Abstract
To evaluate conserved structures of the surface gp120 subunit (SU) of the human immunodeficiency virus type 1 (HIV-1) envelope in gp120-cell interactions, we designed and produced an HIV-1 IIIB (HXB2R) gp120 carrying a deletion of amino acids E61 to S85. This sequence corresponds to a highly conserved predicted amphipathic alpha-helical structure located in the gp120 C1 region. The resultant soluble mutant with a deleted alpha helix 1 (gp120 ΔαHX1) exhibited a strong interaction with CXCR4, although CD4 binding was undetectable. The former interaction was specific since it inhibited the binding of the anti-CXCR4 monoclonal antibody (12G5), as well as SDF1α, the natural ligand of CXCR4. Additionally, the mutant gp120 was able to bind to CXCR4+/CD4− cells but not to CXCR4−/CD4− cells. Although efficiently expressed on cell surface, HIV envelope harboring the deleted gp120 ΔαHX1 associated with wild-type transmembrane gp41 was unable to induce cell-to-cell fusion with HeLa CD4+ cells. Nevertheless, the soluble gp120 ΔαHX1 efficiently inhibited a single round of HIV-1 LAI infection in HeLa P4 cells, with a 50% inhibitory concentration of 100 nM. Our data demonstrate that interaction with the CXCR4 coreceptor was maintained in a SUgp120 HIV envelope lacking αHX1. Moreover, in the absence of CD4 binding, the interaction of gp120 ΔαHX1 with CXCR4 was sufficient to inhibit HIV-1 infection.
Human immunodeficiency virus type 1 (HIV-1) is the etiologic agent of AIDS (3, 33, 52). HIV-1 infection of target cells (monocytes or lymphocytes) is mediated by the viral envelope glycoproteins gp120 and gp41, with gp120 binding primarily to the CD4 receptor (20, 40, 45) with high affinity (44). Deletions and point mutations in gp120 have contributed to the identification of different sites which participate in the association with CD4 (5, 41, 44, 50, 61) and have defined amino acid W432 within the fourth constant (C4) region as being critical in this respect (18). Binding of gp120 to CD4 induces conformational changes in the HIV-1 envelope glycoproteins that are postulated to promote subsequent steps in virus entry (54, 55).
Recently, several members of the seven membrane-spanning chemokine receptor family have been identified as fusogenic coreceptors for HIV-1, HIV-2, and simian immunodeficiency virus (SIV) (1, 15, 23, 24, 28, 30, 38). Distinct tropisms of various HIV strains have been shown to result from their targeting of different chemokine receptors (1, 15, 24, 26, 28, 30), and studies with recombinant HIV-1 envelopes have indicated that the V3 loop of the HIV-1 gp120 protein is central to macrophage tropism and syncytium formation or fusion in CD4+ lymphocytes cultures (11–14, 17, 39). The CXCR4 (fusin) chemokine receptor functions as a coreceptor of T-cell-tropic or T-cell-line-adapted HIV-1 strains (26, 30, 42). It has been proposed that a CD4-induced change in gp120 conformation is necessary for correct HIV binding to chemokine receptors (60, 64), resulting in a trimolecular association between CXCR4 and the gp120/CD4 complex (43, 63).
Structural studies of monomeric and oligomeric forms of gp120 would further our understanding of the interactions between the virus and target cells. The lack of an X-ray crystallographic model of gp120 has resulted in the development of structure-function studies of this protein by different approaches, including computer algorithms, biochemical, mutagenic, and antibody binding analyses (25, 32, 34, 48, 65). These models have provided information about the existence of several beta-strands and five or six highly conserved alpha-helix (αHX) structures among gp120 proteins from different strains of HIV-1 (34). Furthermore, these αHX structures are widely conserved among different members of the retrovirus family including HIV-2, SIV, human T-cell leukemia virus type 1, visna virus, equine infectious anemia virus, bovine leukemia virus, and Rous sarcoma virus (32). Low-stringency antibody screening of a combinatorial peptide library has confirmed that the C1 domain of gp120 contains an αHX (αHX1) (58). This αHX1 is the largest of the αHX structures of gp120 and may be located at the interface between adjacent gp120 molecules in the oligomeric complex, since antibodies directed against this region can bind to monomeric gp120 but do not interact with the native oligomeric protein (48).
In the present study, we introduced a deletion in the surface gp120 subunit (SUgp120) of the HIV-1 IIIB (HXB2) envelope, between amino acids E61 and S85, corresponding to the sequence of the predicted αHX1 structure (34, 58). The resulting gp120 ΔαHX1 mutant envelope did not bind CD4 and had dramatically decreased fusion ability but maintained both the capacity to bind the CXCR4 chemokine receptor and inhibit HIV-1 infection. This may provide the basis for designing CXCR4-specific inhibitors that are CD4 independent in their action.
MATERIALS AND METHODS
Cell lines, viruses, and antibodies.
Cells lines were maintained at 37°C in a 5% CO2 humid atmosphere. HeLa-P4 cells (16) stably expressing the lacZ gene under the control of the HIV-1 long terminal repeat (HeLa CD4 LTR lacZ cells) were a gift from P. Charneau (Pasteur Institute, Paris, France) and were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) (Biomedia), 2 mM l-glutamine, penicillin, streptomycin, and 400 μg of Geneticin (G418) per ml. The HeLa Tat cell line (a gift from O. Schwartz, Pasteur Institute, Paris, France) was transfected to express the HIV-1 envelope and grown in complete DMEM with 2 mM methotrexate. The CEM CD4+ cell line was obtained from the American Type Culture Collection (Rockville, Md.) and grown in RPMI 1640 supplemented with 10% FCS. The CHO-K1 cell line, obtained from the American Type Culture Collection, and CHO-K1 cells transfected with a CXCR4 expression vector (a gift from Marc Parmentier, Euroscreen Co., Brussels, Belgium) were grown in Ham F12 medium (Life Technologies) supplemented with 10% FCS and 400 μg of G418 per ml. Plasmid pHXB2R is an HIV-1 IIIB-derived clone (obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Bethesda, Md.). HIV-1 LAI (obtained from Harvey Holmes, Medical Research Council, AIDS Reagent Project, NIBSC, United Kingdom) was grown in the CEM CD4+ and HeLa CD4 cell lines. The anti-CXCR4 monoclonal antibody (MAb) 12G5 (the kind gift of James Hoxie, University of Pennsylvania, Philadelphia) (29), reacts specifically with the human CXCR4 protein and recognizes a conformational epitope, probably located on the third extramembrane loop of the molecule (7). Samples of pooled HIV-immune immunoglobulin (HIV-Ig) (53) were obtained from the AIDS Reagent Repository, National Institutes of Health, Bethesda, Md. Sheep polyclonal antibody D7324 is an anti-gp120 antibody made against a peptide containing amino acids 497 to 511 of gp120 (Aalto BioReagents). The anti-gp120 110.4 MAb is directed against the GPGR sequence of the V3 loop (Genetic Systems), and the anti-gp120 110-K MAb is directed against the conformational epitope of the CD4 binding site (a gift from F. Traincard, Hybridolab). Rabbit anti-gp120 antiserum was made in our laboratory after immunization of a rabbit with a recombinant HIV-1IIIB gp120 purchased at Intracel Corp. The Anti-CD4 MAb Leu3a was purchased from Becton Dickinson (San Jose, Calif.). The anti-CD4 MAbs 13B8.2/IOKT4A and BL4/IOKT4 were purchased at Immunotech S.A. (Marseilles, France). Anti-CD4 MAb ST4/F101.69, anti-CD4 MAb ST40/F142.63, anti-CD4 MAb BF5, anti-CD100 MAb F93,7G2, and anti-CD5 MAb F145,GF3 were a gift from Sanofi Co. (Montpellier, France). The anti-CD4 MAb OKT4 was purchased from Ortho Diagnostic Systems, Inc. Anti-SDF1α antibodies were purchased from R & D Systems.
