Abstract
Hydrocephalus, characterized by cerebral ventriculomegaly, is the most common disorder requiring brain surgery in children. Recent studies have implicated SMARCC1, a component of the BRG1-associated factor (BAF) chromatin remodelling complex, as a candidate congenital hydrocephalus gene. However, SMARCC1 variants have not been systematically examined in a large patient cohort or conclusively linked with a human syndrome. Moreover, congenital hydrocephalus-associated SMARCC1 variants have not been functionally validated or mechanistically studied in vivo. Here, we aimed to assess the prevalence of SMARCC1 variants in an expanded patient cohort, describe associated clinical and radiographic phenotypes, and assess the impact of Smarcc1 depletion in a novel Xenopus tropicalis model of congenital hydrocephalus.
To do this, we performed a genetic association study using whole-exome sequencing from a cohort consisting of 2697 total ventriculomegalic trios, including patients with neurosurgically-treated congenital hydrocephalus, that total 8091 exomes collected over 7 years (2016–23). A comparison control cohort consisted of 1798 exomes from unaffected siblings of patients with autism spectrum disorder and their unaffected parents were sourced from the Simons Simplex Collection. Enrichment and impact on protein structure were assessed in identified variants. Effects on the human fetal brain transcriptome were examined with RNA-sequencing and Smarcc1 knockdowns were generated in Xenopus and studied using optical coherence tomography imaging, in situ hybridization and immunofluorescence. SMARCC1 surpassed genome-wide significance thresholds, yielding six rare, protein-altering de novo variants localized to highly conserved residues in key functional domains. Patients exhibited hydrocephalus with aqueductal stenosis; corpus callosum abnormalities, developmental delay, and cardiac defects were also common. Xenopus knockdowns recapitulated both aqueductal stenosis and cardiac defects and were rescued by wild-type but not patient-specific variant SMARCC1. Hydrocephalic SMARCC1-variant human fetal brain and Smarcc1-variant Xenopus brain exhibited a similarly altered expression of key genes linked to midgestational neurogenesis, including the transcription factors NEUROD2 and MAB21L2. These results suggest de novo variants in SMARCC1 cause a novel human BAFopathy we term ‘SMARCC1-associated developmental dysgenesis syndrome’, characterized by variable presence of cerebral ventriculomegaly, aqueductal stenosis, developmental delay and a variety of structural brain or cardiac defects. These data underscore the importance of SMARCC1 and the BAF chromatin remodelling complex for human brain morphogenesis and provide evidence for a ‘neural stem cell’ paradigm of congenital hydrocephalus pathogenesis.
These results highlight utility of trio-based whole-exome sequencing for identifying pathogenic variants in sporadic congenital structural brain disorders and suggest whole-exome sequencing may be a valuable adjunct in clinical management of congenital hydrocephalus patients.
Keywords: congenital, neurodevelopment, chromatin, neurosurgery, genetics, genomics
Hydrocephalus is the most common disease requiring neurosurgery in children. Singh et al. show an association between mutations in the chromatin remodelling gene SMARCC1 and congenital hydrocephalus with a variety of syndromic features, and perform functional studies to explore the underlying mechanisms.
Introduction
Epigenetic mechanisms, including methylation, histone modifications and ATP-dependent chromatin remodelling, regulate gene expression by altering chromatin structure.1-4 The SWI/SNF (switch/sucrose non-fermentable) complex [also known as the BRG1-associated factor (BAF) complex] is one of four ATP-dependent chromatin remodelling complexes known in mammals.5-7 The BAF complex mediates nucleosome modification critical to modulating gene expression in multiple essential processes, including cell differentiation and proliferation, and DNA repair.1-5 The combinatorial assembly of numerous gene family paralogues yields many potential types of complex hetero-oligomeric complexes that provide tissue and temporal specificity6-8 for the control of gene transcription that is essential for the development of the brain,9,10 heart11-13 and other organs, as well as the maintenance of embryonic stem cell pluripotency (Table 1).14
Table 1.
De novo mutations in SMARCC1 probands
| Proband ID | Mutation | Class | Position (hg37) | cDNA change | AA change | Domain | MetaSVM rank score | MPC | gnomAD MAF |
|---|---|---|---|---|---|---|---|---|---|
| CHYD115-1a | De novo | Frameshift | 3:47663805:CT:C | c.2672delA | p.(Lys891Argfs*6) | Glu-rich | – | – | 0 |
| CHYD168-1a | De novo | Missense | 3:47718267:T:G | c.1577A>C | p.(His526Pro) | SWIRM | 0.904 | 2.57 | 0 |
| CHYD505-1a | De novo | Splice site | 3:47719687:C:T | c.1571+1G>A | – | SWIRM | – | – | 0 |
| CHYDX1-1 | De novo | Missense | 3:47703958:T:C | c.2204A>G | p.(Asp675Gly) | Myb | 0.13 | 1.22 | 0 |
| CHYDX2-1 | De novo | Splice site | 3:44719687:C:T | c.1571+1G>A | – | SWIRM | – | – | 0 |
| CHYDX3-1 | De novo | Frameshift | 3:47823117:GA:G | c.170delT | p.(Val57Alafs*97) | Chromo | – | – | 0 |
BAFopathies constitute a heterogeneous group of disorders caused by variants in various subunits composing the BAF complex.17 The phenotypic spectrum of BAFopathies includes intellectual disability (ID) and developmental delay (DD), autism, schizophrenia, amyotrophic lateral sclerosis18-20 and other human neurodevelopmental disorders and anatomical congenital defects.21,22 The most recognizable syndrome associated with BAF abnormalities is Coffin-Siris syndrome [CSS (MIM: 135900)]. This is a genetically heterogeneous ID/DD syndrome characterized by speech delay, coarse facial appearance, feeding difficulties, hypoplastic-to-absent fifth fingernails, and fifth distal phalanges.23 This syndrome is associated with variants in multiple BAF complex subunits, including the ATPase subunit SMARCA4 (MIM: 603254), the common core subunit SMARCB1 (MIM: 601607), and BAF accessory subunits such as SMARCE1/BAF57 (MIM: 603111), ARID1A (MIM: 603024), ARID1B (MIM: 614556), ARID2 (MIM: 609539) and DPF2 (MIM: 601671).23,24 Other BAFopathies, such as Nicolaides-Baraitser syndrome (MIM: 601358), have significant phenotypic overlap with CSS and are caused by pathogenic variants in SMARCA2 (MIM: 600014).18-20
SMARCC1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily C member 1) encodes an essential core subunit of the BAF complex highly homologous to SMARCC2.8,25Smarcc1 is highly expressed in the mouse embryonic neuroepithelium and ventricular zone.9,10,14 Similar to other components of the neuroprogenitor-specific BAF complexes, Smarcc1 regulates the proliferation, differentiation and survival of mouse neural progenitors via transcriptional regulation of genes critical for telencephalon development.26-29Smarcc2;Smarcc1 double knockout mice exhibit proteasome-mediated degradation of the entire BAF complex, resulting in impairment of the global epigenetic and gene expression program of cortical development.17,30Smarcc1 knockout causes embryonic lethality in mice.31,32 Approximately 80% of mice homozygous for the Smarcc1msp/msp missense allele exhibit exencephaly due to decreased proliferation and increased apoptosis of neural progenitors in the neural tube.31,33
Recently, whole-exome sequencing (WES) studies in patients with congenital hydrocephalus identified SMARCC1 as a candidate gene, implicating impaired epigenetic regulation of neural progenitor cell (NPC) proliferation and differentiation in the development of ventriculomegaly.15,16 However, despite its significant biological role, SMARCC1 variants have not been conclusively associated with a human syndrome and congenital hydrocephalus-associated SMARCC1 variants have been neither functionally assessed nor mechanistically studied in vivo. The objectives of this study were to: (i) assess the prevalence of rare, damaging de novo variants (DNVs) in SMARCC1 in a large congenital hydrocephalus cohort; (ii) describe the phenotypes of SMARCC1-variant patients; and (iii) functionally validate and assess the cellular and molecular mechanisms of congenital hydrocephalus-associated SMARCC1 variants in a novel animal model of hydrocephalus.
Our findings suggest rare, damaging germline DNVs in SMARCC1 cause a novel human BAFopathy we term ‘SMARCC1-associated developmental dysgenesis syndrome (SaDDS)’ characterized by DD, cerebral ventriculomegaly and other structural brain or cardiac defects. Our data highlight the importance of the ATP-dependent BAF chromatin remodelling complex for human brain morphogenesis and CSF dynamics and further support a ‘neural stem cell paradigm’ of human congenital hydrocephalus.34-36 These data highlight the power of trio-based WES for identifying pathogenic variants in sporadic structural brain disorders and suggest its utility as a prognostic adjunct when evaluating the surgical candidacy and prognosis of congenital hydrocephalus patients.
Materials and methods
Patient cohort
All study procedures and protocols were guided by and in compliance with Human Investigation Committee and Human Research Protection Program at Yale School of Medicine and the Massachusetts General Hospital. All participants provided written, informed consent to participate in accordance with the Declaration of Helsinki. For patients from the clinical laboratory GeneDx, denoted CHYDX, written informed consent for genetic testing was obtained from the guardians of all paediatric individuals undergoing testing. The Western institutional review board waived authorization for the use of de-identified aggregate data for the purposes of this study. Criteria for inclusion into the study was congenital or primary cerebral ventriculomegaly, including congenital hydrocephalus. Patients and participating family members provided buccal swab samples (Isohelix SK-2S DNA buccal swab kits), medical records, neuroimaging studies, operative reports and phenotype data when available. Human phenotype ontology terms were used to aggregate relevant paediatric patients in the GeneDx database. The comparison control cohort consisted of 1798 unaffected siblings of people diagnosed with autism spectrum disorder (ASD) and unaffected parents sourced from the Simons Simplex Collection (SSC).37 Only the unaffected siblings and parents, as designated by SSC, were included in the analysis, and served as controls for this study. Permission to access the genomic data in the SSC on the National Institute of Mental Health Data Repository was obtained. Written and informed consent for all participants was provided by the Simons Foundation Autism Research Initiative.
Kinship analysis
Pedigree information and relationships between proband and parents was confirmed using pairwise PLINK identity-by-descent (IBD) calculation.38 The IBD sharing between the probands and parents in all trios was between 45% and 55%. Pairwise individual relatedness was calculated using KING.39 The ethnicity of each patient from the Yale cohort was determined by single-nucleotide polymorphisms in cases, controls and HapMap samples using EIGENSTRAT, as previously described.40 For the GeneDx cohort, kinship analysis was performed using an internally developed KNN/PCA pipeline.
