Abstract
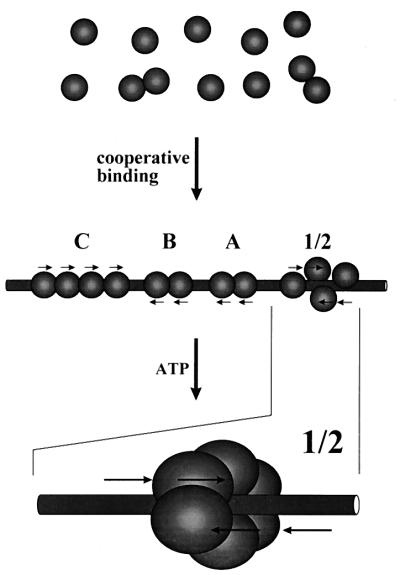
Polyomavirus large T antigen binds to multiple 5′-G(A/G)GGC-3′ pentanucleotide sequences in sites 1/2, A, B, and C within and adjacent to the origin of viral DNA replication on the polyomavirus genome. We asked whether the binding of large T antigen to one of these sites could influence binding to other sites. We discovered that binding to origin DNA is substantially stronger at pH 6 to 7 than at pH 7.4 to 7.8, a range often used in DNA binding assays. Large T antigen-DNA complexes formed at pH 6 to 7 were stable, but a fraction of these complexes dissociated at pH 7.6 and above upon dilution or during electrophoresis. Increased binding at low pH is therefore due at least in part to increased stability of protein-DNA complexes, and binding at higher pH values is reversible. Binding to fragments of origin DNA in which one or more sites were deleted or inactivated by point mutations was measured by nitrocellulose filter binding and DNase I footprinting. The results showed that large T antigen binds cooperatively to its four binding sites in viral DNA, suggesting that the binding of this protein to one of these sites stabilizes its binding to other sites via protein-protein contacts. Sites A, B, and C may therefore augment DNA replication by facilitating the binding of large T antigen to site 1/2 at the replication origin. ATP stabilized large T antigen-DNA complexes against dissociation in the presence, but not the absence, of site 1/2, and ATP specifically enhanced protection against DNase I digestion in the central 10 to 12 bp of site 1/2, at which hexamers are believed to form and begin unwinding DNA. We propose that large T antigen molecules bound to these multiple sites on origin DNA interact with each other to form a compact protein-DNA complex and, furthermore, that ATP stimulates their assembly into hexamers at site 1/2 by a “handover” mechanism mediated by these protein-protein contacts.
Polyomavirus large T antigen initiates DNA unwinding and replication via elaborate interactions with the viral replication origin (2). Specific DNA binding by this 785-amino-acid protein is mediated by a domain that lies between amino acids 282 and 398, as defined by using deletion mutants (52). Large T antigen binds to a target consensus pentanucleotide sequence, 5′-G(G/A)GGC-3′, which is present in multiple copies in the replication origin region between the early transcription start site and the transcriptional enhancer (4, 7, 8, 17, 42). Immunoprecipitation and DNase I protection assays showed that four distinct sites on polyomavirus DNA, denoted 1/2, A, B, and C, are bound by large T antigen in vitro (7, 17, 42). Site 1/2, which is situated within the core origin of DNA replication (23, 26, 28, 34, 43), contains four closely spaced consensus pentanucleotide sequences arranged symmetrically as two partly overlapping pairs on opposite DNA strands (8, 9, 41, 49). Sites A, B, and C are located between the core replication origin and the early transcription unit. These sites contain, respectively, two, two, and four target pentanucleotide sequences in polyomavirus strain A3 and its derivatives (1, 9, 43, 53). Adjacent pentanucleotides are spaced approximately one turn of the DNA helix apart in each of these three sites, implying that large T antigen molecules bound to adjacent pentanucleotides are aligned on one side of the helix. Mutagenesis and methylation interference experiments showed that binding of large T antigen to adjacent pentanucleotides within a given site is cooperative, since removal of one pentanucleotide sequence from a site containing three sequences reduced binding affinity by a factor of 10 (8). Large T antigen of closely related simian virus 40 shares extensive sequence homology with its polyomavirus counterpart and also recognizes G(A/G)GGC pentanucleotide sequences on DNA (40, 41).
Large T antigen molecules can oligomerize; most preparations contain monomers, dimers, trimers, tetramers, and hexamers in solution (6, 10, 19, 44, 55). Incubation with ATP stimulates hexamer formation (10, 44, 55), presumably by inducing a conformational change in large T antigen. In the presence of ATP, two hexamers of simian virus 40 large T antigen assemble on viral DNA at site II in the simian virus 40 replication origin (10, 11, 30, 38, 59); each hexamer is centered on one of the pairs of closely spaced G(A/G)GGC sequences in site II (38). It has been postulated, but not shown directly, that hexamers of polyomavirus large T antigen also assemble at analogous site 1/2 on polyomavirus DNA. Hexamers are circular structures that enclose the DNA like a wheel about an axle (48, 59). Large T antigen hexamers unwind DNA in the replication origin, leading to the initiation of bidirectional DNA replication by cellular DNA polymerase α/primase, which is brought to the origin by interaction with large T antigen (33, 35).
What is the role of polyomavirus large T antigen binding sites A, B, and C in viral DNA replication? Although these sites are not absolutely required to direct large T antigen-mediated DNA replication in vivo (1, 34, 57), their presence augments DNA replication in transfected plasmids (57) and is required for optimal virus replication in permissive mouse cells (1). Large T antigen bound more strongly to sites A, B, and C than to site 1/2 in the absence of ATP (7, 17, 42), but ATP strongly increased the affinity of large T antigen for DNAs containing site 1/2 (27), probably by stimulating the formation of hexamers.
We decided to reexamine the binding of large T antigen to its multiple sites in the replication origin region of polyomavirus DNA by using target DNA fragments with point and deletion mutations. In the course of setting up DNA binding assays, we also studied the effects of pH and of ATP on binding. We found that specific binding to DNA is strong and stable at pH 7 and below but is weaker and reversible above pH 7.4, that ATP stabilizes binding of large T antigen to DNAs that contain site 1/2, and that in the presence of ATP, large T antigen preferentially protects the central 10 to 12 nucleotides (nt) of site 1/2 against DNase I digestion. Using a variety of DNA binding conditions, we found that large T antigen binds cooperatively to its multiple sites in the replication origin. These observations suggest a model in which the assembly of hexamers of large T antigen at the replication origin is facilitated by the “handover” of reversibly bound large T antigen molecules from sites A, B, and C to site 1/2.
MATERIALS AND METHODS
Expression and purification of polyomavirus large T antigen.
Yeast Pichia pastoris transformant E-3 was used to express large T antigen, which was purified by immunoaffinity chromatography as previously described (39). After purification, large T antigen was the predominant protein species when visualized by silver staining of polyacrylamide gels. The quantity of large T antigen was determined both by comparison to protein standards visualized on silver-stained gels and by colorimetric analysis. Purified large T antigen was stored at −70°C in a buffer containing 10 mM potassium phosphate (pH 7.0), 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 20% glycerol. As a control, a baculovirus expression system (45) was also used to produce large T antigen. Recombinant baculovirus vEV51LT stock was generated in Spodoptera frugiperda (Sf9) cells. Large T antigen was expressed in High Five cells in accordance with published methods (45, 46) and was purified as described previously (39).
Construction of plasmids with mutations in the binding region for polyomavirus large T antigen.
Polyomavirus strain AT3-Modori, generated by oligonucleotide-directed mutagenesis from strain AT3 (1), contains four additional restriction endonuclease sites flanking large T antigen binding sites A, B, and C (see Fig. 4A). These sites were chosen to introduce minimal changes to viral DNA; in particular, no G(A/G)GGC consensus sequences were altered, and the distances between sites A, B, C, and 1/2 were unchanged. Plasmid pGEM-Modori contains AT3-Modori DNA cloned into the EcoRI site of plasmid pGEM-3Zf(−) (Promega). Two sets of mutants were generated as follows.
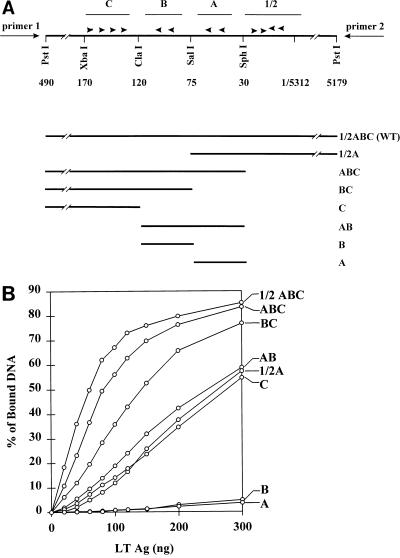
FIG. 4.
