Abstract
While injection molding is becoming the fabrication modality of choice for high-scale production of microfluidic devices, especially those used for in vitro diagnostics, its translation into the growing area of nanofluidics (structures with at least one dimension <100 nm) has not been well established. Another prevailing issue with injection molding is the high startup costs and the relatively long time between device iterations making it in many cases impractical for device prototyping. We report, for the first time, functional nanofluidic devices with dimensions of critical structures below 30 nm fabricated by injection molding for the manipulation, identification, and detection of single molecules. UV-resin molds replicated from Si masters served as mold inserts, negating the need for generating Ni-mold inserts via electroplating. Using assembled devices with a cover plate via hybrid thermal fusion bonding, we demonstrated two functional thermoplastic nanofluidic devices. The first device consisted of dual in-plane nanopores placed at either end of a nanochannel and was used to detect and identify single ribonucleotide monophosphate molecules via resistive pulse sensing and obtain the effective mobility of the molecule through nanoscale electrophoresis to allow its identification. The second device demonstrated selective binding of a single RNA molecule to a solid phase bioreactor decorated with a processive exoribonuclease, XRN1. Our results provide a simple path towards the use of injection molding for device prototyping in the development stage of any nanofluidic or even microfluidic application, through which rapid scale-up is made possible by transitioning from prototyping to high throughput production using conventional Ni mold inserts.
Introduction
Nanofluidics is defined as the transport of fluids through structures in the range of 1 to 100 nm, while microfluidics uses fluid conduits >1000 nm in size. We define nanofluidics as any device that possesses a structure with at least one-dimension <100 nm in size and extended nanofluidics as any device with a structure ranging from 100–1000 nm.2 Because the channel dimensions in nanofluidics are close to molecular dimensions, unique physics occur that can have a dramatic impact on transport phenomena at the nanometer scale that do not occur at the micrometer scale and thus, new applications can be realized using nanofluidics. Examples of unique phenomena occurring in nanofluidics include electrical double layer overlap,1 which can induce co-ion exclusion, transverse electromigration, and parabolic-like flow3 even for electrically-driven transport. In addition, the high surface-to-volume ratio associated with nanochannels can lead to asymmetric ion transport.4 Efforts to fabricate nanofluidic structures <100 nm have also led to the development of devices for DNA sequencing.5,6 Finally, nanofluidics has enabled the ability to stretch dsDNA to near its full contour length when the channel dimensions are close to the persistence length (∼50 nm) of the double-stranded DNA, which can allow for direct reading of sequence variations, such as abasic and methylation sites.7–9 Despite the unique applications of nanofluidics, a challenge with nanofluidics has been the lack of large volume production modalities for devices that would, if realized, broaden the dissemination of nanofluidics into the research and clinical communities and facilitate commercial translation.
Nanofluidic devices usually contain multiscale structures consisting of microfluidic networks to allow interfacing to the macro-world and nanoscale structures such as channels,10–15 pores,16–26 and solid-phase bioreactors27,28 to carry out the intended analysis function. Their fabrication usually requires use of different fabrication methods in series to generate the microfluidic networks and nanostructures. For example, conventional photolithography and wet/dry etching can be used to produce microfluidic networks and that needs to be combined with one or more nanofabrication methods including extreme ultraviolet lithography, electron beam lithography,29,30 proton beam writing,31 or focused ion beam milling (FIB)32,33 to make the pre-requisite nanostructures. Among those methods, FIB milling has been used as a versatile tool for nanofluidic structures with advantages of not requiring resists, photomasks, or a subsequent etching process; the ion beam is used directly to mill structures into the desired substrate.32 Ramsey’s group reported patterning nanochannels in quartz as small as 5 nm using FIB milling,33 which employed a sacrificial conductive layer coated over a quartz substrate. However, the serial processes used for structure generation on different scales and use of high-end nanofabrication tools do not meet the requirements for large-scale production.
To increase the manufacturing throughput, replication methods such as nanoimprint lithography34,35 and hot embossing36,37 have been used for transferring micro- and nano-patterns into polymer substrates. Feature sizes <30 nm have been achieved using NIL.38 In addition, Uba et al. used hot embossing to replicate structures into the thermoplastic PMMA that were ∼18 nm in size both in terms of depth and width.35 While NIL can produce nanofluidic devices forgoing the need for doing direct photolithography and FIB milling to make each device,35,39,40 it is a medium scale production method and as such, may not allow for the wide dissemination of nanofluidic devices into the research and commercial sectors.
As seen in many commercialized microfluidic devices, injection molding of thermoplastics is a strategy with the potential to accommodate industrial-scale manufacturing requirements. Injection molding uses flow of molten polymer into mold cavities under a certain pressure and temperature. While nano-injection molding of structures as small as 25 nm has been reported in a thermoplastic,41 most of the efforts on injection molding of nanostructures focused on fabrication of extended nanostructures for optical applications.42–48 To use injection molding for nanofluidic applications, much tighter tolerances are needed to ensure the continuity of fluid transfer between nanoscale fluidic vias and microfluidic networks upon assembly of devices with a cover plate. Utko et al. reported injection molding devices with arrays of extended nanochannels having 240 nm widths and 150 nm heights using cyclic olefin copolymer (COC) thermoplastic as the substrate and were used for the stretching of DNA molecules.49 To our knowledge, no work has demonstrated functional nanofluidic devices fabricated by injection molding with nanostructures <100 nm in both width and depth.
A drawback of using injection molding in the developmental pipeline of fluidic devices is the high initial startup costs associated with the fabrication of metal mold inserts and the necessary multi-unit die (MUD). Commonly, nickel (Ni) mold inserts are electrodeposited onto Si or photoresist on Si masters, which are used as a sacrificial base.50–53 The need for extensive optimization and the slow pattern filling are time-consuming and the sacrificial Si master or substrate needs to be disposed following electrodeposition. Repeated fabrication of sacrificial Si or photoresist on Si masters with nanofluidic structures using high precision nanofabrication tools is cost prohibitive, which makes it challenging to use injection molding in the prototyping stage of the development of both micro- and nanofluidic devices.
In this work, we report functional nanofluidic devices with dimensions ≤30 nm formed in plastics using nano-injection molding. The need for electrodeposition of Ni molding tools was avoided by using UV-resin mold inserts that were replicated from Si masters by UV nanoimprint lithography, which enabled use of injection molding during the prototyping stage of development for nanofluidic devices. Following injection molding, enclosed nanofluidic devices were made using a hybrid thermal fusion bonding technique we have reported.39 Assembled nanofluidic devices with two different designs to detect and identify single molecules were used to demonstrate the utility of nano-injection molding and assembly processes discussed herein. The first nanofluidic device, consisting of two in-plane nanopores positioned on both ends of a nanochannel, was used for resistive pulse sensing (RPS) and label-free nanoscale electrophoresis of single ribonucleotide monophosphates, rNMPs, molecules.27,54 The second device had multiscale structures that spanned the range from several 10’s of mm to ∼50 nm.27,55 It was used for selectively binding single fluorescently labeled RNA molecules to a processive riboexonuclease (XRN1) that was immobilized on a nanoscale solid phase reactor.
Results and discussion
Nano-injection molding with resin mold inserts
The MUD in our injection molding system consists of movable and stationary platens (Fig. 1A). A blank brass mold insert was positioned on the movable platen while a mold insert with nanofluidic structures was fixed at the stationary platen. As an alternative to Ni mold inserts, we used resin mold inserts replicated from a Si master to forego the need of an electrodeposition step. Our group previously reported the use of resin molds for thermal nanoimprint lithography of thermoplastics, which offers many advantages including low demolding force associated with a low Young’s modulus of the cured UV-resin and reduced thermal expansion coefficient mismatch between the substrate and mold insert, ease of producing molds with positive or negative tones by repetitive replication processes, and extended lifetime of expensive Si master molds.54,56–58
Fig. 1.

