Abstract
Objective:
Perform a scoping review of supervised machine learning in pediatric critical care to identify published applications, methodologies, and implementation frequency to inform best practices for the development, validation, and reporting of predictive models in pediatric critical care.
Design:
Scoping review and expert opinion.
Setting:
We queried CINAHL Plus with Full Text (EBSCO), Cochrane Library (Wiley), Embase (Elsevier), Ovid Medline, and PubMed for articles published between 2000–2022 related to machine learning concepts and pediatric critical illness. Articles were excluded if the majority of patients were adults or neonates, if unsupervised machine learning was the primary methodology, or if information related to the development, validation, and/or implementation of the model was not reported. Article selection and data extraction were performed using dual review in the Covidence tool, with discrepancies resolved by consensus.
Subjects:
Articles reporting on the development, validation, or implementation of supervised machine learning models in the field of pediatric critical care medicine.
Interventions:
None.
Measurements and Main Results:
Of 5075 identified studies, 141 articles were included. Studies were primarily (57%) performed at a single site. The majority took place in the United States (70%). Most were retrospective observational cohort studies. More than three-quarters of the articles were published between 2018–2022. The most common algorithms included logistic regression and random forest. Predicted events were most commonly death, transfer to ICU, and sepsis. Only 14% of articles reported external validation, and only a single model was implemented at publication. Reporting of validation methods, performance assessments, and implementation varied widely. Follow up with authors suggests that implementation remains uncommon after model publication.
Conclusions:
Publication of supervised machine learning models to address clinical challenges in pediatric critical care medicine has increased dramatically in the last five years. While these approaches have the potential to benefit children with critical illness, the literature demonstrates incomplete reporting, absence of external validation, and infrequent clinical implementation.
Keywords: predictive modeling, supervised machine learning, prognostication, critical care
Introduction:
Pediatric critical care medicine requires practitioners to routinely integrate data streams from bedside monitors, laboratory and imaging studies, physical examinations, and numerous other sources. Prediction of patient trajectories and outcomes is a cornerstone of clinical care and is historically based on clinician heuristics. The broad adoption of electronic health records (EHRs) and subsequent development of clinical databases has led to rapid growth in the development of prediction models for pediatric critical care (1). The prediction tasks fundamental to pediatric critical care can now, at least in theory, be enhanced by machine learning-based prediction models. These models may impact the care of individual patients, but also have the potential to fundamentally alter our approach to clinical care and research. As machine learning studies become more common, authors, readers, and reviewers of this journal will require familiarity with prediction modeling and performance assessment.
Supervised learning is a branch of machine learning where computational models are iteratively trained to utilize associations between a set of predictor variables and an outcome of interest to make predictions on new data. To train these models for clinical care, researchers use historical data from a cohort of patients with a labeled outcome (e.g., mortality) and a measured set of candidate predictor variables (e.g., lab results) from a subset of the available cohort. This is in contrast to unsupervised learning, where the data is unlabeled and the goal of the algorithm is to uncover natural groupings in the data based on statistical parameters of similarity. Only studies using supervised machine learning were included in this review.
Developing clinical prediction models using supervised machine learning algorithms is a multistage process, with each stage serving an essential function to the integrity of the model. Although variations may exist, the ideal process is as follows. First, data from the cohort should be partitioned into three non-overlapping sets. A subset of the cohort is used to train the models (i.e., “train set”), with a second subset (i.e., “validation set”) used to initially evaluate the model and make necessary adjustments or “tuning” of the parameters used for model training. A third subset of data from the cohort (i.e., “test set”), which the model has not seen during training or validation, is used to evaluate its performance. A common variation of this set up includes the use of cross-validation to train, validate, and tune models in the train set prior to testing in a separate test set. The complete separation of the test set from other data is important in order to accurately assess the potential performance of the model, including in more recent (and relevant) historical data. Finally, the model’s performance is assessed in other settings (e.g., at a separate institution) through a process of external validation.
Guidelines for the reporting of multivariable prediction models (the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis [TRIPOD] statement) have been developed (2). There are additional guidelines specific to publication of such models in respiratory, sleep, and critical care journals, including Pediatric Critical Care Medicine (3). These guidelines focus on the importance of reporting details about the model’s development, use, and limitations. Prior commentaries in this journal have focused on the characteristics of specific publications, while highlighting that the guidelines may not be directly applicable to models commonly developed in pediatric critical care (4–9). There is a lack of knowledge regarding how models are developed, validated, implemented, and reported in the pediatric critical care literature. The objective of this scoping review was to evaluate the published applications of supervised machine learning in pediatric critical care, with a particular emphasis on methodology and reporting. This evaluation then informed recommendations from our multidisciplinary experts as to developing and disseminating high-quality predictive modeling in our field.
Methods:
This study was performed by members of the Pediatric Data Science and Analytics (PEDAL) subgroup of the Pediatric Acute Lung Injury and Sepsis Investigators (PALISI) Network. As the PALISI subgroup dedicated to data science and predictive modeling, PEDAL represents many of the experts in this multidisciplinary and growing field (10). Membership primarily comprises pediatric critical care clinician-scientists, some of whom have additional training in clinical informatics, and doctoral-trained data scientists. More information about the group can be viewed at https://palisi-pedal.org/. Recent publications by the group have included topics of clinical decision support, model development, and model validation (11–14). This review adhered to the Preferred Reporting Items for Systematic Reviews and Meta Analyses extension for Scoping Reviews guidelines (15, 16) (Supplemental Digital Content 1).