Binding of large T antigen to DNAs containing combinations of adjacent binding sites. (A) The PstI fragment of polyomavirus AT3-Modori DNA that contains the replication origin and large T antigen binding sites 1/2, A, B, and C is shown in the upper panel as part of plasmid pGEM-1/2ABC(+). The arrowheads indicate the G(A/G)GGC consensus sequences within each site. The numbers indicate the positions of the restriction sites shown. The primers used for PCR amplification of fragments within pGEM plasmids are shown at each end. In the lower panel, horizontal lines show sequences retained in deletion mutants made by cutting plasmids and religating them at different restriction sites in the PstI fragment or the pGEM polylinker (pGEM and polylinker sequences are not shown). For the sake of clarity, all deletion mutants are shown in the same orientation; however, mutants BC and B were derived from p1/2ABC(−), and therefore primers 1 and 2 are inverted for those plasmids. WT, wild type. (B) Six femtomoles of labeled DNA fragments containing different combinations of adjacent binding sites (noted at the right) were incubated with various amounts of large T antigen (1 ng = 10 fmol) in potassium phosphate buffer, pH 6, and filter binding assays were carried out as described in Materials and Methods.
(i) Deletion mutants.
Plasmid pGEM-Modori was digested with PstI, and the 623-bp fragment containing the origin region of polyomavirus DNA (nt 5179 to 5312 and 1 to 490) was cloned into the PstI site in the polylinker of pGEM-3Zf(−), resulting in plasmid pGEM-1/2ABC(+) or pGEM-1/2ABC(−), depending on the orientation of the insert. Plasmids containing individual binding sites or combinations of adjacent binding sites were derived from the parent plasmids by restriction cleavage, followed in some instances by blunt ending, and then religation (51). These deletion mutant plasmids were named to describe the sets of binding sites they contain (see Fig. 4A).
(ii) Point mutants.
Point mutations were introduced individually into the consensus G(A/G)GGC binding sequences in binding sites A, B, and C within plasmid pGEM-Modori, as previously described (1). Mutants were named to describe which sites were mutated (see Fig. 5A); e.g., mAmB specifies a mutant in which the two G(A/G)GGC sequences in site A and the two G(A/G)GGC sequences in site B were mutated. Mutant A was named A1A2 by Bertin et al. (1).
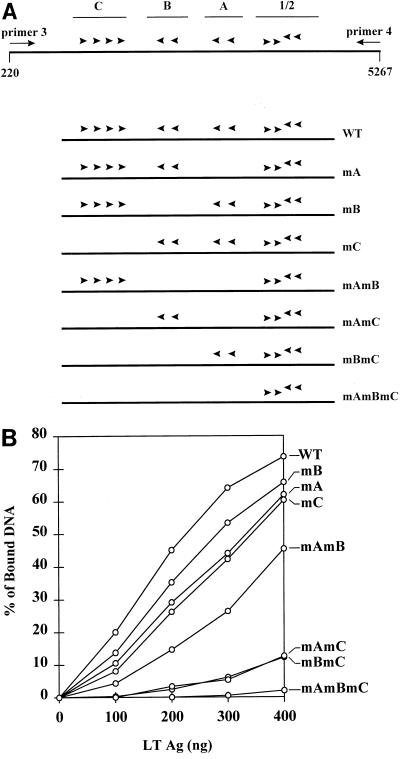
FIG. 5.
Binding of large T antigen to DNAs containing mutated binding sites. (A) The region containing binding sites 1/2, A, B, and C is shown in the upper panel. Below are shown mutants generated by introducing point mutations into consensus pentanucleotide sequences in site A, B, or C or in combinations of these sites (1). Mutants are named for the mutated sites that they contain. Arrowheads show the G(A/G)GGC sequences that remain intact in each plasmid. PCR amplification of the region between nt 220 and 5267 for each mutant was carried out with primers 3 and 4. (B) Six fentomoles of labeled DNA fragments containing mutations in G(A/G)GGC consensus sequences in sites A, B, and C were incubated with various amounts of large T antigen (LTAg) in Tris-HCl buffer, pH 7.0, and filter binding assays were carried out. WT, wild type.
Preparation of 32P-labeled DNA fragments.
Using the plasmids described above as templates, DNA fragments were made by PCR. (i) Fragments containing the wild-type origin or deletion mutants were made from pGEM-1/2ABC and its derivatives by using primer 1 (M13 universal primer), 5′-GTAAAACGACGGCCAGT-3′, and primer 2 (M13 reverse primer), 5′-CAGGAAACAGCTATGAC-3′ (see Fig. 4A). These fragments ranged in size from 736 bp (wild type) to 147 nt (site A alone). DNA products were internally labeled by incorporating (α-32P)dATP during PCR. These DNAs were used in filter binding assays. (ii) A set of 265-bp DNA fragments containing the wild-type origin or mutated binding sites were made from plasmid pGEM-Modori and its derivatives by using primer 3, 5′-GTTCTAGCAGCCTTTCTTTG-3′ (polyomavirus nt 220 to 201), and primer 4, 5′-GTGTGGTTTTGCAAGAGGAAG-3′ (polyomavirus nt 5267 to 5287) (see Fig. 5A). DNAs were either internally labeled as described above or end labeled at nt 5267 by incubating primer 4 with (γ-32P)ATP and T4 polynucleotide kinase before use in PCR. These DNAs were used for filter binding assays, for gel mobility shift assays, and for DNase I footprinting assays. All PCR-generated DNA fragments were purified by agarose gel electrophoresis and were quantitated by measurement of radioactivity.
Filter binding assay.
A procedure, modified from previously published methods (5, 24, 27), for binding of DNA to nitrocellulose filters was utilized. Purified large T antigen was incubated with 32P-labeled DNA fragments in 60 μl of a binding solution containing 50 mM NaCl, 7 mM MgCl2, 83 μg of bovine serum albumin per ml, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 10 ng of aprotinin/ml, and 17 μg of sheared salmon sperm DNA per ml. Buffers used were 50 mM sodium acetate (pH 5.0 to 5.6), 50 to 100 mM potassium phosphate (pH 6.0 to 7.6), and 50 to 100 mM Tris-HCl (pH 7.0 to 8.5). ATP (Boehringer Mannheim or Pharmacia), when used, was dissolved in distilled water, and the pH was adjusted with K2HPO4 or NaOH. After incubation at 37°C for 20 to 25 min, the mixtures were filtered by capillary flow through 13-mm-diameter nitrocellulose filters (BA-85; 0.45-μm pore size; Schleicher & Schuell) that had been boiled in 0.8% sodium dodecyl sulfate, washed in water, and presoaked in washing buffer (5 mM MgCl2, 100 mM NaCl, and 50 mM sodium acetate, potassium phosphate, or Tris-HCl adjusted to the pH values corresponding to those used for binding reactions). The filters were then washed with 2 ml of washing buffer and dried, and bound radioactivity was measured by liquid scintillation counting. Bound DNA was expressed as the percentage of input radioactivity remaining bound to the filter.
DNase I footprinting assay.
Binding reactions were set up as described for the filter binding assays. In a volume of 70 μl, 0.4 to 0.6 μg of large T antigen and 20 fmol of labeled origin DNA were incubated for 20 min at 37°C in Tris-HCl buffer (pH 7.0 to 7.4). ATP (1 mM) was included in the reaction mixture when indicated. The reaction mixture was then added to 5 μl of a solution containing 5 mM CaCl2, 10 mM MgCl2, and 10 μg of sheared salmon sperm DNA/ml. After incubation of the reaction mixture for 1 min at room temperature, 0.02 to 0.05 U of DNase I (GIBCO/BRL) was added. Digestion was allowed to proceed for 1 min at room temperature and was terminated by adding 70 μl of a solution consisting of 100 mM EDTA, 2 M ammonium acetate, 0.2% sodium dodecyl sulfate, and 100 μg of calf thymus DNA/ml. DNA was extracted with phenol-chloroform and subjected to electrophoresis on 12% polyacrylamide–8 M urea gels, which were dried and exposed to X-ray film or to storage phosphor screens that were analyzed in a Molecular Dynamics PhosphorImager.
Gel mobility shift assay.
32P-labeled DNA was incubated with large T antigen at 37°C for 20 min in 60 μl of a binding solution containing 50 mM potassium phosphate (pH 6.0 or 7.6), 7.5% glycerol, and the other components described above. Where indicated, glutaraldehyde was added at a concentration of 0.1% and incubation was continued for an additional 5 min. Samples were directly loaded onto 5% polyacrylamide gels, and electrophoresis was carried out in 50 mM potassium phosphate buffer (pH 6.0 or 7.6) containing 1 mM EDTA for 1.5 to 3 h at 100 V. The gels were dried and exposed to X-ray film.
RESULTS
Specific origin-binding activity of polyomavirus large T antigen is enhanced at pH 7 and below.