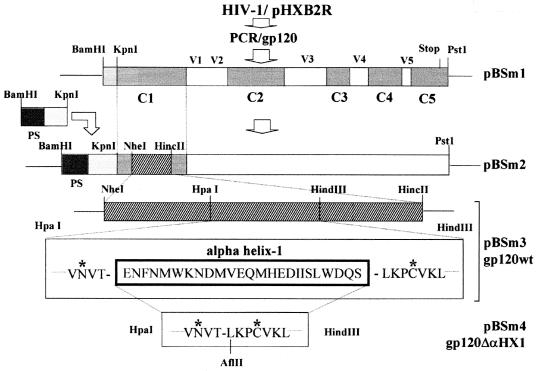
Recombinant gp120.
A 1,414-bp fragment encoding gp120 (from amino acids V12 to R481) was PCR amplified with plasmid pHXB2R as a template and the following two primers: sense primer (5′GCAGGATCCGGTACCTGTGTGGAAGGAAGC3′) and antisense primer (5′GCACTGCAGTTAGCGTTTCTCTCTCTGCACCACTC3′). The generated fragment contained a BamHI site upstream of the gp120 KpnI site (V12) and a stop codon at the end of the gp120 sequence followed by a PstI site. The BamHI-PstI fragment was then cloned into a Bluescript (pBS) vector (Stratagene) in which the KpnI site was eliminated by digestion with KpnI followed by repair with T4 DNA polymerase and religation, yielding the pBSm1 gp120 subclone. The sequence encoding the N-terminal of gp120 (T1 to G11) and a new signal peptide sequence, isolated from ecdysteroid glycosyltransferase gene of the baculovirus of Autographa californica, were added by using overlapping oligonucleotides and inserted into the BamHI-KpnI sites of pBSm1, giving pBSm2.
Two unique restriction sites flanking the αHX1 sequence were introduced by using oligonucleotides which changed codons V57-N58 and K91-L92 without altering their coding ability. The modification GTA→GTT at V57 and AAT→AAC at N58 and the modification AAA→AAG at K91 and TTA→CTT at L92, created HpaI and HindIII sites, respectively. The sequence was then introduced into pBSm2, producing plasmid pBSm3. To delete the αHX1, the wild-type HpaI-HindIII fragment was excised from plasmid pBSm3 and replaced by a mutated HpaI-HindIII fragment. This fragment was reconstituted by using two overlapping oligonucleotides (5′AACGTGACACTTAAGCCATGTGTAA3′ and 5′AGCTTTACACATGGCTTAAGTGTCACGTT3′), and an AflII site was also introduced in codon L86 by changing CTA to CTT to identify the mutated fragment, yielding plasmid pBSm4 (Fig. 1).
FIG. 1.
Schematic structure of HIV-1 gp120 depicting the step-by-step procedure used to delete the amphipathic αHX1 structure within the C1 region of gp120. The signal peptide (ps), conserved regions (C1 to C5), and variable regions (V1 to V5) are indicated. The different transition plasmids, from pBSm1 to pBSm4, through which gp120 wt (pBSm3) and the deleted gp120 (pBSm4) construct were obtained are shown. Asterisks denote conservation of the N-glycosylation and cysteine sites flanking αHX1.
The BamHI-PstI fragment, including the entire coding sequence of gp120, was then excised from pBSm3 or pBSm4 and cloned into the BglII-PstI sites of the P10 baculovirus transfer vector p119P (49a). Sf9 cells were cotransfected with viral DNA purified from the modified baculovirus AcSLP10 (10) and DNA from the recombinant p119P gp120 vector. Recombinant baculoviruses were plaque purified by standard methods (59). Sf9 cells were infected at a density of 5 × 105 cells/ml and at a multiplicity of infection of 5 PFU/cell. Supernatant was collected 6 days postinfection, and gp120 wt or gp120 ΔαHX1 was concentrated and immunopurified by chromatography with the anti-gp120 antibody D7324, linked on bromacetyl-Sepharose. Proteins were separated on a sodium dodecyl sulfate-PhastGel gradient (4 to 15%) in a discontinuous buffer system (PhastSystem; Pharmacia). The resolved protein bands were electrophoretically transferred onto nitrocellulose. After saturation, the blots were incubated with the appropriate labeled antibodies.
The entire gp120 wt or gp120 ΔαHX1 sequences were cloned into the pCEL/E160 HIV-1 envelope expression vector under the control of the cytomegalovirus CMV promoter. pCEL/E160 (a kind gift of Y. Boublik and M. Sitbon, Institut de Génétique Moléculaire, Montpellier, France) was derived from a previously described retroviral envelope expression vector (21, 22) by insertion of the HIV-1 LAI envelope (6). To obtain envelope expression with the desired gp120, the KpnI-NheI fragment was inserted into the original corresponding sequence of pCEL/E160 by using a second NheI restriction site located upstream of the stop codon derived from either pBSm3 or pBSm4.
Transient transfection and HIV-1 envelope-mediated cell fusion.
At 24 h after being plated at a concentration of 8 × 104/well in six-well flat-bottom plates, HeLa Tat cells (27) were transfected with 1 μg of the pCEL/E160-expressing vector by using Lipofectamine reagent (Gibco Life Sciences, Grand Island, N.Y.) as recommended by the manufacturer. The ability of various envelope glycoproteins to induce fusion and syncytium formation was assessed upon coculture with HeLa P4 cells harboring CD4 and HIV-1 long terminal repeat-driven lacZ genes (16). After 24 h, confluent cocultures were washed with phosphate-buffered saline (PBS), fixed with 0.5% glutaraldehyde for 10 min at room temperature, and washed twice with PBS. The cell monolayers were then stained by incubation with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) solution for 2 h at 37°C and washed twice with PBS. Fusion events between HeLa P4 cells and effector cells expressing the HIV-1 envelope and Tat transactivator resulted in induction of the in situ expression of the lacZ reporter gene. For each envelope glycoprotein tested, the total number of blue-stained foci per well was counted and photomicrographs were obtained.