Search Strategy
We conducted a structured scoping review of original research articles published in English between January 2000 and July 2022 that used supervised machine learning methods to develop prediction models of clinical outcomes in the pediatric critical care population. Grey literature, which does not undergo traditional peer review, was not included. Articles were included if children needed, or had the potential need for, critical care services. Regression models can be characterized as those which test for associations between risk factors and outcomes (inference) and those which predict outcomes on new data on the basis of risk factors (prediction); models which were only used for inference purposes were excluded. Articles that only tested a prediction model that used unsupervised machine learning (i.e., unlabeled predictor variables or outcomes) or an expert-generated score (e.g., Pediatric Early Warning Scores) were excluded. Finally, studies were excluded if they took place in a neonatal ICU, if >50% of the study population was >18 years of age or newborns, or if they did not report a clinical outcome (e.g., pharmacokinetic studies).
A comprehensive search strategy was designed by a professional medical librarian (AF) in the following databases: CINAHL Plus with Full Text (EBSCO), Cochrane Library (Wiley), Embase (Elsevier), Ovid Medline, and PubMed. Search queries were adapted to each database to include relevant keywords and controlled vocabulary terms. Representative articles were identified and used to refine the search strategy. The full search strategy is available in Supplemental Digital Content 2.
Review and Extraction Process
Citations were compiled and duplicate records were removed using Endnote (Clarivate, Philadelphia, PA). Abstract, full-text screening, and data extraction were completed in Covidence (Veritas Health Innovation, Melbourne, Australia) and reviewed by two reviewers per article at each phase. Reviewers received training on inclusion/exclusion criteria, study definitions, and other study components through written and real-time training sessions. Conflicts were resolved by a core author group (JAH, MCS, RF, LNS, TDB, ACD, SBW) at screening phases and via consensus between the two reviewers at extraction phase. Variables extracted included site/funding information, study/patient characteristics, model variable selection including outcome measures and feature importance (i.e., scores which demonstrate the relative contribution of a specific variable to the model’s output), results, validation, and implementation. Due to the wide range of included models and methods of performance evaluation, comparisons among the models was not feasible. Data were exported from Covidence and analyzed using Excel (Microsoft, Redmond, WA). Descriptive statistics were performed and reported using n (%) or median (IQR) as applicable.
Follow Up
Articles were assessed for contact information for first and/or corresponding authors. If no contact information was available in the manuscript, online search was undertaken to identify relevant contact information. One author per manuscript was contacted to complete a one-question survey about the current implementation status of the project described in the manuscript. Survey responses were collected using Research Electronic Data Capture Consortium (REDCap; Nashville, TN). Up to two reminder emails were sent via REDCap.
Results:
Review Information
5075 unique studies were identified for initial screening, with 141 studies ultimately included (Supplemental Digital Content 3). Among the 236 articles excluded at the full-text screening stage, the most common reasons for exclusion were incorrect patient population (n=114, 48%) and incorrect methodology (n=71, 30%).
Article Information (Table 1)
Table 1.
Characteristics of included manuscripts. Some characteristics may report more than one answer per category, so totals may be greater than 100%.
| Study Characteristic | N (%) |
|---|---|
| Study Location | |
| United States | 99 (70) |
| Canada | 12 (9) |
| China | 10 (7) |
| Republic of Korea | 5 (4) |
| Other | 22 (16) |
| Number of Sites | |
| 1 | 80 (57) |
| 2 | 9 (6) |
| >2 | 46 (33) |
| Reported Study Design | |
| Retrospective observational cohort | 101 (72) |
| Prospective observational cohort | 31 (22) |
| Randomized control trial | 1 (1) |
| Other (e.g., secondary analyses of cohort studies, case-controls) | 8 (6) |
| Funding Source | |
| Governmental | 72 (51) |
| University | 27 (19) |
| Industry | 10 (7) |
| None | 14 (10) |
| Not reported | 26 (18) |
| Data Source | |
| Electronic health record | 80 (57) |
| Administrative database/registry (e.g. Pediatric Hospital Information System, Virtual Pediatric Systems) | 41 (29) |
| Other | 20 (14) |
| Hospital database | 25 (18) |
| Internal Validation Process | |
| Random split | 42 (30) |
| Temporal split | 14 (10) |
| Cross-validation | 59 (42) |
| Other | 8 (6) |
| None/not reported | 19 (14) |
| External Validation Reported | 20 (14) |
| Methods to Limit Overfitting | |
| Bootstrapping | 21 (15) |
| Cross-validation | 58 (41) |
| Other | 15 (11) |
| None/none reported | 61 (43) |
| Number of Models per Manuscript | Median 3 (IQR 1 – 5) |
Studies were primarily performed at a single site (n=80, 57%). The majority took place in the United States (n=99, 70%). Most were retrospective observational cohort studies (n=101, 72%). Median reported study duration was 4 (IQR 2–8) years. Studies were published between 2000 and 2022 per our inclusion criteria, with 107 (76%) published within the five years prior to the review. Manuscript publication trends by year are shown in Supplemental Digital Content 4. Articles were most commonly published in Pediatric Critical Care Medicine (n=10), Scientific Reports (n=9), Critical Care (n=8), Frontiers in Pediatrics (n=6), and as proceedings of the Annual International Conference of the Institute of Electrical and Electronics Engineers Engineering in Medicine and Biology Society (n=6). For the 123 articles where funding was reported, funding was most often partially governmental (n=72/123, 59%), university (n=27/123, 22%) or industry (n=10/123, 8%). Fourteen articles (11%) reported no funding for the project.
