Abstract
A BHK cell line persistently expressing a Kunjin (KUN) virus replicon RNA (repBHK, similar to our recently described ME/76Neo BHK cell line [A. A. Khromykh and E. G. Westaway, J. Virol. 71:1497–1505, 1997]) was used for rescue and propagation of KUN viruses defective in the RNA polymerase gene (NS5). A new infectious full-length KUN virus cDNA clone, FLSDX, prepared from our previously described cDNA clone pAKUN (A. A. Khromykh and E. G. Westaway, J. Virol. 68:4580–4588, 1994) and possessing ∼105-fold higher specific infectivity than that of pAKUN, was used for preparation of defective mutants. Deletions of the predicted RNA polymerase motif GDD (producing FLdGDD) and of one of the predicted methyltransferase motifs (S-adenosylmethionine [SAM] binding site, producing FLdSAM) were introduced separately into FLSDX. Transcription and transfection of FLdGDD and FLdSAM RNAs into repBHK cells but not into normal BHK cells resulted in their replication and the recovery of defective viruses able to replicate only in repBHK cells. Reverse transcription-PCR and sequencing analyses showed retention of the introduced deletions in the genomes of the recovered viruses. Retention of these deletions, as well as our inability to recover viruses able to replicate in normal BHK cells after prolonged incubation (for 7 days) of FLdGDD- or FLdSAM-transfected repBHK cells, excluded the possibility that recombination had occurred between the deleted defective NS5 genes present in transfected RNAs and the functional NS5 gene present in the repBHK cells. An RNA with a point mutation in the GDD motif (FLGVD) was also complemented in transfected repBHK cells, and defective virus was recovered by day 3 after transfection. However, in contrast to the results with FLdGDD and FLdSAM RNAs, prolonged (4 days or more) incubation of FLGVD RNA in normal BHK cells allowed recovery of a virus in which the GVD mutation had reverted via a single base change to the wild-type GDD sequence. Overall, these results represent the first demonstration of trans-complementation of defective flavivirus RNAs with deleterious deletions in the flavivirus RNA polymerase gene NS5. The complementation system described here may prove to be useful for the in vivo complementation of deletions and mutations affecting functional domains or the essential secondary structure in any of the other flavivirus nonstructural proteins.
The genome of the Australian flavivirus Kunjin (KUN) consists of single-stranded RNA of positive polarity comprising 11,022 nucleotides (14) with one long open reading frame coding for three structural (C, prM, and E) and seven nonstructural (NS; NS1 to NS5) proteins (9). We have been focusing our studies on the components of the flavivirus replication complex using KUN virus as a model for many years (5–7, 34). Previously we partially purified a functionally active KUN replication complex and showed that it was devoid of structural proteins and contained most of the NS proteins (7). Earlier we proposed a model for flavivirus RNA replication based on the recycling role of double-stranded (ds) RNA, the main template for RNA synthesis (5). Colocalization of NS1 protein and ds RNA by immunogold electron microscopy was shown in dengue virus-infected cells (24), suggesting a role for NS1 protein in RNA replication. Recent data on effects of mutations in yellow fever (YF) virus NS1 protein on synthesis of viral RNA also suggest its involvement in RNA replication (25). The involvement of NS3 and NS5 proteins in RNA replication has been implied because of the presence of conserved helicase (NS3) and RNA polymerase (NS5) motifs, experimental in vitro data on the nonspecific RNA-dependent RNA polymerase (RDRP) activity of purified dengue virus NS5 with inhibition of this RDRP activity by anti-NS5 antibodies (31), binding of Japanese encephalitis NS3 and NS5 proteins to the 3′ untranslated region (UTR) (4), and blocking of the exchange of ds RNA templates during in vitro RDRP assays for dengue virus by anti-NS3 and anti-NS5 antibodies (2). Recently we showed colocalization in KUN virus-infected cells of NS1 and NS3 with ds RNA by immunofluorescence (IF) and immunogold electron microscopy analyses and that virtually all the NS but no structural proteins were coprecipitated by antibodies to ds RNA (34). Taken together, these data indicate involvement of nearly all the NS proteins in flavivirus RNA replication.
In extension of our studies of the roles of individual components of the replication complex, we decided to explore the use of our stable full-length KUN virus cDNA clone (14) and our recently developed BHK cell line persistently expressing KUN virus replicon RNA deficient in the structural genes (15) for mutagenesis and complementation analyses of individual KUN virus NS proteins. The NS5 gene was chosen as a first target because it contains several highly conserved domains characteristic of RDRPs of positive-strand RNA viruses (3, 13, 18, 27, 28). Within these domains the sequence motif GDD (KUN virus NS5 residues 665 to 667) (9) was of particular interest because of the demonstrated importance of this sequence for the functional activities of RNA polymerases of other positive-strand RNA viruses, including encephalomyocarditis virus (29), poliovirus (12), and hepatitis C virus (23). In addition to the RNA polymerase domains, two other conserved domains characteristic for methyltransferases were identified at the amino terminus of flavivirus NS5 by computer-assisted analysis (19). No experimental data on the importance of these domains for functional activity and/or viral replication are available for any of the members of Flaviviridae. It was therefore of interest to determine whether mutations or deletions in the described motifs in NS5 would have an effect on virus RNA replication in vivo and furthermore whether they could be complemented by functionally active NS5 supplied in trans. In contrast to the extensive number of complementation studies with RNA polymerase and other NS genes of other positive-stranded RNA viruses, such as poliovirus (reviewed in references 17 and 35; see also references 10, 26, and 32) and alphaviruses (see, for example, references 11 and 21), only one publication describes successful complementation of a defective flavivirus NS gene, the NS1 gene of YF virus (22).
In this report, we present the first direct demonstration in vivo of the deleterious effects on RNA replication of a deletion and a point mutation in the conserved RNA polymerase motif (GDD) and of a deletion in one of the methyltransferase domains (S-adenosylmethionine [SAM] binding site) in the flavivirus NS5 gene. We also show for the first time that the replication of these defective mutated RNAs was complemented in trans by transfection into a BHK cell line persistently expressing KUN virus replicon RNA.
MATERIALS AND METHODS
Cells.
BHK21 cells were grown in Dulbecco’s modification of minimal essential medium (DMEM; Gibco BRL) supplemented with 10% fetal bovine serum (FBS) at 37°C in a CO2 incubator.
RT and PCR amplification.
All reverse transcription (RT) reactions were performed with Superscript II RNase H− reverse transcriptase (Gibco BRL) essentially as described by the manufacturer by using 100 to 200 ng of purified KUN virion RNA or 1 μg of total cell RNA and appropriate primers. PCR amplification after RT of a 6,895-bp DNA fragment was performed with an Expand High Fidelity PCR kit (Boehringer Mannheim) and with a 1/25 to 1/10 volume of RT product as follows. The PCR mixture (50 μl) containing all necessary components except the enzyme mixture (3 parts Taq polymerase and 1 part Pwo polymerase) was preheated at 95°C for 5 min and then the enzyme mixture was added and the following cycles were performed: 10 cycles of 95°C for 15 s and 72°C for 6 min, followed by 6 cycles of 95°C for 15 s and 72°C for 6 min, with an automatic increase in the extension time (at 72°C) of 20 s in each subsequent cycle. All PCRs with Pfu DNA polymerase (Stratagene) were performed essentially as described by the manufacturer.