Flow cytometry analysis of envelope cell surface expression.
The pCEL/E160 expression vector, carrying either wild-type SUgp120 (gp120 wt) or gp120 ΔαHX1, was cotransfected into HeLa-Tat cells with the pMACS-Kk plasmid expressing a truncated mouse H-2Kk membrane molecule (Miltenyi Biotec Inc.). DNA transfection was carried out by particle bombardment delivery with a Biolistic PDS-1000/He apparatus (Bio-Rad) (31). Briefly, 3 mg of 1.6-μm-diameter gold beads was coated with 2.5 μg of the mixed DNA containing 0.5 μg of pMACS-Kk and 2 μg of pCEL/E160. After 24 h of culture, transfected cells were detached, washed, and incubated for 1 h at 4°C with 80 μl of magnetic microbeads coated with anti-H-2Kk MAb. Cells expressing H-2Kk protein were positively selected with RS+ columns by separation on the Vario-MACS magnetic system as recommended by the manufacturer (Miltenyi Biotec Inc, Auburn, Calif.). Selected cells were incubated for 1 h at 4°C with 100 μg of a polyclonal human anti-HIV IgG (HIV-IgG) per ml. Subsequently, washed cells were stained with phycoerythrin (PE)-conjugated goat anti-human IgG (50 μl of a 1/50 dilution [Immunotech S.A.]) for 1 h at 4°C. The cells were then washed three times in PBS–0.3% bovine serum albumin (BSA) before being subjected to flow cytometric analysis with a FACSort apparatus (Becton Dickinson).
Studies of binding of recombinant gp120 wt and gp120 ΔαHX1 proteins to CEM and CHO cells.
All binding experiments were performed with 2 × 105 CEM cells resuspended in 50 μl of PBS–3% BSA containing the desire MAb at the appropriate concentration. After a 1-h incubation with agitation at 37°C, the cells were washed twice in PBS–0.3% BSA before addition of either an anti-mouse IgG-fluorescein isothiocyanate (FITC) conjugate (Sigma), an anti-human IgG-PE conjugate (Immunotech), an anti-human IgG-FITC conjugate (Immunotech), or a streptavidin-PE conjugate (SIGMA) at a 1/50 dilution.
After an additional 1 h of incubation with agitation at room temperature (RT), the cells were washed three times, resuspended in PBS, and analyzed by single color flow cytometry with a FACSort (Becton Dickinson) and LYSIS II software. Each datum point represents the acquisition of 10,000 gated events.
Binding of the anti-CD4 MAbs Leu3a, F101.69, 13B8.2, ST40, BL4, OKT4, and BF5 was also monitored following incubation of CEM cells with soluble gp120 wt or gp120 ΔαHX1 (10 μg/ml) for 1 h at 37°C. The cells were then washed and stained with the appropriate anti-mouse IgG-specific FITC conjugate before being subjected to flow cytometric analysis as described above in the presence or absence of 0.02% sodium azide in the wash and antibody solutions. Various concentrations of soluble gp120 wt or gp120 ΔαHX1 in a volume of 50 μl were evaluated for binding by incubating CEM cells in the presence of sodium azide for 1 h at 37°C with agitation. After two washes in PBS–0.3% BSA, the cells were stained with HIV-Ig (100 μg/ml). Similarly, CHO-K1 CXCR4+/CD4− and CHO-K1 CXCR4−/CD4− cells were incubated for 4 h at 4°C with 2 μg of gp120 proteins and with either 2, 10, or 30 μg of gp120 proteins per ml, respectively. The cells were then washed and stained with an anti-human IgG PE or FITC conjugate and processed for flow cytometric analysis as described above.
Inhibition of stroma-derived factor 1 alpha chemokine (SDF1α) binding was assessed following incubation of CEM cells with various concentrations of gp120 wt and gp120 ΔαHX1 for 1 h at 37°C in PBS–3% BSA. The cells were washed twice, and SDF1α (10 μg/ml) was then added in PBS–3% BSA for 30 min at 37°C. The cells were stained with a biotinylated goat polyclonal anti-human SDF-1α antibody in PBS–3% BSA–0.02% sodium azide for 30 min at RT. Streptavidin-PE conjugate was added for 30 min at RT in PBS–3% BSA–0.02% sodium azide, and the cells were analyzed by flow cytometry.
Inhibition of anti-CXCR4 MAb 12G5 binding was assessed after incubation of CEM and CHO-K1 cells with various concentrations of gp120 wt or gp120 ΔαHX1 or 10 μg of SDF1α per ml for 30 min at 37°C. The cells were washed twice, and CXCR4 accessibility was monitored by addition of anti-CXCR4 MAb 12G5 (10 μg/ml) in the presence of 0.02% sodium azide for 1 h at 4°C. The cells were washed twice with PBS–0.3% BSA–0.02% sodium azide and stained with an anti-mouse IgG FITC-conjugated antibody in PBS–3% BSA–0.02% sodium azide before being subjected to flow cytometric analysis.
Cell-ELISA.
U-shaped Maxisorb microtiter plates (Nunc) were saturated with PBS–3% BSA for 30 min at 37°C and incubated with 25 μl of a 10-μg/ml 1:1 mix of the anti-CD5 MAb (F145,6F3) and anti-CD100 (F93,7G2) for 16 h at 4°C. After being washed, 105 CEM cells were distributed in each well before centrifugation of the plate at 900 × g for 5 min, further incubation for 30 min at 37°C, and two washes with 200 μl of PBS–0.3% BSA per well. Quadruplicate wells were incubated with soluble gp120 proteins for 1 h at 37°C and washed twice in PBS–0.3% BSA. For CD4 inhibition experiments, the cells were further incubated for 30 min at 37°C with a 1/1,000 dilution of the anti-CD4bs MAb F101.69. Alternatively, after incubation with gp120 proteins, SDF1α was added at 10 μg/ml for 30 min at 20°C and the wells were washed twice with PBS–0.3% BSA, incubated with a biotinylated anti-SDF1α goat antibody in PBS–3% BSA–0.02% sodium azide, and then visualized with a streptavidin-biotin-peroxidase complex at 20°C for 30 min. The optical density at 492 nm (OD492) was measured on a Labsystem Multiscan RC spectrophotometer. The enzyme-linked immunosorbent assay (Cell-ELISA) plate included two internal standards with neither gp120 protein nor SDF1α, which served as a reference for the binding capacity of the anti-CD4 MAb F101.69 and the anti-SDF1α biotinylated goat antibodies, respectively. Experimental values were expressed as the percent inhibition of the corresponding reference values. The OD of the references varied between 1.0 and 2.0. Values obtained for wells without cells, saturated with PBS–3% BSA, indicated that nonspecific binding of the recombinant gp120 proteins was less than 5%.