Patient location was primarily the PICU (n=95, 67%) but also included the pediatric cardiac intensive care unit (n=38, 27%), the emergency department (n=27, 19%), the general ward (n=27, 19%), and the operating room (n=6, 4%). The median number of patients included in each manuscript was 982 (IQR 199–8022). While all but one manuscript reported the number of patients, more than a quarter of articles (n=37, 26%) did not report how data were partitioned into training, validation, and test sets. The full list of included articles is found in Supplemental Digital Content 5.
Data and Methods Used
Data were most commonly acquired using the EHR (n=80, 57%) or an administrative database or registry (n=41, 29%). Structured data items (e.g., lab results, intermittent vital signs) were used in the majority of articles (n=130, 92%) with signals (e.g., continuous vital signs) (n=28, 20%) and text (n=20, 14%) less commonly used.
Articles most commonly used logistic regression (n=61, 43%) or random forest (n=44, 31%) modeling methodologies (Figure 1). Nearly one-third (n=41, 29%) of articles reported using at least three supervised machine learning methodologies. Predictor variables were primarily selected by clinician adjudication or expert selection (n=65, 46%), followed by computational or data-driven methods (n=52, 37%), and a combination of clinician and computational selection methodologies (n=17, 12%). A median of 23 predictor variables were assessed per model (IQR 11–60), with the final models including a median of 11 variables (IQR 5–26). Additional details regarding the described models are found in Supplemental Digital Content 6.
Figure 1.
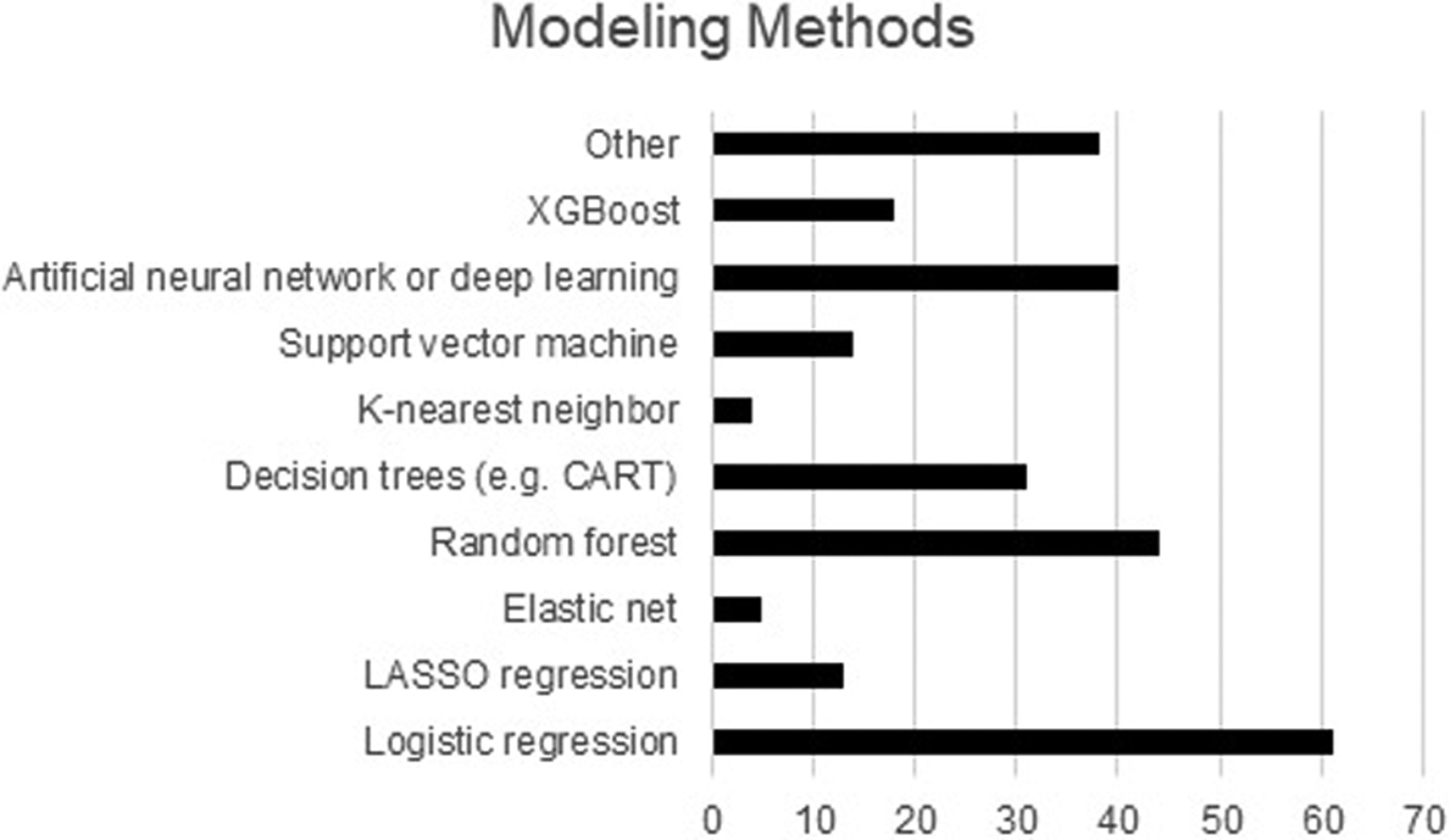
Modeling methods used in the included manuscripts. Articles may have included more than one method, so the total number of methods is greater than the number of articles.
