Abstract
The equine infectious anemia virus (EIAV) often results in lifelong subclinical infection following early episodes of clinical disease. To identify the cellular reservoirs of EIAV during subclinical infection, horses were infected with EIAV and allowed to develop subclinical infections. Horses with acute disease served as a basis for comparison. The tissue distribution, replication status, location of infected cells, and viral load were characterized by PCR for proviral DNA and reverse transcriptase PCR for viral RNA, in situ hybridization, and in situ PCR. Proviral DNA was widespread in tissues regardless of disease status. Viral gag and env RNAs were also detected in tissues of all horses regardless of disease status. Plasma viral RNA (viremia) could be detected in some, but not all, horses with subclinical infections. In situ assays determined that a primary cellular reservoir and site of viral replication during subclinical infection is the macrophage. During subclinical infection, viral load was decreased 4- to 733-fold and there was decreased viral RNA expression within infected cells. These data indicate that viral replication continues at all times, even in horses that are clinically quiescent. Moreover, restricted viral replication at the cellular level is associated with clinical remission.
The factors that control the expression of clinical disease remain a poorly understood aspect of lentiviral pathogenesis. Salient features of lentiviral infections include lifelong persistence and prolonged periods of subclinical infection followed by progressive disease. However, in some proportion of infected domestic animals (6), nonhuman primates (6, 22), and possibly humans (4, 38), clinical disease never develops. The basis for the control of clinical lentiviral disease within a given host has not been fully established. Subclinical human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) infections are generally associated with reductions in viral load relative to initial infection and clinical AIDS, although viremia remains consistently detectable, indicative of continuing viral replication (1, 13, 18, 21, 34). Long-term subclinical HIV-1 infections have been associated with polymorphisms in HIV-1 chemokine coreceptors which affect viral tropism (10, 46), strong HIV-1-specific cell-mediated immune responses (7), and defective viruses (26). However, virus is not eliminated, and the long-term prognosis for these patients has not been determined (4, 38). In contrast, SIV infections of the natural primate host are completely apathogenic, even in the face of continuous viral replication (21). Infections with the ungulate lentiviruses ovine visna virus and caprine arthritis-encephalitis virus do not always result in clinical disease. However, the disease course is progressive if clinical signs do appear (6, 11).
Cellular reservoirs for lentiviruses include cells of the monocyte-macrophage lineage (6). Reservoirs for the immunodeficiency viruses also include latently infected lymphocytes (14) and virus sequestered on the processes of follicular dendritic cells (16). With the non-lymphocyte-tropic ungulate lentiviruses, disease signs are referable to infected macrophages and inflammatory lesions (6). Macrophages are also important as reservoirs, as restricted viral replication in these cells may allow the virus to avoid immunologic detection and may facilitate viral dissemination as “Trojan horses” (6, 17, 39).
Infection of horses with the equine infectious anemia virus (EIAV), a lentivirus, is characterized clinically by recurrent episodes of fever, anemia, and thrombocytopenia (reviewed in reference 43). Some horses die from either acute or chronic clinical disease. However, in most horses, disease episodes progressively decrease in frequency and intensity over about a year, after which the animals remain clinically normal (43). Acute episodes of clinical equine infectious anemia (EIA) are associated with extensive viral replication in tissue macrophages (5, 32) and readily detectable viremia (12, 20, 27, 47). In contrast, during subclinical EIAV infections, more sensitive techniques such as PCR are required to detect viral nucleic acids in tissues (25) and plasma (28), or inoculation of susceptible horses is needed to detect viral infectivity (8, 15, 20, 36). Thus, EIAV is similar to HIV-1 and SIV in that initial extensive viral replication is followed by a reduction in viral load (13, 18, 22). EIAV is distinct, however, in that many horses effectively suppress clinical disease following the onset of clinical signs, even when infected with highly virulent strains (25). This feature makes EIAV a useful model for the examination of a lentiviral infection in which disease can be successfully controlled. Since the sites and mechanisms of EIAV persistence have not been clearly defined, the purpose of this study was to identify the cellular reservoirs of EIAV in vivo during subclinical infection. The presence of viral replication and the amount of virus present were also evaluated.
MATERIALS AND METHODS
Animals, viruses, and clinical parameters.
Seven horses were infected intravenously with EIAV. Four horses (three Arabian foals and one adult pony) were infected with the highly virulent Wyoming strain of EIAV (EIAVWyo); two with 106 and one horse each with 103 and 101 horse infectious doses. The lower doses were used to reduce dose-dependent mortality (23). Three horses (one Arabian foal and two ponies) were infected intravenously with 106 50% tissue culture infectious doses of the WSU5 strain of EIAV (EIAVWSU5). EIAVWSU5 is an equine dermal fibroblast-adapted (29), pony-passaged (37) variant of EIAVWyo that is less virulent than EIAVWyo. Physical examinations, rectal temperatures, hemograms, and platelet counts were performed daily during clinical episodes and intermittently during chronic clinical disease or subclinical infection. Serum, plasma, and peripheral blood mononuclear cell (PBMC) samples were collected, processed, and stored at −80°C until needed. Necropsy tissues were fixed for 48 h in 4% paraformaldehyde and paraffin embedded. Replicate samples of unfixed tissues were snap-frozen in liquid nitrogen and stored at −80°C.
Nucleic acid extractions, PCR, and reverse transcriptase PCR (RT-PCR).
DNA from tissue and PBMC was extracted by using proteinase K digestion and a commercial extraction system (Stratagene DNA extraction kit), phenol-chloroform-isoamyl alcohol, and ethanol precipitation. Total RNA was extracted from tissues homogenized in a commercial guanidinium isothiocyanate-phenol-chloroform solution (Trizol Reagent; Life Technologies), followed by additional phenol-chloroform-isoamyl alcohol extraction and digestion with DNase. Plasma or serum viral RNA was obtained by pelleting the virions from 1 ml of sample at 47,000 × g for 1 h at 4°C, extracted as described above with a polysaccharide gel carrier (Microcarrier Gel-TR; Molecular Research Center).