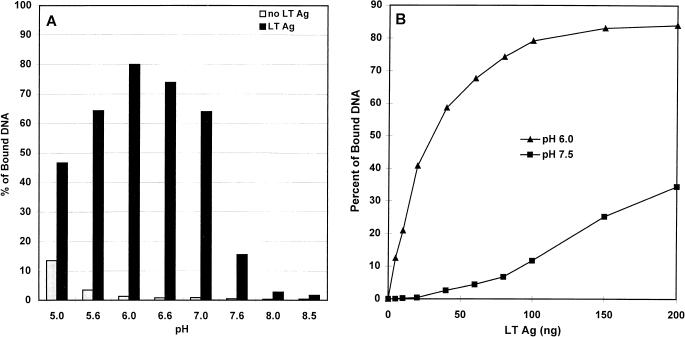
We wanted to quantitate the binding of purified polyomavirus large T antigen to DNA fragments containing the multiple G(A/G)GGC consensus pentanucleotide sequences present in the region of the polyomavirus replication origin. Binding of protein-DNA complexes to nitrocellulose filters is a simple, rapid, and easily quantifiable method of measuring protein-DNA binding (5, 12, 24, 27). We simplified the binding solution such that it included only NaCl, MgCl2, a reducing agent, and a buffer, in addition to protease inhibitors and nonspecific competitor DNA (see Materials and Methods). To characterize our DNA binding assay, we examined the influence of different components on the binding reaction. The pH of the solution was varied between 5.0 and 8.5 by the use of different buffers, and filter binding of a 736-bp 32P-labeled polyomavirus origin DNA fragment containing the four binding sites for large T antigen, 1/2, A, B, and C (1, 7), was determined both in the presence and in the absence of large T antigen (Fig. 1A). Optimal binding was observed between pH 5.6 and 7.0, with a maximum at pH 6.0, and binding activity fell off sharply at pH 7.6 or higher.
FIG. 1.
Effect of pH on DNA-binding activity of polyomavirus large T antigen. (A) Binding to nitrocellulose filters of 3 ng of 32P-labeled 736-bp DNA fragment containing the wild-type polyomavirus origin region was carried out in 50 mM buffers of different pH values (pH 5.0 to 5.6, sodium acetate; pH 6.0 to 7.6, potassium phosphate; pH 8.0 to 8.5, Tris-HCl) after incubation in the presence (black bars) or absence (gray bars) of 100 ng of large T antigen (LT Ag). (B) The same DNA fragment was incubated with increasing amounts of large T antigen in potassium phosphate buffer at pH 6.0 (triangles) or pH 7.5 (squares). Radioactivity bound to filters in the absence of large T antigen was subtracted from the results to give corrected specific binding values.
Previous DNA binding assays of polyomavirus or simian virus 40 large T antigen were carried out at various pH values between 7 and 8 (5, 12, 27, 30, 41, 42, 49, 54). Although differences in binding to nonspecific (18, 32, 36) or specific (12) DNAs were noted, no systematic study of the variation of DNA binding with pH has been published. Since polyomavirus large T antigen purified from P. pastoris had not been previously characterized, we asked whether an increase in DNA binding at a pH below 7 was peculiar to this source. We therefore purified polyomavirus large T antigen made in insect cells by a recombinant baculovirus (45) which has been used extensively in other studies (2, 27, 29, 55, 56) and measured its DNA binding activity as a function of pH; the results (not shown) were similar to those shown in Fig. 1A. To determine whether increased binding at low pH is specific for DNA containing binding sites for large T antigen, we performed filter binding assays with DNAs either containing or lacking G(A/G)GGC consensus sequences. The results showed that binding to nonspecific DNA increased as the pH was lowered but remained less than 2% of the input DNA at pH 7.0 and less than 10% of the input DNA at pH 6.0 under conditions in which 70 to 90% of the specific DNA was bound (data not shown). Furthermore, we carried out DNase I footprinting assays at different pH values and found that binding of large T antigen to origin DNA at pH values between 6.0 and 7.6 gave discrete footprints (see below) similar to those previously reported (7, 27, 29), although much higher concentrations of large T antigen were needed at pH 7.4 and above than at lower pH values.
Figure 1B shows that as little as 5 ng of large T antigen per 60-μl reaction volume was sufficient to bind to a fraction (12%) of a labeled polyomavirus origin DNA fragment when binding was carried out at pH 6.0, and binding was maximal at about 100 ng of large T antigen per reaction. In contrast, binding at pH 7.5 required substantially higher concentrations of large T antigen. The difference in binding affinity at pH 6.0 compared with that at pH 7.5 was estimated from the difference in the initial slopes of the two curves to be 10- to 20-fold. Clearly, the effect of pH on DNA binding by polyomavirus large T antigen is important and must be taken into account when binding studies are carried out.
Large T antigen–DNA complexes are stable at pH 6 to 7 but are unstable at pH 7.6.
In previous reports, DNA binding by polyomavirus or simian virus 40 large T antigen could be detected by gel mobility shift assays only after fixation of protein-DNA complexes with glutaraldehyde (11, 31, 35). The enhanced binding that we found at low pH encouraged us to try DNA band retardation with unfixed complexes by using a pH 6 buffer during electrophoresis. A 265-bp, 32P-labeled DNA fragment containing all four binding sites was incubated with large T antigen in binding solutions containing potassium phosphate buffer at pH 6.0 or 7.6, and reaction mixtures were loaded onto a 5% polyacrylamide gel and subjected to electrophoresis in pH 6 buffer (Fig. 2A). When the binding reaction was carried out at pH 6 (lane 2), all of the radioactive DNA migrated as a band very close to the position of the loading well. Prior fixation of DNA-protein complexes with glutaraldehyde (lane 3) had no effect on the migration of these complexes. When the binding reaction was carried out at pH 7.6 (lane 5), surprisingly, the migration of all of the DNA was also retarded. However, prior fixation of these complexes with glutaraldehyde resulted in a very small proportion of the input DNA being present in the retarded band (lane 6).
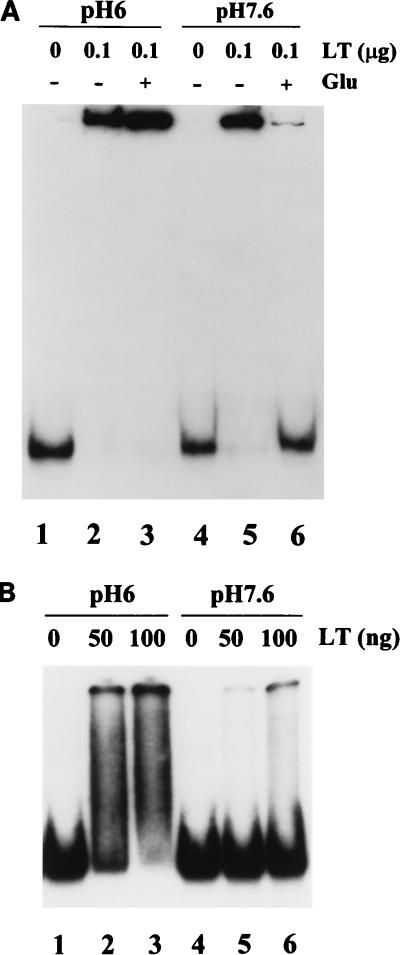
FIG. 2.
Stability of DNA-protein complexes subjected to electrophoresis at pH 6.0 or 7.6. (A) One nanogram of a 32P-labeled 265-bp DNA fragment containing the origin region was incubated without large T antigen (LT) (lanes 1 and 4) or in the presence of 100 ng of large T antigen (lanes 2, 3, 5, and 6) at 37°C for 20 min in potassium phosphate buffer at either pH 6.0 (lanes 1 to 3) or pH 7.6 (lanes 4 to 6). Glutaraldehyde (Glu; 0.1%) was added to reaction mixtures 3 and 6, and incubation was continued for an additional 5 min. All reactions were analyzed on a 5% polyacrylamide gel prepared and run in potassium phosphate buffer, pH 6.0. (B) One nanogram of the same DNA fragment was incubated without large T antigen (lanes 1 and 4) or with 50 ng (lanes 2 and 5) or 100 ng (lanes 3 and 6) of large T antigen at pH 6.0 (lanes 1 to 3) or pH 7.6 (lanes 4 to 6). Electrophoresis was carried out in potassium phosphate buffer, pH 7.6.
We explain these results as follows. Complexes formed at pH 6 between target DNA and polyomavirus large T antigen were stable for the several hours during which they were subjected to electrophoresis at pH 6, resulting in a retarded DNA band. Complexes form less readily at pH 7.6, but when the pH 7.6 binding reaction mixture was loaded onto the pH 6 gel (lane 5), the pH of the reaction dropped before electrophoresis began, allowing stable DNA-protein complexes to form at the lower pH of the loading well. On the other hand, glutaraldehyde fixation of large T antigen that had remained unbound (or not stably bound) to DNA at pH 7.6 eliminated its ability to bind to DNA, leading to reduced binding after the reaction mixture was loaded onto the pH 6 gel (lane 6).
We also prepared binding reaction mixtures at either pH 6 or pH 7.6 and subsequently loaded them onto a 5% polyacrylamide gel run in pH 7.6 buffer (Fig. 2B). When binding was carried out at pH 6, some DNA remained in the retarded band but most of the DNA-protein complexes dissociated during electrophoresis, leading to a smear of radioactive DNA between the positions of the free and the bound DNA (lanes 2 and 3). Very little retarded DNA was seen when binding was carried out at pH 7.6 (lanes 5 and 6). These results show that much of the large T antigen that had initially bound to DNA at pH 6 dissociated when the complexes were exposed to a pH 7.6 environment. Therefore, we conclude that large T antigen-DNA complexes are stable at pH 6 but are unstable at pH 7.6, dissociating on extended incubation at that pH in the absence of free large T antigen. The increased stability of large T antigen-DNA complexes at low pH may explain the more efficient binding of this protein to DNA at these pH values.