Outcomes of Interest
Clinical outcomes predicted by the models, whether individually or as part of a composite outcome, included mortality (n=44, 31%), ICU transfer (n=20, 14%), sepsis diagnosis (n=13, 9%), cardiac arrest (n=11, 8%), respiratory failure (n=11, 8%), and acute kidney injury diagnosis/need for renal replacement therapy (n=6, 4%). Thirty (21%) articles described more than one outcome of interest. Eighty (57%) articles reported the number of events of the outcomes of interest. When reported, the outcomes of interest (which could be more than one per manuscript) occurred in a median of 15.3% of patients per manuscript (IQR 6.3–40.9%, range 0.2–140.9%). Eighty-four articles reported on models that were intended to be used at a single time point, while 43 were intended to provide dynamic prediction over time.
Model Performance Assessments and Validation
Articles reported performance assessments for a median of three models per manuscript (IQR 1–5; range 1–45). The most common metrics included area under the receiver operating curve (AUROC), which was reported in 113 (80%) articles, recall/sensitivity (n=75, 53%) specificity (n=70, 50%) precision/positive predictive value (n=57, 40%), accuracy/percent correctly classified (n=53, 38%), negative predictive value (n=42, 30%). Fewer than one-quarter (n=30, 21%) reported F1 scores (harmonic mean of precision and recall), 13 (9%) reported area under the precision recall curve (AUPRC), 4 (3%) reported Matthew’s correlation coefficients (similar to chi-square testing, and particularly important for imbalanced class models, where the outcome classes have markedly different prevalence), and 4 (3%) reported number needed to alarm/assess. Feature importance was reported in 90 (64%) articles. A majority (87%) of articles reported internal validation, most commonly using a cross-validation approach, whereby the performance of the model is assessed within a subset of the training set (n=59, 42%). External validation was reported in 20 (14%) articles.
Model Implementation and Availability
Only a single model was reported to be used clinically at the time of publication, and 23 additional articles noted planning for implementation. Two articles reported deciding against implementation, one specifically due to poor performance of the model. Implementation status was ascertained on follow up for 64 articles. The majority reported models were never implemented (n=34, 53%). Seventeen models (27%) were actively implemented at either a single or multiple sites, while the balance were either implemented only in a background/silent manner (precluding clinical use) or previously implemented but no longer. Current implementation status as reported by responding authors is listed in Supplemental Digital Content 5. Some degree of public availability was reported for the models in 27 (19%) articles. Models in five (4%) additional articles were proprietary or commercially available.
Discussion:
As machine learning studies become more common, authors, readers, and reviewers will require familiarity with prediction modeling and performance assessment. This scoping review describes the landscape of supervised machine learning in pediatric critical care from 2000 to 2022. We found the following areas for improvement: 1) lack of consistency of terminology and reporting of study methodology across articles; 2) overreliance on AUROC as the sole mechanism to assess the performance of a model; and 3) lack of focus on high-quality, effective implementation of models. Additionally, we found that the studied outcomes were largely high-acuity in nature, with limited focus on low-acuity outcomes which nonetheless may have meaningful impact on unit processes.
One area where a lack of consistency in terminology exists is distinguishing the different data sets (training, validation, testing) utilized in model development, if these numbers were reported at all. The practice of data partitioning is essential for model development and assessment. The test set must be independent, ensuring that no observations are shared with the training or validation sets. In other words, while the data come from a cohort of patients, there should be no overlap on the unit-level analysis (separate patients, patient admissions, etc.). If possible, a longitudinal split between train/validation and test data should be conducted. The independence of the test set is traditionally ensured using a longitudinal train-test split (i.e., using data from different time periods for each component) or testing in an external setting, allowing for assessment of how a model may perform upon deployment on future observations. Any overlap between the training/validation and testing sets could result in a biased, overly-optimistic result that is not an indicator of true future performance. Ideally, this subset of data will also post-date the data used for the training and validation sets, although not always necessary.
We observed relatively consistent use and reporting of the term training set to describe the data used to develop the initial model. The terms validation set and testing set were often used interchangeably to describe the data used to evaluate model performance and their sizes were less commonly reported. While some data science texts (17) endorse the interchangeability of the terms, the traditional convention is that the validation set is distinct from the testing set (18, 19). The validation set is used to select the best combination of variables and model characteristics to fine tune the hyperparameters of the model (i.e., parameters that affect the training of the model), whereas a test set is used to independently assess the performance of the final model. Sharing accurate information about these different data sets, as well as how the partitioning is performed, is both implicitly and explicitly recommended in the current guidelines (2,3).