DNA PCR was performed on 2 or 3 μg of DNA, using a protocol modified from that previously described (19). PCR amplification consisted of 1 cycle of 3 min at 95°C, followed by 35 to 40 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C and then 1 cycle of 7 min at 72°C. For nested PCR, the first round was performed as described above except that the annealing temperature was 50°C, and 1 μl of the first reaction was added to the second reaction and amplified as described above. The oligonucleotide primers used for DNA PCR were from the capsid protein sequence of EIAVWyo gag (40); they included primers 854 (5′ GGCTGGAAACAGAAATTTTA 3′) and 1262 (5′ TAGGTTTTCCAATCATCACT 3′) as internal primers and primers 636 (5′ CCATTGCTGGAAGATGTAAC 3′) and 1399 (5′ TGCGTTCTGAATAGTCAGTG 3′) as flanking primers. RT-PCR for viral RNA was performed on 2 or 3 μg of total RNA from tissues or 2 or 3 μl of RNA extracted from plasma. The assay combined reverse transcription and PCR in a one-tube format using the PCR primers to also initiate reverse transcription (19). RT-PCR and nested RT-PCR for EIAV gag RNA were performed as described above for DNA PCR except that the reaction mixture contained RNase inhibitor and Moloney murine leukemia virus reverse transcriptase (SuperScript II; Life Technologies), and PCR amplification was preceded by a reverse transcription step of 40 min at 42°C. RT-PCR and nested RT-PCR were also performed for the singly spliced message for EIAV env by selecting primers that bridged the mRNA splice site. This protocol was performed as described above, using the following oligonucleotide primers as described previously (3): 297 (5′ CTAGTTTGTCTGTTCGAGATCC 3′) and 5470 (5′ CTTGCTTCCTTCGATTCTGCCATGCTGTTC 3′) as flanking primers, with 297 and 5416 (5′ GGTTGAAACATTGTGTTCTCCTCACACTTAG 3′) as internal primers. The specificity of these assays for RNA was confirmed for positive samples by duplicate reactions without reverse transcriptase.
In situ hybridization (ISH) and immunohistochemistry.
Five-micrometer sections of paraffin-embedded tissues were mounted onto Microprobe-Plus slides (Fisher Scientific). Deparaffinized sections were rehydrated, permeabilized with 0.2 N HCl and 5 μg of proteinase K per ml, acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine buffer, and rinsed in 2× SSC (1× SSC is 150 mM NaCl and 15 mM sodium citrate). Nonspecific nucleic acid binding was blocked by treatment with prehybridization solution [50% formamide with 310 μg of sheared herring sperm DNA, per ml, 310 μg of poly(A) per ml, 31 mM EDTA, 25 mM HEPES, 1 M NaCl, 0.25% sodium dodecyl sulfate, 125 mM dithiothreitol, and 1× Denhardt’s solution). An EIAV-antisense RNA probe was produced by in vitro transcription using T7 RNA polymerase from pEIAp26.1 (pGEM4Z vector [Promega] containing a 450-bp fragment of EIAV gag) and labeled by [35S]dUTP (New England Nuclear-DuPont) or digoxigenin-dUTP (Boehringer Mannheim). Either 5 ng of 35S-labeled probe (approximately 4.5 × 106 cpm) or 100 ng of digoxigenin-labeled probe, diluted in prehybridization solution, was hybridized to each section overnight at 42°C. Following hybridization, unbound probe was digested with 50 μg RNase A per ml, and the sections were washed in decreasing concentrations of SSC. Bound 35S-labeled probe was detected by autoradiography (NTB2 emulsion; Eastman Kodak) for 5 to 14 days at −80°C. Bound digoxigenin-labeled probe was detected with Boehringer Mannheim’s antidigoxigenin Fab fragments conjugated with alkaline phosphatase, 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium chromogen (BCIP-NBT), and the Wash and Block system.
In tissues that have not been subjected to conditions that denature DNA, the antisense gag probe is specific for full-length genomic viral RNA, the presence of which is suggestive of viral replication (5, 41, 44). Controls for specificity included (i) hybridization on nondenatured, EIAV-infected tissues with a complementary-sense probe specific for proviral DNA (unavailable for hybridization), (ii) a nonsense probe from a gene of Trypanosoma cruzi, and (iii) the antisense probe on uninfected tissues.
In situ PCR.
Deparaffinized tissue sections were rehydrated, permeabilized with proteinase K (75 to 200 μg/ml) for 35 min at 37°C, rinsed in water and ethanol, and air dried. In situ PCR for proviral DNA was performed by applying 50 μl of PCR mix (500 pmol each of EIAV gag-specific primers 854 and 1262, 1.2 mM each deoxynucleoside triphosphate, 4.5 mM MgCl2, and 20 U of Taq DNA polymerase in Perkin-Elmer PCR II buffer) to the sections at 70°C, covering the reagents with Amplicover Discs and Clips (Perkin-Elmer), and using the GeneAmp In Situ PCR System 1000 for amplification as follows: 1 cycle of 4 min at 95°C for denaturation and then 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C. Negative controls included tissues from uninfected horses and PCR mix without Taq polymerase on infected tissues. Following thermal cycling, sections were serially rinsed in 2× SSC, phosphate-buffered saline, and ethanol and then air dried. DNA was denatured by heating for 4 min at 92°C, and amplified proviral cDNA was detected by ISH as described above. The probe used for the detection of in situ PCR products was a digoxigenin-labeled, sense-polarity RNA transcript of the same plasmid pEIAp26.1 used in ISH experiments. The sense-orientation probe was generated by transcription with SP6 RNA polymerase, is complementary to EIAV gag, and because of its sense polarity is specific for proviral DNA.