Whole-exome sequencing and variant calling
Patient genomic DNA samples derived from saliva or blood were applied for exon capture using Roche SeqCap EZ MedExome Target Enrichment kit or IDT xGen target capture followed by 101 or 148 base-paired end sequencing on Illumina platforms, as described previously.15,16 BWA-MEM was applied to align sequence reads to the human reference genome GRCh37/hg19. Single nucleotide variants and small indels were called using a combination of GATK HaplotypeCaller and Freebayes41,42 and annotated using ANNOVAR.43 The cDNA change and protein change were accurately annotated using transcript variant NM_003074.4 and protein isoform NP_003065.3, respectively. Allele frequencies were annotated in the Exome Aggregation Consortium, gnomAD (v.2.1.1) and Bravo databases.44 Variant filtration and analysis were conducted following GATK best practices and consensus workflows.45 MetaSVM and MPC algorithms were used to predict the deleteriousness of missense variants (D-Mis, defined as MetaSVM-deleterious or MPC-score ≥2).46 Inferred loss-of-function (LoF) variants consisted of stop-gain, stop-loss, frameshift insertions/deletions, canonical splice site and start-loss. LoF and D-Mis variants were considered ‘damaging’. Analyses were conducted separately for each class of variant—DNVs and rare, heterozygous variants—following previously established analytical methodologies.16,45 First, DNVs from the Harvard-Yale cohort were called from all congenital hydrocephalus parent-offspring trios using the established TrioDeNovo pipeline.47,48 GeneDx DNVs were called as previously defined.49 Candidate DNVs for all samples were further filtered based on whether the variants were called in the exonic or splice-site regions, the variant read depth (DP) was at least 10 in the proband as well as both parents, and the global minor allele frequency was ≤4 × 10−4 in the Exome Aggregation Consortium database. Samples from the Yale cohort were subsequently filtered based on the following criteria: (i) the proband’s alternative read depth was ≥5; (ii) proband alternative allele ratio ≥28% if having <10 alternative reads, or ≤20% if having ≥10 alternative reads; and (iii) the alternative allele ratio in both parents ≤3.5%. Samples from the GeneDx cohort were additionally filtered based on the following criteria: (i) genotype quality (GQ) > 40 for all family members; (ii) variant quality score log odds (VQSLOD) > −10; (iii) Phred-scaled P-value (Fisher's exact test; FS) < 30; (iv) proband alternate allele count >4; (v) proband alternate allele ratio > 0.1; (vi) proband alternate allele ratio >0.15 if REF and ALT calls are of equal length; (vii) proband alternate allele ratio >0.25 if REF and ALT calls are of unequal length; (viii) proband alternate allele ratio < 0.9 in proband if DNV is autosomal; (ix) DNV must be <100 bps in size for both the REF and ALT calls; (x) if the VQSLOD <7 and the alternate allele ratio in the proband <0.3, the variant was omitted; and (xi) DNVs were omitted if they existed in more than two unrelated probands. After filtering as above, in silico visualization was performed, applying in-house software to manually inspect each variant for false-positive calls. Variants found to be false-positive upon manual inspection were removed. SMARCC1 variant annotations were then confirmed through manual cross-reference in the UCSC Genome Browser.43,50 Reported variants passing these filters and manual inspection in SMARCC1 were further confirmed by Sanger sequencing.
Developmental human brain single cell RNA-seq dataset analysis
As described previously,26 the preprocessing and clustering analysis for single cell RNA (scRNA) developmental human brain dataset was completed using Seurat.51 Briefly, cells with fewer than 1000 genes/cell were removed, as were cells with >10% of their individual transcriptome represented in either mitochondrial or ribosomal transcripts. Only genes expressed in at least 30 cells were carried forward in the analysis. The raw counts were normalized and log2 transformed by first calculating ‘size factors’ that represented the extent to which counts should be scaled in each library. Highly variable genes were detected using the proposed workflow of and were subsequently used for unsupervised dimensionality reduction techniques and principal component analysis. Uniform Manifold Approximation and Projection (UMAP) coordinates were calculated using standard Seurat workflow, and clusters were assigned to cells based on previous analysis via a hybrid method using Louvain clustering and weighted gene co-expression network analysis (WGCNA).26 Non-parameteric Wilcoxon rank sum test was used to identify differentially expressed markers across time points, areas and laminar zones by running FindAllMarkers. Heat map expression values were calculated using AverageExpression function and visualization of the heat maps were created using pheatmap package.
Cell type enrichment
Cell type enrichment for the expression of SMARCC1 was tested in scRNA-seq datasets of prenatal human brain using in-house custom-made script in R studio.52,53 Enrichment for each cell type was tested using hypergeometric test, where a gene list was significantly enriched in a cell type if the adjusted P-value was <0.05. Average expression was shown using the DotPlot function from the Seurat package. For control comparison, we used frontal neocortex layer-specific data from the BrainSpan database matched by developmental age.52 Batch correction was applied by quantile normalization in the limma package.54 Median fold changes of gene expression were used to rank the genes. Only protein-coding genes were used for analysis.
SMARCC1 expression in PsychENCODE bulk RNA sequencing
To examine the expression pattern of SMARCC1 during human brain development, we extracted reads per kilobase per million (RPKM) expression from the PsychENCODE bulk tissue RNA sequencing dataset.53 Gene expression was scaled, centred and average values calculated across developmental periods. The expression distributions were visualized in a violin plot.
Gene ontology enrichment analysis
To test for functional enrichment for all modules and the respective genes, we performed gene ontology enrichment analysis (GOEA) for using the gene ontology (GO) set of biological processes. Gene set ‘GO. v5.2.symbols_mouse.gmt’ was obtained from the Molecular Signatures Database. The compareCluster and enrichGo functions from the R package ClusterProfiler (version 3.12.0) were used to determine significant enrichment (q < 0.05) of biological processes. All present genes were used as background (universe). To focus only on neurological gene sets, GO term gene sets were selected for terms including the term ‘neuro’, ‘neural’ and ‘nerv’. Network visualization was performed using the cnetplot function from the R package ClusterProfiler.
Acquisition, preprocessing and differential gene expression analysis of post-mortem tissue
A stillborn male fetus was born at Medstar Washington Hospital Center via induced vaginal delivery at a clinical gestation age of 20 weeks for pregnancy termination due to sonographic and MRI findings of supratentorial ventriculomegaly/and hydrocephalus suggestive of aqueductal stenosis. The mother had a previous history of pregnancy termination at 22 weeks due to a similar fetal anomaly. The patient underwent autopsy assessment at Children’s National Medical Center at the request of the parents, which identified body weight, crown-rump length, crown-heel length, foot length, and organ weights consistent with 20–23 weeks gestation. Representative sections of the cortex, brainstem and cerebellum were retained for analysis at the request of and with the written consent of the patient’s parents. Written informed consent for de-identified genetic testing of samples from the patient and direct family members was obtained. All written consent was obtained in accordance with the Declaration of Helsinki and in compliance with the Human Investigation Committee at Yale University.
Both the patient and the patient’s mother were found to have an amino acid substitution in SMARCC1 c.1723C>T, yielding p.Gln575*. Tissue processing was performed using the Gentra Puregene Tissue Kit. The SMARCC1 bulk-tissue RNA-seq data were then aligned to the hg38 genome assembly and GENCODE v21 gene annotation using STAR, followed by read counting via HTSeq. The count data were then used to compute RPKM in accordance with methods used in the BrainSpan dataset,53 enabling downstream gene expression comparisons between these two datasets.
We used gestational age-matched neocortical samples from the BrainSpan dataset to identify up- and downregulated genes in the SMARCC1-variant cortex. For the downstream analysis, we examined only protein-coding genes, and further removed mitochondria and histone genes as these could bias the analysis. As there was only one cortical sample available, traditional differential gene expression analysis would not be feasible. As an alternative, we performed pairwise comparisons between the SMARCC1 neocortical data and the data of each neocortical area in BrainSpan, and calculated the expression fold changes, with a pseudo-value of 1 added to both numerators and denominators. The median expression fold changes among these pairwise comparisons were compared. To identify the most robust changes, we set the threshold of fold changes at log5 for the upregulated genes and −log5 for the downregulated genes. GOEA was performed on the top 200 up- and downregulated genes using the topGO package in R,55 and the significant terms were selected with a false discovery rate (FDR) cut-off of 0.01.
Xenopus husbandry
Xenopus tropicalis were raised and cared for in our aquatics facility according to protocols approved by the Yale University Institutional Animal Care and Use Committee. Embryos were staged according to Nieuwkoop and Faber.56
Single guide RNA and RNA production
CRISPR
Two non-overlapping CRISPR single guide (sg)RNAs were designed on crisprscan.org for the X. tropicalis smarcc1 gene (Xenbase genome v9.1) and produced using an EnGen sgRNA Synthesis Kit (NEB # E3322). Target sites are located in exon 10 (CRISPR #1: 5ʹ-AGGCTGTGCGCAGTCCCGAGAGG-3ʹ) and exon 1 (CRISPR #2: 5ʹ-CGGCCGGGAAGAGCCCCGCAGGG-3ʹ. CRISPR indels were verified by performing Sanger sequencing on PCR products using genomic DNA from stage 46 embryos. Genomic DNA was extracted from individual anaesthetized embryos at phenotypic stage 46 by 10 min incubation in 50 μl of 50 mM NaOH at 95°C followed by neutralization with 20 μl of 1 M Tris pH 7.4. PCR was performed with either Phusion High-Fidelity DNA Polymerase (NEB #M0530) (CRISPR #1) or Platinum SuperFi II Green PCR Master Mix (Thermo Fisher Scientific #12369050) (CRISPR #2) using primers around the CRISPR cut site (CRISPR #1: 5ʹ-ACATTGGTCCCTGTGCTTTT-3ʹ and 5ʹ-TTCAAGTCCTCGTCTGTTTGG-3ʹ, CRISPR #2: 5ʹ-AACGGCAGCAATAACGGAGA-3ʹ and 5ʹ-AGATACATGTCCCCTCCGCA-3ʹ). PCR sequences were analysed for indels using the online Inference of CRISPR Edits (ICE) tool (Synthego).
Human mRNA
Human SMARCC1 mRNA was produced by cloning a full-length insert (sequence ID NM_003074.4) into a pCS DEST expression plasmid backbone using Gateway recombination techniques. mRNA was synthesized using a mMESSAGE mMACHINE SP6 Transcription Kit (Thermo Fisher Scientific #AM1340). The patient variant 1723T>C (p.Q575*) was produced using inverse PCR. Overlapping primers (forward and reverse) were designed with the base change located in the middle, with 14 bases on either side of the variant. Long range PCR was performed with wild-type plasmid template using Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific # 11304011). DpnI digestion removed methylated template DNA from non-methylated PCR product, which was then used to transform Ca2+-competent Escherichia coli. Individual clones were grown up and plasmid DNA was extracted and sequenced to verify the presence in the insert of only the desired variant. The insert was cloned into a pCS DEST expression vector and mRNA was synthesized as described earlier.
Microinjection, gene expression, knockdown and overexpression
Xenopus tropicalis embryos were microinjected using standard protocols.57-59 Fertilized eggs were injected at either one cell stage with a 2 nl volume, or into one of two cells at two cell stage with a 1 nl volume. Injection mixes for expression knockdown using morpholino oligo (MO) consisted of the fluorescent tracer Dextran Alexa Fluor 488 (Thermo Fisher Scientific #D22910) along with MO targeting the start site of X. tropicalis smarcc1 (gene model XM_002942718.5) (5ʹ-CCTTTGTTTCATGGCTGCTACTCCC-3ʹ, Gene Tools) at a concentration leading to a dose of 0.5–1.0 ng per embryo. A standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′, Gene Tools) was also used at the same dose. We also used CRISPR/Cas9-mediated gene editing for expression knockdown, as previously described.60 CRISPR injection mixes consisted of fluorescent tracer and the following components at a concentration leading to the listed dose per embryo: 1.6 ng Cas9 (CP03, PNA Bio), 400 pg sgRNA. 500 pg mRNA encoding human wild-type or patient variant protein was injected into each embryo to rescue MO knockdown. Injections were verified using a Zeiss SteREO Lumar.V12 microscope to visualize fluorescence at stages 18–46 in the entire embryo (one cell injection) or on only the right or left side (two cell injection).