The majority of articles reported the AUROC as a measure of model performance. The AUROC is a measure of the discriminative ability of a prediction model, quantifying the separation of risk distributions of events and non-events (20). While the AUROC may aid in identifying an optimal model among a group of models trained on the same outcome, it is not a measure of model utility (20, 21). Models developed to predict rare outcomes, where the separation of risk distributions of events and non-events for the outcome are broader, will likely have higher accuracy values than models developed for outcomes with higher incidence. In the case where such unbalanced classes exist, calculation and reporting of the area under the precision-recall curve (AUPRC) may be a better metric by which to assess model performance. Given the relatively low rate of mortality in modern pediatric critical care, a model designed to predict mortality would perform quite well by AUROC if it always predicted a child would survive. However, missed predictions of mortality (i.e., if the model predicted survival and the child died) would significantly impact the recall, thereby reducing AUPRC. Evaluation of a predictive model should include additional metrics such as positive predictive value and the number needed to alarm in order to ensure that the model is clinically, and not just statistically, useful (20, 21).
The outcomes chosen for modeling in this review largely focused on high acuity outcomes. Other outcomes, particularly those which impact unit processes (e.g., normal lab values for avoidance of repeat blood sampling) may benefit from the design and implementation of prediction modeling. However, it is unclear from this review why specific outcomes have been the focus of current model development in pediatric critical care. One possibility is the availability of clinical data and the ability to accurately record the outcome. Information about lower acuity outcomes may be less practical for modeling due to lack of integration of this information in the datasets which served as the source of data for many models in this review. Researchers should also be mindful of the potential ethical implications of our choices of data sources, methodology, or outcomes (22).
The models reported in the identified articles are rarely applied beyond the single center where they were developed. Model performance can vary widely when models are applied to a new population, so external validation is necessary to ascertain the generalizability of a model. Barriers to external validation may include the lack of multi-institutional datasets in pediatric critical care. PhysioNet (the National Institutes of Health’s Research Resource for Complex Physiologic Signals) manages the Medical Information Mart for Intensive Care (MIMIC) and a host of other datasets which primarily contain data related to adult patients (23). These data sets are freely available to researchers, with integrated educational materials, and the PhysioNet community routinely hosts challenges related to addressing clinical questions potentially addressable by the available data. No similar resources exist in pediatric critical care currently, although efforts are underway, such as the PICU Data Collaborative (24). Before implementing prediction models derived from other sites, it is essential to evaluate the performance of the model on local data. The model’s utility and impact in the clinical workflows at that implementation site should be studied, adaptation of the model to the local context and population (including re-fitting the model) should be considered, and new clinical workflows should be designed to maximize the benefit and minimize the possible unintended negative consequences of model implementation (25).
The fact that implementation was rarely attempted prior to model publication and remained relatively rare upon follow up likely reflects the difficulty of this process. Barriers to implementation may include clinician distrust of “black box” models, technical infrastructure challenges, lack of personnel and/or governance mechanisms to guide the implementation and oversight processes, and legal or regulatory concerns (10). Implementation science is a growing field with rigorous methodology; collaboration between these fields may allow us to overcome the barriers to high-quality implementation (26). Additionally, we should be mindful of the potentially evolving ethical and regulatory landscape surrounding all components of model development and implementation.
Guidance for Machine Learning-Based Clinical Prediction Models Studies in PCCM:
As mentioned previously, the TRIPOD guidelines provide key components in several domains (e.g., data source, predictors, model structure, validation, etc.) that should be present for the appropriate evaluation of a prediction model. Guidelines based on the original TRIPOD recommendations for reporting prediction models using machine learning/artificial intelligence algorithms are currently under development (27).
Figure 2 and Tables 2 and 3 provide a summary of our guidance for machine learning-based clinical prediction modeling studies reporting in PCCM based on the results of this scoping review, our assessment of the existing guidelines and recommendation, and our collective expertise in the field. This includes adherence to reporting guidelines, a focus on transparency in reporting model development processes, attention to clinical relevance of performance assessments, and implementation considerations. Exemplars of each guidance item are provided. Additional considerations specific to predictive modeling in pediatrics include consideration of age and developmental status as potential modifier of model performance, the prevalence of chronic critical illness and technology-dependent patients, and patients with immunodeficiencies. Investigators should consider a priori sensitivity analyses of model performance in these categories. Suggested resources and references related to specific aspects of predictive modeling in pediatric critical care are found in Supplemental Digital Content 7.
Figure 2.

Summary of methodology and reporting guidance for predictive modeling work in PCCM
Table 2.