ATP does not affect DNA binding at pH 7 or below but stabilizes a fraction of large T antigen-origin DNA complexes at high pH.
ATP was shown to increase the affinity of polyomavirus large T antigen for DNA fragments within the viral replication origin when binding was done at pH 7.8 (27). Because we found that the affinity of large T antigen for origin DNA was significantly greater below pH 7 than at higher pH values, we decided to test the effect of ATP on DNA binding as a function of pH, using the nitrocellulose filter binding assay. Binding to origin DNA was stimulated twofold by 5 mM ATP in Tris-HCl buffer at pH 7.8 under our binding conditions, in agreement with previous results (27); however, we could detect no stimulation of binding by ATP in Tris-HCl buffer at pH 7.0 or in potassium phosphate buffer at pH 6.0 when using a variety of different concentrations of large T antigen (10 to 100 ng per reaction) or ATP (0.1 to 5 mM) (data not shown).
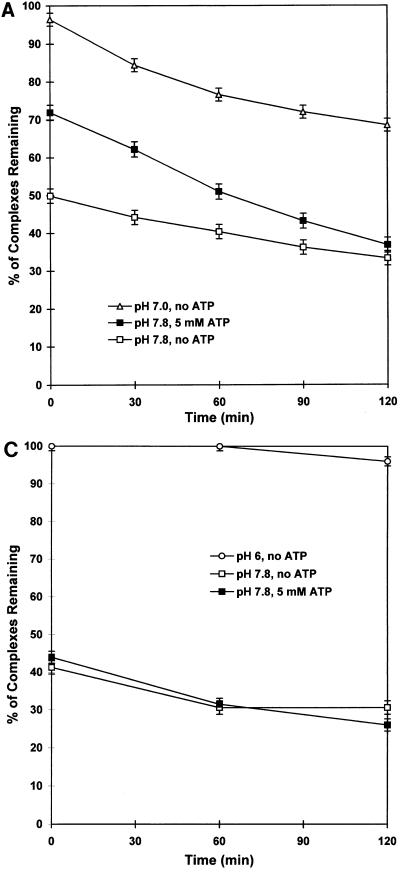
Since we had shown that large T antigen-DNA complexes were unstable at pH 7.6 but stable at pH 6, we asked whether ATP acted by stabilizing protein-DNA complexes at high pH values. We assembled complexes at pH 7.0 and subsequently diluted samples 16-fold into pH 7.0 or pH 7.8 buffer containing a 100-fold excess of unlabeled target DNA, in the presence or absence of 5 mM ATP. Aliquots either were filtered immediately (within 2 min) upon dilution or were incubated at 0°C for various periods of time before filtration to measure the amount of DNA remaining bound by large T antigen. Figure 3A (uppermost curve) shows that large T antigen-DNA complexes that formed at pH 7 were relatively stable after dilution into pH 7 buffer; only 4% of these complexes dissociated within 2 min of dilution, and 29% dissociated during a subsequent 2-h incubation in pH 7 buffer at 0°C.
FIG. 3.
Kinetics of dissociation of protein-DNA complexes on dilution from low to high pH in the presence or absence of ATP. (A) A mixture of 1 ng of a 32P-labeled 265-bp DNA fragment containing sites 1/2, A, B, and C and 100 ng of large T antigen per 60 μl of binding solution in 100 mM Tris-HCl buffer (pH 7.0) was incubated at 37°C for 25 min. Aliquots (60 μl) were then diluted into 1 ml of ice-cold Tris-HCl buffer, pH 7.0 or 7.8, containing all other components of the binding solution (see Materials and Methods), as well as a 100-fold excess of unlabeled 265-bp DNA fragment, in the presence or absence of 5 mM ATP. Mixtures were filtered after incubation at 0°C for the indicated periods of time, and filters were washed with the dilution buffer. Results are expressed as percentages of protein-DNA complexes remaining of those originally present in undiluted 60-μl aliquots of the pH 7.0 binding reaction mixtures. (B) A similar experiment was carried out, except that the initial binding was carried out in 100 mM potassium phosphate buffer, pH 6.0, and dilution was in potassium phosphate buffer at pH 6.0 or in Tris-HCl buffer at pH 7.0 or 7.8. (C) A 32P-labeled 260-bp DNA fragment containing binding sites A, B, and C but lacking site 1/2 was incubated with large T antigen at pH 6.0 as described for panel B and then diluted with pH 6.0 or 7.8 buffer as indicated. Error bars show the maximum difference in radioactivity retained on filters when using duplicate samples. Error bars are omitted from the overlapping curves in panel B for the sake of clarity but are similar to those shown on the lower curves.
Dilution of complexes into pH 7.8 buffer in the absence of ATP (bottommost curve) led to a rapid dissociation of one-half of the complexes formed at pH 7, consistent with the results of the gel mobility shift assays described above. The remaining complexes remained intact but subsequently dissociated, with a half-life of approximately 3 h. When dilution into pH 7.8 buffer was carried out in the presence of ATP (middle curve), a fraction of the large T antigen-DNA complexes were protected from rapid dissociation. In this experiment (Fig. 3A), 50% of the complexes that formed at pH 7 dissociated after dilution to pH 7.8 in the absence of ATP while only 28% of the complexes dissociated in the presence of ATP. Therefore, about two-fifths (22 of 50) of the complexes that dissociated rapidly on dilution to pH 7.8 were protected against dissociation by ATP. This population of ATP-protected complexes, however, subsequently dissociated more rapidly than the remaining complexes and had almost completely disappeared after 2 h at 0°C (half-life, 1 h).
Stabilization of complexes by ATP depends on the presence of site 1/2.
ATP stimulates the formation of hexamers of both polyomavirus and simian virus 40 large T antigens in the absence of DNA (10, 55). The stimulatory effect of ATP on binding of simian virus 40 large T antigen to target DNAs has been ascribed to the formation of hexamers on DNA (5, 12, 27). Hexamer formation is specific to binding site II on simian virus 40 DNA (7, 25, 37), which is analogous to site 1/2 on polyomavirus DNA (16, 21). We therefore postulated that the fraction of protein-DNA complexes protected from dissociation by ATP consists of hexamers formed on the DNA upon dilution of complexes from low to high pH in the presence of ATP. If this were the case, complexes made with a target DNA lacking site 1/2 should not be protected by ATP from dissociation upon dilution to a high pH, since hexamers would not be expected to form on such DNA. The results of such an experiment are shown in Fig. 3B and C. Protein-DNA complexes containing sites A, B, C, and 1/2 formed at pH 6 (Fig. 3B) were very stable when diluted into pH 6 buffer (uppermost two curves) and nearly as stable when diluted into pH 7.0 buffer (middle two curves). ATP had little effect on these complexes when present during dilution to pH 6 or 7. However, ATP protected a fraction of the complexes from rapid dissociation on dilution in a pH 7.8 buffer (bottommost two curves), in agreement with the results shown in Fig. 3A. In a parallel experiment carried out with a target DNA containing only sites A, B, and C, more than one-half of large T antigen-DNA complexes dissociated on dilution to pH 7.8, but ATP did not protect these complexes from dissociation (Fig. 3C). Similar results were obtained when using other DNAs containing various combinations of binding sites A, B, and C but lacking site 1/2 (data not shown). These results suggest that the stabilizing effect of ATP on large T antigen-DNA complexes upon dilution to pH 7.8 is due to the formation of hexamers of large T antigen at site 1/2 on a fraction of the target DNA molecules.
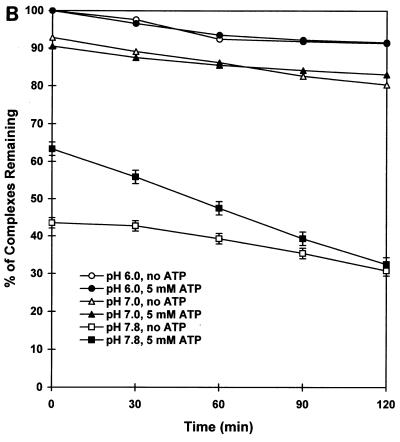
Binding of large T antigen to origin DNA fragments containing one or more adjacent binding sites.