Construction of the plasmids.
Plasmids FLSD and FLSDX, shown in Fig. 1, were obtained from the previously described stable KUN virus full-length cDNA clone pAKUN (14) by replacement of the original cDNA fragments with those obtained by RT and PCR amplification of purified KUN virus RNA (see the previous section) with existing unique restriction sites, which were also incorporated into the primers for PCR amplification. A KUN virus replicon plasmid, C20DXrep, was prepared by replacing SphI at position 2467 and XhoI at position 11021 in C20rep (15) with the fragment from the full-length cDNA clone FLSDX (Fig. 1). The dicistronic replicon construct C20DXrepNeo used for generation of replicon-expressing BHK cells (repBHK) was prepared from C20DXrep by cloning an internal ribosomal entry site-neomycin transferase gene cassette into the 3′ UTR 25 nucleotides downstream of the polyprotein termination codon (similar to ΔME/76Neo [15]).
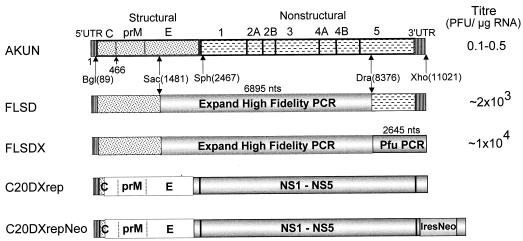
FIG. 1.
Construction and specific infectivities of the full-length KUN virus cDNA clones and the structures of KUN virus replicon RNAs. Schematic representations of the full-length constructs and of the constructs with deletions (replicon) show consecutive replacements of the cDNA fragments in the AKUN clone (stippled boxes) with analogous fragments obtained by RT-PCR from KUN virion RNA (shaded boxes) as described in Materials and Methods. PFU titers on the right-hand side of the figure are averages (of results from three experiments) obtained after electroporation of the transcribed RNAs into BHK21 cells and determined by plaque assay (see Materials and Methods); the titer of purified wild-type KUN virus RNA was ∼105 to ∼106 PFU/μg of RNA. Arrows marked Bgl(89), Sac(1481), Sph(2467), Dra(836), and Xho(11021) indicate restriction enzyme sites used in replacement cloning, with the numbers in parentheses representing nucleotide numbers in the KUN virus sequence (9, 14). An Expand High Fidelity PCR kit (Boehringer Mannheim) was used to obtain the indicated cDNA fragment of 6,895 nucleotides (nts) in the FLSD and FLSDX constructs, and Pfu PCR in FLSDX indicates that the cDNA fragment of 2,645 nucleotides was obtained with Pfu DNA polymerase (Stratagene). C20DXrep and C20DXrepNeo constructs were prepared as described in the Materials and Methods. Open boxes represent the deleted part of the genome (see reference 15). Ires, internal ribosomal entry site of encephalomyelitis virus RNA; Neo, neomycin transferase gene.
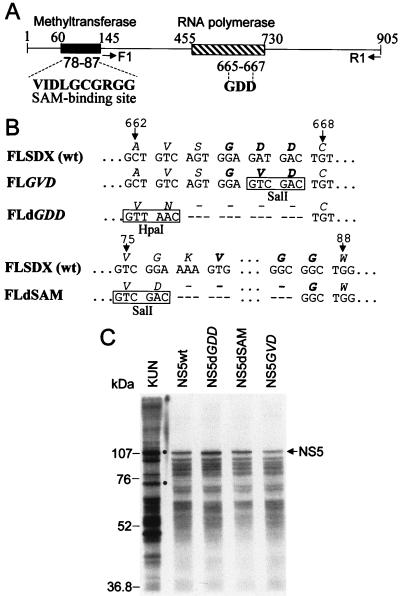
Deletion or mutation of the GDD motif and deletion of the SAM binding motif in the KUN virus NS5 gene (see Fig. 3A and B) were initially introduced into an intermediate plasmid, pBSNS5wt, containing the full-length NS5 gene in the pBluescript IIKS vector (Stratagene) by PCR-directed mutagenesis with high-fidelity Pfu DNA polymerase (8) and appropriate primers (Table 1) to obtain pBSNS5dGDD, pBSNS5GVD, and pBSNS5dSAM, respectively. In order to later distinguish between mutated RNAs and RNAs with deletions in complementation experiments, new restriction sites were incorporated into the individual primers used for the introduced mutation and deletions (Table 1; see Fig. 3B). After confirmation of the introduced mutation and deletions by restriction digestion with appropriate enzymes, fragments of the NS5 gene containing the corresponding mutation or deletions were first transferred into the C20DXrep plasmid and then into the FLSDX plasmid (containing full-length KUN virus cDNA) to obtain FLGVD, FLdGDD, and FLdSAM, respectively (see Fig. 3B). The mutation and deletions in the resulting FLGVD, FLdGDD, and FLdSAM plasmids were confirmed by restriction digest and sequencing analyses.
FIG. 3.
Mutagenesis of KUN virus NS5 gene. (A) Schematic representation of the NS5 gene with motifs indicated; (B) Nucleotide and amino acid sequences of the region with a mutation and the regions with deletions. Numbers represent amino acid positions in the KUN virus NS5 gene (9). The filled box in panel A shows the boundaries of the region encompassing two proposed methyltransferase domains that include the SAM binding site (VIDLLGCGRGGW) (19), which is shown in boldface type in panel A. The hatched box in panel A shows boundaries of the region which includes a number of motifs proposed to be involved in RNA polymerase activity (3, 13, 18, 27, 28), including the RNA polymerase active site GDD shown in boldface type in panels A and B. R1 and F1 indicate the primers used for RT-PCR amplification (Fig. 4B). Dashed regions in panel B represent deleted nucleotides and corresponding amino acids. Boxed letters in panel B show new restriction sites introduced into the sequence during mutagenesis as described in Materials and Methods. (C) Autoradiogram of a sodium dodecyl sulfate–10% polyacrylamide gel containing electrophoresed samples of the [35S]methionine- and [35S]cysteine-labelled proteins translated in rabbit reticulocyte lysates programmed with the native and mutated NS5 RNA transcripts produced by T7 RNA polymerase from the intermediate plasmids containing the corresponding NS5 cDNA sequences in the pBluescript IIKS vector (see Materials and Methods). The arrow shows the position of the full-length NS5 protein. The KUN virus lane represents a [35S]methionine-labeled KUN virus-infected Vero cell lysate; dots identify NS5 and NS3. Numbers on the left represent Bio-Rad low-range prestained-protein standards. wt, wild type.
TABLE 1.
DNA primers used to create mutations in the NS5 gene
| Mutation | Primer | Sequence (5′ to 3′)a | Genomic region (nucleotides) |
|---|---|---|---|
| Deletion of GDD | Forward | ctggttaaCTGTGTGGTAAAGCCCTT | 9688–9704 (+ sense) |
| Reverse | cgggttaaCCATGCGGCTGAGTCTTT | 9654–9670 (− sense) | |
| GDD to GVD | Forward | cctcgtctctTCAGTGGAGtcGACTGT | 9674–9690 (+ sense) |
| Reverse | cctcgtctccCTGACAGCCATGCGG | 9663–9677 (− sense) | |
| Deletion of SAM | Forward | ggggtcgacGGCTGGTGTTATTACAT | 7936–7952 (+ sense) |
| Reverse | ggggtCGACCGGTTCAAGAAA | 7888–7903 (− sense) |
Nucleotides in boldface letters show introduced restriction sites, nucleotides in uppercase letters represent the authenic KUN virus sequence, and nucleotides in lowercase letters represent artificial sequences. Boldface italicized nucleotides in the GVD primers represent complementary free ends released after digestion with type II BsmBI restrictase, which digests beyond the recognition site (underlined).