Reactivities of MAbs with monomeric gp120.
The reactivities of MAbs with native monomeric gp120 were determined as described previously (46, 47). Briefly, either gp120 wt or gp120 ΔαHX1 protein (1 μg/ml) was captured on Maxisorb ELISA plates via its carboxy terminus by using a sheep polyclonal antibody D7324 in the presence of PBS–10% FCS. Anti-gp120 110-K and 110.4 MAbs were bound onto gp120 proteins in PBS–3% BSA–20% sheep serum buffer. After two washes, bound murine MAbs were detected with an anti-mouse IgG–horseradish peroxidase conjugate and the OD492 was measured.
Infectivity assay.
Infections were performed 24 h after seeding 104 HeLa P4 (CD4+ LTR-lacZ) cells (16) per well in 96-microtiter plates. The cells were then preincubated with gentle agitation in serum-free DMEM in the presence of various concentrations of soluble recombinant gp120 wt or gp120 ΔαHX1 for 1 h at 4°C and then continuously incubated for the next 24 h at 37°C with 50 50% tissue culture infective doses (TCID50) of HIV-1 LAI particles. Induction of β-galactosidase activity, the product of the HeLa P4 lacZ gene, reflects Tat transactivation and therefore HIV-1 infection. After 24 h, β-galactosidase activity was measured in cell lysates of quadruplicate wells. For this purpose, cells were lysed in 100 μl of a buffer containing 0.125% Nonidet P-40, 60 mM Na2HPO4, 40 mM NaH2PO4, 50 mM β-mercaptoethanol, 2.5 mM EDTA, 10 mM KCl, 10 mM MgSO4, 100 μl of 80 mM sodium phosphate (pH 7.4), 10 mM MgCl2, and 10 mM β-mercaptoethanol before addition of 6 mM chlorophenol red–β-galactopyranoside monosodium salt. The mixture was incubated for 30 min at 37°C, and the absorbance was measured at 574 nm.
RESULTS
Fusogenic abilities and cell surface expression of envelopes containing gp120 wt and gp120 ΔαHX1.
The cell fusion assay was performed in cocultures of the HeLa P4 cell line, in which lacZ expression is under the control of the HIV-1 LTR, and HeLa Tat cells into which either wild-type or ΔαHX1 envelopes were transfected. Transfection with wild-type envelope yielded a large number of syncytia in which β-galactosidase was expressed (1,000 to 2,000 foci/well). In contrast, no foci were observed following transfection with the ΔαHX1 mutant envelope (Fig. 2) or with an ecotropic Friend murine leukemia virus envelope (21), which is fusogenic only for mouse and rat cells (data not shown).
FIG. 2.
Tat transactivation of an HIV-1 LTR lacZ reporter gene after CD4 envelope-induced cell fusion. lacZ expression was assessed in HeLa P4 cells cocultured overnight with either HeLa Tat cells expressing the wild-type envelope (containing the gp120 wt) (A), HeLa Tat cells expressing the deleted αHX1 envelope (containing the gp120 ΔαHX1) (B), or untransfected HeLa Tat cells (C). The cells were then fixed, and X-Gal staining was performed at 37°C for 2 h. The presence of a blue syncytium is indicated by an arrow.
To determine whether the lack of fusogenic ability of the gp120 ΔαHX1 envelope was due to altered cell surface envelope expression, we monitored the presence of gp120 levels at the surface of transfected HeLa Tat cells by flow cytometry. A unimodal population with low mean fluorescence intensity (MFI = 15.1) was observed following anti-HIV-IgG staining of HeLa Tat cells transfected with the control pMACS-Kk plasmid. However, HeLa Tat cells transfected with either wild-type or ΔαHX1 HIV-1 envelope expression vectors demonstrated similar distributions of negative and positive populations. In both cases, the latter population was detected at an MFI of approximately 170 (Fig. 3).
FIG. 3.
Cell surface expression of gp120 wt and deleted gp120 ΔαHX1 envelopes. At 24 h after cotransfection with pMACS-Kk vector and gp120 wt or gp120 ΔαHX1 expression plasmids, HeLa Tat cells were incubated with an anti-HIV-1 polyclonal antiserum (HIV-Ig) and stained with an anti-human IgG-PE-conjugated antibody. Negative controls were cells transfected with pMACS-Kk (H2Kk) vector alone (stained with HIV-Ig and anti-human IgG-PE) and HeLa Tat cells cotransfected with wt envelope plasmid and pMACS-Kk vector but stained only with the secondary anti-human IgG-PE-conjugated antibody.
Binding properties of soluble recombinant HIV-1 gp120 ΔαHX1.
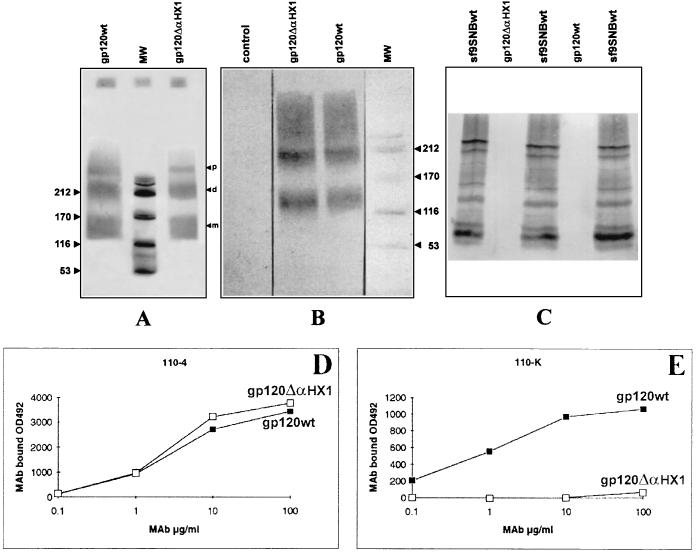
Recombinant baculovirus gp120 proteins were concentrated and immunopurified as described in Materials and Methods. Silver nitrate staining of purified proteins on a nonreducing SDS-PAGE gel revealed two major bands corresponding to monomers and dimers of both soluble gp120 wt and gp120 ΔαHX1, with a purity greater than 95% (Fig. 4A). Immunoblot analysis of the baculovirus-produced proteins with a polyclonal rabbit anti-gp120 antiserum confirmed the presence of gp120 monomeric and oligomeric forms (Fig. 4B). The purity of the gp120 proteins was demonstrated by the lack of reactivity with a rabbit antibody directed against supernatant from Sf9 cells infected with wild-type baculovirus (Fig. 4C).
FIG. 4.