(A) Images of the injection molding machine equipped with a Master Unit Die (MUD) consisting of a movable and a stationary platens. A blank mold insert was placed on the surface of the moving platen while a resin mold insert was fixed to the stationary platen. (B) SEM images of the Si master used for nanoscale electrophoresis, which consist of dual in-plane nanopores flanked into both ends of a nanochannel. (C) SEM images of a nanofluidic structure used for selective binding of a single RNA molecule to a solid phase bioreactor. The structure consist of a nanopillar in the center which is connected to four input/output nanochannel of ∼250 nm × 250 nm in width × depth and one nanochannel of ∼50 nm × 50 nm in width × depth to capture the reaction products from the solid phase reactor. (D) and (E) AFM images of replicated MD700 molds with positive and negative toned structures fabricated by repetitive replication from Si master with the dual in-plane nanopore Time-of-Flight (ToF) sensor structure.
The overall process schematics are shown in Fig. S1,† which broadly consisted of fabricating Si master molds via a combination of photolithography and Si etching for microfluidic networks with FIB milling for generating nanostructures, replication of Si master molds to produce resin mold inserts, and injection molding of thermoplastics with the resin mold inserts. Fig. 1B and C show scanning electron microscopy (SEM) images of the two Si masters used for this study. The first Si master (Fig. 1B) contained dual in-plane nanopore sensors, where two in-plane nanopores with a depth of ∼30 nm were located at both ends of a 5 μm long nanochannel with a cross-section of 100 nm × 100 nm in width and depth. Similar nanosensors formed in polymer substrates via NIL have been used for the detection and identification of the four dNMP molecules via a combination of nanoscale electrophoresis and RPS.54 The second Si master consisted of multiscale structures with a pillar of ∼500 nm in diameter at the center of the device used as a solid phase enzymatic bioreactor, four input/output nanochannels of 250 nm width × 250 nm depth, and a single nanochannel (50 nm width × 50 nm height) to detect and identify the reaction products generated by the bioreactor (Fig. 1C). At the entrance of one of the input nanochannels, an expansion chamber was used to entropically trap RNA or DNA molecules before translocating into the bioreactor. This device was used for selectively surface immobilizing exonuclease enzymes at this solid phase bioreactor and associating to a translocating RNA or DNA with the products generated from this reactor being swept into the flight tube to detect and identify using the dual in-plane nanopore sensor once a cofactor is added to initiate the action of the processive enzyme generating rNMPs or dNMPs, respectively.
The nanofluidic structures in Si were used to produce resin mold inserts via UV-NIL. To determine appropriate UV curable resins that would provide the best performance metrics for injection molding, which included primarily high replication fidelity and high durability to produce a number of replicas, UV-resins with three different resin bases (polyurethane (PUA), tripropyleneglycol diacrylate (TPGDA), and MD700) were evaluated. Their chemical and mechanical properties are shown in Table S1.†59 Among the three UV-resins, TPGDA was known to exhibit the best filling behavior into sharp nanostructures due to its low viscosity, while PUA was the most durable due to its high Young’s modulus while MD700 has the lowest Young’s modulus and surface energy of the three resins.59 Atomic force microscopy (AFM) images of MD700 resin molds with positive and negative toned nanochannels with dual in-plane nanopores are shown in Fig. 1D and E, respectively. The replication fidelity from Si masters in the absence of high aspect ratio nanostructures into the three different resin mold inserts were similar,59 allowing us to compare the performance of different resin mold inserts for injection molding with the same thermoplastic.
Injection molding of cyclic olefin polymer (COP, Tg = 100 °C) was performed with different resin mold inserts under identical injection molding conditions (nozzle temperature, Tnozzle = 260 °C, mold temperature, Tmold = 60 °C, injection speed = 10 cm3 s−1, dosage = 3.7 cm3, and cooling time = 30 s). The corresponding optical micrographs of injection molded COP structures are shown in Fig. S2.† Significant defects were observed for both microfluidic networks and nanostructures for injection molded COP when the PUA resin mold insert was used, while the TPGDA molds produced defects at a portion of the microfluidic network only. Alternatively, MD700 resin resulted in COP devices without any noticeable defects in the devices even after molding >100 devices. The replication fidelity with the three resin molds were in the decreasing order of their Young’s moduli and surface energies, material properties most relevant to adhesion and friction between mold inserts and molded substrates, respectively. Therefore, successful injection molding of nanofluidic structures can be attributed to low demolding force associated with a low Young’s modulus and surface energy of the MD700 mold insert for the resins tested.
The injection molding conditions require optimization for each nanofluidic structure and molding material. While Tnozzle can be set within the range of melting temperatures of the respective polymers, Tmold, which controls the cooling rate of the polymer upon contact with the mold surface after injection, is critical in the optimization process to achieving complete filling of polymer melt into mold cavities and a uniformly smooth surface. When dealing with higher aspect ratios, using elevated mold temperatures becomes essential to ensure complete filling. Using an array of pillars with a diameter of 980 nm shown in Fig. 1C, we studied the effect of Tmold on mold filling for three different thermoplastics with different Tg; COC 8007 (Tg = 78 °C), COC 5013 (Tg = 134 °C) and COP (Tg = 100 °C) (Fig. S3†). Tmold is typically set at a temperature below the Tg of the polymer so that the material can solidify before ejecting it from the mold cavity. The results indicated that Tmold/Tg needs to be higher than ∼0.6 for COP and ∼0.7 for the two COC blends, which also indicated better mold filling with COP. We also observed significant wear of the resin molds when we used Tmold = 100 °C, which can be attributed to the low modulus of the resin molds. Therefore, it is important to use an optimal Tmold to achieve sufficient filling of molten polymers and reduce wear of the resin mold. The injection speed and dosage of the injected polymer are two other important processing parameters to optimize. The dosage of the injected polymer is determined based on the cavity volume, which is a function of the structures’ volume on the molds and the volume of the cavity itself. The injection speed determines how effectively the molten polymer follows the contours of the structures on the mold. Excessive injection speed can lead to distortion of the features on the injection-molded part. The optimized injection molding conditions determined for different thermoplastics and different nanofluidic devices used in this study are summarized in Tables S2 and S3,† respectively.
It should be noted that the viscoelastic property is more important for the filling process, while the Young’s modulus and surface energy are more critical for demolding. We performed dynamic mechanical analysis (DMA) for the three thermoplastics used injection molding (COC 8007, COC 5013, and COP) and the three cured UV-resins used for resin molds (PUA, TPGDA, and MD 700) to obtain their viscoelastic properties. The results are shown in Fig. S4.† The Tg values of three thermoplastics determined by the storage modulus curves (89 °C, 137 °C, and 116 °C for COC 8007, COC 5013, and COP, respectively) were higher than those from the material specification sheets by 3–16 °C (Fig. S4A–C†), which may be associated with dynamic nature of the DMA measurements and re-orientation of polymer chains during sample preparation. The storage modulus values for all three thermoplastics fell to ∼zero after the glass transition was complete. It should be noted that during injection molding the molten polymer at a Tnozzle which is above the melting temperature, is injected through the mold cavities while the temperature at the mold surface (Tmold) is maintained at an elevated temperature to prevent quick solidification of injected polymer melt prior to complete filling. Therefore, at a sufficiently high Tmold and molding time, complete filling should be readily achieved.
On the other hand, the storage modulus curves of three cured UV-resins used for resin molds did not show sharp glass transition behaviors, but gradually decreased with temperature (Fig. S4D–F†). This behavior can be attributed to cross-linked or networked nature of the cured resins. The storage modulus values of cured PUA, TPGDA and MD 700 at Tmold of 60 °C were 316, 1114, and 23 MPa, respectively, which are still significantly larger than the storage modulus of ∼zero for injected molten polymers. Such large difference in the modulus between the cured resin and molten polymer accounts for the utility of resin molds and their high replication fidelity during the molding process of injection molding.
SEM and AFM images for the resulting injection molded COP devices using an MD700 mold insert of two different nanofluidic designs are shown in Fig. 2. The high-resolution AFM image of an in-plane nanopore (Fig. 2A) shows a pore depth of ∼30 nm prior to cover plate bonding. In Fig. 2B, the width and depth of the input/output channels around the solid phase bioreactor determined by SEM and AFM were 250.5 ± 2.7 nm and 282.5 ± 4.8 nm, respectively, while those for the nanochannel were 52.7 ± 1.8 nm and 57 ± 1.5 nm, respectively, indicating good replication fidelity when compared to the dimensions of the Si masters.
Fig. 2.