RNA transcription and transfection and determination of specific infectivity.
RNA transcripts were prepared with SP6 RNA polymerase from the plasmid DNAs FLGVD, FLdGDD, and FLdSAM, linearized with XhoI, and electroporated into BHK21 cells, essentially as described previously (14, 15). Briefly, ∼10 μg of in vitro-transcribed RNAs were electroporated into 2 × 106 BHK21 (normal BHK) or repBHK cells in 400 μl in a 0.2-cm-electrode-gap cuvette (Bio-Rad) with a Bio-Rad Gene Pulser apparatus. To determine specific infectivity, BHK cells were electroporated with 10-μl serial 10-fold dilutions of the RNA transcripts (starting from 1 μg) and incubated in DMEM–10% FBS in 60-mm-diameter culture dishes for 6 h to allow cells to attach. Then cells were overlaid with DMEM–5% FBS in 1.5% agarose and stained with crystal violet after 4 to 5 days of incubation at 37°C.
Preparation of BHK cells persistently expressing the C20DXrepNeo replicon.
BHK21 cells persistently expressing KUN virus replicon RNA C20DXrepNeo (repBHK cells) were established by G418 (Geneticin) selection as described previously for preparation of ΔME/76Neo cells (15).
Immunofluorescence and Northern blot analyses.
Replication of mutated RNAs in transfected cells was monitored by IF analysis with mouse monoclonal antibodies to KUN virus E protein, designated 3.91D, 10A1, and 3.67G (1) (generously provided by Roy Hall, University of Queensland, Brisbane, Australia), as described elsewhere (16). Dual-IF analysis with anti-E and anti-NS3 antibodies was performed essentially as described previously (34). Northern blot hybridization of 2 to 5 μg of total RNA isolated from transfected or infected cells was performed as described previously (15), with (as the hybridization probe) a 32P-labelled cDNA fragment representing 977 nucleotides of the KUN virus prM and E genes (nucleotides 521 to 1498 of KUN virus cDNA) (9, 14).
Treatment of secreted mutant viruses prior to infectivity assays.
The culture fluid recovered from cells after transfection with mutated RNAs was filtered through 0.45-μm-pore-size filters (Sartorius) and treated with RNase A (20 μg per ml) for 20 min at 37°C in order to ensure the absence of particulate cellular material and of free RNA before attempting to transmit virus infections.
RESULTS
Improvement of the specific infectivity of the KUN virus full-length cDNA clone and of the transfection efficacy of the KUN virus replicon RNA.
The specific infectivity of RNA transcribed from our previously described stable full-length KUN virus cDNA clone pAKUN was relatively low (1 to 5 PFU per 10 μg of RNA) (Fig. 1) (14). For mutagenesis and complementation experiments, it was desirable to significantly improve the specific infectivity. Therefore, we replaced the SacII-DraIII (∼7 kb) fragment in the pAKUN clone (Fig. 1) with the corresponding cDNA fragment obtained by RT of purified KUN virion RNA and PCR amplified with an Expand High Fidelity PCR kit (Boehringer Mannheim), using appropriate primers (see Materials and Methods). RNA transcribed from the resulting cDNA clone (FLSD) had a specific infectivity of ∼2 × 103 PFU per 1 μg, compared to only 1 to 5 PFU per 10 μg for AKUN RNA (Fig. 1). We then commenced replacing the rest of the genome using PCR with high-fidelity Pfu DNA polymerase (Stratagene) (8). Thus, a 2,645-nucleotide fragment covering most of the NS5 gene and the 3′ UTR was inserted in FLSD cDNA to produce FLSDX (Fig. 1), which resulted in a total 104- to 105-fold improvement of the original specific infectivity, now equivalent to ∼104 PFU/μg of RNA (Fig. 1). Further replacement of the 1,392-nucleotide fragment covering C, prM, and part of E did not noticeably improve the specific infectivity of the resulting FLBSDX RNA (data not shown). The most infectious FLSDX clone was therefore used in all further mutagenesis experiments.
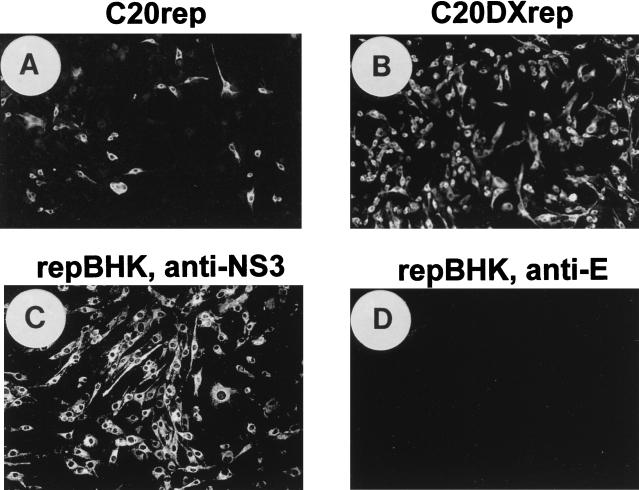
In order to improve the efficiency of transfection of KUN virus replicon RNA, we transferred a fragment from SphI at position 2467 to XhoI at position 11021 from the FLSDX clone into our replicon clone C20rep (15) to obtain the C20DXrep construct (Fig. 1). Electroporation of 5 to 10 μg of C20DXrep RNA resulted in its successful transfection and replication in ∼80% of cells, as judged by IF analysis with anti-NS3 antibodies at 24 h after electroporation (Fig. 2B), which was about eightfold more efficient than transfection with the same amount of C20rep RNA (Fig. 2A).
FIG. 2.
Improvement of transfection efficacy of KUN virus replicon RNA and establishment of a replicon-expressing BHK cell line (repBHK) shown by IF analysis. (A and B) IF-positive BHK21 cells with anti-NS3 antibodies at 24 h after electroporation with the original C20rep RNA and with C20DXrep RNA of improved efficiency, respectively. (C) IF analysis with anti-NS3 antibodies of BHK cells transfected with C20DXNeo RNA (constructed as described in Materials and Methods) and maintained for 38 passages as repBHK cells in medium supplemented with 1 mg of G418 per ml, followed by nine passages in medium without G418. (D) IF analysis of repBHK cells (passage 33) with anti-E monoclonal antibodies. This figure and subsequent figures were prepared by scanning all the original data (slides, autoradiograms, etc.) on an Arcus II scanner (Agfa) with FotoLook software (Agfa) for a Macintosh computer at a resolution of 150 lines per in., followed by adjustment of the brightness and the contrast of some images, assembling of the montages with Microsoft PowerPoint 97 software, and printing of the images on an Epson Stylus Color 400 printer at a resolution of 720 dots per in. on Epson photo-quality ink-jet or Epson photo-quality glossy paper.