Optical coherence tomography imaging
A Thorlabs Ganymede II HR optical coherence tomography (OCT) imaging system using ThorImage OCT version 5.0.1.0 software was used to obtain 2D cross-sectional images and movies. Imaging was obtained as previously described.58
Western blotting
Whole cell lysate was extracted from pooled embryos in RIPA lysis buffer (Millipore #20-188) supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific #78440). Samples were run on Bolt 4–12% Bis-Tris Plus gels (Thermo Fisher Scientific) then transferred to PVDF membranes. Immunoblotting was performed using standard methods. Polyclonal rabbit anti-SMARCC1 antibody (Thermo Fisher Scientific #PA5-96513) was used at 1:1000 and mouse anti-b-Actin (C4) HRP (Santa Cruz Biotechnology #sc47778) was used at 1:100 000 as a loading control.
Whole mount in situ hybridization
Whole mount in situ hybridization (WISH) was performed as previously described.61 Briefly, X. tropicalis embryos were fixed in 4% paraformaldehyde (PFA) in 2 mM EGTA, then dehydrated through methanol washes and stored at −20°C. Embryos were rehydrated through washes in PBS + 0.1% Tween-20, then incubated in 4% hydrogen peroxide in PBS + 0.1% Tween-20 to remove pigment. After post-fixing in 4% PFA in 2 mM EGTA, embryos were hybridized overnight at 60°C with RNA probes. The Digoxigenin-11-UTP (Sigma #11209256910) labelled RNA probes were produced with full-length insert containing expression plasmids using either HiScribe T7 (antisense) or HiScribe SP6 (sense) RNA Synthesis Kits (NEB) according to the manufacturer’s instructions (Table 2).
Table 2.
mRNA synthesis
| Gene | Reference sequence | Expression vector | Linearization restriction enzyme |
|---|---|---|---|
| Smarcc1 | CU075511.1 | pCS 107 | AgeI |
| Neurod2 | NM_001079018.1 | pCMV SPORT 6 | EcoRI |
| Mab21l2 | BC136175.1 | pCMV SPORT 6 | EcoRV |
After overnight hybridization, embryos were washed, blocked then incubated overnight in Anti-Digoxigenin-AP, Fab fragments (Sigma #11093274910). After washes, embryos were incubated in BM Purple (Sigma # 11442074001) until signal was fully visible, then fixed in 4% PFA + 0.1% glutaraldehyde in 2 mM EGTA.
Immunohistochemistry
Uninjected control, one of two cell control MO, or one of two cell smarcc1 MO-injected stage 47 embryos were anaesthetized, then fixed in 4% PFA in PBS for 1 h at room temperature. After washes in PBS, tails, guts and ventral structures of the head including lower jaws and facial cartilage were removed. Pigment was bleached from samples by incubation in 5% formamide + 1.2% H2O2 in PBS while exposed to light. Samples were then washed with PBS + 0.1% Triton X-100 (PTr), blocked in 10% CAS-Block (Thermo Fisher Scientific #008120) in PTr, then incubated in mouse monoclonal anti-PCNA (PC10) (Thermo Fisher Scientific #13-3900) diluted 1:200 in 100% CAS-Block overnight at 4°C. After extensive washes in PTr, samples were blocked in 10% CAS-Block, then incubated overnight at 4°C in Texas Red-conjugated goat anti-mouse IgG (Thermo Fisher Scientific #T-6390) plus Hoechst 33342 (Thermo Fisher Scientific #H3570) diluted 1:200 and 1:5000, respectively in 100% CAS-Block. Samples washed first in PTr, then in PBS, were mounted between two coverslips in ProLong Gold Antifade Mountant (Thermo Fisher Scientific #P36934). Images were obtained using a Zeiss LSM 880 airyscan confocal microscope.
RNA sequencing analysis
Sequenced reads were aligned and quantified using STAR: ultrafast universal RNA-seq aligner (version 2.7.3a) and the murine reference genome, GRCm38p5, from the Genome Reference Consortium. Raw counts were imported using the DESeqDataSetFromHTSeqCount function from DESeq2 (version 1.26.0) and rlog-transformed according to the DESeq2 pipeline. DESeq2 was used for calculation of normalized counts for each transcript using default parameters. All normalized transcripts with maximum overall row mean < 20 were excluded, resulting in 13 284 present protein-coding transcripts. Undesired or hidden causes of variation, such as batch and preparation date, were removed using the sva package. The normalized rlog-transformed expression data were adjusted with four surrogate variables identified by sva using the function removeBatchEffect from the limma package. To determine gene clusters, CoCena (Construction of Co-expression network analysis) was calculated based on Pearson correlation on all present genes. Pearson correlation was performed using the R package Hmisc (version 4.1-1). To increase data quality, only significant (P < 0.05) correlation values were kept. A Pearson correlation coefficient cut-off of 0.803 (present genes; 10 260 nodes and 69 986 edges) was chosen, resulting in networks following the power-law distribution of r2 = 0.934 (scale-free topology). Unbiased clustering was performed using the ‘leiden modularity’ algorithm in igraph (version 1.2.1). Clustering was repeated 100 times. Genes assigned to more than 10 different clusters received no cluster assignment. The mean group fold change expression for each cluster and condition is visualized in the Cluster/Condition heat map. Clusters smaller than 40 genes are not shown.
Single-cell gene expression from age-matched Smarcb1-mutant (n = 44 755) and wild-type (n = 27 230) mouse brains was obtained from a publicly available dataset of cells (Gene Expression Omnibus: GSE212672). Seurat version 4.0.3 was used to cluster cells based upon previously described cell types. The resulting Seurat object was then imported into Monocle 3 version 0.2.3.62 A Monocle 3 cell dataset was constructed and a Moran’s I test was applied to identify differential gene expression in the cell dataset based on low dimensional embedding and the principal graph with neighbour graph = knn, reduction method = UMAP, k = 25, alternative = greater, method = Moran_I and expression_family = quasipoisson. To infer cell–cell interactions based on the expression of known ligand–receptor pairs in different cell types, CellChat63 was applied. The official workflow and databases were implemented. Briefly, the normalized counts were loaded into CellChat, after which the preprocessing functions identifyOverExpressedGenes, identifyOverExpressedInteractions and project Data with standard parameters set were applied. The main analyses were conducted using the functions computeCommunProb, computeCommunProbPathway and aggregateNet with fixed randomization seeds.
Statistics and reproducibility
No power analysis was performed to predetermine sample size, as our sample sizes are similar to those reported in previous publications.15,16,64 Randomization was not relevant to this study as controls and X. tropicalis knockdowns did not receive different treatments and human studies were descriptive studies. All experiments were performed and analysed in a blinded manner. No data were excluded from the analyses. Wilcoxon rank-sum test was used in differential gene expression analysis, as described. Mann-Whitney test was used to analyse experimental data. Elsewhere, data distribution was assumed to be normal, but this was not formally tested.
Results
The total sequenced cohort consisted of a of 8091 exomes (2697 trios) with cerebral ventriculomegaly. This included 2416 new trios from a clinical referral cohort (GeneDx) and 281 trios from an academic neurosurgical cohort (Harvard-Yale) (refer to the ‘Materials and methods’ section). Among the latter, 49 new trios were added to a cohort previously described.15,16 The control cohort consisted of 1798 exomes from unaffected siblings of people diagnosed with ASD and their unaffected parents sourced from the Simons Simplex Collection.15,65 Genomic DNAs were subjected to WES, and variant calling was performed with GATK HaplotypeCaller and Freebayes followed by ANNOVAR annotation and confirmation by the Integrative Genomics Viewer.16,43,50 Reported variants were confirmed by Sanger sequencing.
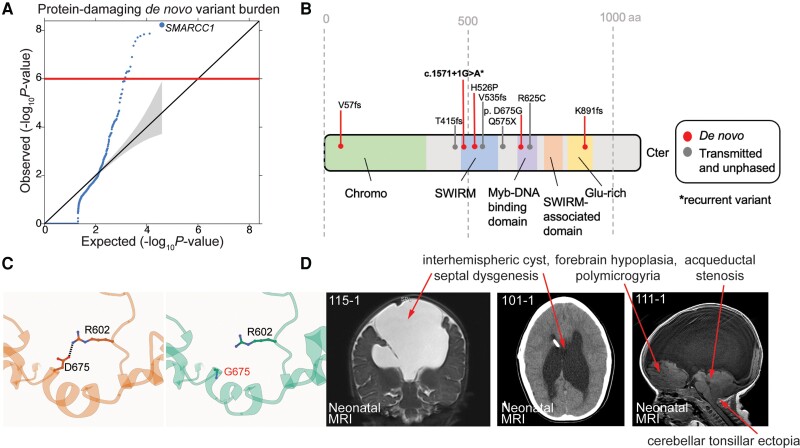
We compared observed and expected numbers of non-synonymous DNVs in all genes in cases and controls. In the Harvard-Yale cohort, three missense or LoF variants in SMARCC1 were identified, yielding a protein-altering DNV burden of 3.92 × 10−8 that surpassed the threshold for exome-wide significance (multiple-testing correction threshold of 8.57 × 10−7 after correction for testing 19 347 RefSeq genes in triplicate using a one-tailed Poisson test). These variants included the previously described variants p.His526Pro, p.Lys891Argfs*6 and c.1571+G>A variants (Table 1).15,16 In the GeneDx cohort, three new protein-altering DNVs in SMARCC1 were identified, including the c.2204A>G (p.Asp675Gly), c.170delT (p.Val57Alafs*97) and recurrent c.1571+1G>A variants, yielding a DNV burden of 7.39 × 10−4. Based on the presence of six total DNVs in the Harvard-Yale and GeneDx cohorts (2697 total patient-parent trios), SMARCC1 carried a protein-altering DNV burden of 5.83 × 10−9, surpassing the threshold for exome-wide significance (Fig. 1A). Among SMARCC1 non-synonymous missense DNVs, p.His526Pro is predicted to abolish interaction with the backbone carbonyl oxygen of p.Leu505 at the end of an adjacent helix in the SWIRM domain mediating BAF complex subunit interactions.66 The p.Asp675Gly variant alters a conserved residue in the Myb domain resulting in an unfavourable loss of an ion pair interaction with p.Arg602 (Fig. 1C). All these DNVs are absent in gnomAD and Bravo databases.
Figure 1.
SMARCC1 mutations are associated with congenital hydrocephalus and cause a novel human BAFopathy featuring cerebral ventriculomegaly. (A) Quantile–quantile (Q-Q) plot of observed versus expected P-values for de novo variants (DNVs) in each gene in 2697 trio cases. P-values were calculated using a one-sided Poisson test (refer to the ‘Materials and methods’ section). For protein-damaging de novo SMARCC1 variants [loss-of-function (LoF), MetaSVM = D and/or MPC > 2], P = 5.83 × 10−9. (B) Schematic diagram showing variant locations in SMARCC1 protein domains. Identified DNVs, transmitted and unknown inheritance variants15,16 are indicated with markers. *Recurrent variant. (C) The p.Asp675Gly variant was predicted to be detrimental to SMARCC1 structure and function by alpha-fold biophysical modelling. Structural protein modelling predicts that p.Asp675Gly alters a conserved residue in the Myb domain resulting in loss of an ion pair interaction with p.Arg602, with a predicted ΔG of 0.73 kcal/mol. (D) Brain MRIs of congenital hydrocephalus patients with SMARCC1 variants demonstrate consistent structural abnormalities. Prenatal imaging is shown for Patients 115-1 (contrast MRI), 101-1 and 111-1. Red asterisks denote ventricular catheter of a ventriculo-peritoneal shunt used to treat obstructive hydrocephalus.