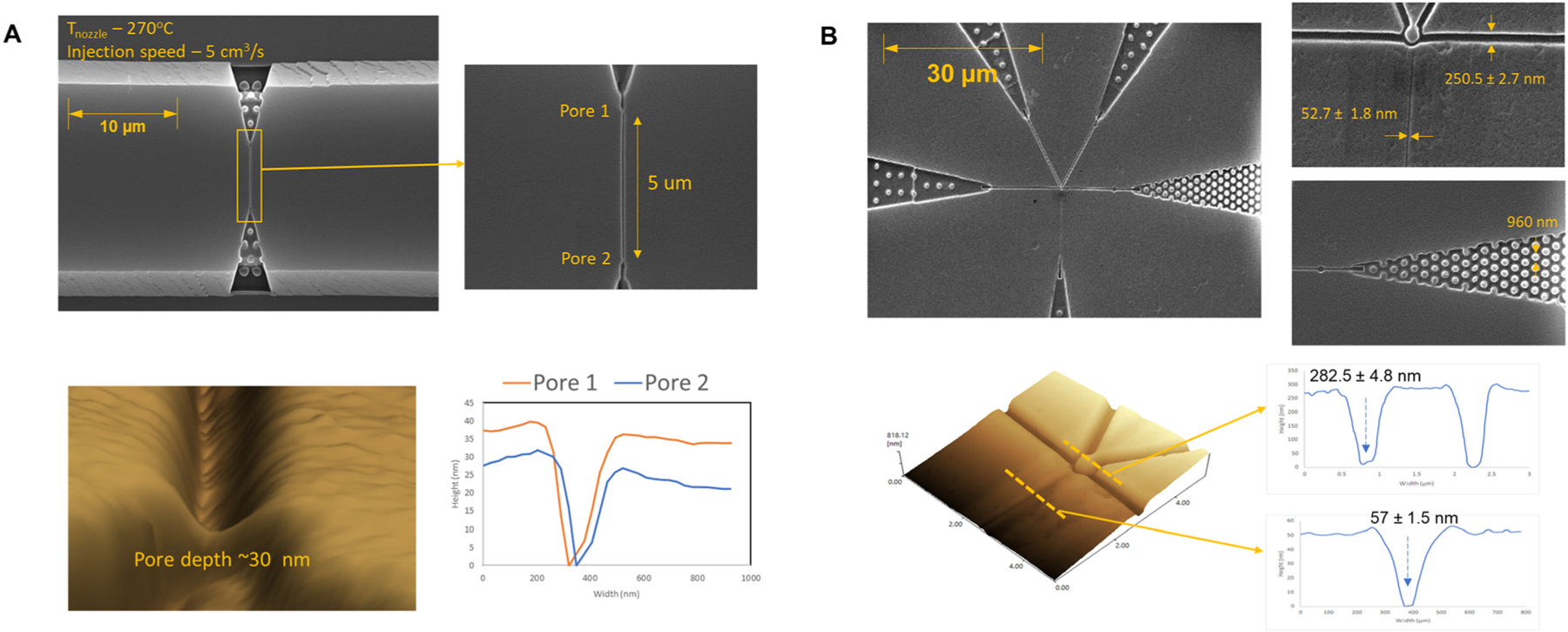
Production of nanofluidic structures by injection molding with COP from MD700 resin molds. (A) Top: Scanning electron microscopy (SEM) images of dual in-plane nanopore time-of-flight (ToF) sensor that consists of dual in-plane nanopores flanked into both ends of a nanochannel. Bottom: Atomic force microscopy (AFM) image of an in-plane nanopore (from resin mold 2, see Fig. 1B) with a ultrasharp AFM tip and cross-sectional profiles of both in-plane nanopores. (B) Top: SEM images of the nanofluidic structure used for selective binding of a single RNA molecule. The structure consists of a solid phase bioreactor located in the center of the image, four input/output nanochannels, and a nanochannel for capturing the reaction products from the solid phase reactor. The input/output channels also contain in-plane sensing pores and entropic trap. Bottom: AFM image near the solid phase bioreactor and cross sectional profiles of the nanochannels at two different locations. Both input/output channels were ∼280 nm in width and depth.
The quantitative analysis of the replication fidelity from the UV-resin mold insert to the injection molded COP chips was not possible because neither of our atomic force microscope or scanning electron microscope could accommodate the large metal block on which the UV-resin mold layer was formed (Fig. 1A). Instead, the durability of the resin mold insert could be indirectly studied by comparing critical dimensions of different iterations of injection molded COP chips. For this, the longevity test of the MD700 UV-resin mold insert was conducted through AFM and SEM measurements on the first and 101st iterations of injection molded chips. The results affirmed consistent dimensions, demonstrating the ability of the MD700 UV-resin mold insert to maintain its shape, structural integrity, and dimensional accuracy for over 100 iterations of the injection molding process. The results are shown in Fig. S5.† Subsequently, we calculated the relative standard deviation (RSD) for these critical dimensions which include the height of the nanopore, the height of the nanochannel flight tube, and the full width at half maximum (FWHM) of the nanopore. The specific values for these critical dimensions and their corresponding RSD percentages are reported in the inset table of Fig. S5.† The RSD values stand at 7.4%, 4.0%, and 3.7% for the height of the nanopore, height of the nanochannel (flight tube), and FWHM of the nanopore, respectively. These findings highlight the high replication fidelity achieved by the injection molding process for a large number of iterations, which also indicate the durability of the resin mold insert used. The number of times a resin mold can be used before starting to fail could not be determined experimentally, but it strongly depends on the structures in the mold as well as process parameters. Once successful injection molding was achieved with optimal parameters for certain mold structures, then reproducible results could be achieved for more than hundreds of iterations.
Following injection molding, functional nanofluidic devices were made via a hybrid thermal fusion bonding technique,41 which uses a low Tg cover plate bonded to a higher Tg substrate at a temperature slightly above the Tg of the cover plate and produces devices at high yield rates (>90%). After the COP thermoplastic device and COC 8007 cover plate were exposed to an O2 plasma at 50 W for 1 min, the cover plate was then placed over the device and thermally fusion bonded to the molded COP substrate at 74 °C and 170 psi for 15 min. Under these specific conditions, we expect from our previous work that the pore depth would decrease by more than 57%, leading to a cross-sectional area reduction of over 77%.38 However, this decrease is primarily caused by the deformation of the COC8007 cover sheet.
Single-molecule detection and identification of rCMP using a dual in-plane nanopore sensor
Using the assembled dual in-plane nanopore sensor device, we sought to detect and identify single rCMP molecules using a time-of-flight (ToF) method. The ToF is a molecular dependent property and is associated with the molecule’s electrophoretic mobility. For this application, the nanofluidic device consisted of two in-plane pores (12 nm in effective diameter after cover plate bonding) that were located on either end of a nanochannel serving as the flight tube (Fig. 2A). The ToF is associated with the apparent electrophoretic mobility of the single molecule electrokinetically translocating through the sensor and is measured from the time difference between the appearances of two resistive pulses (see Fig. 3B). The sensor also included a tapered input funnel to allow for high sampling efficiency of single molecules from the microchannels.57 We have demonstrated the use of similar devices fabricated in polyethylene glycol diacrylate (PEGDA) by UV-NIL for detection and identification of dNMP molecules by their apparent mobilities.54 In that study, the identification accuracy increased as the length of the nanochannel was increased from 0.5 μm to 5 μm. An identification accuracy of 94% was achieved when a 5 μm long nanochannel flight tube was used.54
Fig. 3.