Summary of methodology guidance for predictive modeling work in PCCM
| Guidance | Example |
|---|---|
| Data and methods used | |
| Select methodology that allows for balancing the factors of precision, parsimony, and transparency appropriately for the clinical scenario | “Clinically important confounding variables and statistically significant risk factors found in univariate analysis were evaluated in multivariable regression modeling….Independent risk factors identified upon multivariable analysis were multiplexed in a predictive risk algorithm for the probability of HA-VTE.” (31) |
| Model performance assessments and validation | |
| Include performance assessments which include measures of discrimination (e.g., AUROC), calibration (e.g., scaled Brier score), and clinical performance relevant to the use case (e.g., positive predictive value at different sensitivity thresholds for rare events). Report criteria for choosing the final model(s). Highlight feature importance. | “In Fig. 4a,b, we list the top 15 features by mean absolute SHAP value for both the binary and multi-label classification, and different colored circles in Fig. 4b represent the feature importance of each category in multi-label classification.” (33) |
| Perform (when possible) and report the results of external validation. Recognize that performance will vary across institutions with different demographic and clinical populations. | “Each model was then retrained using the combined derivation and validation data from Site 1 and the retrained models were used for the validation in Site 2. Data from Site 2 constituted the external test dataset… On the external test dataset from Site 2, pCART also significantly outperformed all models, including the modified BedsidePEWS (AUC 0.80 vs. 0.74, P < 0.001).” (34) |
| Model implementation and availability | |
| Understand the sociotechnical environment and develop a plan for implementation, ideally based on known frameworks. Consider phased implementation with prospective “silent” testing and usability testing. Report stage of implementation, usability outcome, and future plans for clinical use. | “Implementation would also have to be considered in the greater context of regional resources, disproportionately affected populations, and in close collaborations with public health departments, regional healthcare partners, and healthcare coalitions rather than on an individual hospital or provider basis.” (36) |
| Support open science by providing public access to the data preparation scripts and model code, as well as the data set if feasible. | “Publicly available datasets were analyzed in this study. This data can be found here: GitHub, https://gitbub.com/fouticus/hptbi-hackathon.” (37) |
Table 3.
Summary of reporting guidance for predictive modeling work in PCCM
| Guidance | Example |
|---|---|
| Article information | |
| Follow appropriate reporting guidelines such as Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) and Reporting of studies Conducted using Observational Routinely-collected Data (RECORD). | “We designed our analysis to follow the recommendations of the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) statement, a recently released international guideline endorsed by 11 major journals.” (28) |
| Report the characteristics of the patients in the cohort being used to derive the model, comparing patients with the outcome against those without and those in the various data set partitions. | “Table 2. Clinical outcomes of patients with and without [serious bacterial infection], stratified by documented antibiotic-exposure in the 48-hours prior to PICU admission.” (29) |
| Data and Methods Used | |
| Report key elements of model development including: approach to partitioning of the data sets into non-overlapping training, validation, and testing data sets; prevalence and handling of missing data; outcome definition (and validation, if needed), approach to variable/feature selection; and model parameters and tuning. | “Three unique datasets were created from the registry, based on two different non-random data splits…. 1. A training set, from 5 sites, April 1, 2013 - December 31, 2016. 2. A temporal holdout set from the 5 original sites, January 1, 2017 - June 30, 2018. 3. A geographical holdout set from a small pediatric community-based ED site, April 1, 2013 - June 30, 2018. The temporal test set was intended as the primary test set, while the geographic test set was designed to be a small, preliminary evaluation of the performance of the model in a non-tertiary setting.” (30) |
| Outcomes of interest | |
| Report the number of outcome events in the data set. Recognize that the size of the data set, source of the model’s features, and number of outcome events will determine the number of predictor variables (or features) that can be included in the model. Articulate how these relationships may impact the model being reported. | “Because we sought to identify early AKI, analysis of candidate variables was limited to data available within the first 12 hours of ICU admission…. Of the [9,396] patients analyzed, 4% had early AKI…. Of the 33 candidate variables tested in each of the nine pathophysiologic groups, 17 had an association with early AKI….” (32) |
| Model implementation and availability | |
| Explicitly describe the model’s intended purpose (e.g., trigger-based, trajectory-based) and how it might be operationalized for use in clinical care | “For example, when a laboratory test is ordered, the clinician may be alerted that the likelihood of identifying a normal value is 95%, which may prompt a consideration of foregoing testing.” (35) |
Conclusion:
Prediction of a patient’s outcome will continue to be a cornerstone of pediatric critical care, whether done solely through clinical means or through the use of machine learning approaches. While these approaches have demonstrated the ability to create predictive models, there is a clear need for consistency in the content and quality of model development and reporting as well as standardization of relevant terminology to maximize clinical utility. An early focus on implementation, preferably in coordination with experts in implementation to ensure a user-centered design approach, will be particularly important for the ability of accurate models to lead to improved clinical care.
Supplementary Material
Research in Context:
Prediction of patient trajectories and outcomes is a core aspect of critical care and may be enhanced by machine learning-based prediction models.
General guidelines about the development and reporting of prediction models have been adopted by respiratory, sleep, and critical care journals.
Little is known about the current use of predictive modeling in pediatric critical care.
What This Study Means:
The publication of prediction models in pediatric critical care has substantially increased over time.
There is a lack of consistency of terminology and reporting of study methodology across articles, which must be overcome for the field to benefit from this important work.
An early focus on implementation may lead to improved success in the ability of clinical prediction models to improve clinical care.