We examined the clinical phenotypes of probands harbouring SMARCC1 DNVs and other published rare, damaging transmitted or unknown inheritance congenital hydrocephalus-associated SMARCC1 variants16 (Table 3). The latter included two transmitted LoF variants (p.Gln575* and p.Val535Serfs*29), one unknown inheritance rare LoF variant (p.Thr415Lysfs*29) and one transmitted rare damaging missense (D-Mis) variant (p.Arg652Cys). Strikingly, 10/10 had perinatally diagnosed cerebral ventriculomegaly, and at least seven required neurosurgical CSF diversion by endoscopic third ventriculostomy or ventriculoperitoneal shunting. Nine of 10 had aqueductal stenosis, 9/10 had partial or complete corpus callosum abnormalities, including septal agenesis, 9/10 exhibited moderate-to-profound DD and 9/10 had cardiac defects including atrial septal defect, ventricular septal defect, double outlet right ventricle and cardiac hypoplasia. Other neurodevelopmental phenotypes, such as seizures, structural brain defects like cerebellar tonsillar ectopia, and craniofacial defects including cleft palate, microtia and auditory canal atresia were variably present (Table 3 and Fig. 1D). These data suggest that SMARCC1 variants, in addition to conferring congenital hydrocephalus risk, lead to a novel human syndrome with phenotypes that resemble other BAFopathies.18,19,67,68
Table 3.
Phenotypic characteristics of de novo and transmitted SMARCC1 probands
| Proband ID | Type | cDNA change | AA change | CNS structural | CNS Functional | Other | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aqueductal stenosis | Corpus callosum abnormalities | Septal agenesis | Cerebellar tonsillar ectopia | Macrocephaly | Polymicrogyria | Developmental delay | Seizures | Cardiac defects | Craniofacial defects | ||||
| CHYD115-1a | De novo | c.2672delA | p.(Lys891Argfs*6) | + | + | + | + | + | + | + | + | + | − |
| CHYD168-1a | De novo | c.1577A>C | p.(His526Pro) | + | + | + | + | − | − | + | − | − | − |
| CHYD505-1a | De novo | c.1571+1G>A | - | + | + | + | − | + | − | + | + | + | − |
| CHYD451-1a | Transmitted | c.1954C>T | p.(Arg652Cys) | + | N/A | N/A | N/A | − | − | + | − | + | − |
| CHYD364-1a | Transmitted | c.1723C>T | p.(Gln575*) | + | + | + | − | − | − | − | − | + | − |
| CHYD111-1a | Transmitted | c.1602_1603 insAGTGGGGACTC |
p.(Val535Serfs*29) | + | + | + | + | + | + | + | − | + | + |
| CHYD101-1a | Unphased | c.1243_1244insAA | p.(Thr415Lysfs*29) | + | + | + | + | + | − | + | + | − | + |
| CHYDX1-1b | De novo | c.2204A>G | p.(Asp675Gly) | ++ | +++ | +++ | + | + | − | +++ | + | ++ | + |
| CHYDX2-1b | De novo | c.1571+1G>A | – | ||||||||||
| CHYDX3-1b | De novo | c.170delT | p.(Val57Alafs*97) | ||||||||||
| Total | − | − | – | 9/10 | 9/10 | 9/10 | 5/10 | 5/10 | 2/10 | 9/10 | 4/10 | 7/10 | 3/10 |
We also examined the clinical phenotypes of 13 individuals reported in the literature with rare, damaging SMARCC1 DNVs or transmitted variants (Supplementary Table 3), including four damaging de novo variants (p.Gln742Arg, p.Trp279*, p.Trp279*, c.2782-1G),69-71 four inherited damaging variants (p.Arg912*, p.Gln972Sfs*19, p.Gln956*, p.Lys615Ilefs*49),72,73 two damaging variants (p.Gln1005*, p.Asp821Glufs*4) of unknown inheritance and two exon deletions (deletion of exon 4 and deletion of exon 4–6)68 unknown inheritance. Interestingly, the patient with the de novo c.2782-1G variant had congenital hydrocephalus and aqueductal stenosis along with appendicular skeletal defects. Among the patients, 6/12 patients had DD, 4/12 had ASD and 3/12 patients had craniofacial defects and appendicular skeletal defects (as well as scoliosis and vertebral defects). Other developmental abnormalities included polymicrogyria, attention deficit hyperactivity disorder and seizures. These findings are consistent with the phenotypes of our cohort and further expand SMARCC1 phenotypic spectrum.
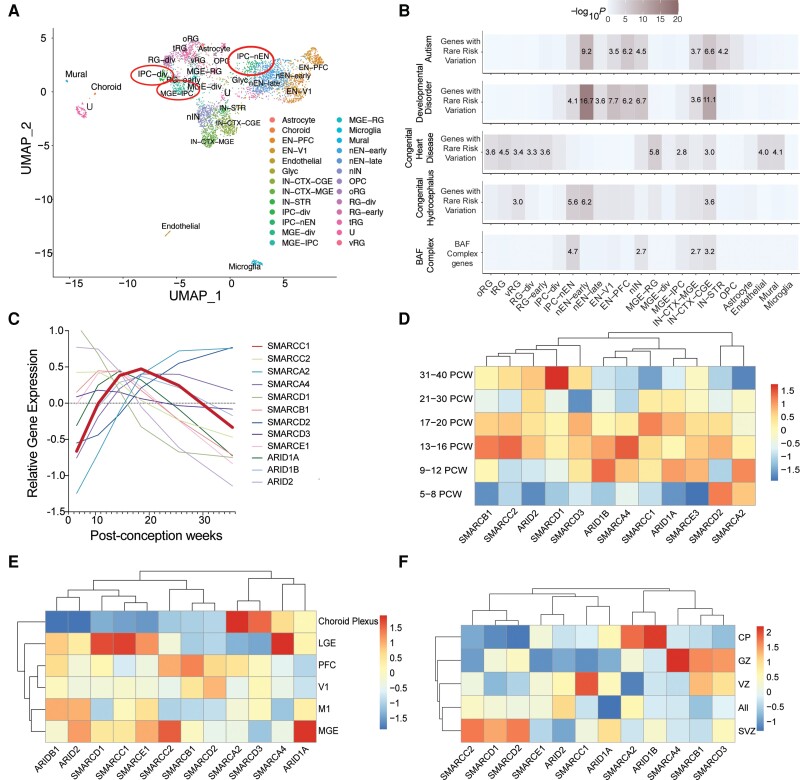
Smarcc1 is expressed in mouse ventricular zone neuroepithelial and neural progenitor cells during midgestation,10,15,74,75 a key epoch during which neurogenesis contributes to the development of the diencephalon and telencephalon.9,26,31,75-78 We studied the expression of SMARCC1 in the human brain during development using scRNA sequencing database of 4261 cells from developmental human whole brain tissue during post-conception weeks (PCW) 6–40.26 We found that SMARCC1 and other BAF complex genes are expressed highly in intermediate progenitor cells (IPCs) between PCW 13–20. (Fig. 2A–D). In addition, SMARCC1 is highly expressed in the lateral ganglionic eminence, an NPC niche within the ventral telencephalon that harbours NPCs destined for cortical and striatal interneurons and oligodendrocyte precursor cells (OPCs) (Fig. 2E).79 When investigating ventricular lamina expression, we found that BAF complex members are also expressed throughout the ventricular laminae. Further, SMARCC1 is most highly expressed in the ventricular zone (Fig. 2F). These data show that SMARCC1 is highly expressed in human fetal periventricular NPCs.
Figure 2.
SMARCC1 is expressed in intermediate progenitors of the cortical lamina during human brain development. (A) Analysed transcriptomic dataset26 showing uniform manifold approximation and projection (UMAP) clustering of developmental human brain cells, coloured by cell type. EN-PFC = early and late born excitatory neuron prefrontal cortex; EN-V1 = early and late born excitatory neuron V1; IN-CTX-CGE = caudal ganglionic eminence (CGE)/lateral ganglionic eminence (LGE)-derived inhibitory neurons; IN-CTX-MGE = medial ganglionic eminence (MGE)-derived cortex inhibitory neuron; IN-STR = striatal neurons; IPC-div = dividing intermediate progenitor cells radial glia (RG)-like; IPC-nEN = intermediate progenitor cells excitatory neuron (EN)-like; MGE-div = dividing MGE progenitors; MGE-IPC = MGE progenitors; MGE-RG = MGE radial glia; Mural = mural/pericyte; Nen-early = newborn excitatory neuron-early born; nEN-late = newborn excitatory neuron-late born; nIN = MGE newborn neurons; OPC = oligodendrocyte progenitor cell; oRG = outer radial glia; RG-div = dividing radial glia; RG-early = earlyvRG; tRG = truncated radial glia; U = unknown; vRG = ventricular radial glia. Expression in neural progenitors is featured in red circles. (B) Analysed transcriptomic dataset26 showing enrichment analysis across cell type markers of the developmental human brain for genes with rare risk variation in autism, developmental disorders, congenital heart disease and congenital hydrocephalus compared to BAF complex genes. Tiles labelled with −log10(P-value) and an asterisk represent significant enrichment at the Bonferroni multiple-testing cut-off (α = 0.05/23 = 2.17 × 10−3). (C) Temporal gene expression profiles for SMARCC1 and other BAF complex genes between post-conception weeks (PCW) 5–36. (D) Analysed transcriptomic dataset26 showing heat map of gene expression levels for SMARCC1 and other BAF complex genes across different developmental time points. Vertical axis shows time points in post-conception weeks. (E) Analysed transcriptomic dataset26 showing heat map of gene expression levels for SMARCC1 and other BAF complex genes across different brain regions. V1 = primary visual cortex; M1 = primary motor cortex. (F) Analysed transcriptomic dataset26 showing heat map of gene expression levels for SMARCC1 and other BAF complex genes across cortical lamina, PCW 5–40. CP = cortical plate; GZ = germinal zone; VZ = ventricular zone; SVZ = subventricular zone.
To functionally validate SMARCC1 as a novel disease gene, we generated Smarcc1 mutant X. tropicalis tadpoles since mice with Smarcc1 deletion are embryonic lethal.31-33 Additionally, brain morphogenesis and CSF circulation can be studied in live Xenopus tadpoles using CT (Supplementary Fig. 1).80,81 We knocked down Xenopus Smarcc1 using CRISPR/CAS9 targeting exon1 and exon10, as well as by using a morpholino oligo targeting the Smarcc1 transcription start site (Fig. 3A and B and Supplementary Fig. 2).
Figure 3.