Having defined the optimal conditions for DNA binding assays, we proceeded to ask whether polyomavirus large T antigen binds independently or cooperatively to its multiple target sites in the polyomavirus origin region. In a first set of experiments, we prepared target DNAs from which one or more of sites 1/2, A, B, and C were deleted (Fig. 4A) and measured binding as a function of the concentration of large T antigen by using the filter binding assay at pH 6.0, in the absence of ATP. Under these conditions, binding to the wild-type origin region reached a maximal value of 85% (Fig. 4B); half-maximal binding was achieved with 50 ng of large T antigen per 60-μl reaction volume. Binding to DNA containing only site A or site B was barely detectable; each of these sites contains only two G(A/G)GGC consensus binding sequences, and previous studies using such sites also showed inefficient binding (4, 7, 60). However, fragment AB, containing both site A and site B, had about a 10-fold-higher affinity for large T antigen than A or B separately. Fragment C, which contains four G(A/G)GGC consensus sequences, showed about the same affinity for large T antigen as fragment AB. Addition of site B, or both A and B, to site C increased binding by a factor of about 2 (BC) or 4 (ABC), respectively, as determined by the concentration of large T antigen required for half-maximal binding of wild-type DNA. Thus, two inherently weak binding sites (A and B) strengthened binding of large T antigen to DNA containing a moderately strong binding site (C) when positioned adjacent to that site. Furthermore, fragment 1/2A bound about as well as fragment AB or C, but combining 1/2A and BC increased binding by a factor of about 2.5 (determined by comparison of the half-maximal binding of 1/2ABC with that of BC).
These results show that large T antigen binds cooperatively to its multiple binding sites in the region of the replication origin. Binding of large T antigen is some 50-fold stronger to DNA containing all four sites than to DNA containing only site A or B and is fivefold stronger to the former than to DNA containing only site C. We also measured DNA binding at pH 7.0 in Tris-HCl buffer for most of the deletion mutants described in Fig. 4A (data not shown). Although the overall level of binding was lower, the relative affinities of binding to the different DNAs were the same as those shown in Fig. 4B.
Binding of large T antigen to DNA targets containing mutated binding sites.
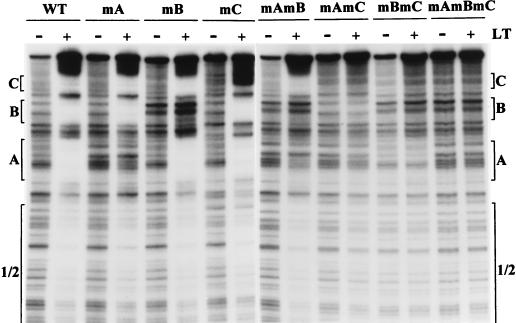
One drawback of the use of deletion mutants as shown in Fig. 4 is that the DNA fragments used have different sizes (ranging from 147 to 736 bp) and contain different flanking sequences; these differences could affect binding affinities. We tested binding to PCR-generated DNA fragments of different sizes and detected no major differences (data not shown). However, the availability of DNAs containing point mutations in the G(A/G)GGC consensus sequences within sites A, B, and C (1) allowed us to determine the binding strengths of a set of DNA targets identical in size and sequence except for a small number of nucleotide substitutions (Fig. 5A). This set of DNAs also allowed the determination of the binding strengths of combinations of sites different from those of the collection of deletion mutants; all of these DNA fragments contain an intact site 1/2.
Mutation of either site A, B, or C (mA, mB, or mC) reduced binding of large T antigen to the origin DNA fragment by only 1.3- to 1.6-fold (Fig. 5B), as measured by the ratios of large T-antigen concentration at half-maximal binding. It is of interest that mA and mC had nearly identical binding strengths; only two G(A/G)GGC sequences are mutated in mA, but four are mutated in mC, and the results in Fig. 4B showed that site C alone binds much more strongly than site A. This suggests that the proximity of sites affects the overall binding strength of the DNA fragment. Fragment mA contains a gap of some 70 nt between sites 1/2 and B, while the three sites (B, A, and 1/2) in mC are separated by only 25 to 30 nt each.
The double mutant mAmB bound significantly less strongly than either mA or mB. Fragment mAmB required a twofold-higher concentration of large T antigen for half-maximal binding than did wild-type DNA; however, at a concentration of 100 ng of large T antigen per 60 μl, fivefold more wild-type DNA than mAmB DNA bound. Thus, at low concentrations of large T antigen, the differences in binding strengths between wild-type DNA and either mAmB, mAmC, or mBmC were greater than at higher concentrations of large T antigen. Fragment mAmB also bound less strongly than did mC, although both mutants are missing four G(A/G)GGC sequences, and DNA fragments containing both sites A and B or site C alone had very similar binding strengths (Fig. 4B). This difference may be due to a 100-nt gap between site C and site 1/2 in mutant mAmB that may render cooperative interactions less efficient.
DNAs containing only site A or site B in addition to site 1/2 (mBmC and mAmC) bound about sevenfold less strongly than DNA containing both sites A and B (mC). Thus, mutation of a weak binding site (B or A) strongly affected binding of large T antigen to the remaining sites on the target DNA. Finally, a DNA target containing only site 1/2 (mAmBmC) barely showed any binding to large T antigen at the highest concentration used. This experiment was carried out at pH 7.0 in Tris-HCl buffer, in the absence of ATP; similar results were obtained at pH 6, except that higher overall levels of binding were detected.
DNase I footprinting of mutant DNAs in the presence of large T antigen.
Filter binding and gel retardation assays do not provide information on the occupancy of each of the binding sites on the various DNAs that we used. To understand more precisely the nature of the cooperative interactions between large T antigen and its binding sites, we carried out DNase I footprinting experiments (Fig. 6) with the mutant DNAs shown in Fig. 5A. This experiment was carried out at pH 7, using 400 ng of large T antigen per binding reaction, but essentially identical results were obtained at pH 7.4 with a higher concentration of antigen and at pH 6 with a lower concentration. The left half of Fig. 6 shows the footprints of three single mutants (mA, mB, and mC) in comparison to that of the wild-type origin. As expected, there was no protection against DNase I digestion in the mutated region of each DNA, but all other sites were protected to approximately the same degree as in the wild-type DNA. This agrees with the results presented in Fig. 5B, which showed that single mutations in these sites reduced the overall binding affinity for large T antigen by only a small amount.
FIG. 6.
DNase I footprinting of DNAs containing mutated binding sites. Twenty femtomoles of 265-bp end-labeled DNA fragments containing binding sites mutated as shown at the top were incubated in the absence (−) or presence (+) of 400 ng of large T antigen (LT) in Tris-HCl buffer, pH 7, and then incubated with DNase I, as described in Materials and Methods. The DNAs were analyzed on 12% polyacrylamide–8 M urea gels. Regions protected against DNase I digestion are indicated by brackets. WT, wild type.
The right half of Fig. 6 shows the footprints of double and triple mutants. It is instructive to examine the protection of site 1/2 in these four mutants and to compare it with that of wild-type DNA. There was little binding of large T antigen to site 1/2 in DNA of the triple mutant mAmBmC, as could be expected from examination of Fig. 5B. Protection of site 1/2 was strongly increased by the presence of site C, some 100 nt distant from site 1/2, in DNA of mutant mAmB. Mutants mAmC and mBmC showed only partial protection of site 1/2 and, furthermore, showed little protection of site B (in mAmC) or site A (in mBmC), even though these sites were intact in the target DNA. If we compare these patterns to the patterns for single mutants mA, mB, and mC, it is clear that binding of large T antigen to site B requires the presence of either site C or site A, and that binding to site A requires the presence of either site B or site C, in addition to site 1/2, which is present on all of these DNAs. These results further document the cooperative nature of the binding of large T antigen to polyomavirus origin DNA. In particular, they emphasize that optimal binding to site 1/2 requires the presence of either site C or sites A and B.
ATP specifically enhances protection against DNase I digestion of the central 10 to 12 bp of site 1/2.
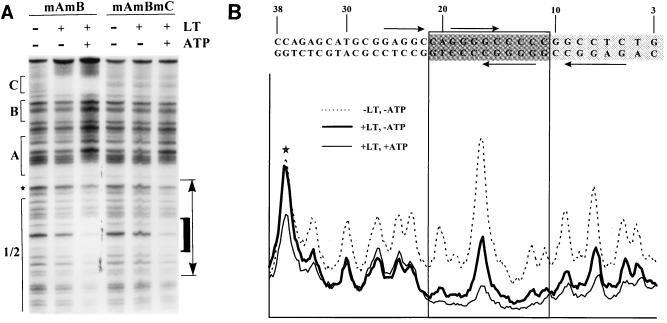
We then tested the effect of ATP on footprint patterns at pH 7.4, using the different mutant DNAs shown in Fig. 5A. In our initial experiments, we observed what appeared to be increased protection against DNase I digestion by 4 mM ATP throughout site 1/2 and decreased protection in sites A, B, and C. However, we found that 4 mM ATP reduced overall DNase I activity (perhaps by sequestering free Mg2+), leading to increased levels of uncleaved DNA and decreased levels of cleaved DNA, particularly in regions close to the labeled end of the DNA (smaller DNA fragments). We therefore reduced the ATP concentration from 4 to 1 mM and titrated the amounts of DNase used in all reactions, so that similar levels of cleavage took place in the presence and absence of ATP, by making sure that the amounts of uncleaved, full-length DNA in all digested reaction mixtures were similar. Under these conditions, ATP had little or no effect outside of site 1/2 on wild-type DNA or any of the mutant DNAs. Sample results with mutants mAmB and mAmBmC are shown in Fig. 7A, and a PhosphorImager tracing of a gel lane containing mutant mC is shown in Fig. 7B. Precise alignment of the footprint pattern with the DNA sequence was achieved by digesting end-labeled DNA with different restriction endonucleases and comparing the migrations of those DNAs with the footprint patterns of the corresponding DNAs run in the same gel (results not shown); HpaII cuts between nt 9 and 10 on the bottom (labeled) strand, and SphI cuts between nt 32 and 33 on the same strand. Therefore, determination of the sites at which ATP altered protection of DNA by large T antigen was accurate to within 1 or 2 nt in this region.
