J, et al. . Deep learning-accelerated t2-weighted imaging of the prostate: reduction of acquisition time and improvement of image quality . Eur J Radiol 2021. ; 137: 109600 . doi: 10.1016/j.ejrad.2021.109600 [DOI] [PubMed] [Google Scholar]
- 47. Johnson PM, Tong A, Donthireddy A, Melamud K, Petrocelli R, et al. . Deep learning reconstruction enables highly accelerated biparametric mr imaging of the prostate . J Magn Reson Imaging 2021. . doi: 10.1002/jmri.28024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeelani H, Martin J, Vasquez F, Salerno M, Weller DS . Image quality affects deep learning reconstruction of MRI . 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) ; Washington, DC . ; April 2018. 10.1109/ISBI.2018.8363592 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.