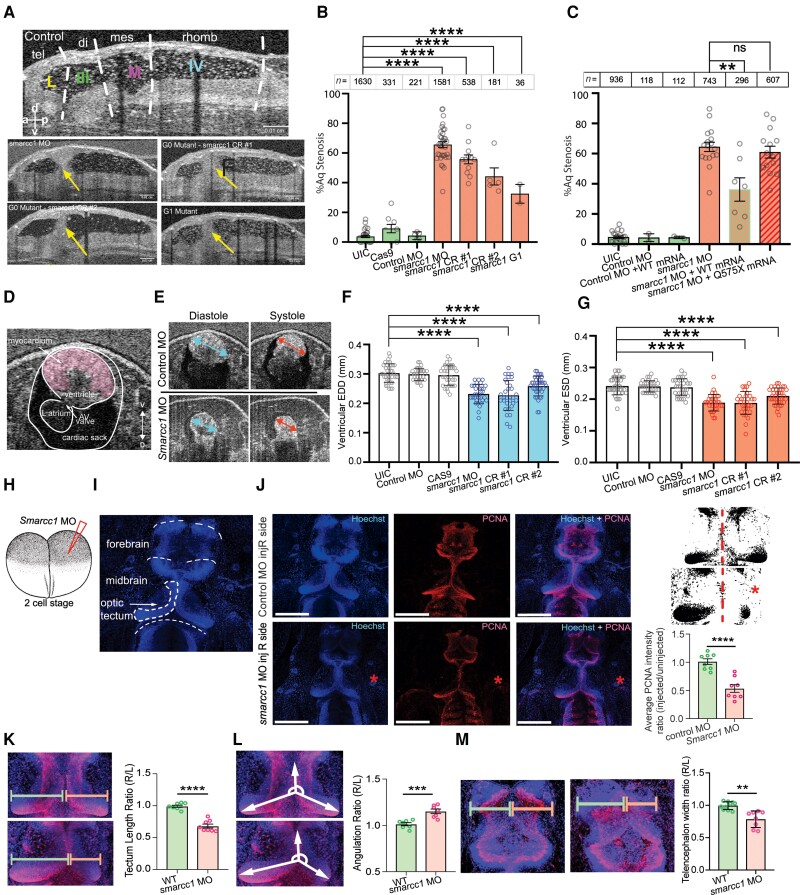
SMARCC1 mutation causes hydrocephalus by disrupting midbrain architecture in Xenopus tropicalis. (A) Mid-sagittal view of the Xenopus ventricular system. Dotted white lines indicate boundaries between labelled regions: tel = telencephalon; di = diencephalon; mes = mesencephalon; rhomb = rhombencephalon; L = lateral ventricle; III = third ventricle; M = midbrain ventricle; IV = fourth ventricle. Representative mid-sagittal views for experimental conditions (G0 variant from morpholino oligo, G0 variant from CRISPR #1, G0 variant from CRISPR #2, and G1 variant progeny from Smarcc1 MO animals) are shown with aqueductal occlusion marked by arrows. (B) Quantification of per cent aqueductal stenosis in uninjected controls (UIC); Cas9 control and control MO, as well as in the experimental conditions Smarcc1 MO, Smarcc1 CRISPR #1, Smarcc1 CRISPR #2 and Smarcc1 G1 variant. Data are shown as mean ± standard error of the mean (SEM). Open circles indicate the number of experiments, with animal counts indicated above each column. Significance was calculated by one-way ANOVA; ****P ≤ 0.0001. (C) Quantification of rescue of aqueductal stenosis phenotype with Smarcc1 MO + WT mRNA (P = 0.0024) with recapitulation of phenotype by pathogenic mRNA from p.Q575* variant (P = 0.4888). Data are shown as mean ± SEM. Open circles indicate number of experiments, with animal counts indicated above each column. Significance was calculated by Mann-Whitney test. (D) Representative X. tropicalis cardiac OCT image on ventral-dorsal axis, the ventral three chamber view (VTCV). Labelled structures are myocardium, ventricle, left atrium, AV valve and cardiac sack. (E) Representative cardiac measurements by OCT shown for UIC and smarcc1 MO. EDD = end diastolic diameter; ESD = end systolic diameter. (F) Quantification of EDD in UIC, Cas9 control and control MO, as well as experimental conditions smarcc1 MO, smarcc1 CRISPR #1 and smarcc1 CRISPR #2. Data are shown as mean ± SEM. Significance was calculated by one-way ANOVA using GraphPad Prism. ****P ≤ 0.0001. (G) Quantification of ESD in UIC, Cas9 control and control MO, as well as experimental conditions smarcc1 MO, smarcc1 CRISPR #1 and smarcc1 CRISPR #2. Data are shown as mean ± SEM. Significance was calculated by one-way ANOVA using GraphPad Prism. ****P ≤ 0.0001. (H) Schematic of two-cell injection protocol in X. tropicalis. (I) Labelled representative fluorescence microscopy of wild-type (WT) stage 46 X. tropicalis stained with Hoechst. Olfactory bulb, forebrain, midbrain, optic tectum and cerebellum are indicated. (J) Representative immunofluorescence images of right side-injected control MO and right side-injected Smarcc1 MO stage 46 X. tropicalis for PCNA (red) and merged images (with Hoechst, blue). Scale bar = 500 um. Schematic and chart for quantification of average PCNA intensity ratio for control and smarcc1 MO injected on the right side with left side uninjected, P ≤ 0.0001 with unpaired t-test. Data are shown as mean ± SEM. (K) Schematic and chart for quantification of optic tectum length ratio for wild-type control and Smarcc1 MO injected on the right side with left side uninjected, P ≤ 0.0001 with unpaired t-test. Data are shown as mean ± SEM. (L) Schematic and chart for quantification of optic tectum angulation ratio for wild-type control and Smarcc1 MO injected on the right side with left side uninjected, P = 0.0008 with unpaired t-test. Data are shown as mean ± SEM. (M) Schematic and chart for quantification of telencephalon width ratio for wild-type control and Smarcc1 MO injected on the right side with left side uninjected, P = 0.0019 with unpaired t-test. Data are shown as mean ± SEM. ns = not significant; OCT = optical coherence tomography.
All three resulting Smarcc1 mutant and morphant tadpoles exhibited highly penetrant aqueductal stenosis that was transmitted to G1 mutant progeny (Fig. 3A and B). Despite this, OCT imaging demonstrated intact ependymal cilia-driven CSF circulation (Supplementary Fig. 3). Overexpression of human wild-type SMARCC1 but not human CH-variant SMARCC1 p.Gln575* in Smarcc1-depleted X. tropicalis variants (refer to the ‘Materials and methods’ section) rescued aqueductal stenosis (Fig. 3C andSupplementary Fig. 4). Smarcc1-depleted variants also exhibited decreased cardiac function resembling hypoplastic cardiomyopathy (e.g. see proband CHYD111-1), as quantified by end-diastolic diameter and end-systolic diameter (Fig. 3D–G). These results show congenital hydrocephalus-associated SMARCC1 variants or Smarcc1 depletion in Xenopus phenocopies the core brain and cardiac pathology of humans with SMARCC1 variants.
To begin to elucidate the cellular pathogenesis of SMARCC1-variant hydrocephalus, we leveraged the fate patterning of Xenopus, in which embryos at the two-cell stage can be selectively injected with MO on one side of the organism and then compared with the opposite side injected with nonsense MO as an isogenic control (Fig. 2H). PCNA immunostaining (a marker of cellular proliferation) at stage 46 showed Smarcc1 variants have significantly fewer PCNA+ periventricular cells on the MO-injected side compared to the control side (Fig. 3I–L). This was particularly evident in the midbrain and tectum, structures situated dorsal and ventral to the aqueduct, respectively. The length of the tectum in Smarcc1-depleted tissue was markedly reduced and its angulation with the anterior portion of the midbrain was altered, indicating significant dysmorphology or atresia. Forebrain thickness was also significantly reduced on the MO-injected versus the control side (Fig. 3M). These data are consistent with cortical and midbrain dysgenesis secondary to the impaired proliferation of SMARCC1-variant NPCs.
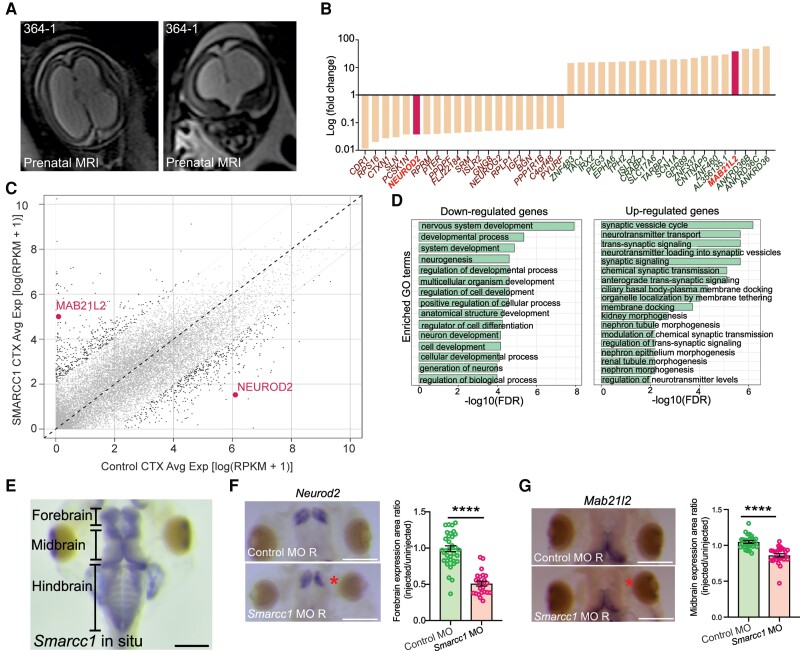
To gain insight into the molecular impact of SMARCC1 variants in humans, we performed bulk RNA-seq analysis on human frontal and motor-sensory cortex tissue from severely hydrocephalic proband CHYD364-1 (p.Gln575*), who unfortunately underwent fetal demise at PCW 20 (Fig. 4A). We compared these results to a spatial and developmental time-matched RNA-seq control dataset from the BrainSpan database.53,82 Analysis of differentially expressed genes (DEGs) identified several genes with significantly higher or lower expression compared to control samples (Fig. 4B and C). GO analysis of significantly downregulated DEGs showed enrichment in multiple terms related to structural neurodevelopment, including ‘nervous system development’, ‘neurogenesis’ and ‘regulation of cell development’, whereas significantly upregulated DEGs were enriched for terms related to neural transport and signalling (Fig. 4D).
Figure 4.
SMARCC1 mutation dysregulates transcription factors involved in intermediate progenitor biology in human and Xenopus tropicalis. (A) Prenatal ultrasound imaging for Patient CHYD364-1 demonstrates severe cerebral ventriculomegaly and aqueductal stenosis. (B) Median fold-change of top 20 differentially expressed genes. NEUROD2 and MAB21L2 are highlighted. (C) Dot plot showing differentially expressed genes between SMARCC1 variant and control CTX samples. Each dot represents a gene. The x- and y-axes represent average gene expression in control and variant samples, respectively. Genes with a fold change >5 between samples are in black; others are in grey. MAB21L2 and NEUROD2 are highlighted with text. (D) Gene ontology (GO) analysis of congenital hydrocephalus risk genes, including ranked and selected terms. Significance was calculated by two-sided Fisher’s exact test. Scale bar = 500 μm. (E) Representative photomicrograph of DNA in situ hybridization showing Smarcc1 expression in wild-type stage 46 of X. tropicalis. The forebrain, midbrain and hindbrain are indicated. Scale bar = 500 μm. (F) Representative photomicrographs of DNA in situ hybridization showing Neurod2 expression in stage 46 X. tropicalis, control and Smarcc1 MO-injected on the right side, with left side uninjected. Quantification of forebrain expression area is shown, P ≤ 0.0001 with unpaired t-test. Data are shown as means ± SEM. Scale bar = 500 μm. (G) Representative photomicrographs of DNA in situ hybridization showing Mab21l2 expression and quantification as in F. Scale bar = 500 μm.
Among the most significant DEGs were NEUROD2 and MAB21L2, transcription factors with human orthologues in Xenopus, which have been shown in both mice and Xenopus to play critical roles in brain morphogenesis via NPC regulation83-86 (Supplementary Fig. 5). Examination of the expression profiles of NEUROD2 and MAB21L2 in human prenatal scRNA-seq datasets revealed highly enriched expression in intermediate progenitor cells (IPC1), the same cell type with robust SMARCC1 expression during early brain development (Supplementary Fig. 6). WISH showed MO-mediated Smarcc1 depletion in the two-cell model caused a significant reduction of Neurod2 and Mab21l2 on the MO-injected versus control side (Fig. 4E–G). These results suggest SMARCC1 variants in congenital hydrocephalus may cause cortical and midbrain dysgenesis by altering the expression of key transcription factors, including Neurod2 and Mab21l2, that are involved in the regulation of the growth and proliferation of NPCs.