Preparation of the KUN virus replicon-expressing BHK cell line.
Recently we described the preparation of a BHK cell line persistently expressing the KUN virus replicon RNA ME/76Neo suitable for use in complementation experiments (15). To ensure maximum complementation efficiency, we established a new BHK cell line (repBHK) persistently expressing the replicon RNA C20DXrepNeo, a derivative of C20Dxrep that was constructed as described in Materials and Methods. Continuous passaging of these cells in the presence of 1 mg of G418 per ml showed persistent replication of replicon RNA in virtually 100% cells for at least 6 months (68 passages) after transfection, as judged by IF analysis with anti-NS3 antibodies (data not shown). Removing the selection pressure for at least nine passages did not have any noticeable effect on the proportion of positive cells and the intensity of fluorescence (Fig. 2C), although the cells grew better in the absence of G418 (data not shown). Importantly for the subsequent complementation experiments (see below), repBHK cells were completely negative in IF with anti-E antibodies (Fig. 2D).
In order to detect possible interference of KUN virus replication in repBHK cells by the replicating C20DXrepNeo RNA, we compared the levels of production of virus after infection of normal BHK cells and of repBHK cells (passage 30) using ∼1 multiplicity of infection per cell of wild-type KUN virus. Some delay or inhibition of KUN virus replication was observed in the first 25 h after infection in repBHK cells compared with the rate of replication in normal BHK cells (yields, 1 × 107 and 7 × 107 PFU/ml, respectively), but by 45 h KUN virus production in repBHK cells (1.4 × 108 PFU/ml) was apparently similar to if not better than that in normal BHK cells (6 × 107 PFU/ml). Encouraged by the efficient replication of C20DXrepNeo RNA and the lack of any continuing interference with KUN virus replication, we commenced the complementation experiments described below using this newly prepared repBHK cell line.
Complementation of the mutated virion RNA FLGVD by replicon RNA.
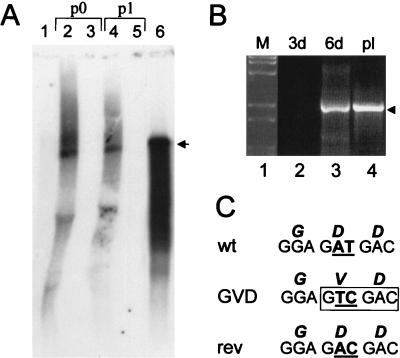
The GDD RNA polymerase motif in NS5 was mutated to GVD in the KUN virus full-length cDNA clone in plasmid FLSDX (see the previous section) as described in Materials and Methods (see Fig. 3A and B). In order to ensure that the introduced GVD mutation had not affected the open reading frame or the efficiency of translation, the mutated NS5 (NS5GVD) and native NS5 (NS5wt) mRNAs (prepared from the intermediate pBS plasmids) (see Materials and Methods and Fig. 1) were translated in rabbit reticulocyte lysates. We observed synthesis of similar amounts of predominantly full-size NS5 protein from both NS5GVD and NS5wt RNAs (Fig. 3C) plus some smaller products detected previously in similar assays which appeared to result from internal initiation of translation (33). Replication of FLGVD RNA was observed by 3 days posttransfection in repBHK cells but not in normal BHK cells, as judged by IF analysis with anti-E antibodies (results not shown) or by Northern blot analysis with a prM- and E-specific probe (Fig. 4A, lanes 2 and 3). When filtered and RNase-treated culture fluid from repBHK cells collected at 3 days after transfection was used for infection, replication of FLGVD RNA was again observed only in repBHK and not in normal BHK cells by 2 days after infection (Fig. 4A, lanes 4 and 5). Thus, an apparently lethal mutation in FLGVD RNA was successfully complemented in repBHK cells. However, a longer incubation of normal BHK cells after transfection of FLGVD RNA resulted in accumulation in culture fluid by 6 days posttransfection of a virus able to replicate after infection of fresh normal BHK cells (data not shown). RT-PCR analysis of RNA isolated from these transfected cells confirmed the presence of KUN virus-specific RNA at day 6 but not at day 3 (Fig. 4B, lanes 2 and 3). Sequencing analysis of a PCR fragment showed that one of the changed bases (T) in the mutant Val codon (GTC) had back mutated, resulting in restoration of the wild-type GDD amino acid sequence (Fig. 4C). Interestingly, the adjacent second mutated nucleotide (C) remained unchanged (Fig. 4C), thus confirming that the recovered RNA was indeed derived from the initially transfected FLGVD RNA.
FIG. 4.
Complementation in KUN virus replicon-expressing BHK (repBHK) cells of full-length KUN virus RNA with a GDD-to-GVD mutation in the NS5 gene. (A) Northern blot analysis of total RNA isolated from repBHK cells (lanes 2 and 4) or normal BHK cells (lanes 3 and 5) at 3 days after transfection with FLGVD RNA (p0) and at 2 days after infection with 3-day-posttransfection culture fluid (p1), with a radiolabelled cDNA probe representing 977 nucleotides of KUN virus prM and E genes (see Materials and Methods). Lane 1 contains mock-transfected repBHK cells, and lane 6 contains 10 ng of control FLGVD RNA transcribed in vitro. An arrowhead indicates the position of RNA of about 11 kb, determined relative to migration in the same gels of ethidium bromide-stained λ DNA digested with BstEII (New England Biolabs). (B) RT-PCR analysis with F1 and R1 primers (Fig. 3A) of total RNA isolated from FLGVD RNA-transfected normal BHK cells. Lane 1 contains λ DNA digested with BstEII (New England Biolabs). Lanes 2 and 3 contain RNA samples isolated from normal BHK cells at 3 and 6 days (3d and 6d), respectively, after electroporation with FLGVD RNA. Lane 4 contains the control DNA fragment of 2.5 kb obtained by PCR amplification of FLGVD cDNA. (C) Comparison of the nucleotide and deduced amino acid (boldface italic letters above the nucleotides) sequences in the GDD motif of wild-type (wt) and GVD mutated cDNAs with that of revertant (rev) cDNA. The sequence of the rev cDNA was obtained by automatic cycle sequencing of the purified 2.5-kb RT-PCR fragment shown in lane 3 in panel B with appropriate primers and by using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Brisbane, Australia). Boldface underlined letters represent nucleotides that either were targeted for mutation (AT in the wt sequence), mutated (TC in the GVD sequence), or reverted after replication of FLGVD RNA in BHK cells (AC in the rev sequence). Boxed letters show the SalI recognition site introduced by the GVD mutation.
Complementation of the KUN virus genome with a deletion of the GDD motif in the NS5 gene.
In order to eliminate the possibility of reversion of the mutated GDD motif to a wild-type sequence, as was observed with the FLGVD RNA (see the previous section), we prepared RNA with a coding deletion of 4 amino acids including the GDD motif (FLdGDD) (Fig. 3A and B). In vitro translation analysis of NS5dGDD mRNA transcribed from the intermediate pBSNS5dGDD plasmid (see Materials and Methods) showed efficient translation of full-length NS5dGDD protein (Fig. 3C), indicating that the introduced deletion did not affect either the open reading frame or the translational efficiency of the resulting mRNA.