FIG. 7.
ATP enhances protection of the central portion of site 1/2 against DNase I digestion. (A) Twenty femtomoles of 265-bp labeled DNA fragments containing binding sites mutated as shown at the top were incubated with (+) or without (−) 600 ng of large T antigen (LT) in Tris-HCl binding buffer, pH 7.4, in the presence (+) or absence (−) of 1 mM ATP. DNase I footprinting was then carried out. A DNase-sensitive band that corresponds to nt 37 in polyomavirus DNA, between sites A and 1/2, is noted by a star. The domain whose protection is enhanced in the presence of ATP is indicated by a thick black bracket on the right side. (B) PhosphorImager profile of relative band intensities in the region denoted by the two-headed arrow in panel A, from a DNase I footprint prepared with mutant DNA mC. The DNA sequence in this region is shown above the density profile; nucleotide numbers refer to the polyomavirus strain A3 genome. G(A/G)GGC sequences on each DNA strand are designated by arrows. The darker box indicates the region whose protection is strongly enhanced in the presence of ATP; the lighter box indicates a region whose protection is weakly enhanced in the presence of ATP. The star denotes the same band as that marked by a star in panel A.
Figures 7A and B show that ATP had distinct effects on different parts of site 1/2. At the early (left) side of site 1/2, a major DNase-sensitive site at nt 37 was not protected by large T antigen at this concentration in the absence of ATP. Binding in the presence of ATP led to protection at this site, but little or no enhancement of protection was evident in the adjacent region of site 1/2, between nt 35 and 22 (compare the thick and thin continuous lines in Fig. 7B). This region contains one of the four GAGGC sequences in site 1/2. However, ATP strongly enhanced protection of the central region between nt 21 and 11, shown by the heavy bracket to the right of Fig. 7A and the more darkly shaded sequence in Fig. 7B. Furthermore, protection of the region from nt 10 and beyond (the more lightly shaded sequence in Fig. 7B) was also enhanced, but to a lesser extent than in the central region. The central region contains two overlapping GGGGC consensus binding sequences and represents approximately one turn of the DNA helix. Similar results were obtained when the experiment was carried out at pH 7, but ATP had no detectable effects on footprint patterns at pH 6.0 (data not shown).
DISCUSSION
The results presented in this paper show that polyomavirus large T antigen binds in a cooperative fashion to its multiple target sites within and adjacent to the origin of DNA replication. Binding of large T antigen to sites A, B, and C facilitates binding to site 1/2, in the core replication origin, where hexamers presumably form to initiate unwinding and DNA replication. The presence of these auxiliary sites (1, 15, 57) near the origin may therefore allow initiation of DNA replication at substantially lower concentrations of large T antigen than would be possible in their absence.
No previous studies have attempted to quantitate binding of polyomavirus large T antigen to origin DNA in which individual binding sites were deleted or mutated. Binding to DNA fragments containing isolated site A, B, or C, or some combination of these sites, was first shown by immunoprecipitation of large T antigen-DNA complexes (7, 8, 42). Site A in polyomavirus strain A2 contains three GAGGC pentanucleotide sequences (in strain A3 and derivatives used here, there are two pentanucleotides [1, 9, 43, 53]); inactivation of any one of these three sequences by methylation at a single G residue reduced binding by large T antigen by a factor of about 10 (8). This led Cowie and Kamen (8) to propose that large T antigen molecules bind cooperatively to the adjacent G(A/G)GGC sequences, approximately one helical turn apart, within an individual site (A, B, or C). These results, as well as those of DNase footprinting studies (7, 27, 39), also suggested that each large T antigen molecule occupies approximately one helical turn when bound to target DNA and that the mutual interaction of these closely packed large T antigen molecules helps to stabilize their binding to DNA. The cooperative binding described in this paper involves molecules of large T antigen bound to sites that lie between 20 and 100 nt apart and in which G(A/G)GGC sequences are not always on the same DNA strand. Our results (Fig. 4 to 6) suggest that large T antigen bound to site A, B, C, or 1/2 can interact with large T antigen bound to any of the other sites.
We propose a model (Fig. 8) to account for these results. This model allows interactions between DNA-bound large T antigen molecules by virtue of folding of the DNA in this 170-nt region into a compact protein-DNA complex. Interactions between sites C and 1/2, A and B, A and 1/2, and B and 1/2 are suggested by contacts between spheres representing molecules of large T antigen bound to each set of sites. Interactions between site A or B and site C can be imagined by folding the top and bottom parts of the complex vertically above the plane of the page. DNase I footprinting (references 7, 27, and 39 and this paper) showed that there are unprotected regions between each of the four sites, shown as DNA not covered by protein in the model. In addition, on binding of large T antigen, there is increased DNase I sensitivity at positions located between sites A, B, and C (Fig. 6) (2, 30), consistent with the bending or distortion of the DNA helix that would be necessary to bring the various large T antigen molecules into contact with each other. However, we do not claim to know either the exact path of the DNA through this multiprotein complex or how stable such a complex would be in the cell.
FIG. 8.
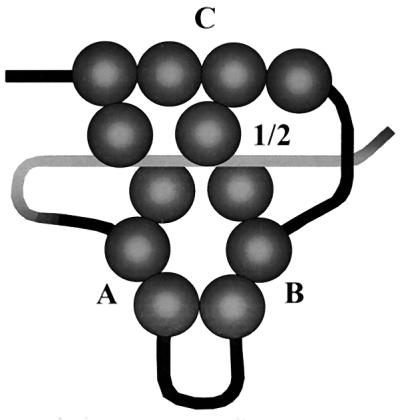
Model for cooperative binding of large T antigen to origin DNA. Monomers of large T antigen (spheres) are shown bound to the multiple G(A/G)GGC sequences within sites C, B, A, and 1/2 on origin DNA in such a way that they can interact cooperatively with each other. The structure containing sites A and B could fold upward to enable contact with site C as well as site 1/2.
We observed a strong pH dependence for the binding by large T antigen to specific DNA, from a maximum at pH 6 to nearly no detectable binding at pH 8.5. Previous in vitro binding studies were carried out at pH values between 7 and 8 (5, 12, 27, 30, 42, 49, 54); however, no quantitative data on the pH dependence of specific DNA binding by either polyomavirus or simian virus 40 large T antigen have been reported. Binding over the range of pH values tested (6.0 to 7.8) was specific for G(A/G)GGC sequences on DNA, and the patterns of DNase I-protected regions on origin DNA were similar (in the absence of ATP) across this pH range. Moreover, cooperative binding to the multiple binding sites on origin DNA was observed at all pH values tested. Stronger binding at low pH can be explained by increased stability of large T antigen-DNA complexes at low pH values, as shown by resistance to dissociation on dilution or during gel electrophoresis. Increased binding strength and increased stability of protein-DNA complexes at low pH allow more sensitive detection of large T antigen by nitrocellulose filter binding or gel retardation assays than was previously possible. Purification of large T antigen by binding to DNA at low pH and release from the DNA at high pH might also be possible. The pH effect should also be considered when interpreting any studies on biochemical activities of large T antigen mediated by DNA binding, since the decrease in binding strength as the pH rises is particularly steep in the range between pH 7.4 and 8.0, which is used in many binding and replication assays (5, 12, 27, 30, 49, 54).
What might be the biological significance of the variation in binding as a function of pH? Most mammalian cells have an intracellular pH of 7.1 to 7.3, although values ranging from pH 6.8 to 7.5 have been measured (13, 47, 50). Intracellular pH is regulated by Na+/H+ antiport and by HCO3− exchange (20, 47, 50). Regulation of a number of cellular processes, including cell spreading and attachment, DNA replication, and cellular proliferation, has been correlated with changes in intracellular pH or in the Na+/H+ antiport activity (14, 20, 50). At pH 7.2, the binding affinity of large T antigen for origin DNA is intermediate between its maximum, at pH 6.0, and its minimum, at pH 8 and above (Fig. 1A). It is possible that polyomavirus DNA replication is responsive to changes in intracellular pH as a result of the pH dependence of the affinity of large T antigen for its DNA target. In particular, a lower intracellular pH may favor binding to origin DNA and therefore accumulation of a sufficient number of large T antigen molecules near the replication origin, and a higher pH may favor mobilization of these bound protein molecules and the formation of hexamers by a handover mechanism (see below).