Acknowledgements:
We thank Matthew P. Malone, MD of University of Arkansas for Medical Sciences/Arkansas Children’s Hospital and Sanjiv D. Mehta, MD, MBE of Children’s Hospital of Philadelphia for their participation in data collection for this project. We additionally appreciate the input of the PALISI Scientific Committee in reviewing this manuscript. We thank William Sveen, MD of University of Minnesota for his assistance with the study infographic (Figure 2); images used in the study infographic are “Paper” by Throwaway Icons, “Model” by Adrien Coquet, “Chart” by Alice Design, “Checked Box” by Abdullah Al Noman, and “Implementation” by Daisy, all from Noun Project. REDCap at the University of Minnesota is supported by grant Number UL1TR002494 from the National Institutes of Health’s National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Copyright Form Disclosure:
Drs. Bennett and Mayampurath’ s institution received funding from the National Heart, Lung, and Blood Institute. Drs. Bennett, Dziorny, Kamaleswaran and Mayampurath received support for article research from the National Institutes of Health (NIH). Dr. Bennett’s institution received funding from the National Institute of Child Health and Human Development and the National Center for Advancing Translational Sciences. Dr. Sanchez-Pinto received funding from Celldom, Inc, Allyx, Inc., and Saccharo, Inc. Dr. Martin’s institution received funding from the Children’s Hospital Colorado Research Institute and the Thrasher Research Fund. Dr. Kamaleswaran’s institution received funding from the NIH. The remaining authors have disclosed that they do not have any potential conflicts of interest.
References:
- 1.Shah N, Arshad A, Mazer MB, Carroll CL, Shein SL, Remy KE. The use of machine learning and artificial intelligence within pediatric critical care. Pediatr Res. 2023;93(2):405–412. doi: 10.1038/s41390-022-02380-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594 [DOI] [PubMed] [Google Scholar]
- 3.Leisman DE, Harhay MO, Lederer DJ, et al. Development and Reporting of Prediction Models: Guidance for Authors From Editors of Respiratory, Sleep, and Critical Care Journals. Crit Care Med. 2020;48(5):623–633. doi: 10.1097/CCM.0000000000004246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiens J, Fackler J. Striking the Right Balance-Applying Machine Learning to Pediatric Critical Care Data. Pediatr Crit Care Med. 2018;19(7):672–673. doi: 10.1097/PCC.0000000000001578 [DOI] [PubMed] [Google Scholar]
- 5.Fackler JC, Rehman M, Winslow RL. Please Welcome the New Team Member: The Algorithm. Pediatr Crit Care Med. 2019;20(12):1200–1201. doi: 10.1097/PCC.0000000000002149 [DOI] [PubMed] [Google Scholar]
- 6.O’Brien CE, Noguchi A, Fackler JC. Machine Learning to Support Organ Donation After Cardiac Death: Is the Time Now?. Pediatr Crit Care Med. 2021;22(2):219–220. doi: 10.1097/PCC.0000000000002639 [DOI] [PubMed] [Google Scholar]
- 7.Sanchez-Pinto LN, Bennett TD. Evaluation of Machine Learning Models for Clinical Prediction Problems. Pediatr Crit Care Med. 2022;23(5):405–408. doi: 10.1097/PCC.0000000000002942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pittman E, Bernier M, Fackler J. Data, Anomalies; a Call to Cease and Desist. Pediatr Crit Care Med. 2023;24(1):72–74. doi: 10.1097/PCC.0000000000003133 [DOI] [PubMed] [Google Scholar]
- 9.Bennett TD. Pediatric Deterioration Detection Using Machine Learning. Pediatr Crit Care Med. 2023;24(4):347–349. doi: 10.1097/PCC.0000000000003222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randolph AG, Bembea MM, Cheifetz IM, et al. Pediatric Acute Lung Injury and Sepsis Investigators (PALISI): Evolution of an Investigator-Initiated Research Network. Pediatr Crit Care Med. 2022;23(12):1056–1066. doi: 10.1097/PCC.0000000000003100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Pinto LN, Bennet TD, Stroup EK, et al. Derivation, Validation, and Clinical Relevance of a Pediatric Sepsis Phenotype With Persistent Hypoxemia, Encephalopathy, and Shock. Pediatr Crit Care Med. 2023;10.1097/PCC.0000000000003292. doi: 10.1097/PCC.0000000000003292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winter MC, Day TE, Ledbetter DR, et al. Machine Learning to Predict Cardiac Death Within 1 Hour After Terminal Extubation. Pediatr Crit Care Med. 2021;22(2):161–171. doi: 10.1097/PCC.0000000000002612 [DOI] [PubMed] [Google Scholar]
- 13.Typpo K, Watson RS, Bennett TD, et al. Outcomes of Day 1 Multiple Organ Dysfunction Syndrome in the PICU. Pediatr Crit Care Med. 2019;20(10):914–922. doi: 10.1097/PCC.0000000000002044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziorny AC, Heneghan JA, Bhat MA, et al. Clinical Decision Support in the PICU: Implications for Design and Evaluation. Pediatr Crit Care Med. 2022;23(8):e392–e396. doi: 10.1097/PCC.0000000000002973 [DOI] [PubMed] [Google Scholar]
- 15.Tricco AC, Lillie E, Zarin W, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann Intern Med. 2018;169(7):467–473. doi: 10.7326/M18-0850 [DOI] [PubMed] [Google Scholar]
- 16.Toh TSW, Lee JH. Statistical Note: Using Scoping and Systematic Reviews. Pediatr Crit Care Med. 2021;22(6):572–575. doi: 10.1097/PCC.0000000000002738 [DOI] [PubMed] [Google Scholar]