Quantitative PCR.
The copy number of proviral DNA in tissue was determined by adaptation of a method previously described for the quantitation of HIV-1 DNA (34). PCRs (primers 854 and 1262) were performed in triplicate and repeated nine times for each sample. Reaction products were visualized in ethidium bromide-stained agarose gels, and their densities were quantitated by a commercial digital imaging system (IS1000; Alpha Innotech). The copy number of provirus in 2 to 3 μg of tissue DNA was determined from a standard curve constructed by performing PCR in triplicate on known copy numbers of an EIAV-containing plasmid (33) and reported as copies per 10,000 cell-equivalents of tissue DNA. The plasmid copy number was calculated from its mass; mass was determined by comparing band intensities of the plasmid to dilutions of a DNA-mass standard (Life Technologies) in an ethidium bromide-stained agarose gel. PCRs for the test samples and plasmid were performed simultaneously, using a reagent-master mix, and were analyzed simultaneously on a single agarose gel.
RESULTS
Clinical disease.
All seven EIAV-infected horses initially developed clinical disease and detectable plasma viral RNA (viremia) (Table 1). Disease severity was related to the virulence of the infecting viral strain and, in contrast to a previous report (23), did not appear to be dose dependent. Signs in all horses included fever (up to 40.8°C) and thrombocytopenia; anemia was present in horses 2084, 2092, 2079, and 2085. Three horses (2079, 2084, and 2092) were euthanized with acute disease; two of these (2084 and 2092) developed hemorrhagic diathesis and were euthanized in extremis. Horse 2085 exhibited chronic EIA (43), characterized by prolonged clinical signs (30 days), followed by resolution of fever but persistent thrombocytopenia, anemia, and poor body condition until necropsy at day 124 postinfection. Horse 524 experienced an initial disease episode of 14 days, followed by six episodes of recrudescent disease (fever up to 41°C and thrombocytopenia but never anemia) lasting 2 to 5 days each. Plasma viral RNA was readily detected by RT-PCR during clinical episodes and the intervening subclinical periods. Following recrudescent disease, this horse remained afebrile until necropsy at day 878. During most of the afebrile period, this horse was borderline thrombocytopenic; however, at the time of necropsy, the platelet count had normalized and this horse was classified as subclinically infected. The remaining two horses (489 and 498) were necropsied at days 665 and 726, at which time they were clinically normal.
TABLE 1.
Summary of EIAV-infected horses, clinical parameters, and plasma viremia
| Horse no. | Virus strain | Virus dosea | Initial clinical diseaseb | Days postinfection at necropsy | Clinical status at necropsy | Viremia during acute diseasec |
|---|---|---|---|---|---|---|
| 2084 | Wyo | 106 | Severe | 15 | Acute clinical | Yes |
| 2092 | Wyo | 101 | Severe | 22 | Acute clinical | Yes |
| 2079 | WSU5 | 106 | Mild | 29 | Acute clinical | Yes |
| 2085 | Wyo | 106 | Severe | 124 | Chronic clinical | Yes |
| 524 | Wyo | 103 | Severe | 879 | Subclinical | Yes |
| 489 | WSU5 | 106 | Mild | 665 | Subclinical | Yes |
| 498 | WSU5 | 106 | Mild | 726 | Subclinical | Yes |
EIAVWyo, horse infectious doses; EIAVWSU5, 50% tissue culture infectious doses.
Severe disease characterized by evidence of hemorrhagic diathesis.
Virus was detected by RT-PCR on plasma and/or virus isolation on neonatal equine kidney cells.
Tissue proviral DNA and viral RNA.
PCR for proviral DNA and RT-PCR for viral RNA was performed on nucleic acids extracted from a panel of tissues to identify the tissue reservoirs of EIAV during acute disease and subclinical infection. In subclinically infected horses, this was to identify infected tissues for subsequent in situ studies. Amplicons were shown to be derived from EIAV gag or env by sequencing. Tissues from an uninfected control horse were negative (data not shown).
In the subclinically infected horses, not all tissues contained sufficient proviral DNA to be detected with a single round of PCR, although all tissues tested (except PBMC) were positive by nested PCR (Table 2). RT-PCR detected viral gag and/or env RNA in either spleen, bone marrow, or plasma of all three subclinically infected horses, although levels were sufficiently low in these horses to require nested RT-PCR (Table 3). In the two horses with subclinical EIAVWSU5 infections, plasma viral RNA could not be detected, even by using nested RT-PCR on as much as 4 ml of plasma. In contrast, the horses with acute and chronic clinical disease contained levels of proviral DNA that were readily detected with a single round of PCR in PBMC, lung, spleen, lymph node, and bone marrow (Table 2); also strongly positive were kidney, ileum, colon, heart, and brain (data not shown). Viral gag or env RNA was readily detected in either spleen, bone marrow, and plasma of these horses (Table 3). The widespread tissue distribution of EIAV is consistent with a number of previous reports (5, 24, 32, 42) for horses with acute disease and the single report for a subclinically infected horse examined by PCR (25). The presence of viral gag and env RNA indicates viral replication in the subclinically infected horses, even when EIAV is undetectable in plasma or PBMC.
TABLE 2.