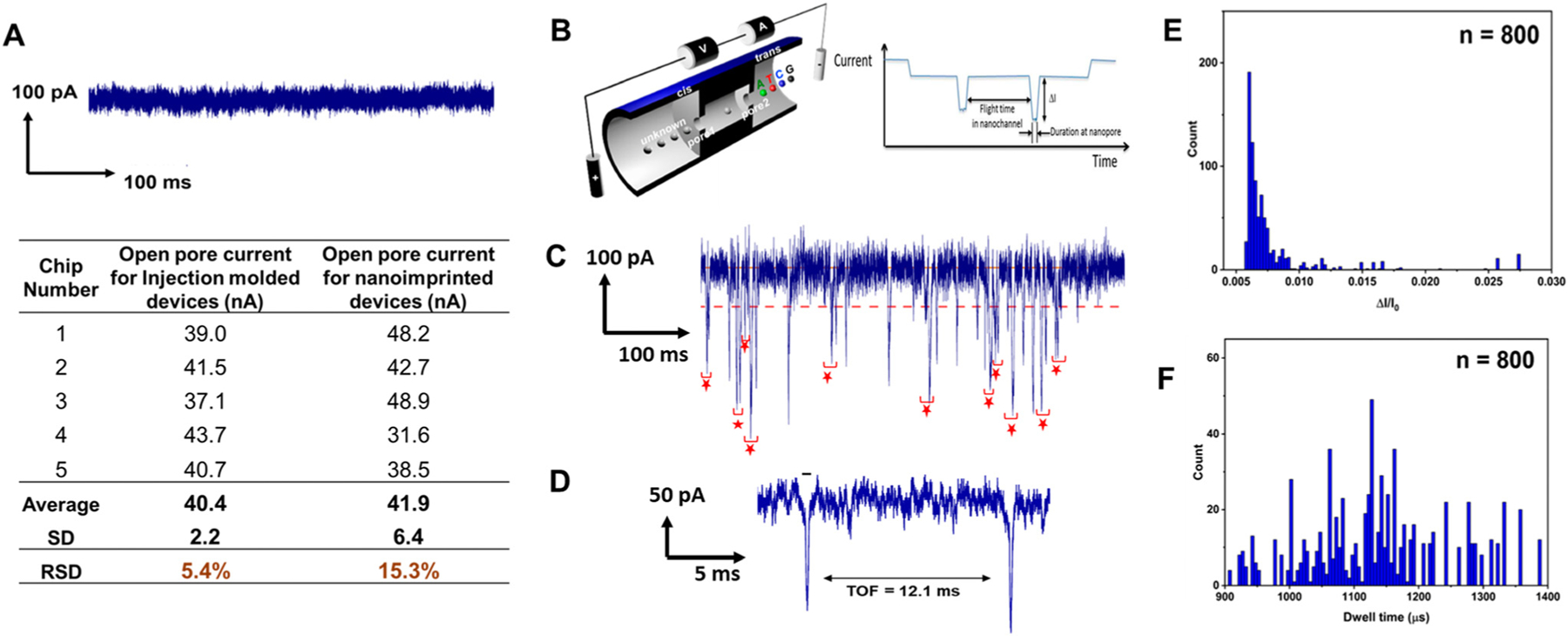
Dual in-plane nanopore sensor for measuring the Time-of-Flight (ToF) of single molecules, such as ribonucleotide monophosphates. (A) A 0.5 s current transient trace obtained prior to introducing rCMPs. The inset table shows open pore currents for five randomly picked assembled nanosensors produced by injection molding and nanoimprint lithography. (B) Diagram illustrating the dual-nanopore ToF sensor configuration, featuring a pair of in-plane nanopores positioned on opposite sides of a nanochannel serving as the nanochannel column for nanoscale electrophoresis. The schematic was taken from ref. 54. (C) A 0.5 s current transient trace of signal amplitudes obtained after injecting rCMPs (10 nM) in 1× NEBuffer 3 at pH.7.9 into the nanosensor. (D) Example current transient data showing a peak pair from the two in-plane nanopores for translocation of a single rCMP molecule. (E) Histogram of the normalized peak amplitude (ΔI/Io) data from the current transient data shown in Fig. 3C. (F) Histogram of the dwell time data from the current transient data shown in Fig. 3C.
Prior to nanoscale electrophoresis experiments, the size of in-plane nanopores in the assembled device was determined. The thermal fusion bonding process inevitably involves mixing of polymer chains between the nanofluidic substrate and cover plate and thus changes the effective dimensions of the assembled nanostructures from those of injection molded nanostructures particularly for the in-plane nanopores (Fig. 2A), which are the smallest structures contained within this nanofluidic device. Previously, we demonstrated controlled reduction of in-plane nanopores made in thermoplastics via the thermal fusion bonding process.55 Due to inaccessibility to the enclosed nanostructures after cover plate bonding, it was difficult to directly measure the pore dimensions. Instead, the open pore current measured after filling the device with an electrolyte could provide an indirect estimate of the effective diameters of the in-plane nanopores.54 Therefore, the open pore current for assembled nanofluidic devices can be used to study the reproducibility of our manufacturing protocol by injection molding and thermal fusion bonding processes (Fig. 3A).
We randomly picked five injection molded COP substrates out of more than 100, assembled them with COC 8007 cover plates at 74 °C and 170 psi for 15 min, and measured open pore currents after filling the devices with 1× PBS buffer containing 1 M KCl. For comparison, we also produced the same nanofluidic devices via thermal NIL with the MD700 resin mold and fusion bonded the COC cover plate to imprinted COP substrates under identical conditions as used for injection molded devices. The average open pore current for injection molded devices was 40.4 ± 2.2 nA with the relative standard deviation (RSD) being 5.4%, which is three times smaller than the RSD of NIL produced devices (15.3%) (Fig. 3A). During injection molding, demolding of the injection molded part was done spontaneously when the movable MUD platen was retracted during each injection molding cycle. On the other hand, demolding for NIL was done manually by peeling the resin mold from the imprinted substrate, generating more lateral motion during demolding. The improved uniformity of injection molded devices is attributed to the uniform demolding process in conjunction with low adhesion (or demolding force) associated with the use of resin mold inserts. The results are significant in that increased manufacturing throughput and precision can be achieved simultaneously even for nanofluidic structures by using injection molding over the conventional prototyping tool of NIL.
The effective diameter of the in-plane nanopores in the assembled nanofluidic device as estimated by the open pore current was 12 nm, which was determined from the physical length of the pore (10 nm), the bias voltage (2.5 V), and the open pore current (40.4 nA), and is within the range of pore size used for resistive pulse sensing of dNMPs from our previous study.54 Assembled COP/COC dual nanopore devices were primed with 50% v/v methanol followed by introduction of 1× NEBuffer 3 (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH = 7.9). Then, a rCMP solution (10 nM) in 1× NEBuffer 3 at pH 7.9 was introduced into the device with an applied potential of 2.5 V and RPS detection was carried out using an Axopatch current amplifier. Fig. 3C and D shows a 0.5 s current transient trace and an example peak pair from the dual in-plane nanopores. The criteria to determine peak pairs from our previous study54 was used. For the RPS data trace, 41 peak pairs were identified and the average ToF, which was the time required for a molecule to travel between the two pores, was 12 ± 2.1 ms (RSD = 18%). The average current transient amplitudes of rCMP events was 467 ± 72 pA (Fig. 3E) and the average dwell time was determined to be 925 ± 270 μs (Fig. 3F). The values are comparable to those reported in.54 An in-depth analysis of the identification and detection of all rNMPs will be forthcoming in a paper currently in preparation by our group using injection molded thermoplastic devices.
Immobilization of XRN1 and selective binding of a single RNA molecule
The second nanofluidic device tested was designed to bind a single RNA molecule by a processive XRN1 enzyme that was immobilized to a solid phase bioreactor with the injection molded COP-COC device as shown in Fig. 1C and 2B.
For selective binding of a single RNA molecule to the central solid phase bioreactor, it was hypothesized that when immobilized XRN1 was present within the solid phase bioreactor the translocating RNA would enter the immobilized enzyme’s active site through electrostatic interactions and effectively complex with the enzyme causing it to be held in place.60 XRN1 is a processive exonuclease and clips single RNA molecules into their constituent rNMP nucleotides when activated by a Mg2+ cofactor. Without the Mg2+ cofactor, XRN1 can be used to bind electrostatically through its active site to a single RNA molecule without dissociation into nucleotides.
We first used two input/output nanochannels in the upper left and right sides in Fig. 2B to electrokinetically introduce the EDC/NHS and XRN1 solutions sequentially (thoroughly rinsed after each step) to covalently attach the XRN1 enzymes to the central pillar surface using EDC/NHS coupling chemistry following O2 plasma activation to generate surface –COOH groups.27
After immobilizing the enzyme to the central solid phase bioreactor, the input/output nanochannels in the right and left sides of Fig. 2B were used to electrokinetically drive SYTO82 labelled CAS9 RNA molecules (4.5 kb) to the bioreactor so that the translocating RNA molecules could potentially associate with the XRN1 enzyme immobilized around the solid phase bioreactor (Fig. 4A). A bright field image of the device used for these experiments is shown in Fig. 4B. These experiments were performed using NEBuffer 3, because XRN1 shows high enzymatic activity in this buffer system. Representative translocation dynamics of the RNA molecule in the device without XRN1 immobilization is shown in Fig. 4C. This RNA molecule translocated past the nanoscale bioreactor without remaining stationary due to the absence of XRN1 at the solid phase bioreactor. When the bioreactor had undergone XRN1 immobilization, RNA would move into the nanoreactor and become stationary even under the presence of a driving electric field, producing a fluorescent area of high intensity (see Fig. 4D). This was indicative of association between the solution-phase CAS9 RNA and the immobilized XRN1 enzyme.