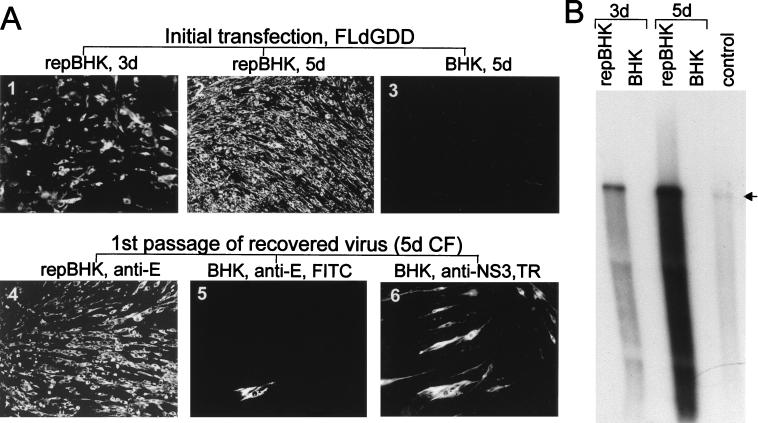
When FLdGDD RNA was transfected into repBHK and normal BHK cells in parallel experiments, replication of FLdGDD RNA was detected in repBHK cells but not in normal BHK cells; approximately 50 and 100% of repBHK cells were positive by IF analysis with anti-E antibodies at 3 and 5 days, respectively (Fig. 5A, photos 1 and 2), indicating replication and secondary spread of the mutated genomic RNA after 3 days. Replication and secondary spread were confirmed by the observed accumulation of FLdGDD RNA in transfected repBHK cells detected by Northern blot analysis with a radiolabelled prM- and E-specific probe (Fig. 5B, lanes repBHK). Importantly, no evidence of FLdGDD replication in normal BHK cells was detected at 5 days (Fig. 5A, photo 3, 5B, lane 5d BHK) or as late as 7 days (data not shown) after transfection. Infection of fresh repBHK cells with culture fluid collected at 5 days after productive transfection with FLdGDD RNA resulted in replication of defective FLdGDD virus in 100% of repBHK cells by 42 h after infection, as detected by IF with anti-E antibodies (Fig. 5A, photo 4). The infectious titer was determined by similar IF analysis with anti-E antibodies by using serial dilutions of the day 5 culture fluid assayed on repBHK cells. The number of IF foci decreased linearly with the dilutions of culture fluid, and the titer was ∼5 × 105 infectious units per ml. Recently we showed that KUN virus replicon RNA can be packaged by KUN virus structural proteins expressed in trans from another expression vector (16). Therefore, it was possible for the KUN virus replicon RNA present in repBHK cells to be packaged by structural proteins expressed from the complemented FLdGDD RNA. Furthermore, it was theoretically possible for FLdGDD RNA to be able to replicate in normal BHK cells, if single cells were simultaneously infected with two particles, one containing replicon RNA and the other containing FLdGDD RNA. We therefore performed dual-IF analysis at 42 h after infection of normal BHK cells with the virus recovered as described above at 5 days after transfection of FLdGDD RNA into repBHK cells, using anti-NS3 antibodies (able to detect replication of both replicon and FLdGDD RNAs) and anti-E antibodies (able to detect replication of only FLdGDD RNA). The results showed that ∼1% of infected normal BHK cells were positive for NS3 expression and that, of these NS3 positive cells, only ∼1 in 200 were also positive for E expression (Fig. 5A, photos 5 and 6). The implication of these results is that only those very rare (normal) BHK cells determined by IF to be positive for expression of both NS3 and E proteins were able to support replication of defective FLdGDD RNA. Significantly, because of cell division, a minor increase only in the number of cells positive for both E and NS3 was observed when normal BHK cells infected with FLdGDD virus were incubated longer (4 days) (data not shown), indicating that no spread of the defective FLdGDD virus had occurred. Importantly, virus able to replicate and spread in normal BHK cells was never detected in the culture fluid of FLdGDD-transfected repBHK cells even after prolonged incubation, or after two to three passages of secreted FLdGDD virus on repBHK cells (data not shown). These results exclude the presence of any replication-competent (recombinant) virus in the stock of defective FLdGDD virus and thus strongly indicate that no detectable recombination between the deleted NS5 gene (coded in FLdGDD RNA) and the functional native NS5 gene (present in repBHK cells) ever occurred in these experiments. Our results are comparable to those of Lindenbach and Rice (22), who also did not find any evidence for recombination between the functional YF NS1 gene expressed from a Sindbis vector and a deleted YF NS1 gene in YF genomic RNA in their trans-complementation experiments. Taken together, these results demonstrate that the deletion of the putative RNA polymerase GDD motif in the NS5 gene of genomic KUN virus RNA can be efficiently complemented in trans by wild-type NS5 expressed from KUN virus replicon RNA persistently replicating in repBHK cells.
FIG. 5.
Complementation of full-length KUN virus RNA with a GDD deletion in the NS5 gene (construct FLdGDD). Results of IF analysis with anti-E antibodies (A) and Northern blot analysis with the E-specific probe (B) for detection of replicating FLdGDD RNA are shown. Photos 1 and 2 in panel A and the corresponding repBHK lanes in panel B demonstrate replication of FLdGDD RNA in repBHK cells at 3 and 5 days (3d and 5d), respectively, after electroporation. Photo 3 in panel A and the BHK lanes in panel B indicate the absence of replication of FLdGDD RNA as late as 5 days after transfection into normal BHK cells. Photo 4 in panel A shows results of IF analysis with anti-E antibodies of repBHK cells at 2 days after infection with the complemented FLdGDD virus recovered at 5 days after transfection of repBHK cells with FLdGDD RNA. Photos 5 and 6 show results of dual-IF analysis with anti-E (fluorescein isothiocyanate [FITC] stain; photo 5) and anti-NS3 (Texas Red [TR] stain; photo 6) antibodies of normal BHK cells infected for 2 days with complemented FLdGDD virus recovered at 5 days after transfection with FLdGDD RNA. CF, culture fluid. The control lane in panel B contains 10 ng of in vitro-transcribed FLdGDD RNA. The arrow indicates the position of RNA of about 11 kb, determined as described in the legend to Fig. 4. The Northern blot was exposed to X-ray film for 3 h.
Complementation of the KUN virus genome with a deletion in the methyl transferase motif in the NS5 gene.