Recombinant gp120 proteins expressed from a baculovirus vector were resolved by gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4 to 15% polyacrylamide). (A to C) Proteins (4 μg) were stained with silver nitrate or transferred to a nitrocellulose membrane (A) and subsequently immunoblotted with rabbit anti-gp120 polyclonal antiserum (B) or rabbit polyclonal antiserum generated against supernatant of baculovirus wt infected Sf9 cells (SF9SNBwt) (C). Lanes with gp120 wt, gp120 ΔαHX1, and SF9SNBwt supernatants are indicated, and molecular weight (MW) markers are shown in thousands. The positions of gp120 monomers (m), dimers (d), and polymers (p) are marked by arrows. (D and E) The reactivities of various concentrations of anti-gp120 110.4 (D) and 110-K (E) MAbs with native gp120 wt (shaded squares) and gp120 ΔαHX1 (open squares) were assessed. All data are corrected for background antibody absorption in the absence of gp120 (usually <0.100 OD492 unit).
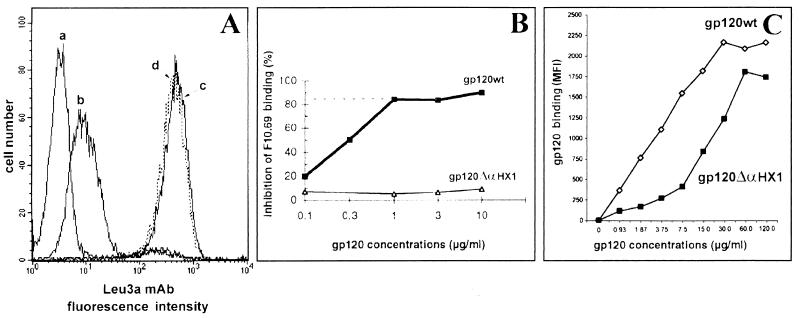
We then examined the ability of the gp120 ΔαHX1 protein to be recognized by the 110.4 and 110-K anti-gp120 MAbs, which react with the V3 loop and the C4 region through the conformational CD4bs epitope, respectively. We found that whereas the V3 loop was recognized equivalently in gp120 wt and gp120 ΔαHX1 (Fig. 4D), the conformational CD4bs MAb was able to interact only with gp120 wt (Fig. 4E). The ability of soluble gp120 ΔαHX1 to associate with CD4 was assessed by determining the binding level of the CD4-specific MAbs Leu3a and F101.69-PE, which interact with the CDR2 loop in the first D1 domain of CD4, after preincubation of the cells with gp120 wt or gp120 ΔαHX1 (10 μg/ml). As demonstrated by fluorescence-activated cell sorter (FACS) analysis (Fig. 5A), gp120 wt inhibited more than 98% of Leu3a binding whereas gp120 ΔαHX1 did not alter antibody binding (<5.6%). Similar results were obtained with the anti-CD4 F101.69-PE conjugate antibody (data not shown) and confirmed by the Cell-ELISA method (Fig. 5B). Furthermore, equivalent studies performed with various anti-CD4 MAbs, 13B8.2, ST40, BL4, OKT4, and BF5 directed against sites distinct from the CDR2 loop of D1 in CD4, showed that gp120 ΔαHX1 did not interfere with the ability of any of these MAbs to bind CD4 (Table 1). Therefore, the gp120 ΔαHX1 appeared unable to bind the CD4 receptor.
FIG. 5.
Effect of gp120 wt and gp120 ΔαHX1 on cell surface binding of the anti-CD4 MAbs Leu3a and F101.69. (A) Binding by the anti-CD4 MAb Leu3a was assessed in CEM cells in the presence of 0.02% sodium azide by FACS analysis: curve a, incubation of CEM cells with a goat anti-mouse IgG-FITC conjugate (negative control), curve b, binding of the Leu3a anti-CD4 MAb after incubation with gp120 wt; curve c, binding of Leu3a after incubation with gp120 ΔαHX1; curve d, total binding of Leu3a to CEM cells. (B) The effect of gp120 wt and gp120 ΔαHX1 on F101.69 MAb binding was examined by Cell ELISA in the absence of sodium azide. Similar results were obtained in the presence of sodium azide (0.02%) (data not shown). (C) Direct binding of gp120 wt or gp120 ΔαHX1 was assessed following incubation with CEM cells for 1 h at 37°C in the presence of sodium azide (0.02%).
TABLE 1.
Effect of gp120 on anti-CD4 MAb bindinga
| Antibody | MFIb after preincubation with:
|
||
|---|---|---|---|
| Medium | gp120 wt | gp120 ΔαHX1 | |
| 13B8.2 | 1054.78 | 73.13 | 940.74 |
| ST40 | 416.00 | 87.73 | 360.16 |
| BL4 | 489.38 | 93.40 | 418.96 |
| OKT4 | 526.53 | 481.65 | 530.00 |
| BF5 | 430.00 | 410.00 | 424.31 |
Binding of anti-CD4 MAbs directed against sites distinct from the CDR2 loop was assessed following a preincubation of CEM cells with medium alone, gp120 wt or gp120 ΔαHX1.
All the anti-CD4 MAbs were detected with an FITC-labeled goat anti-mouse IgG, with the exception of the 13B8.2 MAb, which was PE conjugated.
Since gp120 ΔαHX1 did not appear to bind CD4, we next assessed whether the deleted gp120 was able to bind to the surface of CEM cells. FACS analysis was performed on cells incubated with gp120 proteins for 1 h at 37°C in the presence of sodium azide (0.02%) to prevent possible internalization of target receptors. Under these conditions, gp120 ΔαHX1 bound to the cell surface of CEM cells in a dose-dependent manner, albeit at lower levels than gp120 wt (Fig. 5C). When the latter experiments were performed in the absence of sodium azide, no binding of gp120 ΔαHX1 could be observed on CEM cells (data not shown). Interestingly, the absence of sodium azide did not alter the ability of gp120 wt to inhibit the binding of anti-CD4 MAbs (data not shown).
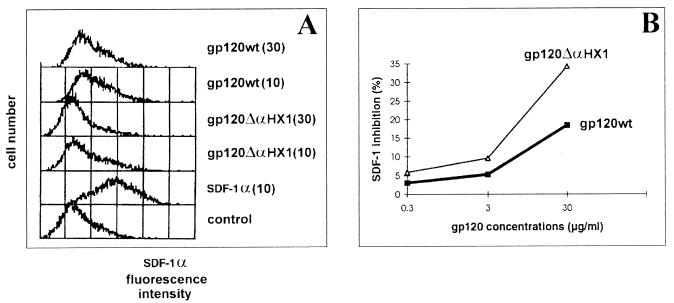
Since gp120 ΔαHX1 bound to CEM cells, we determined whether association with the CXCR4-chemokine receptor might be responsible for this phenomenon. For this purpose, we examined whether either gp120 wt or gp120 ΔαHX1 could inhibit the binding of SDF1α, the natural ligand of CXCR4 (4, 49). We observed that the binding of SDF1α (10 μg/ml) decreased to 44.4 and 36% of the control value in the presence of 10 and 30 μg of gp120 wt per ml, respectively (Fig. 6A). Following preincubation with gp120 ΔαHX1 at 10 and 30 μg/ml, significant decreases (to 35.3 and 25% of the control value, respectively) were also observed. Similar results were obtained by using the Cell-ELISA method, as represented by respective levels of inhibition (Fig. 6B). Therefore, a specific interaction between CXCR4 and both wild-type and deleted gp120 molecules was observed. We also evaluated the ability of gp120 wt and gp120 ΔαHX1 to inhibit the binding of the anti-CXCR4 specific MAb, 12G5. gp120 wt and gp120 ΔαHX1 concentrations of 10 to 40 μg/ml allowed us to observe 12G5 MAb binding levels of approximately 60 and 80% of control levels, respectively. Moreover, a gp120 ΔαHX1 concentration of 90 μg/ml decreased the 12G5 MAb binding to 45% of the control level. Preincubation with SDF1α decreased binding to 20% of the control level of binding to CEM cells. Similar experiments assessing inhibition of anti-CXCR4 MAb binding on CHO-K1 CXCR4+/CD4− cells were also performed. We observed that both wild-type and mutant gp120 proteins inhibited 12G5 MAb binding by approximately 25 and 36% at concentrations of 2 and 10 μg/ml, respectively (Table 2).