Tissue distribution of EIAV gag DNA by PCR
| Horse no. | Clinical status | Virus strain | PCR (nested PCR) resulta
|
|||||
|---|---|---|---|---|---|---|---|---|
| PBMC | Lung | Spleen | Node | Bone marrow | Liver | |||
| 2084 | Acute clinical | Wyo | + | + | + | + | + | + |
| 2079 | Acute clinical | WSU5 | + | + | + | + | + | + |
| 2085 | Chronic clinical | Wyo | + | + | + | + | + | + |
| 524 | Subclinical | Wyo | + | + | + | + | + | + |
| 489 | Subclinical | WSU5 | − (−) | ± (+) | + | − (+) | − (+) | NT |
| 498 | Subclinical | WSU5 | − (−) | + | + | + | + | NT |
+, positive PCR result detected in ethidium bromide-stained agarose gels; −, negative result; ±, positive result weak and inconsistent; NT, not tested (tissue extracts inhibitory to PCR).
TABLE 3.
EIAV RNA in tissues and plasma, detected by RT-PCR and ISH
| Horse no. | Clinical status | Virus strain | RT-PCR (nested PCR)a
|
ISH, gag in spleen | ||||
|---|---|---|---|---|---|---|---|---|
| Spleen
|
Bone marrow
|
gag in plasma | ||||||
| gag | env | gag | env | |||||
| 2084 | Acute clinical | Wyo | + | + | ND | ND | + | + |
| 2079 | Acute clinical | WSU5 | + | + | ND | ND | + | + |
| 2085 | Chronic clinical | Wyo | + | + | + | − (+) | + | + |
| 524 | Subclinical | Wyo | + | − (+) | − (+) | − (+) | − (+) | + |
| 489 | Subclinical | WSU5 | − (+) | − (+) | ND | ND | −b | − |
| 498 | Subclinical | WSU5 | − (+) | − (+) | ND | ND | −b | − |
+, positive PCR result detected in ethidium bromide-stained agarose gels; −, negative result; ND, not done.
Ultracentrifuge pellets from 4 ml of plasma were pooled for nested RT-PCR.
ISH and in situ PCR.
ISH for viral gag RNA and in situ PCR for proviral gag DNA were performed to determine the cellular tropism of EIAV. Spleens were used for in situ experiments since this tissue, based on solution-phase PCR results, contained readily detected levels of viral DNA and RNA in all horses. Viral RNA-expressing cells were identified in spleens from horses with chronic clinical disease and subclinical infection with the virulent strain of EIAV (EIAVWyo) (Tables 3 and 4; Fig. 1a). The locations of these infected cells in the splenic sinusoids suggested that they were macrophages. Viral RNA-expressing cells could not be detected in spleens from the horses subclinically infected with the less virulent strain (EIAVWSU5), despite numerous attempts with both isotopic and nonisotopic ISH. Proviral DNA also could not be detected by ISH in any tissues from horses with subclinical infections. Proviral DNA was also undetectable by ISH in tissues from acutely infected horses, including tissues in which viral RNA was easily detected by ISH, indicating that the copy number of proviral DNA per cell was below the threshold of detection for the ISH assay. In situ PCR, however, did detect cells containing proviral DNA in the spleens of subclinically EIAVWSU5-infected horses; the presence of hemosiderin in some of these cells suggested that they were macrophages (Fig. 1b). Varying the amount of protease digestion used in experiments to identify proviral DNA in macrophages, in order to control for cell type variation in protease digestion optima, did not reveal additional types of infected cells. The ability to detect viral RNA (both gag and env) by solution-phase RT-PCR, but not by ISH, in these splenic macrophages (cells that contain levels of viral RNA readily detected by ISH during acute disease) indicates relative restriction of viral transcription at the cellular level. In situ PCR on spleens of horses with acute or chronic clinical disease did not identify proviral DNA in cells that differed in location, morphology, or numbers from the cells expressing viral RNA (Fig. 2), suggesting unrestricted replication during acute disease.
TABLE 4.
Amount of virus in spleen of EIAV-infected horses
| Horse no. | Virus strain | Clinical status | Mean copy no. of viral DNAa | % Infected cellsc |
|---|---|---|---|---|
| 2084 | Wyo | Acute clinical | 1,224 (189–4,154b) | 4.6 |
| 2092 | Wyo | Acute clinical | 2,200 (677–6,028) | 2.6 |
| 2079 | WSU5 | Acute clinical | 328 (50–589) | 0.77 |
| 2085 | Wyo | Chronic clinical | 273 (67–596) | 0.12 |
| 524 | Wyo | Subclinical | 85 (17–217) | 0.0025 |
| 489 | WSU5 | Subclinical | 3 (1–6) | NS |
| 498 | WSU5 | Subclinical | 15 (3–31) | NS |
Copy number in 10,000 cell-equivalents of spleen DNA. Each value is the mean for nine independent experiments.
Range of values for different experiments.
Ratio of infected cells, detected by ISH for viral RNA, to total cells in 120 4,500-μm2 fields. NS, viral RNA-containing cells could not be detected by ISH.
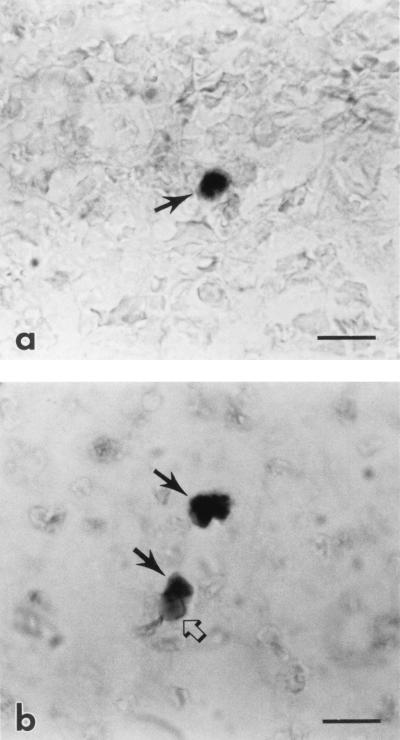
FIG. 1.