Fig. 4.
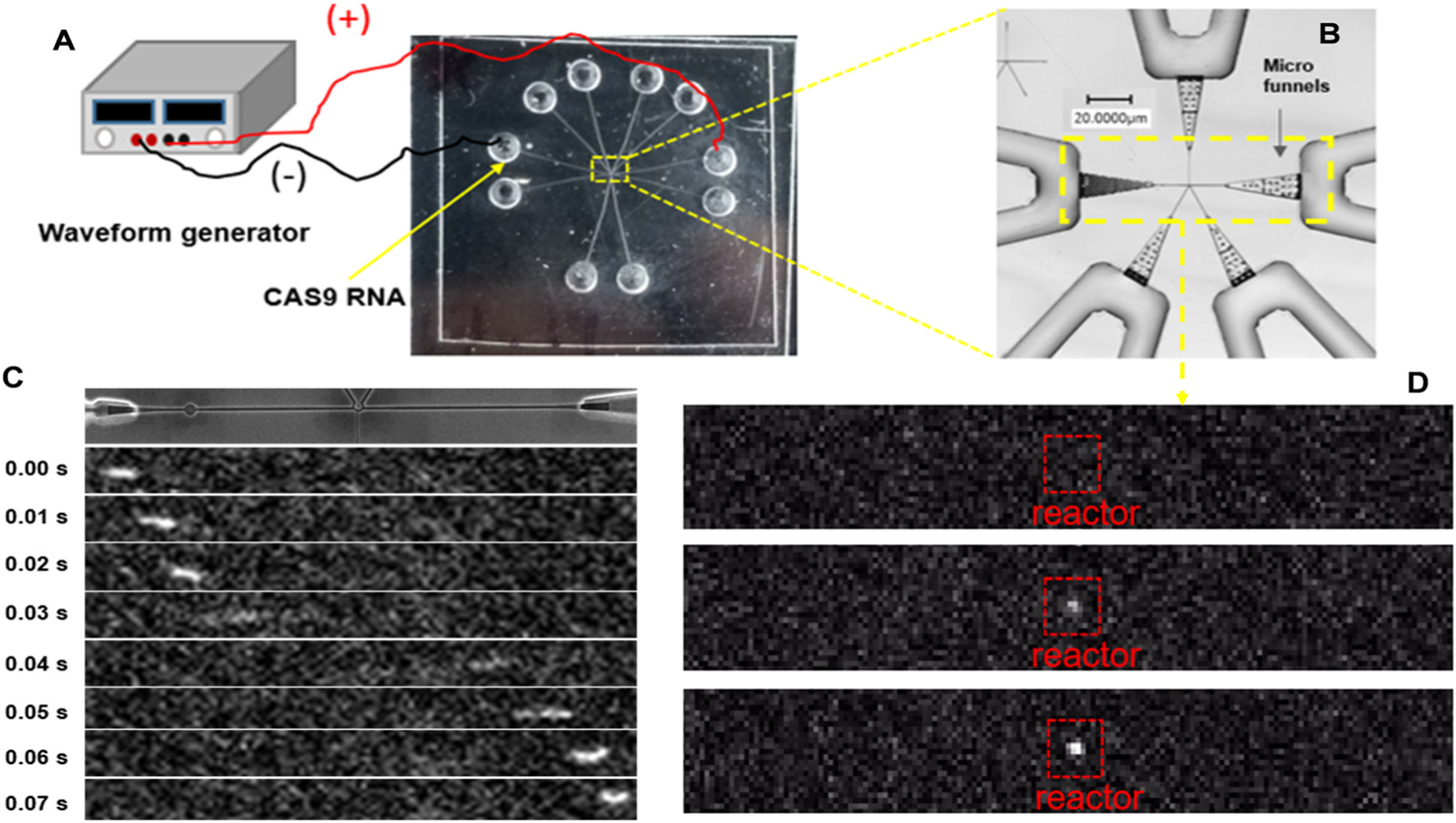
RNA translocation through an injection molded single-molecule sequencing device. (A) Experimental setup showing the electrical connections to the chip with a waveform generator for supplying the electrical field for driving the RNA (CAS9) through the chip. (B) Rapid scanning confocal image of the single-molecule sequencing device with the yellow box showing the area that is imaged with the single-molecule laser-induced fluorescence tracking microscope. (C) Fluorescence image Syto 82 labeled RNA electrically translocating through the input/output channels of the mixed-scale sequencing device. In this case, there was no ribo-exonuclease covalently attached to the solid-phase bioreactor portion of the device. Also, this device did not contain the in-plane nanopores within the input/output channel network. (D) Same conditions as shown and discussed in (C), but in this case, there was XRN1 ribo-exonuclease attached to the solid-phase bioreactor, which associates to the translocating RNA molecule causing it to remain stationary.
Materials and methods
Fabrication of multiscale micro/nanofluidic silicon masters
Silicon masters with multiscale micro/nanofluidic structures were prepared using a combination of photolithography and wet-chemical or dry etching for microstructuring, followed by focused ion beam (FIB) milling for nanostructuring (Fig. 1B and C). Photolithography was carried out by first spin coating an AZ 9260 (Microchemicals GmbH, Ulm, Germany) positive photoresist onto a silicon/silicon dioxide (Si/SiO2) wafer (UniversityWafer, Inc., South Boston, MA) with the thermal oxide film thickness of 300 nm at 4000 rpm for 60 s followed by resist baking at 100 °C for 5 min. After the bake, the microstructure patterns defined by a photomask were transferred onto the resist layer by exposing it to UV light for 24 s and developing the resist with AZ 300 MIF developer (Microchemicals GmbH, Ulm, Germany). The top oxide layer was removed by etching in a BOE 10 : 1 solution (Sigma-Aldrich, St. Louis, MO) followed by either wet or dry etching the underlying Si to create structures with a depth of 8 μm. The necessary nanostructures were fabricated using FIB milling (Quanta 3D Dual Beam system, FEI, Hillsboro, OR). The milling was performed at a beam voltage and current of 30 kV and 10–50 pA, respectively, using the bitmap mode.
Generating resin molds on a stainless-steel block to serve as a mold insert
Resin mold inserts were mounted in a master unit die (MUD) of the injection machine. Resin molds on a stainless-steel block were prepared using the following procedure (Fig. 1A). The patterns in the Si master were transferred to a UV-curable resin (mixture of 97 wt% MD700 (Solvay Solexis, Italy) and 3 wt% Darocure 1173 (Sigma Aldrich, St. Louis, MO)). Drops of the UV-resin were dispensed onto the surface of the Si master followed by placing a flexible polyethylene terephthalate (PET) sheet coated with an adhesive layer (NOA72, Norland Products, Jamesburg, NJ) gently against the liquid drop and used as a backbone substrate. During the curing process, the sample was exposed to UV light (365 nm) for 3 min at an intensity of 30 mW cm−2 using an ELC-500 UV curing system (Electro-Lite, Bethel, CT). This created features on the first MD700 resin mold that were protrusive (positive tone). These patterns were used to form a second MD700 resin mold with negative toned features using another PET backbone through the same UV curing process. To create the final resin mold insert, the top surface of a stainless-steel block underwent sanding followed by treatment with O2 plasma at 50 W for 2 min and coating with an NOA72 adhesive layer. Then, UV-resin molds with different UV-resin bases (polyurethane acrylate (PUA), tripropyleneglycol diacrylate (TPGDA), and MD700) were produced on the metal block using the second MD700 resin mold with negative toned structures. After demolding, positive toned UV-resin mold inserts were formed on the stainless-steel block and used for nano-injection molding. It should be noted that the final resin mold insert on a stainless-steel block with positive toned structures could be produced directly from the Si master without intermediate replication steps. However, we performed two additional replication processes to prevent any potential damage of the Si master during its repetitive use in producing the final resin mold inserts.