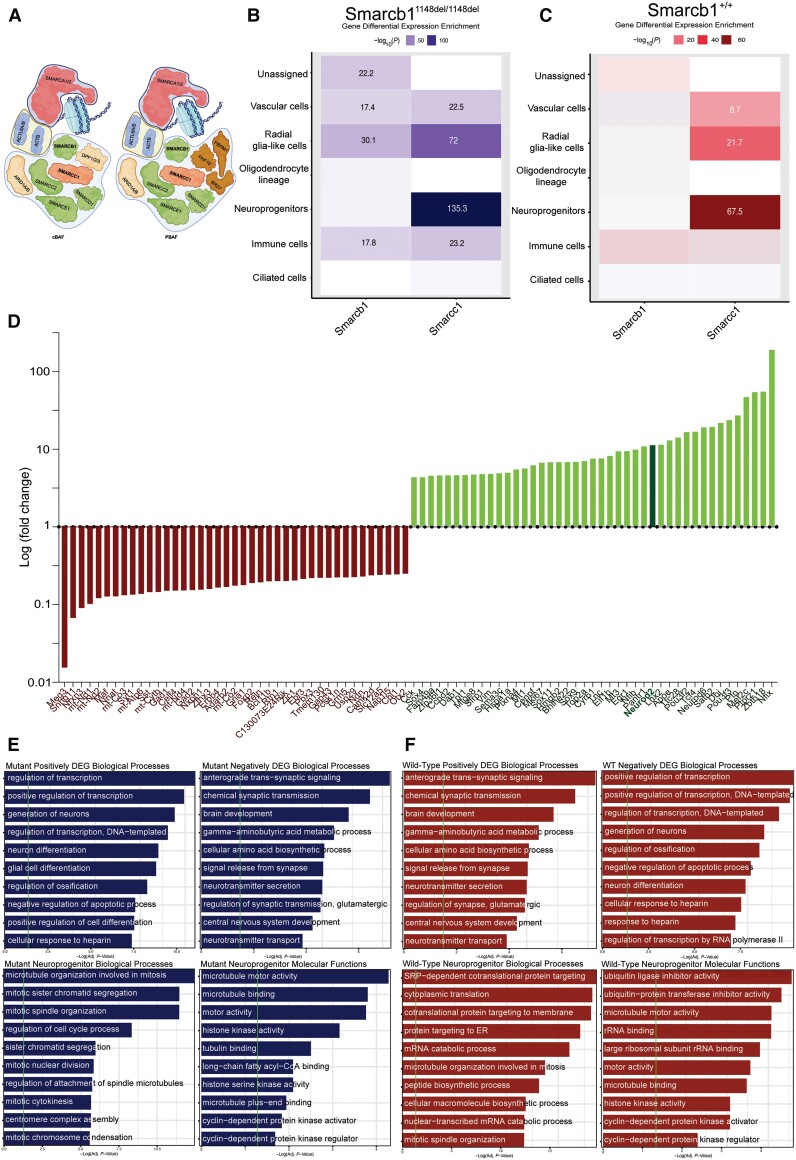
Given the limitations of studying the brain transcriptome in the very rare scenario of a single human fetus with SMARCC1 mutation above, we studied a scRNA-seq atlas of 71 985 individual cells from the brains of age-matched wild-type and Smarcb1-mutant mice.87 The non-truncating variant in Smarcb1, encoding a core subunit of the SWI/SNF complex that interacts with Smarcc1 (Fig. 5A), leads to an elongated Smarcb1 protein product that causes severe congenital hydrocephalus with high penetrance during fetal development.
Figure 5.
Altered Smarcc1 expression in NPCs and disrupted neurogenesis in Smarcb1-mutant hydrocephalic mice. (A) Schematic of the human SWItch/Sucrose Non-Fermentable (SWI/SNF complex), highlighting SMARCC1 and SMARCB1, core subunits of the complex. (B) Differential expression of Smarcc1 and Smarcb1 by cell type in Smarcb1-mutant mice. Significant values are labelled numerically. (C) Representation of positively and negatively differentially expressed genes (DEGs) by log(fold-change) in hydrocephalic Smarcb1-mutant mice relative to wild-type. Neurod2 is highlighted. (D) Smarcb1-mutant positively and negatively DEGs gene ontology (GO) biological processes. Vertical green line indicates statistical significance corrected by the Benjamini-Hochberg method. (E) Smarcb1-mutant NPC-type marker GO biological processes and molecular functions. Vertical green line indicates statistical significance corrected by the Benjamini-Hochberg method. NPC = neural progenitor cell.
Consistent with our findings from the patient above, in this model we found Smarcc1 expression was highly enriched in NPCs in both Smarcb1-mutant and wild-type mice. Interestingly, Smarcc1 had significantly higher expression in NPCs of Smarcb1-mutant mice (P = 1.99 × 10−135) compared to NPCs in their wild-type counterparts of the same age (P = 3.16 × 10−67) (Fig. 5B and C). Analysis of DEGs identified genes with significantly higher or lower expression in Smarcb1-mutant mice compared to wild-type; among the highest of these Neurod2, as in the human patient (Fig. 5D). GO analysis of significantly downregulated DEGs showed enrichment in multiple terms related to structural neurodevelopment as in the human patient, including ‘brain development’ (Padj. = 3.70 × 10−5) and ‘axon guidance’ (Padj. = 8.49 × 10−4) whereas significantly upregulated DEGs were enriched for terms related to neural precursor replication and differentiation including ‘regulation of stem cell proliferation’ (Padj. = 1.83 × 10−4) and ‘positive regulation of cellular differentiation’ (Padj. = 1.67 × 10−7) (Fig. 5E and F). Together, these findings suggest Smarcc1 and its interacting SWI/SNF complex components are key regulators of the NPC transcriptome during fetal brain development, and mutational disruption of this pathway in NPCs results in impaired neurogenesis and the development of aqueductal stenosis and cerebral ventriculomegaly and other structural brain defects.
Discussion
Our data provide evidence that DNVs in SMARCC1 cause a novel human syndrome characterized by cerebral ventriculomegaly and aqueductal stenosis, DD and other associated structural brain and cardiac defects. We propose the name ‘SMARCC1-associated developmental dysgenesis syndrome (SaDDS)’ to describe this novel BAFopathy. These results highlight the importance of SMARCC1 and the BAF chromatin remodelling complex in human brain morphogenesis and provide further support for a ‘neural stem cell’ paradigm of human congenital hydrocephalus.34,35,88,89 These data also demonstrate the power of trio-based WES for identifying pathogenic variants in sporadic structural brain disorders and suggest WES may be a useful prognostic adjunct when evaluating the surgical candidacy of congenital hydrocephalus patients.
The variant spectrum of SMARCC1 involving its BAF complex-interacting SWIRM, Chromo and Myb-DNA-binding domains suggests that these variants lead to functional impairment of the BAF complex.15,16,66 Indeed, neurodevelopmental disorders (NDDs) with overlapping clinical phenotypes are associated with pathogenic variants in other subunits of the BAF chromatin remodelling complex. Thus, the SMARCC1-associated condition presented here partially overlaps with BAFopathies such as CSS and Nicolaides-Baraitser-like syndrome, characterized by ID/DD and neurobehavioural abnormalities. In support of this, several individuals with deleterious SMARCC1 variants have been reported in other cohorts ascertained on the basis of congenital anomalies, including neural tube defects,70 autism72 or congenital heart disease (CHD)73 and others68,69,71 (Supplementary Table 3). However, a defining phenotype of patients with SMARCC1 variation appears to be cerebral ventriculomegaly associated with aqueductal stenosis that often requires neurosurgical intervention. This phenotype is likely over-represented in our patients, insofar as recruitment for our cohort was based on the presence of cerebral ventriculomegaly, including those treated with neurosurgery. Nonetheless, other SWI/SNF complex members including SMARCB190 and ARID1A91 have been implicated in cerebral ventriculomegaly, and knockout mouse models of Smarcb1 and Smarca4, encoding binding partners of Smarcc1, also exhibit congenital hydrocephalus,87,92 similar to the new Smarcc1 mutant frogs reported here. These findings support the pathogenicity of the reported SMARCC1 variants in the pathogenesis of cerebral ventriculomegaly. These findings are in line with the known phenotypic heterogeneity and incomplete penetrance of other BAFopathy genes.20,67,68,93 As more patients are collected, the full phenotypic spectrum of the human SMARCC1 syndrome will be clarified and genotype-phenotype correlations may arise.
Our results suggest incomplete penetrance and variable expressivity for some SMARCC1 variants, a phenomenon well recognized for other congenital hydrocephalus and ASD risk genes,15,16 as well as for other BAF complex genes.20,67,94,95 Mechanistic drivers of incomplete penetrance and variable expressivity in this specific context remain unclear but may include common genetic or environmental modifiers, such as inflammatory or oxidative triggers,96,97 as well as stochastic components resulting in mosaicism.98 As the paralogous SMARCC1 and SMARCC2 gene products form hetero- or homodimers17 and share functional scaffolding properties,30 upregulation of SMARCC2 or other BAF members could compensate for the loss of SMARCC1, possibly leading to less severe phenotypes. As different SMARCC1 isoforms are expressed in different tissues and at different developmental stages,15,26,82 both the spatio-temporal profile and isoform of the protein containing the variant could affect the phenotype. Further, the wide phenotypic variety and lack of clear genotype-phenotype correlation characterizing conditions associated with BAF-complex variants suggest possible gene dosage-dependent mechanisms affecting variants of the ATP-dependent chromatin remodelling machinery.22
The well orchestrated spatiotemporal regulation of BAF complex subunit assembly and activity is essential for the development and function of the CNS.28,99,100 BAF complexes specific to NPCs control cell proliferation, differentiation and survival through the epigenetic regulation of gene expression that is essential for telencephalon development.26,29 We showed that SMARCC1 is highly expressed in fetal human NPCs, and our analysis of a human SMARCC1-mutant hydrocephalic fetus suggests that SMARCC1 variants dysregulate the expression of genes critical for neurogenesis, including transcription factors NEUROD2 and MAB21L2. Similar results were derived from our analysis of the scRNA-seq atlas of Smarcb1-mutant hydrocephalic fetal brains. Interestingly, Smarcc1 expression was significantly increased in Smarcb1-mutant NPCs, perhaps compensating for the depletion of Smarcb1. Together, these data suggest SMARCC1 variants, possibly by altering the epigenetic regulation of gene expression, impair NPC growth and proliferation. Resultant attenuation of neurogenesis and gliogenesis could then lead to non-obstructive (ex vacuo) ventriculomegaly from a thinned cortical mantle, as has been shown with other congenital hydrocephalus-associated gene variants,88 and obstructive hydrocephalus secondary to midbrain dysgenesis and aqueductal stenosis. The latter is consistent with the fact that normal cerebral aqueduct development requires the precise regulation of the proliferation and differentiation of NPCs and the development of their associated fibre tracts in the mesencephalon following the prenatal closure of the neural tube.101-104 Notably, the BAF complex also modulates the expression of cardiac progenitor cells and regulates cardiac remodelling during development and repair.11-13,105 Inactivation of the complex has been implicated in cardiac hypertrophy, shortened or incomplete separation of outflow tracts, and persistent truncus arteriosus in rodents.11,106 In particular, a p.Lys615Ile frameshift variant in SMARCC1 has been associated with hypoplastic right or left heart syndrome in humans.73 Recent studies indicate that CHD patients exhibit increased prevalence of neurodevelopmental disabilities, and CHD patients with neurodevelopmental outcomes have a higher burden of damaging DNVs, particularly in genes important to both heart and brain development.107
Multiple ‘impaired brain plumbing’ mechanisms have been proposed to account for hydrocephalus, including increased CSF secretion, decreased intraventricular CSF transit from cilia dysfunction, and decreased CSF reabsorption associated with elevated venous pressure, arachnoid granulation immaturity and lymphatic dysplasia.108 However, accumulating genetic data15,16,88,109 suggest that impaired neurogenesis rather than overactive CSF accumulation may underlie some forms of congenital hydrocephalus. Our findings with SMARCC1 support such a ‘neural stem cell’ paradigm of disease.15,16,88 In the case of patients with variants in TRIM71 (another congenital hydrocephalus-associated gene), impaired post-transcriptional silencing of RNA targets leads to decreased NPC proliferation at even earlier time points, resulting in severe cortical hypoplasia and secondary ventriculomegaly.88 The opposite phenotype of NPC hyperproliferation due to PTEN variant and constitutive activation of the PI3K pathway may also occur.110
The diversity of genetic aetiologies and underlying biochemical pathways in congenital hydrocephalus supports the implementation of routine clinical WES for newly diagnosed patients. Current recommendations for work-up of fetal and neonatal ventriculomegaly include rapid commercial microarray testing for known chromosomal and copy-number abnormalities.111 However, this strategy does not address congenital hydrocephalus cases explained by recently detected variants in many new congenital hydrocephalus genes.15,16,88 Application of routine WES or whole genome sequencing could improve the management of children with congenital hydrocephalus by aiding prognostication and treatment stratification (including when or when not to operate); increasing vigilance for medical screening of variant-associated conditions (such as cancer surveillance for congenital hydrocephaluspatients with variants in PIK3CA or PTEN); and providing recurrence rates to increase reproductive confidence.