(a) Photomicrograph of a cell containing viral RNA (arrow) in the spleen from a horse subclinically infected with EIAVWyo. The cell is labeled by ISH (darkly staining cytoplasm) for EIAV gag RNA with the digoxigenin-labeled antisense probe and BCIP-NBT. Bar = 10 μm. (b) Photomicrograph of two cells containing proviral DNA (solid arrows) in the spleen from a horse subclinically infected with EIAVWSU5. The cell is labeled by in situ PCR (darkly staining nucleus) followed by ISH for EIAV gag cDNA with the digoxigenin-labeled sense probe and BCIP-NBT. Note the presence of hemosiderin (open arrow) in the cytoplasm of an infected cell, indicating that it is a macrophage. Bar = 10 μm.
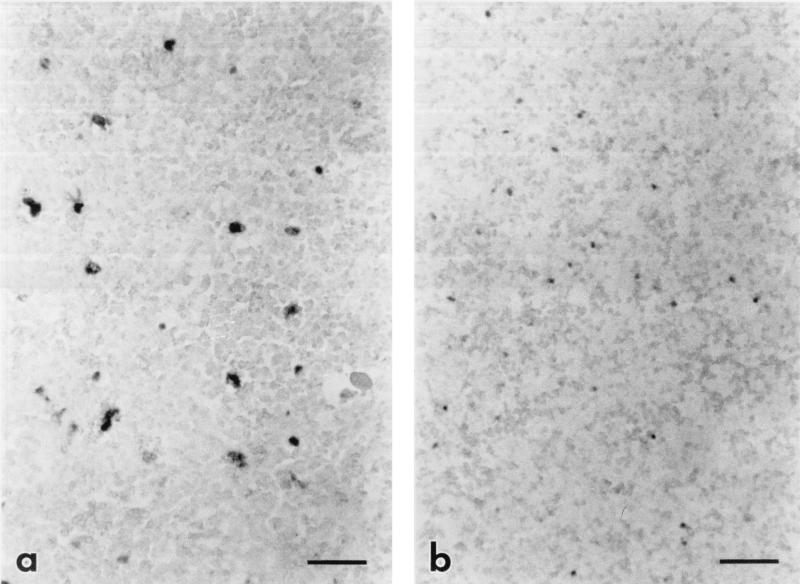
FIG. 2.
Photomicrograph of subadjacent sections from the spleen of a horse with acute clinical EIAVWSU5 infection, comparing the number cells containing viral RNA to the number of cells containing viral DNA. (a) Cells containing viral RNA (darkly staining cells) are labeled by ISH for EIAV gag RNA with the digoxigenin-labeled antisense probe and BCIP-NBT. Bar = 50 μm. (b) Cells containing viral DNA (darkly staining cells) are labeled by in situ PCR followed by ISH for EIAV gag cDNA with the digoxigenin-labeled sense probe and BCIP-NBT. Bar = 50 μm. Note the similar numbers of labeled cells in the sections, indicating that most or all infected cells are replicating virus during clinical disease.
During acute disease, viral RNA-containing cells were present in tissues from all major organ systems in all infected horses. ISH with control (sense and nonsense) probes, and with the antisense probe on tissues from an uninfected horse, did not detect significant nonspecific hybridization. Consistent with previous reports (5, 32), viral RNA-containing cells were predominantly macrophages, based on their location, morphology, and coexpression of lysozyme in dual-labeling experiments (35). Viral RNA-containing endothelial cells were also identified (35). Spleens contained large numbers of infected macrophages (Table 4) in the sinusoids and lymphoid germinal centers. A diffuse hybridization pattern in germinal centers suggested viral trapping on the processes of dendritic cells (Fig. 3a) (2, 14, 16). Lymph nodes, in contrast, did not contain large numbers of infected cells or have preferential hybridization in lymphoid follicles (Fig. 3b).
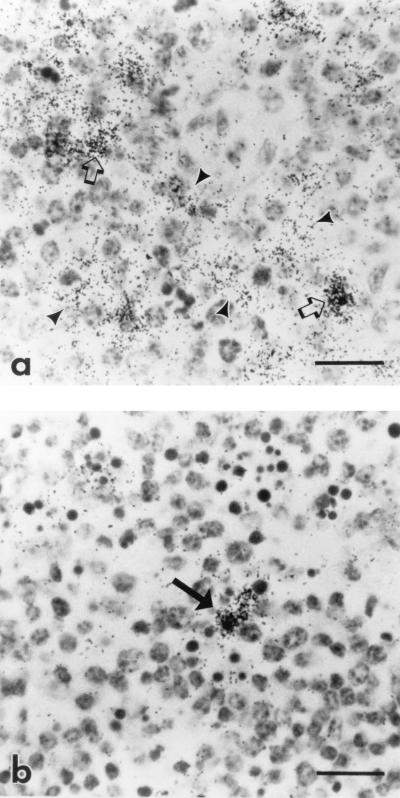
FIG. 3.
Photomicrograph of spleen and lymph node from a horse with acute clinical EIAV infection. Cells containing viral RNA (dark silver grains) are labeled by ISH for EIAV gag with the 35S-labeled antisense probe. (a) Germinal center in the spleen. Note the reticular pattern of silver grains indicating the presence of virus on the processes of follicular dendritic cells (arrowheads), in addition to the discrete pattern of silver grains indicating infection of individual cells (arrows). Bar = 20 μm. (b) Germinal center in the lymph node. Note the discrete pattern of silver grains indicating infection of individual cells (arrow) and absence of trapping of EIAV by dendritic cells. Bar = 20 μm.
Quantitation of viral load.