Previous reports showed that ATP, in the presence of Mg2+, stimulated binding of both polyomavirus and simian virus 40 large T antigens to their respective origin DNAs when binding reactions were carried out at 37°C in pH 7.5 to 7.8 buffers (5, 11, 12, 27). We confirmed that ATP stimulates binding of polyomavirus large T antigen to an origin DNA fragment at pH 7.6, when measured by the nitrocellulose filter binding assay, but found no effect of ATP on binding at pH 7 or below. We further found that a fraction of the protein-DNA complexes formed at pH 6 or 7 were protected from dissociation on dilution to pH 7.6 when ATP was present. This protective effect was seen when a DNA fragment containing the entire origin region was used, but it was not seen with fragments lacking site 1/2. ATP is required for generation of hexameric forms of simian virus 40 and polyomavirus large T antigen in solution or at the replication origin (30, 34, 55); hexamers of simian virus 40 large T antigen have been shown to form at site II (10, 59), which is homologous to polyomavirus site 1/2 (16). It is likely that the effect of ATP on DNA binding results from its ability to stimulate hexamer formation at site 1/2. Hexamers may not form on DNA when the pH is below 7, either because of a changed protein conformation at low pH or because molecules of large T antigen are too tightly bound to G(A/G)GGC pentanucleotide target sequences on DNA and therefore cannot be released to form hexamers. Hexamer formation on shifting to a higher pH in the presence of ATP could stabilize protein-DNA complexes. Hexamers probably bind to DNA by topologically enclosing the DNA double helix within the central hole formed by the circular hexamer (48, 59) rather than by recognizing and interacting with specific nucleotides. The ATP-stabilized complexes generated on dilution to high pH (Fig. 3A and B) dissociated relatively rapidly, with a half-life of about 1 h. This could be due to instability of hexamers, but hexamers of simian virus 40 large T antigen were shown to be stable for several hours in vitro (10, 59). Alternatively, hexamers may simply fall off the ends of the linear DNA fragments to which they are bound in our in vitro assay.
At pH 7.4, ATP specifically enhanced protection of the central 10 to 12 bp of site 1/2 when less-than-saturating concentrations of large T antigen were used (Fig. 7). A previous study of the effect of ATP on the DNase I footprint pattern of wild-type polyomavirus origin DNA (27) showed increased protection of a region including sites A and 1/2 but did not detect specific enhancement in this part of site 1/2. However, those experiments were carried out under different conditions (a higher concentration of large T antigen; pH 7.8), and the opposite DNA strand was labeled, making it difficult to detect closely spaced DNase-sensitive bands in site 1/2.
Studies of the structure of polyomavirus origin DNA bound by large T antigen in the presence of 5′-adenylyl imidodiphosphate (AMPPNP), a nonhydrolyzable analog of ATP (2), detected KMnO4-sensitive nucleotides at positions 10, 11, and 20 to 22. These sites are located at the borders of the 10- to 12-bp region in the center of site 1/2 whose protection against DNase I digestion is enhanced in the presence of ATP. These KMnO4-sensitive sites may reveal distortion of the DNA helix at the edges of bound protein molecules. Taken together, our data and those of Bhattacharyya et al. (2) suggest that ATP, or AMPPNP, induces the formation of a complex of large T antigen that covers the central part of site 1/2 over a single turn of the DNA helix. In contrast, simian virus 40 large T antigen protects a larger region of the homologous site II against DNase I digestion (5, 38), and there are no KMnO4-sensitive sites generated at the equivalent positions in site II (2). Simian virus 40 large T antigen has been shown to form double hexamers at site II via a cooperative process directed by the two halves of site II (38). A single hexamer of polyomavirus large T antigen may form at the center of site 1/2. This is consistent with the different structures of these sites: in simian virus 40 site II, the central GAGGC pentanucleotides are separated by 1 nt, perhaps allowing the formation of a hexamer on each half of site II; however, in polyomavirus site 1/2, the central GGGGC pentanucleotides overlap by 2 nt (the 3′-terminal GC on each strand). Furthermore, simian virus 40 DNA replication begins at approximately the same site on each DNA strand (22), while polyomavirus DNA replication begins near nt 30 on the early strand but some 16 nt beyond (nt 46) on the late strand (23). Therefore, it is possible that a single hexamer forms at site 1/2 and begins DNA unwinding and replication in the early direction, with the formation of a second hexamer occurring once replication has begun. This would also account for the unidirectional rolling-circle replication known to take place in polyomavirus (3). Such unidirectional replication could result from the progression of a single replication fork if the second hexamer were not assembled.
The cooperative binding of large T antigen to its multiple target sites on origin DNA, coupled with the reversibility of binding at intracellular pH values, suggests a model for the pathway of assembly of hexamers of large T antigen leading to the initiation of DNA replication at the origin (Fig. 9). This model proposes that monomers of large T antigen bind cooperatively to DNA via interactions with the G(A/G)GGC pentanucleotides in all four sites, forming a complex similar to that shown in Fig. 8. Such cooperative binding would tend to concentrate large T antigen on one or a few DNA molecules, avoiding nonfunctional binding of a small number of monomers among several DNA molecules. This could be important to the efficiency of DNA replication in the beginning of the replication cycle, when small amounts of large T antigen are present. Large T antigen molecules bound to DNA then assemble into hexamers at site 1/2 in the presence of ATP, which presumably induces a conformational change in large T antigen favoring hexamer formation (27, 48, 59). Hexamer assembly would be favored by the proximity of large T antigen molecules and their mutual interaction in the protein-DNA complex; they could therefore be “handed over” from sites A, B, and C, to which they are reversibly bound, to site 1/2, where hexamer formation occurs. When large T antigen is present at high concentrations, it could alternatively assemble directly from solution onto hexamers forming at site 1/2. A single hexamer is shown in Fig. 9, reflecting our observation of enhanced DNase I protection by ATP at the center of site 1/2. It is not known whether a second hexamer can subsequently form adjacent to this hexamer, as with simian virus 40 (38, 58, 59), or forms only after the displacement of this hexamer during unwinding and initiation of DNA replication.
FIG. 9.
“Handover” model for assembly of hexamers at the replication origin. Monomers of large T antigen (spheres) bind cooperatively to the four binding sites in origin DNA to form a complex as shown in Fig. 9. In the presence of ATP, bound monomers in contact with each other dissociate from the G(A/G)GGC sequences to which they are bound and rearrange to form a hexamer at the center of site 1/2, which can unwind DNA and allow initiation of DNA replication. A second hexamer may subsequently form by a similar rearrangement.
ACKNOWLEDGMENTS
Noelle-Ann Sunstrom constructed the deletion mutant plasmids which were used as PCR templates to make DNA fragments containing single or multiple binding sites. Spodoptera frugiperda (Sf9) insect cells and the polyomavirus large T antigen-producing recombinant baculovirus vEV51LT were kindly provided by Marcel Bastin. High Five cells were a gift from Fernando Congote. Cells producing F5 monoclonal antibody were kindly provided by Carol Prives and by Marcel Bastin.
Yu-Cai Peng was supported by a McGill Max Stern recruitment fellowship, an F. C. Harrison fellowship, and the Medical Research Council of Canada. This research was supported by the Medical Research Council of Canada (grant MT-7281).