Injection molding
An all-electric hydraulic injection molding machine (Arburg 307A, Rocky Hill, Ct) was equipped with a master unit die (MUD), which had two frames where the platens with the slots for holding the mold inserts were attached. As can be seen in Fig. 1A, one side of the frame was movable, and fitted with a brass blank while the opposite side of the frame was stationary and contained the resin mold inserts; the molten polymer fills the mold cavity formed by the blank and resin mold inserts and generates the desired device. The mold inserts was maintained at a temperature (mold temperature) during injection molding, as controlled by a thermolator (Advantage Engineering, Greenwood, IN). In a general sequence of injection molding, the process starts with the mold cavity closing followed by injecting molten polymer into the mold cavity through a nozzle. After packing the molten polymer under a holding pressure of 800 bar for 2 s, the thermoplastic is allowed to cool down for a fixed cooling time via conduction through the blank and resin mold inserts that are maintained at the mold temperature. Finally, the movable frame was removed from the stationary frame, when the injection molded device is ejected from the mold cavity. Each injection molding cycle took less than 1 min, which mostly consists of times for injection, holding, and cooling.
Metrology of micro- and nanostructures
The Si master and injection molded devices containing micro/nano structures were analyzed using a variable pressure scanning electron microscope (SEM, Hitachi FlexSEM 1000 II, Hitachi High Tech, Schaumburg, IL) and atomic force microscope (AFM). The SEM can image non-conductive surfaces, such as thermoplastics, with a thin metal layer coating to interrogate the surface profile of the molded parts and was operated at 20 kV using a working distance of 6.0 mm. The widths of micro-pillars, nanochannels, and in-plane nanopores were determined using SEM images. High-resolution AFM scans (7.5 μm × 7.5 μm, 256 × 256 pixels, scan rate 0.5 Hz) were secured using a SPM HT-9700 (Shimadzu Scientific Instruments, Columbia, MO) tapping mode AFM with super sharp Si tips (curvature radius 2 nm, Nanosensors AG, Neuchatel, Switzerland). The depths of nanochannels and the dimensions of in-plane nanopores were determined using AFM imaging.
Characterization of viscoelastic properties of polymer
In the context of injection molding, the way that molten material fills the mold is significantly influenced by the inherent viscoelastic properties of the polymer matrix. A solid understanding of these material traits is vital for fine-tuning injection molding processes and achieving high-quality end products. To deepen this understanding, we conducted Dynamic Mechanical Analysis (DMA) using a Q-800 dynamic mechanical analyzer from TA Instruments, Inc. (New Castle, DE, USA). The DMA analysis involved a tensile mode setup with a fixed frequency of 1 Hz, a nominal strain amplitude of 0.1%, and a controlled temperature ramp of 3 °C per minute, ranging from room temperature to 180 °C. The specimens used for testing maintained consistent dimensions of 1–2 mm in thickness, 2–3 mm in width, and 20 mm in length. Before DMA testing, the samples underwent 10 minutes of UV exposure at a wavelength of 365 nm using the ELC-500 UV curing system from Electro-Lite in Bethel, CT, at an intensity of 30 mW cm−2.
RPS of single rCMP molecules
Translocation experiments were performed for rNMPs in dual in-plane nanopore devices bonded at 200 psi for 5 min. Briefly, after priming, 1× NEBuffer 3 (New England Biolabs, Ipswich, MA) was introduced into the device. Finally, 10 nM of rCMP seeded into 1× NEBuffer 3 was introduced into one of the reservoirs of the device. The devices were placed in a Faraday cage and Ag/AgCl electrodes were immersed in the reservoirs of the device. A potential of −2.5 V was applied between two electrodes by serially connecting 1.5 V battery to the Axopatch Digidata 1440B circuit (Molecular Devices, San Jose, CA) and the data was acquired at a sampling frequency of 250 kHz, a head stage configuration of β = 0.1, gain = 1, and a low pass filter of 10 kHz. Data were collected for a period of 10 min and Clampfit 11.1 software (Molecular Devices, San Jose, CA) was used for data acquisition and analysis.
Selective immobilization of XRN1 at bioreactor
Attachment of XRN1 to the enzymatic nanobioreactor was achieved using EDC/NHS coupling chemistry.61 To ensure delivery of reagents to the bioreactor, a waveform generator was used to apply a bias voltage. This was done by immersing electrodes into reservoirs of the microchannels that connect to the bioreactor. Both the COC 8007 (Topas Polyplastics, Farmington Hills, MI) cover plate and molded COP substrate were activated with an O2 plasma before thermal fusion bonding. The plasma treatment generated carboxyl groups on the thermoplastic surface.62 After initial aqueous buffer primer, two microchannels were used to deliver reagents to the nanobioreactor. These microchannels were filled with 0.1 M MES buffer (Sigma Aldrich, St. Louis, MO) containing 10 mg of EDC and 1 mg of NHS. The waveform generator was used to apply a bias voltage of 5 V across these microchannels and the nanobioreactor for 30 min. This drove the solution of EDC and NHS through the nanoreactor to ensure that these reagents reached and reacted with the carboxylic acid groups on the thermoplastic surfaces. This solution was removed and replaced with a 737 nM XRN1 enzyme solution. The waveform generator was again used with a bias voltage of 5 V to drive the enzyme solution through the reactor for 30 min. The waveform generator was removed and the device was left for an additional 90 min to ensure enzyme immobilization. This was followed by a buffer wash (1× NEBuffer 3).
RNA translocation
After enzyme immobilization, the devices were filled with NEBuffer 3 (100 mM NaCl, 50 mM Tris-HCl, 1 mM DTT, pH = 7.9) (New England Biolabs, Ipswich, MA). CAS9 RNA (4245 nt) (ThermoFisher Scientific, Waltham, MA) was labelled with SYTO-82 (RNA staining dye) (ThermoFisher Scientific, Waltham, MA) and added to access microchannels that connected to the input funnel. Electrodes were connected to a waveform generator and immersed in reservoirs on either side of the nanobioreactor. The entire device was mounted onto the stage of an epifluorescence microscope, which consisted of a Nikon TE 2000 microscope (Nikon Instruments, Melville, NY), 100×/1.4 NA oil-immersion objective, and an Andor iXon3 EMCCD camera (Oxford Instruments, Belfast, UK). The laser source was a Coherent (Santa Clara, CA) Nd:YAG; λex = 532 nm; P = 0.01–5 W; 2.2 mm beam diameter. With the application of the DC driving voltage, CAS9 RNA was electrokinetically driven through the device. Images were collected using the EMCCD camera with Metamorph advanced v7.5.6.0 software (Nashville, TN). ImageJ (National Institutes for Health, Bethesda, MD) software later used to subtract the background from recorded videos for further imaging processing. Fluorescence intensity data was processed using Origin 2021b SR2 version 9.85.212 (OriginLab, Northampton, MA) software with a 50% percentile filter applied to reduce noise.
Conclusions
For the first time, we demonstrated functional nanofluidic devices produced by injection molding with nanostructures ≤30 nm in depth. Injection molding was performed using UV-resin mold inserts consisting of a metal block bearing a patterned UV-resin layer. Among various UV-resins and thermoplastics tested, the combination of the MD700 resin mold insert and COP substrate resulted in the best replication fidelity for injection molding, which was attributed to the low demolding force associated with the low Young’s modulus and surface energy of the MD700 resin. While Young’s modulus and surface energy of the resin mold are key physical and chemical factors in determining ease of demolding, the Tg of the molding thermoplastics is the most critical material factor for the filling process together with important process parameters such as Tmold, injection speed, and dosage of the injected polymer. We have uniquely shown the ability to injection mold multiscale micro/nanofluidic thermoplastic devices, bond cover plates to the molded substrates, and utilize the devices in two different applications.
The functional devices were realized by assembling cover plates to the injection molded substrate with high process yield rates using thermal fusion bonding of a low Tg cover plate to a higher Tg substrate.63 The smallest functional nanostructures achieved in the enclosed nanofluidic devices were in-plane nanopores with an effective diameter of ∼12 nm, which were positioned at both ends of a nanochannel that was 50 × 50 nm in width × depth. We demonstrated label-less detection of single rCMP molecules by RPS from dual in-plane nanopores and obtained the molecular ToF from peak pairs, which produced signatures that could be used as a means for identifying molecules.38 We also demonstrated selective binding of a single CAS9 RNA molecule to a solid phase bioreactor with the processive enzyme XRN1 surface immobilized via EDC/NHS coupling chemistry to the single sub-micron polymer pillar.