The copy number of provirus in 10,000 cell-equivalents of spleen DNA was determined by PCR to compare viral load during acute disease and subclinical infection (Table 4). An example of a standard curve used to determine copy number of proviral DNA is shown in Fig. 4. During subclinical infection, the copy number of EIAV provirus was decreased 4- to 733-fold in comparison to acute disease (3 to 85 copies for subclinical infection, versus 328 to 2,200 copies for acute disease). The horse with chronic clinical disease had a level of proviral DNA (273 copies) that overlapped levels for horses with both acute disease and subclinical infections. Although large standard deviations (ranging from 52 to 120%) and the small number of horses in each group do not allow statistical comparison, the data suggest that subclinical EIAV infections are associated with reductions in viral load.
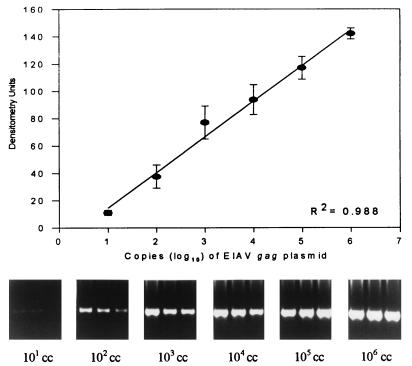
FIG. 4.
Example of a standard curve used to quantitate EIAV gag DNA in tissues by PCR. Plasmid copy number is plotted against densitometry units. R2, regression coefficient. Portions of agarose gel, stained with ethidium bromide, show PCR bands in triplicate from serial dilutions of EIAV gag-containing plasmid. cc, copies of plasmid.
Viral load was also evaluated by using the percentage of viral RNA-expressing spleen cells, detected by ISH, in 120 4,500-μm2 microscope fields (Table 4). During subclinical infection, the percentage of infected cells was decreased 308- to 1,840-fold in comparison to acute disease (0.77 to 4.6% for acute disease, versus 0.0025% for subclinical infection). The horse with chronic clinical disease had an intermediate percentage of infected cells (0.12%).
Viral load also appears to be influenced by the virulence of the infecting viral strain. Horses with acute EIAVWyo infection (horses 2084 and 2092) had a greater viral load (based on either copies of provirus or RNA-expressing cells) than the horse with acute EIAVWSU5 infection (horse 2079). Similarly, the horse with subclinical EIAVWyo infection (horse 524) had a greater viral load than the horses with subclinical EIAVWSU5 infections (horses 489 and 498). These differences indicate that EIAV virulence is related to viral load and viral replication.
DISCUSSION
The results of this study indicate that subclinical EIAV infection is associated with (i) reductions in viral load and (ii) relative transcriptional restriction in the same cell population (tissue macrophages) that is highly permissive during acute disease. Cell types other than the macrophage do not appear to serve as reservoirs of persistence for EIAV. Infected endothelial cells, detected during acute disease (35), were not apparent during subclinical infection.
Consistent with previous reports (5, 24, 25, 32, 42), solution-phase PCR for proviral DNA in this study showed that EIAV is present in most tissues, even after as long as 2 years of clinical remission. Detection of viral RNA in subclinically infected horses indicates that viral replication occurs during subclinical infection. The spleen appears to be a predominant site of both viral replication and persistence. However, the spleen is not the sole site of viral persistence during subclinical infection, as EIAV replication also occurs in other tissues such as bone marrow. Detection of viral RNA may not correlate with productive infection if there are posttranscriptional blocks to viral replication, as has been reported for visna virus (17). The detection of plasma viral RNA in one of the subclinically infected horses (horse 524) indicates that even if there are posttranscriptional blocks to productive infection, they are not absolute. Thus, virologic latency is not a requirement for EIAV persistence or subclinical infection.
Decreased viral load was also a feature of subclinical infection in this study. The amounts of proviral DNA in 10,000 cell-equivalents of spleen DNA, and the amount of plasma viral RNA, were markedly lower during subclinical infection than during acute disease. These reductions in viral load are similar to those reported between asymptomatic and immunosuppressed HIV-1-infected patients (9, 34) but are less than the 105-fold reduction reported for EIAV by others (25). This discrepancy may be related to the other study’s examination of liver (not analyzed in this study due to the consistent presence of PCR inhibitors) or the longer period of subclinical infection (23 years). Spleens from horses acutely infected with the virulent EIAVWyo contained higher levels of proviral DNA per unit of spleen, and higher numbers of RNA-containing cells, than spleens from horses acutely infected with the less virulent EIAVWSU5. This finding suggests that a determinant of virulence for EIAVWyo is enhanced viral replication during acute disease and may be related to differences in the long terminal repeat enhancer regions (31).
Macrophages appear to be highly permissive in vivo during acute disease (5) and therefore likely to be rapidly eliminated by viral cytopathology and/or the immune system. Thus, it was anticipated that another, less permissive cell type may be a reservoir of persistence. This, however, does not seem to be the case; the site of viral persistence and replication during subclinical infection are apparently macrophages. Trapping of virus by follicular dendritic cells in spleen or lymph node was not present in horses with chronic clinical disease or subclinical infections; thus, while EIAV appeared to be collected on the processes of dendritic cells during high-titered viremia, these cells are not an extracellular reservoir of virus as in HIV-1 and SIV infections (2, 16). PCR in situ was necessary to detect infected cells (proviral DNA containing) during subclinical infection; ISH was insufficiently sensitive. ISH with the sense-polarity probe for proviral DNA, on tissue sections with denatured DNA, was consistently negative, even in tissue sections with readily detectable viral RNA. Since only the spleen was examined in situ, the possibility that other, nonmonocyte/macrophage cell types in other tissues may be sites of persistence cannot be entirely excluded.