Our study is limited by the fact our cohort was ascertained on the basis of congenital cerebral ventriculomegaly, including treated hydrocephalus, given previous work in much smaller cohorts suggested a role for SMARCC1in these conditions.15,16 Continued identification of variant patients will expand our knowledge of the SMARCC1 phenotypic spectrum, including those patients without ventriculomegaly. Additional patients may also permit genotype-phenotype correlations. In addition, brain tissue for bulk RNA-seq was available from only one human subject, as access to SMARCC1-mutated fetal brain is such an exceedingly rare event. Using scRNA-seq on multiple human subjects and inclusion of biological replicates would be a goal of future investigations. Another limitation is the embryonic lethality phenotype of Smarcc1 knockout mice,31,32 which necessitated our use of the non-mammalian Xenopus model for disease modelling and mechanistic study.
These findings have clinical implications. These data suggest that detection of a de novo SMARCC1 variant should trigger a brain MRI in that patient (if not already done) as well as a referral to a clinical geneticist for a systematic phenotypic assessment. However, the risk for future pregnancies for that proband is uncertain given the poorly understood incomplete penetrance and variable expressivity of SMARCC1 variants. In affected probands found to have a transmitted SMARCC1 variant from an apparently unaffected carrier parent, a screening brain MRI and a heart ultrasound for the carrier parent and genetic testing for other children, with screening imaging reserved for variant-positive children, could be entertained. Thus, variants in SMARCC1 should be evaluated on a case-by-case basis for potential monogenic congenital hydrocephalus pathogenicity, considering individual variant characteristics, patient alignment to the described phenotypic spectrum and family history of disease. We further encourage the use of and—where applicable—the contribution of data to publicly available resources, such as ClinVar, gnomAD and dbGaP for the evaluation of SMARCC1 variant pathogenicity at the individual patient level.
In conclusion, our data suggest pathogenic DNVs in SMARCC1 variants cause a novel human BAFopathy characterized by DD, cerebral ventriculomegaly and a variety of structural brain or cardiac defects. These data underscore the importance of SMARCC1 and the BAF chromatin remodelling complex for human brain morphogenesis and support a neural stem cell paradigm of human congenital hydrocephalus pathogenesis. These results highlight the power of trio-based WES for identifying risk genes for congenital structural brain disorders and suggest WES may be a valuable adjunct in the management of patients with congenital hydrocephalus.
Supplementary Material
Acknowledgements
We are grateful to the patients and families who participated in this research.
Contributor Information
Amrita K Singh, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA.
Garrett Allington, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA; Department of Genetics, Yale University, New Haven, CT 06510, USA.
Stephen Viviano, Department of Pediatrics, Yale University, New Haven, CT 06510, USA.
Stephen McGee, GeneDx, Gaithersburg, MD 20877, USA.
Emre Kiziltug, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA.
Shaojie Ma, Department of Genetics, Yale University, New Haven, CT 06510, USA; Department of Neuroscience, Yale University, New Haven, CT 06510, USA.
Shujuan Zhao, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA; Departments of Genetics and Pediatrics, Washington University School of Medicine, St Louis, MO 63110, USA.
Kedous Y Mekbib, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA.
John P Shohfi, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA.
Phan Q Duy, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA; Department of Neuroscience, Yale University, New Haven, CT 06510, USA.
Tyrone DeSpenza, Jr, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA; Department of Neuroscience, Yale University, New Haven, CT 06510, USA.
Charuta G Furey, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA.
Benjamin C Reeves, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA.
Hannah Smith, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA; Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA.
André M M Sousa, Waisman Center, University of Wisconsin-Madison, Madison, WI 53705, USA.
Adriana Cherskov, Department of Neuroscience, Yale University, New Haven, CT 06510, USA.
August Allocco, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA.
Carol Nelson-Williams, Department of Genetics, Yale University, New Haven, CT 06510, USA.
Shozeb Haider, Department of Pharmaceutical and Biological Chemistry, University College London School of Pharmacy, London, WC1N 1AX, UK; UCL Centre for Advanced Research Computing, University College London, London, WC1H 9RN, UK.
Syed R A Rizvi, Department of Pharmaceutical and Biological Chemistry, University College London School of Pharmacy, London, WC1N 1AX, UK.
Seth L Alper, Division of Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Division of Nephrology and Vascular Biology Research Center, Beth Israel Deaconess Medical Center, Boston, MA 02215, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA.
Nenad Sestan, Department of Genetics, Yale University, New Haven, CT 06510, USA; Department of Pediatrics, Yale University, New Haven, CT 06510, USA.
Hermela Shimelis, Department of Radiology, Neuroradiology section, Kingston Health Sciences Centre, Queen's University Faculty of Health Sciences, Kingston, Ontario, Canada.
Lauren K Walsh, Department of Radiology, Neuroradiology section, Kingston Health Sciences Centre, Queen's University Faculty of Health Sciences, Kingston, Ontario, Canada.
Richard P Lifton, Laboratory of Human Genetics and Genomics, The Rockefeller University, New York, NY 10065, USA.
Andres Moreno-De-Luca, Department of Radiology, Neuroradiology section, Kingston Health Sciences Centre, Queen's University Faculty of Health Sciences, Kingston, Ontario, Canada; Department of Radiology, Diagnostic Medicine Institute, Geisinger, Danville, PA, 17822, USA.
Sheng Chih Jin, Departments of Genetics and Pediatrics, Washington University School of Medicine, St Louis, MO 63110, USA.
Paul Kruszka, GeneDx, Gaithersburg, MD 20877, USA.
Engin Deniz, Department of Pediatrics, Yale University, New Haven, CT 06510, USA.
Kristopher T Kahle, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115, USA; Division of Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Division of Genetics and Genomics, Boston Children's Hospital, Boston, MA 02115, USA.
Data availability
The data that support the findings of this study will be made available upon reasonable request from kahle.kristopher@mgh.harvard.edu
Funding
This work was supported by the Yale-NIH Center for Mendelian Genomics (grant 5U54HG006504) and the University of Washington Center for Mendelian Genomics (grants UW-CMG, UM1HG006493, U24HG008956), National Institutes of Health (grants R01 NS111029-01A1, R01 NS109358, R01 NS127879-01, R01HL109942, R21 NS116484-02, and K12 228168) (K.K., E.D, S.C.J); the Rudi Schulte Research Institute (K.K.), the National Heart, Lung, and Blood Institute (R00HL143036-02) (S.C.J), the Hydrocephalus Association Innovator Award (E.D., K.K., S.C.J.), the Clinical & Translational Research Funding Program Award (CTSA1405; (S.C.J.), the Children’s Discovery Institute Faculty Scholar Award (CDI-FR-2021-926) (S.C.J), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (R01HD104938-01A1) (A.M.D).
