Abstract
Background
N‐acetyltransferase 10 (NAT10) serves as a critical enzyme in mediating the N4‐acetylcytidine (ac4C) that ensures RNA stability and effective translation processes. The role of NAT10 in driving the advancement of breast cancer remains uninvestigated.
Methods
We observed an increase in NAT10 expression, both at mRNA level through the analysis of the Cancer Genome Atlas (TCGA) database and at the protein level of tumor tissues from breast cancer patients. We determined that a heightened expression of NAT10 served as a predictor of an unfavorable clinical outcome. By screening the Cancer Cell Line Encyclopedia (CCLE) cell bank, this expression pattern of NAT10 was consistency found across almost all the classic breast cancer cell lines.
Results
Functionally, interference of NAT10 expression exerts an inhibitory effect on proliferation and invasion of breast cancer cells. By using ac4C RNA immunoprecipitation (ac4c‐RIP) and acRIP‐qPCR assays, we identified a reduction of ac4C enrichment within the ATP binding cassette (ABC) transporters, multidrug resistance protein 1 (MDR1) and breast cancer resistance protein (BCRP), consequent to NAT10 suppression. Expressions of MDR1 and BCRP exhibited a positive correlation with NAT10 expression in tumor tissues, and the inhibition of NAT10 in breast cancer cells resulted in a decrease of MDR1 and BCRP expression. Therefore, the overexpressing of MDR1 and BCRP could partially rescue the adverse consequences of NAT10 depletion. In addition, we found that, remodelin, a NAT10 inhibitor, reinstated the susceptibility of capecitabine‐resistant breast cancer cells to the chemotherapy, both in vitro and in vivo.
Conclusion
The results of our study demonstrated the essential role of NAT10‐mediated ac4c‐modification in breast cancer progression and provide a novel strategy for overcoming chemoresistance challenges.
Keywords: ABC transporters, ac4C‐modification, breast cancer, NAT10, remodelin
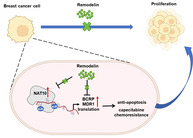
Increased expression of N‐acetyltransferase 10 (NAT10) was observed in breast cancer patients and cell lines as a predictor of an unfavorable clinical outcome. Both multidrug resistance protein 1 (MDR1) and breast cancer resistance protein (BCRP) are N4‐acetylcytidine (ac4C)‐modified mRNA targets regulated by NAT10. Remodelin, a NAT10 inhibitor, can inhibit the expression of MDR1 and BCRP overcome capecitabine resistance, and attenuate breast cancer cell proliferation, in vitro and in vivo.
INTRODUCTION
Breast cancer is a widespread malignancy that affects women worldwide. 1 Despite the advancement in early detection and effective treatment, the incidence and mortality rates are on the increase due to lifestyle changes and the frequent development of formidable resistance to chemotherapy and radiotherapy. 2 While some target genes exhibit potential in treating breast cancer, their clinical applicability remains limited. As a result, identifying novel genes is necessary for devising innovative new strategies to improve clinical outcomes.
The landscape of mRNA epigenetic modifications encompasses over 100 variants, including N6‐adenosine methylation (m6A), cytosine hydroxylation (m5C), and N1‐adenosine methylation (m1A), which have been revealed to mediate the stability, functionality, and splicing process of specific mRNAs. 3 , 4 N4‐acetylcytidine (ac4C) emerges as a conserved chemical RNA modification. Several studies have shown that RNA ac4C modulates mRNA stability and translation efficiency. 5 It is noteworthy that ac4C acetylation, the first acetylation modification identified in mRNA, is intricately associated with the occurrence and prognosis of cancer progression. 6 , 7 N‐acetyltransferase 10 (NAT10), a catalytic enzyme involved in the acetylation modification of tRNA, rRNA, and mRNA, has been implicated in various tumors. 8 , 9 Previous studies showed the critical involvement of NAT10 in fostering gastric cancer metastasis and epithelial‐mesenchymal transition (EMT) by regulating the mRNA ac4C writing pathway. 10 Similarly, NAT10 is recognized for acetylating KIF23 mRNA, thereby regulating colorectal cancer progression and metastasis. 7 However, the precise role of NAT10‐mediated ac4C modification in the trajectory of breast cancer progression remains unclear.
In this study, we investigated NAT10 expression in both breast cancer patients and cell lines, delving into the role of NAT10 in steering disease progression. Furthermore, we identified the catalyzation of multidrug resistance protein 1 (MDR1) and breast cancer resistance protein (BCRP) mRNAs by the acetyltransferase NAT10 for N4‐acetylcytidine (ac4C) modification. In a translational context, we evaluated the significance of targeting NAT10, encompassing its potential to suppress MDR1 and BCRP expressions and to overcome capecitabine resistance through a series of in vitro and in vivo experiments.