Restricted viral transcription in macrophages was present during subclinical infection. During acute disease, the majority of provirus-containing cells also expressed viral RNA, suggesting productive infection and lack of transcriptional restriction in these cells. Viral RNA was undetectable by ISH in two of the horses with subclinical infections. It could, however, be detected by solution-phase RT-PCR. Most or all of these macrophages express levels of viral RNA detectable by ISH during acute disease, which suggests relative restriction of viral transcription during subclinical infection. The significance of restricted viral replication to viral persistence, or to the genesis of antigenic variants which may initiate recrudescent disease (27), has not been determined. The mechanisms that downregulate transcription in vivo also have not been determined but are likely to be important in the pathogenesis of EIA. In vitro, EIAV can be transcriptionally upregulated during differentiation of monocytes into macrophages (30, 44) and downregulated by cytokines expressed by macrophages (45). In vivo, differences in long terminal repeat enhancer region motifs which may affect viral transcription and virulence have also been identified (31). Identification of the host and/or viral factors that influence the biology of reservoir macrophages and differentiate them from the macrophages that are highly permissive to viral replication during acute disease will be important in the further characterization of the mechanisms of persistence of this equine lentivirus.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grants AI01255 (J.L.O.) and AI24291 (T.C.M.) from the National Institute of Allergy and Infectious Diseases and grant HL46651 from the National Heart Lung and Blood Institute (T.B.C.).
We acknowledge and thank Lori Fuller, Emma Karel, Brett Graham, and Jessica Kinney for excellent technical assistance and Susan Tornquist for assistance with hematology.
REFERENCES
- 1.Bagnarelli P, Menzo S, Valenza A, Manzin A, Giacca M, Ancarani F, Scalise G, Varaldo P E, Clementi M. Molecular profile of human immunodeficiency virus type 1 infection in symptomless patients and in patients with AIDS. J Virol. 1992;66:7328–7335. doi: 10.1128/jvi.66.12.7328-7335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskin G B, Martin L N, Murphey-Corb M, Hu F S, Kuebler D, Davison B. Distribution of SIV in lymph nodes of serially sacrificed rhesus monkeys. AIDS Res Hum Retroviruses. 1995;11:273–285. doi: 10.1089/aid.1995.11.273. [DOI] [PubMed] [Google Scholar]
- 3.Beisel C E, Edwards J F, Dunn L L, Rice N R. Analysis of multiple mRNAs from pathogenic equine infectious anemia virus (EIAV) in an acutely infected horse reveals a novel protein, Ttm, derived from the carboxy terminus of the EIAV transmembrane protein. J Virol. 1993;67:832–842. doi: 10.1128/jvi.67.2.832-842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 5.Clabough-Sellon D C, Perry S T, Coggins L, Fuller F J. Wild-type equine infectious anemia virus replicates in vivo predominantly in tissue macrophages, not in peripheral blood monocytes. J Virol. 1992;66:5906–5913. doi: 10.1128/jvi.66.10.5906-5913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements J E, Zink M C. Molecular biology and pathogenesis of animal lentivirus infections. Clin Microbiol Rev. 1996;9:100–117. doi: 10.1128/cmr.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clerici M, Shearer G M. Correlates of protection in HIV infection and the progression of HIV infection to AIDS. Immunol Lett. 1996;51:69–73. doi: 10.1016/0165-2478(96)02557-6. [DOI] [PubMed] [Google Scholar]
- 8.Coggins L. Carriers of equine infectious anemia virus. J Am Vet Med Assoc. 1984;184:279–281. [PubMed] [Google Scholar]
- 9.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford T B, Adams D S. Caprine arthritis-encephalitis: clinical features and presence of antibody in selected goat populations. J Am Vet Med Assoc. 1981;178:713–719. [PubMed] [Google Scholar]
- 12.Crawford T B, Wardrop K J, Tornquist S J, Reilich E, Meyers K M, McGuire T C. A primary production deficit in the thrombocytopenia of equine infectious anemia. J Virol. 1996;70:7842–7850. doi: 10.1128/jvi.70.11.7842-7850.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daar E S, Moudgil T, Meyer R D, Ho D D. Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324:961–964. doi: 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- 14.Embretson J, Zupancic M, Ribas J L, Burke A, Racz P, Tenner-Racz K, Haase A T. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 15.Evans K S, Carpenter S L, Sevoian M. Detection of equine infectious anemia virus in horse leukocyte cultures derived from horses in various stages of equine infectious anemia viral infection. Am J Vet Res. 1984;45:20–25. [PubMed] [Google Scholar]
- 16.Fox C H, Tenner-Racz K, Racz P, Firpo A, Pizzo P A, Fauci A S. Lymphoid germinal centers are reservoirs of human immunodeficiency virus type 1 RNA. J Infect Dis. 1991;164:1051–1057. doi: 10.1093/infdis/164.6.1051. [DOI] [PubMed] [Google Scholar]
- 17.Gendelman H E, Narayan O, Molineaux S, Clements J E, Ghotbi Z. Slow, persistent replication of lentiviruses: role of tissue macrophages and macrophage precursors in bone marrow. Proc Natl Acad Sci USA. 1985;82:7086–7090. doi: 10.1073/pnas.82.20.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant R M, Kaur A, Johnson R P, McClure H, Staprans S, Feniberg M B. Abstracts of the XI International Conference on AIDS. 1996. Host-virus relationships in pathogenic and nonpathogenic SIV infections, abstr. We.A; p. 400. [Google Scholar]