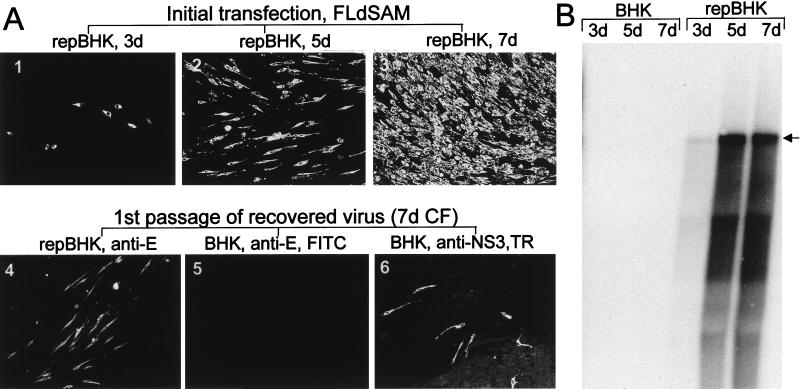
The methyltransferase motif of flaviviruses identified by Koonin (19) by computer-assisted analysis consists of two conserved domains in NS5: domain 1, containing the SAM binding site (VIDLGCGRGG, KUN virus NS5 amino acids 78 to 87) (Fig. 3A), and domain 2, containing DTLLCD (KUN virus NS5 amino acids 150 to 155). There are no published experimental data showing a requirement of these motifs in flavivirus replication. To address this issue, we deleted the more conserved domain, the SAM binding site, from the KUN virus full-length clone FLSDX without disrupting the NS5 open reading frame (FLdSAM) (Fig. 3B) and examined the effect of this deletion on the replication of transcribed RNA after transfection into normal BHK cells. No replication of FLdSAM RNA was detected either by IF analysis with anti-E antibodies (data not shown) or by Northern blotting with an E-specific probe (Fig. 6B, BHK lanes) up to 7 days after transfection of normal BHK cells. Furthermore, no replicated RNA was detected after transfer of culture fluids collected at 5 and 7 days after transfection into fresh normal BHK cells (data not shown). We showed that the deletion did not affect translation of NS5 protein, because when we translated NS5dSAM in a rabbit reticulocyte lysate using RNA prepared from the intermediate plasmid pBSNS5dSAM (see Materials and Methods), there were no significant differences in the size and in the amount of translated NS5dSAM protein from those of NS5wt protein (Fig. 3B). Thus, the absence of replication of FLdSAM RNA in transfected normal BHK cells implies an important role for the SAM binding motif in viral RNA replication and indicates that it may be associated with the RNA capping reaction (19).
FIG. 6.
Complementation of full-length KUN virus RNA with a deletion of the SAM binding site in the NS5 gene (construct FLdSAM). Results of IF analysis with anti-E antibodies (A) and Northern blot analysis with the E-specific probe (B) for detection of replicating FLdSAM RNA are shown. Photos 1 to 3 in panel A and the corresponding repBHK lanes in panel B demonstrate replication of electroporated FLdSAM RNA in repBHK cells at 3, 5, and 7 days (3d, 5d, and 7d, respectively). BHK lanes in panel B show the absence of replication of FLdSAM RNA at 3, 5, and 7 days after transfection into normal BHK cells. Photo 4 in panel A shows results of IF analysis with anti-E antibodies of repBHK cells at 2 days after infection with complemented FLdSAM virus recovered at 7 days after transfection of repBHK cells with FLdSAM RNA. Photos 5 and 6 show results of dual-IF analysis with anti-E (fluorescein isothiocyanate [FITC] stain; photo 5) and anti-NS3 (Texas Red [TR] stain; photo 6) antibodies of normal BHK cells infected with complemented FLdSAM virus recovered at 7 days after transfection of repBHK cells with FLdSAM RNA and immunostained 2 days later. CF, culture fluid. The arrow in panel B indicates the position of RNA of about 11 kb, determined as described in the legend to Fig. 4. The Northern blot was exposed to X-ray film for 24 h, compared to 3 h for the blot in Fig. 5B.
In order to study whether the defective NS5 protein with the deleted SAM binding site could be complemented in trans by functional NS5 protein, FLdSAM RNA was transfected into repBHK cells. Replication of FLdSAM RNA was detected but at a significantly lower rate than in the complementation experiments with FLdGDD RNA. Only ∼1 to ∼2% of cells were positive by IF with anti-E antibodies at 3 days after transfection, ∼40% were positive at 5 days, and 100% were positive at 7 days (Fig. 6A, photos 1 to 3). Northern blot results also showed that accumulation of FLdSAM RNA in repBHK cells was slower than that of FLdGDD RNA (compare exposure times and lanes 1 and 3 in Fig. 5B with lanes 4 and 5 in Fig. 6B). The difference in levels of efficiency of complementation and replication between these two RNAs became even more evident when we compared the proportion of anti-E-positive repBHK cells assayed at 42 h after infection with that of repBHK cell culture fluids collected at 5 days after transfection with FLdGDD RNA (∼100%) (Fig. 5A, photo 4) and at 7 days after transfection with FLdSAM RNA (∼10%) (Fig. 6A, photo 4). Likewise, the titer of defective FLdSAM virus in the day 7 culture fluid, determined by IF assay of infected fresh repBHK cells with anti-E antibodies (as described in the previous section), was ∼2 × 104 infectious units per ml, about 25 times lower than the corresponding titer of the day 5 FLdGDD culture fluid (∼5 × 105 infectious units per ml) (see the previous section). When the day 7 culture fluid from FLdSAM-transfected repBHK cells was used to infect normal BHK cells, dual-IF analysis with anti-NS3 and anti-E antibodies detected a few anti-NS3-positive and no anti-E-positive cells after 2 days in culture (Fig. 6A, photos 5 and 6). When these infected cells were incubated longer (4 days), some single isolated anti-E-positive cells were detected (results not shown), probably arising from the slowly replicating FLdSAM RNA in cells doubly infected with both FLdSAM and C20DXrepNeo particles (see the previous section). Moreover, no virus able to replicate and spread was detected by IF analysis of normal BHK cells infected with secreted defective virus recovered after two passages on repBHK cells (data not shown). The possibility of recombination occurring between replicon RNA and FLdSAM RNA was thus apparently excluded.
Structures of FLdGDD and FLdSAM virion RNAs.