- 17.Kuhn M, Johnson K: Applied Predictive Modeling. First Edition. Philadelphia, Springer, 2013, p. 67 [Google Scholar]
- 18.Russell SJ, Norvig P: Artificial Intelligence - A Modern Approach. Third Edition. London, Pearson, 2009, p. 709 [Google Scholar]
- 19.Ripley B: Pattern Recognition and Neural Networks. First Edition. Cambridge, Cambridge University Press, 1996, p 354 [Google Scholar]
- 20.Janssens ACJW Martens FK. Reflection on modern methods: Revisiting the area under the ROC Curve. Int J Epidemiol. 2020;49(4):1397–1403. doi: 10.1093/ije/dyz274 [DOI] [PubMed] [Google Scholar]
- 21.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128–138. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michelson KN, Klugman CM, Kho AN, Gerke S. Ethical Considerations Related to Using Machine Learning-Based Prediction of Mortality in the Pediatric Intensive Care Unit. J Pediatr. 2022;247:125–128. doi: 10.1016/j.jpeds.2021.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberger AL, Amaral LA, Glass L, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):E215–E220. doi: 10.1161/01.cir.101.23.e215 [DOI] [PubMed] [Google Scholar]
- 24.Bennett T, Flynn A, Sanchez-Pinto LN, et al. 539: The Pediatric ICU Data Collaborative. Crit Care Med. 2021;49(1)33148950 [Google Scholar]
- 25.Trinkley KE, Kroehl ME, Kahn MG, et al. Applying Clinical Decision Support Design Best Practices With the Practical Robust Implementation and Sustainability Model Versus Reliance on Commercially Available Clinical Decision Support Tools: Randomized Controlled Trial. JMIR Med Inform. 2021;9(3):e24359. Published 2021 Mar 22. doi: 10.2196/24359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods-Hill CZ, Wolfe H, Malone S, et al. on behalf of the Excellence in Pediatric Implementation Science (ECLIPSE) authorship group for the Pediatric Acute Lung Injury and Sepsis Investigators (PALISI) Network. Implementation Science Research in Pediatric Critical Care Medicine. Ped Crit Care Med. In press. [Google Scholar]
- 27.Collins GS, Dhiman P, Andaur Navarro CL, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open. 2021;11(7):e048008. Published 2021 Jul 9. doi: 10.1136/bmjopen-2020-048008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett TD, DeWitt PE, Dixon RR, et al. Development and Prospective Validation of Tools to Accurately Identify Neurosurgical and Critical Care Events in Children With Traumatic Brain Injury. Pediatr Crit Care Med. 2017;18(5):442–451. doi: 10.1097/PCC.0000000000001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin B, DeWitt PE, Scott HF, Parker S, Bennett TD. Machine Learning Approach to Predicting Absence of Serious Bacterial Infection at PICU Admission. Hosp Pediatr. 2022;12(6):590–603. doi: 10.1542/hpeds.2021-005998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott HF, Colborn KL, Sevick CJ, Bajaj L, Deakyne Davies SJ, Fairclough D, Kissoon N, Kempe A. Development and Validation of a Model to Predict Pediatric Septic Shock Using Data Known 2 Hours After Hospital Arrival. Pediatr Crit Care Med. 2021. Jan 1;22(1):16–26. doi: 10.1097/PCC.0000000000002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerris EWJ, Sharron M, Zurakowski D, Staffa SJ, Yurasek G, Diab Y. Hospital-Associated Venous Thromboembolism in a Pediatric Cardiac ICU: A Multivariable Predictive Algorithm to Identify Children at High Risk. Pediatr Crit Care Med. 2020. Jun;21(6):e362–e368. doi: 10.1097/PCC.0000000000002293. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Pinto LN, Khemani RG. Development of a Prediction Model of Early Acute Kidney Injury in Critically Ill Children Using Electronic Health Record Data. Pediatr Crit Care Med. 2016. Jun;17(6):508–15. doi: 10.1097/PCC.0000000000000750. [DOI] [PubMed] [Google Scholar]
- 33.Zeng X, Hu Y, Shu L, Li J, Duan H, Shu Q, Li H. Explainable machine-learning predictions for complications after pediatric congenital heart surgery. Sci Rep. 2021. Aug 26;11(1):17244. doi: 10.1038/s41598-021-96721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayampurath A, Sanchez-Pinto LN, Hegermiller E, et al. Development and External Validation of a Machine Learning Model for Prediction of Potential Transfer to the PICU. Pediatr Crit Care Med. 2022;23(7):514–523. doi: 10.1097/PCC.0000000000002965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel BB, Sperotto F, Molina M, et al. Avoidable Serum Potassium Testing in the Cardiac ICU: Development and Testing of a Machine-Learning Model. Pediatr Crit Care Med. 2021;22(4):392–400. doi: 10.1097/PCC.0000000000002626 [DOI] [PubMed] [Google Scholar]
- 36.Killien EY, Mills B, Errett NA, Sakata V, Vavilala MS, Rivara FP, Kissoon N, King MA. Prediction of Pediatric Critical Care Resource Utilization for Disaster Triage. Pediatr Crit Care Med. 2020. Aug;21(8):e491–e501. doi: 10.1097/PCC.0000000000002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonseca J, Liu X, Oliveira HP, Pereira T. Learning Models for Traumatic Brain Injury Mortality Prediction on Pediatric Electronic Health Records. Front Neurol. 2022. Jun 10;13:859068. doi: 10.3389/fneur.2022.859068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