FIG. 6.
Inhibition of SDF1α binding to CXCR4 by soluble gp120 wt and gp120 ΔαHX1. (A) FACS analysis. SDF1α binding was monitored by staining with a biotinylated anti-SDF1α polyclonal antibody revealed with streptavidin-PE. CEM cells were incubated either with SDF1α alone (10 μg/ml) or following a preincubation with gp120 wt or gp120 ΔαHX1 at 10 and 30 μg/ml. Stained CEM cells which were not exposed to SDF1α were used as a negative control. (B) SDF1α binding was monitored in a Cell ELISA test in the presence of various concentrations of gp120 wt and gp120 ΔαHX1.
TABLE 2.
Effect of gp120 on the cell binding capacity of an anti-CXCR4 MAb
| Preincubation | Binding capacity (%) of 12G5 to cells at following ligand concn (μg/ml)
|
||||
|---|---|---|---|---|---|
| CEM CD4+a
|
CHO-K1 CXCR4+/CD4−b
|
||||
| 10 | 40 | 90 | 2 | 10 | |
| SDF1α | 20.7 | NDc | ND | ND | ND |
| gp120 wt | 59.5 | 58.0 | ND | 75.0 | 64.0 |
| gp120 ΔαHX1 | 80.0 | 77.2 | 44.8 | 74.0 | 61.4 |
Binding capacity of the 12G5 anti-CXCR4 MAb (10 μg/ml) to CEM cells was assessed after preincubation with either soluble gp120 wt, gp120 ΔαHX1, or SDF1α and is presented as the percentage of total binding (100%).
Binding capacity of the 12G5 anti-CXCR4 MAb (10 μg/ml) to CHO-K1 CXCR4+/CD4− cells was assessed after preincubation with either soluble gp120 wt or gp120 ΔαHX1 and is presented as the percentage of total binding (100%).
ND, not determined.
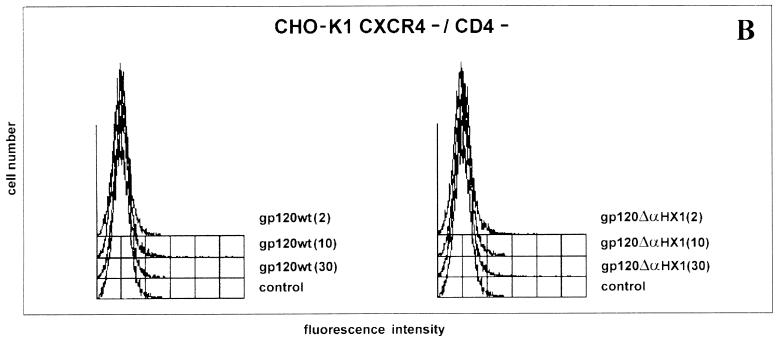
Since gp120 ΔαHX1 did not appear to bind CD4 and could inhibit the binding of several CXCR4 ligands, we next assessed whether the mutant gp120 protein bound to the surface of the CD4-negative CHO-K1 cell line expressing the CXCR4 receptor (the CHO-K1 CXCR4+/CD4− cell line) (29). The binding of gp120 wt and gp120 ΔαHX1 (2 μg/ml) to CHO-K1 cells was assessed by using an anti-HIV-1 IgG and revealed with a PE-conjugated anti-human IgG. Indeed, binding was observed with MFI for gp120 wt and gp120 ΔαHX1 reaching 11.92, and 13.92 respectively, whereas the control MFI, obtained in the absence of gp120 proteins, was 3.35 (Fig. 7A). Similar low MFIs were obtained after incubation of CHO-K1 CXCR4−/CD4− cells with concentrations up to 30 μg of either wt or mutant gp120 proteins per ml (Fig. 7B).
FIG. 7.
Direct binding of gp120 proteins to the CD4− CHO-K1 cell line in the presence of sodium azide (0.02%) for 4 h at 4°C. (A) Binding of gp120 wt (2 μg/ml) (curve c) and gp120 ΔαHX1 (2 μg/ml) (curve b) to the CD4-negative CHO-K1 cell line expressing the recombinant CXCR4 chemokine receptor background fluorescence (curve a). (B) Binding of different concentrations of gp120 wt and gp120 ΔαHX1 (2, 10 and 30 μg/ml) to CHO-K1 CXCXR4−/CD4− cells. The control is the background fluorescence.
Inhibition of HIV-1 infectivity by soluble recombinant gp120 ΔαHX1.
The ability of recombinant gp120 ΔαHX1 to inhibit HIV-1 infection was assessed by preincubating 104 target HeLa P4 cells with this soluble protein at 4°C for 1 h and then exposing them to 50 TCID50 of HIV-1 LAI for 24 h at 37°C in the presence of the gp120 proteins. Interestingly, 50% viral inhibitory concentration (IC50) was obtained following incubation and was maintained throughout the infection in the presence of either gp120 wt or gp120 ΔαHX1 at a concentration of 100 nM (Fig. 8). Therefore, despite a lack of interaction of gp120 ΔαHX1 with CD4, this molecule was capable of inhibiting HIV-1 infection, probably through its interaction with the CXCR4 secondary receptor.
FIG. 8.
gp120 wt and gp120 ΔαHX1 both inhibit HIV-1 infection of HeLa P4 cells. HeLa P4 cells were preincubated for 1 h at 4°C with various concentrations of gp120 wt or gp120 ΔαHX1. Cells (104 cells) were then incubated for 24 h at 37°C in the presence of 50 TCID50 of HIV-1 LAI and either gp120 wt or gp120 ΔαHX1. The IC50 was obtained after incubation of cells with 100 nM either gp120 protein.