REFERENCES
- 1.Bertin J, Sunstrom N-A, Acheson N H. Mutation of large T-antigen-binding site A, but not site B or C, eliminates stalling by RNA polymerase II in the intergenic region of polyomavirus DNA. J Virol. 1993;67:5766–5775. doi: 10.1128/jvi.67.10.5766-5775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya S, Lorimer H E, Prives C. Murine polyomavirus and simian virus 40 large T antigens produce different structural alterations in viral origin DNA. J Virol. 1995;69:7579–7585. doi: 10.1128/jvi.69.12.7579-7585.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjursell G. Effects of 2′-deoxy-2′-azidocytidine on polyoma virus DNA replication: evidence for rolling cycle-type mechanism. J Virol. 1978;26:136–142. doi: 10.1128/jvi.26.1.136-142.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondeson K, Ronn O, Magnusson G. Preferred DNA-binding sites of polyomavirus large T antigen. Eur J Biochem. 1995;227:359–366. doi: 10.1111/j.1432-1033.1995.tb20397.x. [DOI] [PubMed] [Google Scholar]
- 5.Borowiec J A, Hurwitz J. ATP stimulates the binding of simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc Natl Acad Sci USA. 1988;85:64–68. doi: 10.1073/pnas.85.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley M K, Griffin J D, Livingston D M. Relationship of oligomerization to enzymatic and DNA-binding properties of the SV40 large T antigen. Cell. 1982;28:125–134. doi: 10.1016/0092-8674(82)90382-8. [DOI] [PubMed] [Google Scholar]
- 7.Cowie A, Kamen R. Multiple binding sites for polyomavirus large T antigen within regulatory sequences of polyomavirus DNA. J Virol. 1984;52:750–760. doi: 10.1128/jvi.52.3.750-760.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowie A, Kamen R. Guanine nucleotide contacts within viral DNA sequences bound by polyomavirus large T antigen. J Virol. 1986;57:505–514. doi: 10.1128/jvi.57.2.505-514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dailey L, Basilico C. Sequences in the polyomavirus DNA regulatory region involved in viral DNA replication and early gene expression. J Virol. 1985;54:739–749. doi: 10.1128/jvi.54.3.739-749.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 11.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deb S P, Tegtmeyer P. ATP enhances the binding of simian virus 40 large T antigen to the origin of replication. J Virol. 1987;61:3649–3654. doi: 10.1128/jvi.61.12.3649-3654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deitmer J W, Rose C R. pH regulation and proton signalling by glial cells. Prog Neurobiol. 1996;48:73–103. doi: 10.1016/0301-0082(95)00039-9. [DOI] [PubMed] [Google Scholar]
- 14.Demaurex N, Downey G P, Waddell T K, Grinstein S. Intracellular pH regulation during spreading of human neutrophils. J Cell Biol. 1996;133:1391–1402. doi: 10.1083/jcb.133.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 16.DePamphilis M L, Martinez-Salas E, Cupo D Y, Hendrickson E A, Fritze C E, Folk W R, Heine U. Initiation of polyomavirus and SV40 DNA replication, and the requirements for DNA replication during mammalian development. Cancer Cells. 1988;6:165–175. [Google Scholar]
- 17.Dilworth S M, Cowie A, Kamen R, Griffin B E. DNA binding activity of polyoma virus large tumor antigen. Proc Natl Acad Sci USA. 1984;81:1941–1945. doi: 10.1073/pnas.81.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorn A, Brauer D, Otto B, Fanning E, Knippers R. Subclasses of simian virus 40 large tumor antigen (partial purification and DNA-binding properties of two subclasses of tumor antigen from productively infected cells) Eur J Biochem. 1982;128:53–62. [PubMed] [Google Scholar]
- 19.Gidoni D, Scheller A, Barnet B, Hantzopoulos P, Oren M, Prives C. Different forms of simian virus 40 large tumor antigen varying in their affinities for DNA. J Virol. 1982;42:456–466. doi: 10.1128/jvi.42.2.456-466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinstein S, Rotin D, Mason M J. Na+/H+ exchange and growth factor-induced cytosolic pH changes. Role in cellular proliferation. Biochim Biophys Acta. 1989;988:73–97. doi: 10.1016/0304-4157(89)90004-x. [DOI] [PubMed] [Google Scholar]
- 21.Hassell J A, Brinton B T. SV40 and polyomavirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 639–677. [Google Scholar]
- 22.Hay R T, Hendrickson E A, DePamphilis M L. Sequence specificity for the initiation of RNA-primed simian virus 40 DNA synthesis in vivo. J Mol Biol. 1984;175:131–157. doi: 10.1016/0022-2836(84)90471-6. [DOI] [PubMed] [Google Scholar]
- 23.Hendrickson E A, Fritze C E, Folk W R, DePamphilis M L. The origin of bidirectional DNA replication in polyoma virus. EMBO J. 1987;6:2011–2018. doi: 10.1002/j.1460-2075.1987.tb02465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinkle D C, Chamberlin M J. Studies of the binding of Escherichia coli RNA polymerase to DNA. J Mol Biol. 1972;70:157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- 25.Joo W S, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the simian virus 40 (SV40) T antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katinka M, Yaniv M. DNA replication origin of polyoma virus: early proximal boundary. J Virol. 1983;47:244–248. doi: 10.1128/jvi.47.1.244-248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorimer H E, Wang E H, Prives C. The DNA-binding properties of polyomavirus large T antigen are altered by ATP and other nucleotides. J Virol. 1991;65:687–699. doi: 10.1128/jvi.65.2.687-699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luthman H, Nilsson M G, Magnusson G. Non-contiguous segments of the polyoma genome required in cis for DNA replication. J Mol Biol. 1982;161:533–550. doi: 10.1016/0022-2836(82)90406-5. [DOI] [PubMed] [Google Scholar]
- 29.Marton A, Marko B, Delbecchi L, Bourgaux P. Topoisomerase activity associated with polyoma virus large tumor antigen. Biochim Biophys Acta. 1995;1262:59–63. doi: 10.1016/0167-4781(95)00050-q. [DOI] [PubMed] [Google Scholar]
- 30.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 31.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montenarh M, Henning R. The binding of simian virus 40 large T antigen to the polyphosphate backbone of nucleic acids. Biochim Biophys Acta. 1982;697:322–329. doi: 10.1016/0167-4781(82)90095-1. [DOI] [PubMed] [Google Scholar]
- 33.Moses K, Prives C. A unique subpopulation of murine DNA polymerase α/primase specifically interacts with polyomavirus T antigen and stimulates DNA replication. Mol Cell Biol. 1994;14:2767–2776. doi: 10.1128/mcb.14.4.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller W J, Mueller C R, Mes A-M, Hassell J A. Polyomavirus origin for DNA replication comprises multiple genetic elements. J Virol. 1983;47:586–599. doi: 10.1128/jvi.47.3.586-599.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami Y, Hurwitz J. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 36.Oren M, Winocour E, Prives C. Differential affinities of simian virus 40 large tumor antigen for DNA. Proc Natl Acad Sci USA. 1980;77:220–224. doi: 10.1073/pnas.77.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons R, Tegtmeyer P. Spacing is crucial for coordination of domain functions within the simian virus 40 core origin of replication. J Virol. 1992;66:1933–1942. doi: 10.1128/jvi.66.4.1933-1942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons R E, Stenger J E, Ray S, Welker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Y C, Acheson N H. Production of active polyomavirus large T antigen in yeast Pichia pastoris. Virus Res. 1997;49:41–47. doi: 10.1016/s0168-1702(97)01455-x. [DOI] [PubMed] [Google Scholar]
- 40.Pipas J M. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz B J, Hassell J A. Polyomavirus and simian virus 40 large T antigens bind to common DNA sequences. J Virol. 1984;49:925–937. doi: 10.1128/jvi.49.3.925-937.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pomerantz B J, Mueller C R, Hassell J A. Polyomavirus large T antigen binds independently to multiple, unique regions on the viral genome. J Virol. 1983;47:600–610. doi: 10.1128/jvi.47.3.600-610.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prives C, Murakami Y, Kern F G, Folk W, Basilico C, Hurwitz J. DNA sequence requirements for replication of polyomavirus DNA in vivo and in vitro. Mol Cell Biol. 1987;7:3694–3704. doi: 10.1128/mcb.7.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynisdottir I, Lorimer H E, Friedman P N, Wang E H, Prives C. Phosphorylation and active ATP hydrolysis are not required for SV40 T antigen hexamer formation. J Biol Chem. 1993;268:24647–24654. [PubMed] [Google Scholar]
- 45.Rice W C, Lorimer H E, Prives C, Miller L K. Expression of polyomavirus large T antigen by using a baculovirus vector. J Virol. 1987;61:1712–1716. doi: 10.1128/jvi.61.5.1712-1716.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson C D. Baculovirus expression protocols. Methods in Mol. Biol. Vol. 39. Totowa, N.J: Humana Press; 1995. [Google Scholar]
- 47.Roos A, Boron W F. Intracellular pH. Physiol Rev. 1981;61:296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- 48.San Martin M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 49.Scheller A, Prives C. Simian virus 40 and polyomavirus large tumor antigens have different requirements for high-affinity sequence-specific DNA binding. J Virol. 1985;54:532–545. doi: 10.1128/jvi.54.2.532-545.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strazzabosco M, Boyer J L. Regulation of intracellular pH in the hepatocyte. J Hepatol. 1996;24:631–644. doi: 10.1016/s0168-8278(96)80153-x. [DOI] [PubMed] [Google Scholar]
- 51.Sunstrom N-A. Ph.D. thesis. Montreal, Quebec, Canada: McGill University; 1991. [Google Scholar]
- 52.Sunstrom N-A, Acheson N H, Hassell J A. Determination of the origin-specific DNA-binding domain of polyomavirus large T antigen. J Virol. 1991;65:6998–7003. doi: 10.1128/jvi.65.12.6998-7003.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Triezenberg S J, Folk W R. Essential nucleotides in the polyomavirus origin region. J Virol. 1984;51:437–444. doi: 10.1128/jvi.51.2.437-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogt B, Vakalopoulou E, Fanning E. Allosteric control of simian virus 40 T-antigen binding to viral origin DNA. J Virol. 1986;58:765–772. doi: 10.1128/jvi.58.3.765-772.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang E H, Prives C. ATP induces the assembly of polyoma large tumor antigen into hexamers. Virology. 1991;184:399–403. doi: 10.1016/0042-6822(91)90858-9. [DOI] [PubMed] [Google Scholar]
- 56.Wang E H, Prives C. DNA helicase and duplex DNA fragment unwinding activities of polyoma and simian virus 40 large T antigen display similarities and differences. J Biol Chem. 1991;266:12668–12675. [PubMed] [Google Scholar]
- 57.Weichselbraun I, Haider G, Wintersberger E. Optimal replication of plasmids carrying polyomavirus origin regions requires two high-affinity binding sites for large T antigen. J Virol. 1989;63:961–964. doi: 10.1128/jvi.63.2.961-964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weisshart K, Fanning E. Roles of phosphorylation in DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 295–330. [Google Scholar]
- 59.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright P J, DeLucia A L, Tegtmeyer P. Sequence-specific binding of simian virus 40 A protein to nonorigin and cellular DNA. Mol Cell Biol. 1984;4:2631–2638. doi: 10.1128/mcb.4.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]