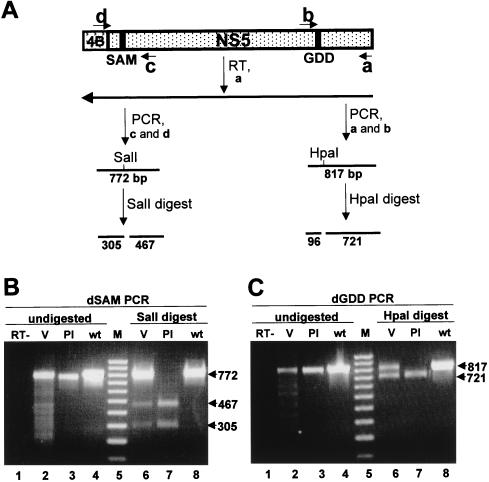
In order to confirm that the viruses recovered after transfection of FLdGDD and FLdSAM RNAs in repBHK cells retained the introduced deletions, we isolated RNAs from filtered and RNase A-treated culture fluids (collected at 5 days for FLdGDD and 7 days for FLdSAM) for restriction mapping and sequence analysis. The RNAs were reverse transcribed with a primer complementary to the C-terminal sequence of the NS5 gene (primer a) (Fig. 7A), and the resulting cDNAs were then PCR amplified with pairs of primers in the SAM and GDD regions (primer pairs c and d and a and b, respectively) (Fig. 7A). The predicted products were 772 bp for the FLdSAM RT-PCR and 817 bp for the FLdGDD RT-PCR (lanes 2 in Fig. 7B and C, respectively), as was found for the fragments amplified with the same primers from plasmids containing FLdSAM and FLdGDD cDNAs (lanes 3 in Fig. 7B and C, respectively) or wild-type FLSDX cDNA (Fig. 7B and C, lanes 4). PCR amplification from the parallel control reactions mixtures lacking reverse transcriptase produced no products (Fig. 7B and C, lanes 1). Because of the presence in culture fluid from complementation experiments of two types of particles with either encapsidated wild-type (replicon) or deleted (FLdGDD or FLdSAM) RNA (Fig. 5A and 6A), and because the primer used for RT did not distinguish between these two RNAs (primer a) (Fig. 7A), amplification of both deleted and wild-type fragments was anticipated. Thus, restriction digest of gel-purified PCR fragments with SalI and HpaI restrictases demonstrated that indeed a mixed population of RNAs with partially deleted and wild-type NS5 genes was present in the RNA isolated from the culture fluid collected after transfection with FLdSAM or FLdGDD RNA in repBHK cells (Fig. 7A, B, lanes 6 to 8, and C, respectively). A noticeably high percentage of undigested RT-PCR fragment (∼50% in FLdGDD particles and ∼80% in FLdSAM particles [lanes 6 in Fig. 7B and C, respectively]) probably represents a higher proportion of encapsidated replicon RNA in the particles secreted from transfected repBHK cells than was expected from the results of dual-IF analysis with anti-NS3 antibodies (detecting both mutated full-length and nonmutated replicon RNAs) and anti-E antibodies (detecting only mutated full-length RNA) (Fig. 5A and 6A, photos 5 and 6). However, it must be noted that the IF assay was performed at 2 days after infection with recovered complemented viruses (see legend to Fig. 5 and 6). This period allowed the defective viruses to spread in repBHK cells, which resulted in detection of expressed FLdGDD RNA by anti-E antibodies in ∼100% (FLdGDD) or ∼20% (FLdSAM) of infected repBHK cells (photos 4 in Fig. 5A and 6A, respectively), while replicon RNA could not escape from normal cells. Thus, in FLdGDD- and FLdSAM-infected normal BHK cells, only ∼1% or fewer cells were replicon positive (anti-NS3 positive) (photos 6 in Fig. 5A and 6A, respectively). In addition, a high percentage of undigested RT-PCR fragment may have been due to the preferential RT-PCR amplification of replicon (wild-type) cDNA over mutated (FLdGDD and FLdSAM) full-length RNAs because of the possible effects of introduced deletions on the RNA secondary structure. In order to demonstrate the presence of the introduced deletions in the defective FLdSAM and FLdGDD genomes by sequencing analysis, we cloned their PCR fragments separately into the pGEM-T vector (Promega) and recombinant plasmids containing the inserts with the appropriate restriction digest patterns were sequenced across the deleted regions. The results confirmed the retention of the deleted sequences (Fig. 3B) in the genomes of the defective viruses (data not shown).
FIG. 7.
Determination of the structure of the defective genomes. (A) Schematic representation of the KUN virus genome in the vicinity of the NS5 gene and the details of the RT-PCR protocol. SAM and GDD represent deleted motifs (Fig. 3). a, b, c, and d represent primers used for RT and PCR and correspond to the published KUN virus plus-sense sequence (9, 14). a, nucleotides 10378 to 10398 (minus sense); b, nucleotides 9576 to 9597 (plus sense); c, nucleotides 8372 to 8400 (minus sense); d, nucleotides 7606 to 7621 (plus sense). Lines marked SalI and HpaI indicate the positions of new sites in the defective genomes introduced into their cDNAs during construction (Fig. 3B). Numbers indicate the predicted sizes of the fragments obtained by PCR amplification and restriction digestion. 4B, NS4B. (B and C) Results of RT-PCR and restriction digest analyses of the RNAs isolated from the culture fluid collected at 7 days after transfection of FLdSAM RNA and at 5 days after transfection of FLdGDD RNA, respectively. The primer for RT was a for both reactions; the primer pairs for PCR were a and b for FLdGDD samples and c and d for FLdSAM samples (see panel A). Lanes 1 in both panels B and C contain PCR products from the parallel control reaction lacking reverse transcriptase (RT−). Lanes 2 contain the PCR products obtained from the RT reactions performed with RNAs from the defective viruses (V) FLdSAM (B) and FLdGDD (C). Lanes 3 contain the PCR products obtained after amplification of the plasmid DNAs (Pl) FLdSAM (B) and FLdGDD (C). Lanes 4 contain the PCR products obtained from the parental FLSDX plasmid DNA with primers specific for SAM (B) and GDD (C). Lanes 6 and 7 contain restriction digests of the corresponding purified PCR fragments shown in lanes 2 to 4 with SalI (B) or HpaI (C) restrictases. Lanes 5 in both panels B and C contain a 100-bp molecular size marker (M) (Progene, Brisbane, Australia). Arrows show the sizes (in thousands) of the undigested and digested DNA fragments. wt, wild type.
Overall, the results described in the last four sections clearly demonstrate that our established repBHK cell line, which expresses KUN virus NS proteins from the persistently replicating KUN virus replicon RNA, was successfully used to complement defective KUN virus genomes with deletions or a mutation in NS5, the putative RNA polymerase gene. It is also reasonable to assume from these results that this repBHK cell line can be used with a high probability of success for trans-complementation of KUN virus (or other flavivirus?) genomes with lethal deletions and mutations in any of the other NS genes.
DISCUSSION
A complementation system allowing trans rescue of defective KUN virus RNAs with deleterious deletions in an NS protein has been developed and involves transfection of these RNAs into a BHK cell line persistently expressing a KUN virus replicon (repBHK cells). Complementation by the replicon permits recovery of defective viruses able to replicate only in repBHK cells and not in normal BHK cells. Thus, deletions in an RNA polymerase motif (GDD) or a methyltransferase motif (SAM binding site) in the NS5 gene were rescued and corresponding defective viruses were recovered. This is the first report on successful trans-complementation of defined functional motifs in any of the flavivirus NS proteins. However, a successful trans-complementation of a large deletion with no assigned function in the flavivirus NS1 gene was recently demonstrated by the recovery of defective YF virus after transfection of YF RNA with the corresponding deletion in cells expressing NS1 protein from a noncytopathic Sindbis replicon (22).
In order to facilitate detection of possibly inefficient complementation of mutated full-length KUN virus RNAs in repBHK cells, we improved the specific infectivity of RNA transcribed from the parental cDNA by ∼105-fold by replacing 87% of the genome in the FLSDX clone with the cDNA fragments obtained after RT-PCR of purified virion RNA with high-fidelity polymerases. We then also prepared a new replicon construct, C20DXrep, for use as a helper in complementation by transferring a fragment coding for the NS region and the 3′ UTR from the FLSDX plasmid into our recently prepared C20rep replicon plasmid (15). About 80% of cells were successfully transfected by C20DXrep RNA, representing an ∼5- to ∼10-fold improvement in transfection efficiency of the parental C20rep RNA (see Results). The neomycin resistance gene inserted in C20DXrep allowed establishment of the repBHK cell line persistently expressing C20DXrepNeo RNA for complementation assays (see Results and Fig. 2).