In addition to the demonstration of two types of injection molded, functional nanofluidic devices, our work significantly advances the injection molding technology in that we were able to demonstrate the use of injection molding as a prototyping tool by avoiding the electrodeposition process used to produce Ni mold inserts, thus allowing accommodation of design changes with reduced development time and also rapid transition to a production line after prototyping. Once a final design is determined, high scale production can be realized using a conventional Ni shim to allow for producing a large number of parts.
Supplementary Material
Acknowledgements
The authors thank the NIH for financial support of this work (NIBIB R01-EB031579; NIBIB P41-EB020594; R21CA272351, and P20-GM130423). The authors also acknowledge the KU Nanofabrication Facility (KUNF, P20GM103638), LSU Shared Instrumentation Facilities (SIF), and LSU Center for Advanced Materials and Devices (CAMD). The authors would also like to thank Bionano Genomics for partial financial support of this project.
Footnotes
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00543g
Conflicts of interest
The authors have declared no conflict of interest.
References
- 1.Haywood DG, Saha-Shah A, Baker LA and Jacobson SC, Fundamental Studies of Nanofluidics: Nanopores, Nanochannels, and Nanopipets, Anal. Chem., 2015, 87, 172–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mawatari K, Kazoe Y, Shimizu H, Pihosh Y and Kitamori T, Extended-Nanofluidics: Fundamental Technologies, Unique Liquid Properties, and Application in Chemical and Bio Analysis Methods and Devices, Anal. Chem., 2014, 86, 4068–4077. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Yu H, Fan J, Wang F, Lu D, Liu H and Wu H, Channel-width dependent pressure-driven flow characteristics of shale gas in nanopores, AIP Adv., 2017, 7, 045217. [Google Scholar]
- 4.Nanofluidics is on the rise, Nat. Mater., 2020, 19, 253. [DOI] [PubMed] [Google Scholar]
- 5.Riehn R, Lu M, Wang Y-M, Lim SF, Cox EC and Austin RH, Restriction mapping in nanofluidic devices, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10012–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riehn R, Austin RH and Sturm JC, A Nanofluidic Railroad Switch for DNA, Nano Lett., 2006, 6, 1973–1976. [DOI] [PubMed] [Google Scholar]
- 7.Tegenfeldt JO, Prinz C, Cao H, Huang RL, Austin RH, Chou SY, Cox EC and Sturm JC, Micro- and nanofluidics for DNA analysis, Anal. Bioanal. Chem., 2004, 378, 1678–1692. [DOI] [PubMed] [Google Scholar]
- 8.Douville N, Huh D and Takayama S, DNA linearization through confinement in nanofluidic channels, Anal. Bioanal. Chem., 2008, 391, 2395–2409. [DOI] [PubMed] [Google Scholar]
- 9.Eijkel JCT and Berg A.v.d., Nanofluidics: what is it and what can we expect from it?, Microfluid. Nanofluid., 2005, 1, 249–267. [Google Scholar]
- 10.Guo R, Qi L, Xu L and Zou H, Fabrication of sub-50 nm nanochannel array by an angle forming lift-off method, J. Manuf. Process., 2022, 75, 584–592. [Google Scholar]
- 11.Zangle TA, Mani A and Santiago JG, Theory and experiments of concentration polarization and ion focusing at microchannel and nanochannel interfaces, Chem. Soc. Rev., 2010, 39, 1014–1035. [DOI] [PubMed] [Google Scholar]
- 12.Keyser UF, van Dorp S and Lemay SG, Tether forces in DNA electrophoresis, Chem. Soc. Rev., 2010, 39, 939–947. [DOI] [PubMed] [Google Scholar]
- 13.Pennathur S, Baldessari F, Santiago JG, Kattah MG, Steinman JB and Utz PJ, Free-Solution Oligonucleotide Separation in Nanoscale Channels, Anal. Chem., 2007, 79, 8316–8322. [DOI] [PubMed] [Google Scholar]
- 14.Menard LD and Ramsey JM, Electrokinetically-Driven Transport of DNA through Focused Ion Beam Milled Nanofluidic Channels, Anal. Chem., 2013, 85, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng L-J and Guo LJ, Nanofluidic diodes, Chem. Soc. Rev., 2010, 39, 923–938. [DOI] [PubMed] [Google Scholar]
- 16.Storm AJ, Chen JH, Ling XS, Zandbergen HW and Dekker C, Fabrication of solid-state nanopores with single-nanometre precision, Nat. Mater., 2003, 2, 537–540. [DOI] [PubMed] [Google Scholar]
- 17.Wu M-Y, Krapf D, Zandbergen M, Zandbergen H and Batson PE, Formation of nanopores in a SiN/SiO[sub 2] membrane with an electron beam, Appl. Phys. Lett., 2005, 87, 113106. [Google Scholar]
- 18.Iqbal SM, Akin D and Bashir R, Solid-state nanopore channels with DNA selectivity, Nat. Nanotechnol., 2007, 2, 243–248. [DOI] [PubMed] [Google Scholar]
- 19.Uddin A, Yemenicioglu S, Chen CH, Corigliano E, Milaninia K and Theogarajan L, Integration of solid-state nanopores in a 0.5 mum CMOS foundry process, Nanotechnology, 2013, 24, 155501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Stein D, McMullan C, Branton D, Aziz MJ and Golovchenko JA, Ion-beam sculpting at nanometre length scales, Nature, 2001, 412, 166–169. [DOI] [PubMed] [Google Scholar]
- 21.Stein D, Li J and Golovchenko JA, Ion-Beam Sculpting Time Scales, Phys. Rev. Lett., 2002, 89, 276106. [DOI] [PubMed] [Google Scholar]
- 22.Keyser UF, Krapf D, Koeleman BN, Smeets RMM, Dekker NH and Dekker C, Nanopore Tomography of a Laser Focus, Nano Lett., 2005, 5, 2253–2256. [DOI] [PubMed] [Google Scholar]
- 23.Krapf D, Wu M-Y, Smeets RMM, Zandbergen HW, Dekker C and Lemay SG, Fabrication and Characterization of Nanopore-Based Electrodes with Radii down to 2 nm, Nano Lett., 2005, 6, 105–109. [DOI] [PubMed] [Google Scholar]
- 24.Kim MJ, Wanunu M, Bell DC and Meller A, Rapid Fabrication of Uniformly Sized Nanopores and Nanopore Arrays for Parallel DNA Analysis, Adv. Mater., 2006, 18, 3149–3153. [Google Scholar]
- 25.Fischbein MD and Drndiƒá M, Sub-10 nm Device Fabrication in a Transmission Electron Microscope, Nano Lett., 2007, 7, 1329–1337. [DOI] [PubMed] [Google Scholar]
- 26.Mussi V, Fanzio P, Repetto L, Firpo G, Scaruffi P, Stigliani S, Tonini GP and Valbusa U, DNA-functionalized solid state nanopore for biosensing, Nanotechnology, 2010, 21, 145102. [DOI] [PubMed] [Google Scholar]
- 27.Athapattu US, Amarasekara CA, Nagel AC, Immel JR, Bloom S, Barany F and Soper SA, Solid-Phase XRN1 Reactions for RNA Cleavage: Application in Single-Molecule Sequencing, Nucleic Acids Res., 2021, 40, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver-Calixte NJ, Uba FI, Battle KN, Weerakoon-Ratnayake KM and Soper SA, Immobilization of Lambda Exonuclease onto Polymer Micropillar Arrays for the Solid-Phase Digestion of dsDNAs, Anal. Chem., 2014, 86, 4447–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stormonth-Darling JM, Pedersen RH, How C and Gadegaard N, Injection moulding of ultra high aspect ratio nanostructures using coated polymer tooling, J. Micromech. Microeng., 2014, 24, 075019. [Google Scholar]
- 30.Maximov I, Sarwe EL, Beck M, Deppert K, Graczyk M, Magnusson MH and Montelius L, Fabrication of Si-based nanoimprint stamps with sub-20 nm features, Microelectron. Eng., 2002, 61–62, 449–454. [Google Scholar]
- 31.Wang YH, Malar P and van Kan JA, Resist evaluation for proton beam writing, Ni mold fabrication and nano-replication, Microsyst. Technol., 2014, 20, 2079–2088. [Google Scholar]