DISCUSSION
We describe a deletion of an amphipatic αHX structure in a highly conserved region of the HIV-1 SUgp120 that abolished CD4 binding while maintaining CXCR4 recognition and binding. In our mutant, amino acids N58 to T60 and L86 to K91 were preserved in order to maintain the potential N58 glycosylation site and C89-linked disulfide bond. This mutant envelope displayed an ability to inhibit HIV-1 infection, probably through its binding to CXCR4. By using a highly sensitive assay for HIV-1 envelope cell-to-cell fusion, we were able to demonstrate a complete lack of fusion by an HIV-1 envelope harboring this mutation despite its efficient expression at the cell surface. This is in agreement with previous reports concluding that CD4 is required for fusion and syncytium formation by gp120/HIV-1 LAI/IIIB (8).
Several groups have shown that the CD4 recognition and binding properties of gp120 are maintained upon deletion of either specific variable loops (51, 66) or the N-terminal C1 and C-terminal C5 constant regions, which are implicated in the noncovalent association with gp41 (9, 36). Recently, smaller deletions within the N-terminal domain of C1, including the 30 amino acids from the signal peptide, have demonstrated that CD4 binding is preserved upon deletion of the first 85 amino acids but is lost upon elimination of 93 amino acids (65). Our results extend the latter finding and indicate that the region between amino acids E61 and S85 in the C1 region, which does not include the first 30 amino acids from the signal peptide, either (i) harbors essential amino acids or structures which are required for binding to the CD4 receptor or (ii) participates in the correct folding of the gp120 binding site for CD4.
The finding that gp120 undergoes a conformational change upon binding to CD4 (54, 55) led to the hypothesis that this interaction might play a role in chemokine receptor binding and subsequent HIV-1 entry (60, 64). However, more recently, it has been reported that gp120 wt is likely to interact with CXCR4 in a CD4-independent manner (37). Our results with gp120 ΔαHX1 unambiguously demonstrate that CD4 binding is indeed not required for CXCR4 association. This was established by our observation that wt and deleted forms of gp120 competed equivalently with the binding of SDF1α, the CXCR4 natural ligand, and 12G5, a MAb probably directed against the third extramembrane loop of CXCR4 (7). This was also clearly demonstrated by the ability of the mutant gp120 protein to bind to CHO-K1 CXCXR4+/CD4− cells but not to the parental CHO-K1 CXCXR4−/CD4− cells. Further topological mapping with MAbs probing for global and local conformation will be necessary to more precisely assess the folding of our mutant protein.
The levels of CCR5 and CXCR4 chemokine receptors are known to be down modulated following interaction with their natural ligands. Thus, after binding of SDF1α via the N-terminal segments of the second and third CXCR4 extramembrane receptor loops (19), CXCR4 is internalized rapidly (57) and reexpressed at the cell surface after recycling (2, 35). In the experiments described here, under conditions where internalization could occur, i.e., in the absence of sodium azide, binding of gp120 ΔαHX1 to the cell surface could not be detected while gp120 wt remained efficiently associated at the surface. In contrast, upon inhibition of internalization with sodium azide, both gp120 wt and gp120 ΔαHX1 were strongly associated at the cell surface. Thus, binding of gp120 to CXCR4 but not to CD4 probably results in rapid receptor down modulation. Accordingly, binding of SDF1α and SDF1α analogs at 100 nM has been shown to inhibit HIV-1 infection, most probably by down regulating CXCR4 levels (4, 49, 56, 62).
Incubation of cells with either gp120 wt or gp120 ΔαHX1 resulted in an inhibition of HIV-1 infection with an IC50 of 100 nM. It will be of interest to determine whether gp120 ΔαHX1 inhibited HIV-1 infection by competing for HIV-1 binding sites and/or by down regulating of CXCR4 receptor surface expression. Site-directed mutagenesis of gp120 within the highly conserved αHX1 structure will allow a more precise definition of the amino acids and structure required for interactions with CD4 and/or CXCR4 receptors. This may provide the basis for the design of CXCR4-specific inhibitors that are CD4 independent in their action.
ACKNOWLEDGMENTS
This study would have been impossible without the generosity and kindness of our colleagues, to whom we are indebted for scientific and technical input. We especially thank Marc Sitbon, and Quentin Sattentau for providing reagents and for insightful discussions; Naomi Taylor for critical reading of the manuscript; Pierre Charneau, Olivier Schwartz, and Marc Parmentier for generously providing plasmids, antibodies, and cell lines; James Hoxie for the 12G5 MAb; Ian Clark Lewis for SDF1α; François Traincard for anti-gp120 MAbs; Christophe Duperay, Bernard Geoffroy, Claudine Franche, Michel Secondy, Yvan Boublik, Arnaud Dupuis D’Angeac, and Ilias Stefas for helpful discussions and technical assistance; and Jeanne Anne Ville for her continuous encouragement.
This work was supported by the Institute for Scientific Cooperation and Development, CNRS, the World Health Organization, and Sidaction-France.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Murphy P M, Berger E. CC-CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Amara A, Le Gall S, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J L, Arenzana-Seisdedos F. HIV coreceptor down regulation as antiviral principle: SDF-1α-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:1139–1146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barré-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vézinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphocyte retrovirus from a patient at risk for acquired immunodeficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 4.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 5.Bolmstedt A, Hemming A, Flodby P, Berntsson P, Travis B, Lin J P C, Ledbetter J, Tsu T, Wigzell H, Hu S L, Olofsson S. Effects of mutations in glycosylation sites and disulfides bonds on processing CD4-binding and fusion activity of human immunodeficiency virus envelope glycoproteins. J Gen Virol. 1991;71:1269–1277. doi: 10.1099/0022-1317-72-6-1269. [DOI] [PubMed] [Google Scholar]
- 6.Boublik, Y., C. Denesvre, and M. Sitbon. Unpublished data.