Use of a single cell line (repBHK) with persistently replicating KUN virus replicon RNA should have an advantage for a large number of proposed trans-complementation experiments, compared with the alternative use of a number of cell lines or expression constructs, each expressing an individual NS protein. Persistent replication of replicon RNA should ensure continuous expression and correct processing of all the seven NS proteins and their intermediates in a functionally active form and therefore provide a quick, reliable, and universal system for complementing any defective NS genes. Replication of complemented defective full-length genomes in repBHK cells can be easily distinguished from the helper replicon RNA either by IF analysis with anti-E antibodies or by Northern blot hybridization with the prM- and E-specific probe. We showed previously that IF analysis can detect KUN virus protein products only if transfected KUN virus RNA is amplified and that the results of IF analysis correlate well with the results of detection of RNA accumulation by Northern blot analysis (15). As a first step in exploring our complementation system, we successfully complemented in repBHK cells two nonreplicating full-length KUN virus RNAs with deletions of the putative RNA polymerase (GDD; FLdGDD) and methyltransferase (SAM binding site; FLdSAM) motifs in the NS5 gene (see Results and Fig. 5 and 6). Moreover, this complementation resulted in the accumulation and secretion of defective viruses which could be passaged further in repBHK cells but not in normal BHK cells (Fig. 5 and 6, photos 4 and 5). Importantly, either deletion in the KUN virus NS5 gene resulted in the complete loss of ability of the mutated full-length RNAs to replicate in transfected normal BHK cells.
Although replicon RNA present in repBHK cells was encapsidated into a small proportion of secreted transmissible particles (Fig. 5A and 6A, photos 6), interpretation of complementation results was not compromised. Double infection of the same normal BHK cell with both types of encapsidated particles occurred with very low frequency (compare results in photos 5 and 6 in Fig. 5A and 6A) and was associated with a relatively high titer of defective viruses accumulated in the culture fluid. Moreover, no further spread of defective viruses in these cells (except by division of cells containing both defective and replicon RNAs) was detected even after prolonged (4 days) incubation. Importantly, no recombination apparently occurred between NS5 genes with deletions (in FLdGDD and FLdSAM RNAs) and the functional NS5 gene (in replicon RNA). This conclusion is based on (i) the absence of free virus spread in normal BHK cells after infection with the defective viruses recovered either at 5 to 7 days posttransfection of repBHK cells or after two to three passages on repBHK cells, as detected by IF analysis; (ii) a linear decrease in the number of IF-positive foci in repBHK cells infected with serial dilutions of the defective viruses; and (iii) retention of the introduced deletions in the recovered defective viral genomes as confirmed by RT-PCR, restriction digestion, and sequencing analysis. Significantly, although it occurs in other positive-strand RNA viruses of vertebrates such as alphaviruses, picornaviruses, and coronaviruses (for a review, see reference 20), to the best of our knowledge, recombination has never been reported for any member of the Flavivirus genus. Moreover, in the similar study reporting the trans-complementation of the YF NS1 protein (22), no recombination was detected even after three serial passages of defective virus.
Attempted complementation of the point mutation in FLGVD appeared to be successful early after transfection in repBHK cells, but after more than 3 days of incubation in normal BHK cells, a revertant (GDD) virus was recovered. The reversion was unexpected, because lethal point mutations involving more conservative substitutions of the first D in the GDD motif (e.g., with E, H, N, or Q) of poliovirus 3Dpol were stable for at least 5 days after transfection of the mutated full-length cDNAs (12). The emergence of the viable revertant virus from the nonviable mutant must mean that limited replication which was sufficient to produce a back mutation occurred early after transfection. Similar results relating to the emergence of viable viruses from nonviable mutants were recently described for N-terminal mutants of the Sindbis virus RNA polymerase nsP4 (30). Alternatively, an error producing a rare miscopied (wild-type) RNA molecule which finally gave rise to a revertant virus in transfected normal cells may have been introduced during its transcription from cDNA by SP6 RNA polymerase. A particular advantage of our complementation assay is that reversion or recombination, if it occurs in transfected repBHK cells, can be readily detected by transferring secreted (complemented) virus particles onto normal BHK cells and by monitoring by IF analysis the formation of any spreading foci of infection.
Taken together, these results clearly demonstrate that the repBHK cell line persistently expressing a KUN virus replicon can successfully be used for trans-complementation of lethal mutations in the KUN virus NS5 gene. We are exploiting this system by introducing mutations and deletions in the other NS proteins in the FLSDX KUN virus cDNA clone, examining their effects on RNA synthesis or virus replication, and testing for trans-complementation using the repBHK cell line. We believe that this efficient, quick, reliable, and generally applicable complementation system represents a major advance in flavivirus molecular genetics and should provide a powerful tool for studying the functional roles of flavivirus NS proteins in RNA and virus replication.
ACKNOWLEDGMENTS
We are grateful to Roy Hall for providing KUN virus anti-E monoclonal antibodies.
This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Adams S C, Broom A K, Sammels L M, Hartnett A C, Howard M J, Coelen R J, Mackenzie J S, Hall R A. Glycosylation and antigenic variation among Kunjin virus isolates. Virology. 1995;206:49–56. doi: 10.1016/s0042-6822(95)80018-2. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomeusz A I, Wright P J. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch Virol. 1993;128:111–121. doi: 10.1007/BF01309792. [DOI] [PubMed] [Google Scholar]
- 3.Bruenn J A. Relationships among the positive-strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991;19:217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C-J, Kuo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu P W, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 6.Chu P W, Westaway E G. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology. 1987;157:330–337. doi: 10.1016/0042-6822(87)90275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu P W, Westaway E G. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125:177–191. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- 8.Cline J, Braman J C, Hogrefe H H. PCR fidelity of Pfu polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 10.Collis P S, O’Donnell B J, Barton D J, Rogers J A, Flanegan J B. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J Virol. 1992;66:6480–6488. doi: 10.1128/jvi.66.11.6480-6488.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De I, Sawicki S G, Sawicki D L. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J Virol. 1996;70:2706–2719. doi: 10.1128/jvi.70.5.2706-2719.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jablonski S A, Morrow C D. Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J Virol. 1995;69:1532–1539. doi: 10.1128/jvi.69.3.1532-1539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7283. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh A A, Westaway E G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khromykh A A, Varnavski A N, Westaway E G. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkegaard K. Genetic analysis of picornaviruses. Curr Opin Genet Dev. 1992;2:64–70. doi: 10.1016/S0959-437X(05)80324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 19.Koonin E V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 20.Lai M C. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M-L, Wang H-L, Stollar V. Complementation of and interference with Sindbis virus replication by full-length and deleted forms of the nonstructural protein, nsP1, expressed in stable transfectants of HeLa cells. Virology. 1997;227:361–369. doi: 10.1006/viro.1996.8342. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach B D, Rice C M. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohman V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie J M, Jones M K, Young P R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 25.Muylaert I R, Galler R, Rice C M. Genetic analysis of the yellow fever NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 27.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice C M, Lenches E M, Eddy S R, Shin S J, Sheets R L, Strauss J H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 29.Sankar S, Porter A G. Point mutations which drastically affect the polymerisation activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J Biol Chem. 1992;267:10168–10175. [PubMed] [Google Scholar]
- 30.Shirako Y, Strauss J H. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J Virol. 1998;72:2310–2315. doi: 10.1128/jvi.72.3.2310-2315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan B-H, Fu J, Sugrue R J, Yap E-H, Chan Y-C, Tan Y H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 32.Teterina N L, Zhou W D, Cho M W, Enrenfeld E. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J Virol. 1995;69:4245–4254. doi: 10.1128/jvi.69.7.4245-4254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westaway, E. G., and A. P. Schrader. Unpublished data.
- 34.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]