Competing interests
S.M. and P.K. are employees of GeneDx. All other co-authors have no conflicts of interest to disclose.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–171. [DOI] [PubMed] [Google Scholar]
- 2. Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. [DOI] [PubMed] [Google Scholar]
- 3. Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. [DOI] [PubMed] [Google Scholar]
- 5. Alfert A, Moreno N, Kerl K. The BAF complex in development and disease. Epigenetics Chromatin. 2019;12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao K, Wang W, Rando OJ, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. [DOI] [PubMed] [Google Scholar]
- 8. Yan L, Xie S, Du Y, Qian C. Structural insights into BAF47 and BAF155 Complex formation. J Mol Biol. 2017;429:1650–1660. [DOI] [PubMed] [Google Scholar]
- 9. Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17:537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan Z, Wang Z, Sharova L, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bevilacqua A, Willis MS, Bultman SJ. SWI/SNF chromatin-remodeling complexes in cardiovascular development and disease. Cardiovasc Pathol. 2014;23:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lei I, Liu L, Sham MH, Wang Z. SWI/SNF in cardiac progenitor cell differentiation. J Cell Biochem. 2013;114:2437–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vieira JM, Howard S, Villa Del Campo C, et al. BRG1-SWI/SNF-dependent regulation of the wt1 transcriptional landscape mediates epicardial activity during heart development and disease. Nat Commun. 2017;8:16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furey CG, Choi J, Jin SC, et al. De novo mutation in genes regulating neural stem cell fate in human congenital hydrocephalus. Neuron. 2018;99:302–314.e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin SC, Dong W, Kundishora AJ, et al. Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus. Nat Med. 2020;26:1754–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Machol K, Rousseau J, Ehresmann S, et al. Expanding the Spectrum of BAF-related disorders: De novo variants in SMARCC2 cause a syndrome with intellectual disability and developmental delay. Am J Hum Genet. 2019;104:164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sousa SB, Abdul-Rahman OA, Bottani A, et al. Nicolaides-Baraitser syndrome: Delineation of the phenotype. Am J Med Genet A. 2009;149A:1628–1640. [DOI] [PubMed] [Google Scholar]
- 19. Mari F, Marozza A, Mencarelli MA, et al. Coffin-Siris and Nicolaides-Baraitser syndromes are a common well recognizable cause of intellectual disability. Brain Dev. 2015;37:527–536. [DOI] [PubMed] [Google Scholar]
- 20. Van Houdt JKJ, Nowakowska BA, Sousa SB,et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–449. S1. [DOI] [PubMed] [Google Scholar]
- 21. Hoyer J, Ekici AB, Endele S, et al. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012;90:565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bogershausen N, Wollnik B. Mutational landscapes and phenotypic Spectrum of SWI/SNF-related intellectual disability disorders. Front Mol Neurosci. 2018;11:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Santen GWE, Aten E, Sun Y, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause coffin-siris syndrome. Nat Genet. 2012;44:379–380. [DOI] [PubMed] [Google Scholar]
- 24. Wieczorek D, Bögershausen N, Beleggia F, et al. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 2013;22:5121–5135. [DOI] [PubMed] [Google Scholar]
- 25. DelBove J, Rosson G, Strobeck M, et al. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics. 2011;6:1444–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nowakowski TJ, Bhaduri A, Pollen AA, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen H, Sokpor G, Pham L, et al. Epigenetic regulation by BAF (mSWI/SNF) chromatin remodeling complexes is indispensable for embryonic development. Cell Cycle. 2016;15:1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front Mol Neurosci. 2017;10:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Narayanan R, Pham L, Kerimoglu C, et al. Chromatin remodeling BAF155 subunit regulates the genesis of basal progenitors in developing Cortex. iScience. 2018;4:109–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Narayanan R, Pirouz M, Kerimoglu C, et al. Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 2015;13:1842–1854. [DOI] [PubMed] [Google Scholar]
- 31. Harmacek L, Watkins-Chow DE, Chen J, et al. A unique missense allele of BAF155, a core BAF chromatin remodeling complex protein, causes neural tube closure defects in mice. Dev Neurobiol. 2014;74:483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Panamarova M, Cox A, Wicher KB, et al. The BAF chromatin remodelling complex is an epigenetic regulator of lineage specification in the early mouse embryo. Development. 2016;143:1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim JK, Huh SO, Choi H, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001; 21:7787–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duy PQ, Rakic P, Alper SL, et al. Brain ventricles as windows into brain development and disease. Neuron. 2022;110:12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodríguez EM, Guerra MM. Neural stem cells and fetal-onset hydrocephalus. Pediatr Neurosurg. 2017;52:446–461. [DOI] [PubMed] [Google Scholar]
- 36. Rodríguez EM, Guerra MM, Vío K, et al. A cell junction pathology of neural stem cells leads to abnormal neurogenesis and hydrocephalus. Biol Res. 2012;45:231–242. [DOI] [PubMed] [Google Scholar]
- 37. Krumm N, Turner TN, Baker C, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stram DO. Software for tag single nucleotide polymorphism selection. Hum Genomics. 2005;2:144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.11–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang YC, Wu Y, Choi J, et al. Computational genomics in the era of precision medicine: Applications to variant analysis and gene therapy. J Pers Med. 2022;12:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wei Q, Zhan X, Zhong X, et al. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics. 2015;31:1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Diab NS, King S, Dong W, et al. Analysis workflow to assess de novo genetic variants from human whole-exome sequencing. STAR Protoc. 2021;2:100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaplanis J, Samocha KE, Wiel L, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020; 586:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller JA, Ding SL, Sunkin SM, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li M, Santpere G, Imamura Kawasawa Y, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-Sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. [DOI] [PubMed] [Google Scholar]
- 56. Gerhart JKM. Normal table of Xenopus laevis (daudin): A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. 1st ed. Garland Science; 2020. [Google Scholar]
- 57. Lane M, Mis EK, Khokha MK. Obtaining Xenopus tropicalis eggs. Cold Spring Harb Protoc. 2022;2022(4):Pdb.prot106344. [DOI] [PubMed] [Google Scholar]
- 58. Deniz E, Mis EK, Lane M, Khokha MK. Xenopus tadpole craniocardiac imaging using optical coherence tomography. Cold Spring Harb Protoc. 2022;2022:Pdb.prot105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Deniz E, Mis EK, Lane M, Khokha MK. CRISPR/cas9 F0 screening of congenital heart disease genes in Xenopus tropicalis. Methods Mol Biol. 2018;1865:163–174. [DOI] [PubMed] [Google Scholar]
- 60. Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK. CRISPR/cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol. 2015;408:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. [DOI] [PubMed] [Google Scholar]
- 62. Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barish S, Barakat TS, Michel BC, et al. BICRA, a SWI/SNF Complex member, is associated with BAF-disorder related phenotypes in humans and model organisms. Am J Hum Genet. 2020;107:1096–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duran D, Zeng X, Jin SC, et al. Mutations in chromatin modifier and ephrin signaling genes in vein of galen malformation. Neuron. 2019;101:429–443 e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006;103:2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Santen GWE, Aten E, Vulto-van Silfhout AT, et al. Coffin-Siris syndrome and the BAF complex: Genotype-phenotype study in 63 patients. Hum Mutat. 2013;34:1519–1528. [DOI] [PubMed] [Google Scholar]
- 68. Chen CA, Lattier J, Zhu W, et al. Retrospective analysis of a clinical exome sequencing cohort reveals the mutational spectrum and identifies candidate disease-associated loci for BAFopathies. Genet Med. 2022;24:364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kosmicki JA, Samocha KE, Howrigan DP, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Al Mutairi F, Alzahrani F, Ababneh F, Kashgari AA, Alkuraya FS. A Mendelian form of neural tube defect caused by a de novo null variant in SMARCC1 in an identical twin. Ann Neurol. 2018;83:433–436. [DOI] [PubMed] [Google Scholar]
- 71. Lefebvre M, Bruel AL, Tisserant E, et al. Genotype-first in a cohort of 95 fetuses with multiple congenital abnormalities: When exome sequencing reveals unexpected fetal phenotype-genotype correlations. J Med Genet. 2021;58:400–413. [DOI] [PubMed] [Google Scholar]
- 72. Wilfert AB, Turner TN, Murali SC, et al. Recent ultra-rare inherited variants implicate new autism candidate risk genes. Nat Genet. 2021;53:1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reuter MS, Chaturvedi RR, Liston E, et al. The cardiac genome clinic: Implementing genome sequencing in pediatric heart disease. Genet Med. 2020;22:1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tuoc TC, Boretius S, Sansom SN, et al. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. [DOI] [PubMed] [Google Scholar]
- 76. Ninkovic J, Steiner-Mezzadri A, Jawerka M, et al. The BAF complex interacts with pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. [DOI] [PubMed] [Google Scholar]
- 78. Chandler RL, Magnuson T. The SWI/SNF BAF-A complex is essential for neural crest development. Dev Biol. 2016;411:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Encha-Razavi F, Sonigo P. Features of the developing brain. Childs Nerv Syst. 2003;19(7–8):426–428. [DOI] [PubMed] [Google Scholar]
- 80. Dur AH, Tang T, Viviano S, et al. In Xenopus ependymal cilia drive embryonic CSF circulation and brain development independently of cardiac pulsatile forces. Fluids Barriers CNS. 2020;17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Date P, Ackermann P, Furey C, et al. Visualizing flow in an intact CSF network using optical coherence tomography: Implications for human congenital hydrocephalus. Sci Rep. 2019;9:6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. [DOI] [PubMed] [Google Scholar]
- 84. Wullimann MF, Rink E, Vernier P, Schlosser G. Secondary neurogenesis in the brain of the African clawed frog, Xenopus laevis, as revealed by PCNA, Delta-1, neurogenin-related-1, and NeuroD expression. J Comp Neurol. 2005;489:387–402. [DOI] [PubMed] [Google Scholar]
- 85. Olson JM, Asakura A, Snider L, et al. Neurod2 is necessary for development and survival of central nervous system neurons. Dev Biol. 2001;234:174–187. [DOI] [PubMed] [Google Scholar]
- 86. Wong RL, Chan KK, Chow KL. Developmental expression of Mab21l2 during mouse embryogenesis. Mech Dev. 1999;87(1–2):185–188. [DOI] [PubMed] [Google Scholar]
- 87. Brugmans AK, Walter C, Moreno N, et al. A carboxy-terminal smarcb1 point mutation induces hydrocephalus formation and affects AP-1 and neuronal signalling pathways in mice. Cell Mol Neurobiol. 2023;43:3511–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Duy PQ, Weise SC, Marini C, et al. Impaired neurogenesis alters brain biomechanics in a neuroprogenitor-based genetic subtype of congenital hydrocephalus. Nat Neurosci. 2022;25:458–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Duy PQ, Rakic P, Alper SL, et al. A neural stem cell paradigm of pediatric hydrocephalus. Cereb Cortex. 2023;33:4262–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Diets IJ, Prescott T, Champaigne NL, et al. A recurrent de novo missense pathogenic variant in SMARCB1 causes severe intellectual disability and choroid plexus hyperplasia with resultant hydrocephalus. Genet Med. 2019;21:572–579. [DOI] [PubMed] [Google Scholar]
- 91. Slavotinek A, Lefebvre M, Brehin AC, et al. Prenatal presentation of multiple anomalies associated with haploinsufficiency for ARID1A. Eur J Med Genet. 2022;65:104407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cao M, Wu JI. Camk2a-Cre-mediated conditional deletion of chromatin remodeler brg1 causes perinatal hydrocephalus. Neurosci Lett. 2015;597:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rive Le Gouard N, Nicolle R, Lefebvre M, et al. First reports of fetal SMARCC1 related hydrocephalus. Eur J Med Genet. 2023;66:104797. [DOI] [PubMed] [Google Scholar]
- 94. Cappuccio G, Sayou C, Tanno PL, et al. De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. Genet Med. 2020;22:1838–1850. [DOI] [PubMed] [Google Scholar]
- 95. Kaufman L, Ayub M, Vincent JB. The genetic basis of non-syndromic intellectual disability: A review. J Neurodev Disord. 2010;2:182–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Morrison AJ. Chromatin-remodeling links metabolic signaling to gene expression. Mol Metab. 2020;38:100973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kenneth NS, Mudie S, van Uden P, Rocha S. SWI/SNF regulates the cellular response to hypoxia. J Biol Chem. 2009;284:4123–4131. [DOI] [PubMed] [Google Scholar]
- 98. D'Gama AM, Walsh CA. Somatic mosaicism and neurodevelopmental disease. Nat Neurosci. 2018;21:1504–1514. [DOI] [PubMed] [Google Scholar]
- 99. Ronan JL, Wu W, Crabtree GR. From neural development to cognition: Unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166c:333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jellinger G. Anatomopathology of non-tumoral aqueductal stenosis. J Neurosurg Sci. 1986;30(1–2):1–16. [PubMed] [Google Scholar]
- 102. Takashima S, Fukumizu M. Pathology of congenital aqueductal stenosis and posthemorrhagic hydrocephalus. No To Hattatsu. 1994;26:216–221. [PubMed] [Google Scholar]
- 103. Cinalli G, Spennato P, Nastro A, et al. Hydrocephalus in aqueductal stenosis. Child's Nervous System. 2011;27:1621. [DOI] [PubMed] [Google Scholar]
- 104. Tully HM, Dobyns WB. Infantile hydrocephalus: A review of epidemiology, classification and causes. Eur J Med Genet. 2014;57:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wiley MM, Muthukumar V, Griffin TM, Griffin CT. SWI/SNF chromatin-remodeling enzymes brahma-related gene 1 (BRG1) and brahma (BRM) are dispensable in multiple models of postnatal angiogenesis but are required for vascular integrity in infant mice. J Am Heart Assoc. 2015;4:e001972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mehrotra A, Joe B, de la Serna IL. SWI/SNF chromatin remodeling enzymes are associated with cardiac hypertrophy in a genetic rat model of hypertension. J Cell Physiol. 2013;228:2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Govaert P, Oostra A, Matthys D, Vanhaesebrouck P, Leroy J. How idiopathic is idiopathic external hydrocephalus? Dev Med Child Neurol. 1991;33:274–276. [DOI] [PubMed] [Google Scholar]
- 109. Kundishora AJ, Singh AK, Allington G, et al. Genomics of human congenital hydrocephalus. Childs Nerv Syst. 2021; 37:3325–3340. [DOI] [PubMed] [Google Scholar]
- 110. DeSpenza T Jr, Carlson M, Panchagnula S, et al. PTEN mutations in autism spectrum disorder and congenital hydrocephalus: Developmental pleiotropy and therapeutic targets. Trends Neurosci. 2021;44:961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Etchegaray A, Juarez-Peñalva S, Petracchi F, Igarzabal L. Prenatal genetic considerations in congenital ventriculomegaly and hydrocephalus. Childs Nerv Syst. 2020;36:1645–1660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study will be made available upon reasonable request from kahle.kristopher@mgh.harvard.edu