- 19.Hamel A L, Wasylyshen M D, Nayar G P S. Rapid detection of bovine viral diarrhea virus by using RNA extracted directly from assorted specimens and a one-tube reverse transcription PCR assay. J Clin Microbiol. 1995;33:287–291. doi: 10.1128/jcm.33.2.287-291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond S A, Cook S J, Lichtenstein D L, Issel C J, Montelaro R C. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartung S, Boller K, Cichutek K, Norley S G, Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemeny L J, Mott L O, Pearson J E. Titration of equine infectious anemia virus. Effect of dosage on incubation time and clinical signs. Cornell Vet. 1971;61:687–695. [PubMed] [Google Scholar]
- 24.Kim C H, Casey J W. Genomic variation and segregation of equine infectious anemia virus during acute infection. J Virol. 1992;66:3879–3882. doi: 10.1128/jvi.66.6.3879-3882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim C H, Casey J W. In vivo replicative status and envelope heterogeneity of equine infectious anemia virus in an inapparent carrier. J Virol. 1994;68:2777–2780. doi: 10.1128/jvi.68.4.2777-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 27.Kono Y. Recurrences of equine infectious anemia. Approaches to an understanding of the mechanisms. In: Bryans J T, Gerber H, editors. Equine infectious diseases III. S. Basel, Switzerland: Karger; 1973. pp. 175–186. [Google Scholar]
- 28.Langemeier J L, Cook S J, Cook R F, Rushlow K E, Montelaro R C, Issel C J. Detection of equine infectious anemia viral RNA in plasma samples from recently infected and long-term inapparent carrier animals by PCR. J Clin Microbiol. 1996;34:1481–1487. doi: 10.1128/jcm.34.6.1481-1487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmquist W A, Barnett D, Becvar C S. Production of equine infectious anemia antigen in a persistently infected cell line. Arch Gesamte Virusforschung. 1973;42:361–370. doi: 10.1007/BF01250717. [DOI] [PubMed] [Google Scholar]
- 30.Maury W. Monocyte maturation controls expression of equine infectious anemia virus. J Virol. 1994;68:6270–6279. doi: 10.1128/jvi.68.10.6270-6279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maury W, Perryman S, Oaks J L, Seid B K, Crawford T, McGuire T, Carpenter S. Localized sequence heterogeneity in the long terminal repeats of in vivo isolates of equine infectious anemia virus. J Virol. 1997;71:4929–4937. doi: 10.1128/jvi.71.7.4929-4937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuire T C, Crawford T B, Henson J B. Immunofluorescence localization of equine infectious anemia virus. Am J Pathol. 1971;62:283–292. [PMC free article] [PubMed] [Google Scholar]
- 33.McGuire T C, O’Rourke K I, Baszler T V, Leib S R, Brassfield A L, Davis W C. Expression of functional protease and subviral particles by vaccinia virus containing equine infectious anaemia virus gag and 5′ pol genes. J Gen Virol. 1994;75:895–900. doi: 10.1099/0022-1317-75-4-895. [DOI] [PubMed] [Google Scholar]
- 34.Michael N L, Vahey M, Burke D S, Redfield R R. Viral DNA and mRNA expression correlate with the stage of human immunodeficiency virus (HIV) type 1 infection in humans: evidence for viral replication in all stages of HIV disease. J Virol. 1992;66:310–316. doi: 10.1128/jvi.66.1.310-316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oaks, J. L., C. Ulibarri, and T. B. Crawford. Endothelial cell infection by equine infectious anemia virus. Submitted for publication.
- 36.O’Rourke K I, Besola M L, McGuire T C. Proviral sequence detected by polymerase chain reaction in peripheral blood cells of horses with equine infectious anemia lentivirus. Arch Virol. 1991;117:109–119. doi: 10.1007/BF01310496. [DOI] [PubMed] [Google Scholar]
- 37.O’Rourke K I, Perryman L E, McGuire T C. Antiviral, anti-glycoprotein and neutralizing antibodies in foals with equine infectious anaemia virus. J Gen Virol. 1988;69:667–674. doi: 10.1099/0022-1317-69-3-667. [DOI] [PubMed] [Google Scholar]
- 38.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori S D, Orenstein J M, Fox C, Schrager L K, Margolick J B, Buchbinder S, Giorgi J V, Fauci A S. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 39.Peluso R, Haase A, Strowring L, Edwards M, Ventura P. A trojan horse mechanism for the spread of visna virus in monocytes. Virology. 1985;147:231–236. doi: 10.1016/0042-6822(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 40.Perry S T, Flaherty M T, Kelley M J, Clabough D L, Tronick S R, Coggins L, Whetter L, Lengel C R, Fuller F. The surface envelope protein gene region of equine infectious anemia virus is not an important determinant of tropism in vitro. J Virol. 1992;66:4085–4097. doi: 10.1128/jvi.66.7.4085-4097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz R J, Trono D, Feinberg M B, Baltimore D. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell. 1990;61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- 42.Rice N R, Lequarre A S, Casey J W, Lahn S, Stephens R M, Edwards J. Viral DNA in horses infected with equine infectious anemia virus. J Virol. 1989;63:5194–5200. doi: 10.1128/jvi.63.12.5194-5200.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sellon D C, Fuller F J, McGuire T C. The immunopathogenesis of equine infectious anemia virus. Virus Res. 1994;32:111–138. doi: 10.1016/0168-1702(94)90038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sellon D C, Walker K M, Russell K E, Perry S T, Covington P, Fuller F J. Equine infectious anemia virus replication is upregulated during differentiation of blood monocytes from acutely infected horses. J Virol. 1996;70:590–594. doi: 10.1128/jvi.70.1.590-594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sellon D C, Walker K M, Russell K E, Perry S T, Fuller F J. Phorbol ester stimulation of equine macrophage cultures alters expression of equine infectious anemia virus. Vet Microbiol. 1996;52:209–221. doi: 10.1016/s0378-1135(96)00071-5. [DOI] [PubMed] [Google Scholar]
- 46.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 47.Tornquist S J, Oaks J L, Crawford T B. Elevation of cytokines associated with the thrombocytopenia of equine infectious anemia. J Gen Virol. 1997;78:2541–2548. doi: 10.1099/0022-1317-78-10-2541. [DOI] [PubMed] [Google Scholar]