- 7.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broder C C, Berger E A. CD4 molecules with a diversity of mutations encompassing the CDR3 region efficiently support human immunodeficiency virus type 1 envelope glycoprotein-mediated cell fusion. J Virol. 1993;67:913–926. doi: 10.1128/jvi.67.2.913-926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acids changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaabihi H, Ogliastro M H, Martin M, Giraud C, Devauchelle G, Cérutti M. Competition between baculovirus polyhedrin and P10 gene expression during infection of insect cells. J Virol. 1993;67:2664–2671. doi: 10.1128/jvi.67.5.2664-2671.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesebro B, Wehrly K, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho M W, Lee M, Carney M, Breson J, Doms R, Martin M. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, MacKay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1–20. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 16.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 18.Cordonnier A, Montagnier L, Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature. 1989;340:571–574. doi: 10.1038/340571a0. [DOI] [PubMed] [Google Scholar]
- 19.Crump M P, Gong J H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J L, Baggiolini M, Sykes B D, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1α dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalgeish A G, Beverly P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–766. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 21.Denesvre C, Carrington C, Corbin A, Takeuchi Y, Cosset F-L, Schulz T, Sitbon M, Sonigo P. TM domain swapping of murine leukemia virus and human T-cell leukemia virus envelopes confers different infectious abilities despite similar incorporation into virions. J Virol. 1996;70:4380–4386. doi: 10.1128/jvi.70.7.4380-4386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denesvre C, Sonigo P, Corbin A, Ellerbrok H, Sitbon M. Influence of transmembrane domains on the fusogenic abilities of human and murine leukemia retrovirus envelopes. J Virol. 1995;69:4149–4157. doi: 10.1128/jvi.69.7.4149-4157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng H, Unutmaz D, Kewal Ramani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 24.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R, Hill M, Davis C, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 25.Ditzel H J, Parren P W, Binley J M, Sodroski J, Moore J P, Barbas C F. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267:684–695. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- 26.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Nature. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 27.Dragic T, Alizon M. Different requirements for membrane fusion mediated by the envelopes of human immunodeficiency virus types 1 and 2. J Virol. 1993;67:2355–2359. doi: 10.1128/jvi.67.4.2355-2359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dragic T, Litwin V, Allaway G P, Martin S, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 29.Endres M J P R, Clapham M, Marsh M, Ahuja J, Davis-Turner A, Mcknight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T C N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR-4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 31.Fitzpatrick-McElligott S. Gene transfer to tumor-infiltrating lymphocytes and other mammalian somatic cells by microprojectile bombardment. Bio/Technology. 1992;10:1036–1040. doi: 10.1038/nbt0992-1036. [DOI] [PubMed] [Google Scholar]
- 32.Gallaher W R, Ball J M, Garry R F, Martin-Amedee A M, Montelaro R C. A general model for the surface glycoproteins of HIV and others retroviruses. AIDS Res Hum Retroviruses. 1995;11:191–202. doi: 10.1089/aid.1995.11.191. [DOI] [PubMed] [Google Scholar]
- 33.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Hayer B F, Palker T J, Redfield R, Oleske J, Safai G, White G, Foster P, Markham P D. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 34.Hansen J E, Lund O, Nielsen J O, Brunak S, Hansen J E S. Prediction of the secondary structure of HIV-1 gp120. Proteins. 1996;25:1–11. doi: 10.1002/(SICI)1097-0134(199605)25:1<1::AID-PROT1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 35.Haribabu B, Ricardo M, Fisher I, Sozzano S, Peiper S C, Horuk R, Ali H, Snyderman R. Regulation of phosphorylation in desensitization and internalization. J Biol Chem. 1997;272:28726–28731. doi: 10.1074/jbc.272.45.28726. [DOI] [PubMed] [Google Scholar]
- 36.Helseth E, Olshevsky U, Furman C, Sodrosky J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein important for association with gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 38.Horuk R, Hesselgesser J, Zhou Y, Faulds D, Halks-Miller M, Harvey S, Taub D, Samson M, Parmentier M, Rucker J, Doranz B J, Doms R W. The CC chemokine I-309 inhibits CCR8-dependent infection by diverse HIV-1 strains. J Biol Chem. 1998;273:386–391. doi: 10.1074/jbc.273.1.386. [DOI] [PubMed] [Google Scholar]
- 39.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 40.Klatzmann D, Champagne E, Chamaret S, Gruest J, Gruetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 41.Kowalski M, Poltz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 42.Kozak S L, Platt E J, Mandani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR5 dependencies for infection by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp 120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 44.Lasky L A, Nakamura G, Smith D H, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, Capon D. Delineation of a region of the human immunodeficiency virus type1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–985. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- 45.Maddon P, Dalgleish A, McDougal J S, Clapham P, Weiss R, Axel R. The T4 gene encodes the AIDS receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 46.Moore J. Simple methods for monitoring HIV-1 and HIV-2 gp120 binding to sCD4 by ELISA: HIV-2 has a 25-fold lower affinity than HIV-1 for soluble CD4. AIDS. 1990;3:297–305. doi: 10.1097/00002030-199004000-00003. [DOI] [PubMed] [Google Scholar]
- 47.Moore J, Thali M, Jameson B A, Vignaux F, Lewis G K, Poon S W, Charles M, Fung M S, Sun B, Durda P J, Akerblom L, Wahren B, Ho D D, Sattentau Q, Sodroski J. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J Virol. 1993;67:4785–4796. doi: 10.1128/jvi.67.8.4785-4796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore J, Sattentau Q, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legier D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 49a.Ogliastro, M. H. Unpublished data.
- 50.Olsevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollard S, Rosa M D, Rosa J, Willey D. Truncated variants of gp120 bind CD4 with high affinity and suggest a minimum CD4 binding region. EMBO J. 1992;11:4325–4332. doi: 10.1002/j.1460-2075.1992.tb05090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 53.Prince A M, Reesink H, Pascual D, Horowitz B, Hewlett I, Murthy K K, Cobb K E, Eichberg J W. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res Hum Retroviruses. 1991;7:971–973. doi: 10.1089/aid.1991.7.971. [DOI] [PubMed] [Google Scholar]
- 54.Sattentau Q, Moore J. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sattentau Q, Moore J, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schols D, Struyf S, Van Damme J, Esté J A, Henson G, De Clerq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Signoret N, Oldridge J, Pelchen-Matthews A, Klasse P J, Tran T, Brass L F, Rosenkilde M M, Schwartz T W, Holmes W, Dallas W, Luther M A, Wells T N C, Hoxie J A, Marsh M. Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol. 1997;139:651–664. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stern B, Denisova D G, Buyaner D, Raviv D, Gershoni J M. Helical epitopes determined by low-stringency antibody screening of a combinatorial peptide library. FASEB J. 1997;11:147–153. doi: 10.1096/fasebj.11.2.9039957. [DOI] [PubMed] [Google Scholar]
- 59.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station bulletin 1555. College Station: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 60.Trkola A, Dragic T, Arthos J, Binley J, Olson W, Allaway G, Cheng-Mayer C, Robinson J, Maddon P, Moore J. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 61.Tschachler E, Buchow H, Gallo R C, Reitz M S. Functional contribution of cysteine residues to the human immunodeficiency virus type 1 gp120 envelope. J Virol. 1990;64:2250–2259. doi: 10.1128/jvi.64.5.2250-2259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ueda H, Siani M A, Gong W, Thompson D A, Brown G G, Wang J M. Chemically synthesized SDF-1α analogue, N33A, is a potent chemotractive agent for CXCR4/Fusin/LESTR-expressing human leukocytes. J Biol Chem. 1997;272:24966–24970. doi: 10.1074/jbc.272.40.24966. [DOI] [PubMed] [Google Scholar]
- 63.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q J. HIV-1 gp120 induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 64.Wu L, Gerard N, Wyatt R, Choe H, Parolin C, Ruffing N, Boresetti A, Cardoso A, Desjardins E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 65.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevky V, Sodroski J. Analysis of interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71:9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the of human immunodeficiency virus type 1 gp120 envelope epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]