- 32.Mabe TL, Ryan JG and Wei J, in Fundamentals of Nanoparticles, ed. Barhoum A and Hamdy Makhlouf AS, Elsevier, 2018, pp. 169 –213. [Google Scholar]
- 33.Menard LD and Ramsey JM, Fabrication of Sub-5 nm Nanochannels in Insulating Substrates Using Focused Ion Beam Milling, Nano Lett., 2011, 11, 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Chantiwas R, Amirsadeghi A, Soper SA and Park S, Complete plastic nanofluidic devices for DNA analysis via direct imprinting with polymer stamps, Lab Chip, 2011, 11, 2984–2989. [DOI] [PubMed] [Google Scholar]
- 35.Uba FI, Pullagurla SR, Sirasunthorn N, Wu J, Park S, Chantiwas R, Cho Y-K, Shin H and Soper SA, Surface charge, electroosmotic flow and DNA extension in chemically modified thermoplastic nanoslits and nanochannels, Analyst, 2015, 140, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W and Soper SA, Bio-MEMS: Technologies and Applications, Boca Raton, 1st edn, 2006. [Google Scholar]
- 37.Qi S, Liu X, Ford S, Barrows J, Thomas G, Kelly K, McCandless A, Lian K, Goettert J and Soper SA, Microfluidic devices fabricated in poly(methyl methacrylate) using hot-embossing with integrated sampling capillary and fiber optics for fluorescence detection, Lab Chip, 2002, 2, 88–95. [DOI] [PubMed] [Google Scholar]
- 38.Athapattu US, Rathnayaka C, Vaidyanathan S, Gamage SST, Choi J, Riahipour R, Manoharan A, Hall AR, Park S and Soper SA, Tailoring Thermoplastic In-Plane Nanopore Size by Thermal Fusion Bonding for the Analysis of Single Molecules, ACS Sens., 2021, 6, 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uba FI, Hu B, Weerakoon-Ratnayake K, Oliver-Calixte N and Soper SA, High process yield rates of thermoplastic nanofluidic devices using a hybrid thermal assembly technique, Lab Chip, 2015, 15, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weerakoon-Ratnayake KM, O’Neil CE, Uba FI and Soper SA, Thermoplastic nanofluidic devices for biomedical applications, Lab Chip, 2017, 17, 362–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schift H, David C, Gabriel M, Gobrecht J, Heyderman LJ, Kaiser W, Köppel S and Scandella L, Nanoreplication in polymers using hot embossing and injection molding, Microelectron. Eng., 2000, 53, 171–174. [Google Scholar]
- 42.Dundar Arisoy F, Czolkos I, Johansson A, Nielsen T and Watkins JJ, Low-cost, durable master molds for thermal-NIL, UV-NIL, and injection molding, Nanotechnology, 2020, 31, 015302. [DOI] [PubMed] [Google Scholar]
- 43.Hobæk TC, PhD thesis, Technical University of Denmark, 2014. [Google Scholar]
- 44.Calaon M, Tosello G, Garnaes J and Hansen HN, Injection and injection-compression moulding replication capability for the production of polymer lab-on-a-chip with nano structures, J. Micromech. Microeng., 2017, 27, 105001. [Google Scholar]
- 45.Mönkkönen K, Pakkanen TT, Hietala J, Pääkkönen EJ, Pääkkönen P, Jääskeläinen T and Kaikuranta T, Replication of sub-micron features using amorphous thermoplastics, Polym. Eng. Sci., 2002, 42, 1600–1608. [Google Scholar]
- 46.Zhang N, Byrne CJ, Browne DJ and Gilchrist MD, Towards nano-injection molding, Mater. Today, 2012, 15, 216–221. [Google Scholar]
- 47.Matschuk M, Bruus H and Larsen NB, Nanostructures for all-polymer microfluidic systems, Microelectron. Eng., 2010, 87, 1379–1382. [Google Scholar]
- 48.Miller J, Hemmati H, Lechuga O, Claytor N, Claytor R, Fraelich M and Hoppe M, Injection molding of subwavelength structures over a large surface area, SPIE, 2022. [Google Scholar]
- 49.Utko P, Persson F, Kristensen A and Larsen NB, Injection molded nanofluidic chips: Fabrication method and functional tests using single-molecule DNA experiments, Lab Chip, 2011, 11, 303–308. [DOI] [PubMed] [Google Scholar]
- 50.Park DS, Singh V, You BH, Kim N, Chen P-C, Soper SA, Nikitopoulos DE, Goettert J and Murphy MC, ASME 2009 International Mechanical Engineering Congress and Exposition, 2009, vol. 12: Micro and Nano Systems, Parts A and B, pp. 263–269. [Google Scholar]
- 51.Kim K, Park S, Lee JB, Manohara H, Desta Y, Murphy M and Ahn CH, Rapid replication of polymeric and metallic high aspect ratio microstructures using PDMS and LIGA technology, Microsyst. Technol., 2002, 9, 5–10. [Google Scholar]
- 52.Tang PT, Christensen TR and Jensen MF, Electroforming of Tool Inserts for Injection Molding of Optical or Microfluidic Components, AESF SUR/FIN Conference, Navy Pier, Chicago, 2004, pp. 763–774. [Google Scholar]
- 53.Ren J, Zhu Z, Xia C, Qu N and Zhu D, Abrasive-assisted Nickel Electroforming Process with Moving Cathode, Chin. J. Mech. Eng., 2017, 30, 294–300. [Google Scholar]
- 54.Choi J, Jia Z, Riahipour R, McKinney CJ, Amarasekara CA, Weerakoon-Ratnayake KM, Soper SA and Park S, Label-Free Identification of Single Mononucleotides by Nanoscale Electrophoresis, Small, 2021, 17, 2102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Athapattu US, Rathnayaka C, Vaidyanathan S, Gamage SST, Choi J, Riahipour R, Manoharan A, Hall AR, Park S and Soper SA, Tailoring Thermoplastic In-Plane Nanopore Size by Thermal Fusion Bonding for the Analysis of Single Molecules, ACS Sens., 2021, 6, 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Neil C, Amarasekara CA, Weerakoon-Ratnayake KM, Gross B, Jia Z, Singh V, Park S and Soper SA, Electrokinetic transport properties of deoxynucleotide monophosphates (dNMPs) through thermoplastic nanochannels, Anal. Chim. Acta, 2018, 1027, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaidyanathan S, Weerakoon-Ratnayake KM, Uba FI, Hu B, Kaufman D, Choi J, Park S and Soper SA, Thermoplastic nanofluidic devices for identifying abasic sites in single DNA molecules, Lab Chip, 2021, 21, 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu JH, Chantiwas R, Amirsadeghi A, Soper SA and Park S, Complete plastic nanofluidic devices for DNA analysis via direct imprinting with polymer stamps, Lab Chip, 2011, 11, 2984–2989. [DOI] [PubMed] [Google Scholar]
- 59.Jia Z, Choi J and Park S, Selection of UV-resins for nanostructured molds for thermal-NIL, Nanotechnology, 2018, 29, 365302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang JH, Xiang S, Xiang K, Manley JL and Tong L, Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1, Nat. Struct. Mol. Biol., 2011, 18, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Athapattu US, Amarasekara CA, Immel JR, Bloom S, Barany F, Nagel AC and Soper SA, Solid-phase XRN1 reactions for RNA cleavage: application in single-molecule sequencing, Nucleic Acids Res., 2021, 49, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amarasekara CA, Athapattu US, Rathnayaka C, Choi J, Park S and Soper SA, Open-tubular nanoelectrochromatography (OT-NEC): gel-free separation of single stranded DNAs (ssDNAs) in thermoplastic nanochannels, Electrophoresis, 2020, 41, 1627–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uba FI, Hu B, Weerakoon-Ratnayake K, Oliver-Calixte N and Soper SA, High process yield rates of thermoplastic nanofluidic devices using a hybrid thermal assembly technique, Lab Chip, 2015, 15, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