METHODS
Patient sample collection and ethical approvals
Patient samples were sourced from Tianjin Medical University Cancer Institute and Hospital, following the approval of the institutional ethics committee. All individuals enrolled in the study provided signed informed consent before their inclusion in the research.
Cell cultures and treatments
Breast cancer cell lines (MCF‐7, MDA‐MB‐231, MDA‐MB‐468) and HEK293T cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences. These cell lines were maintained in Dulbecco's modified Eagle medium (DMEM: Gibco) media supplemented with 10% fetal bovine serum (FBS: Hyclone). All cells were cultured at 37°C in a 5% CO2 incubator, and the experiments were conducted using cultures that achieved a confluence of 70%–80%. The breast cancer cell lines were infected with lentivirus carrying NAT10 shRNA or the NAT10‐coding sequences in six‐well plates, following the manufacturer's instructions. The medium was replaced with the complete medium on the following day. Stably infected cells were selected by incubation with puromycin for 2 weeks. The efficiency of NAT10 knockdown or overexpression was examined by quantitative polymerase chain reaction (qPCR) and western blotting.
Subcutaneous transplantation tumor model
All animal experiments complied with the Guide for the Care and Use of the Animal Ethics Committee of Institute of Radiation Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College. First, 1 × 106 MCF‐7 cells were resuspended in Matrigel (Becton Dickinson) and subsequently subcutaneously implanted into lateral flanks of adult female NOD/SCID mice. The mice were treated with capecitabine (0.55 mmol/kg/day) and remodelin (100 mg/kg) starting on day 7, twice a week. Mice were randomly allocated into two experimental groups (n = 12 per group) and maintained in specific pathogen‐free conditions throughout the study. Tumor dimensions, including length (L) and width (W), were measured every other day; tumor volumes were calculated using the formula (L × W2)/2. Treatment was initiated once the tumor volume reached about 100 mm3. The mice were housed for 4–8 weeks before being euthanized and sampled.
Western blotting
For protein extraction, cells were lysed with the RNA immunoprecipitation assay (RIPA) buffer (Invitrogen) with protease and phosphatase inhibitors (Roche). Total cellular protein was extracted and the protein concentrations were determined using bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific). The protein samples were separated by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis and transferred to NC membranes. The NC membranes were sectioned based on predicted molecular weight and then subjected to blocking with 5% nonfat milk for 1 h, followed by incubation with primary antibodies against NAT10 (1:1000; Abcam, ab194297), MDR1 (1:1000; CST, #13978), BCRP (1:1000; Abcam, ab130244), or β‐actin (1:50000; abclonal, AC026), respectively. The NC membranes were incubated with HRP‐labeled secondary antibodies (1: 2000, CST), and the protein bands were visualized using chemiluminescent reagents.
Immunohistochemistry staining
Breast cancer and corresponding nontumor tissues were sectioned into slices measuring 4 μm in thickness. Sections from each block were deparaffinized in xylene and rehydrated through a descending series of alcohol. Antigen retrieval was performed by heating a pressure cooker in 10 mmol/L citrate buffer (pH 6.0). The quenching of endogenous peroxidases occurred by incubation in 3% H2O2 for 10 min, followed by incubation with 5% serum to reduce nonspecific binding. Subsequently, the sections were incubated with NAT10 antibody, BCRP antibody, or MDR1 antibody (1:200; Abcam) at 4°C overnight. Then, the slides were incubated with an anti‐rabbit secondary antibody in a humidified box for 30 min at room temperature. After washing in phosphate‐buffered saline (PBS), slides were developed using 3,3‐diaminobenzidine (DAB) chromogen solution and counterstained with Mayer's hematoxylin. Negative controls were performed in parallel by replacing the primary antibody with nonspecific serum. Regarding quantifying the immunohistochemical (IHC) results, IHC intensity was scored as follows: 0 for negative staining; 1 for weak staining, 2 for moderate staining, and 3 for strong staining. The percentage of positive cells was classified into the subsequent categories: 0 for 0%, 1 for 1% to 10%, 2 for 11% to 50%, 3 for 51% to 80%, and 4 for >80%. The final scoring, ranging from 0 to 12, was achieved by multiplying the scores of staining intensity and extent.
Tunel staining
Cells were fixed with 4% methanol‐free formaldehyde for 20 min and permeated with 0.4% Triton X‐100 for 20 min at 4°C. Then, cells were washed with PBS three times, and stained with a Tunel staining kit (Beyotime, C1086) following the manufacturer's instructions.
Ac4C dot blot
Total RNA was heated at 65°C for 5 min and spotted to the Hybond‐N+ membrane after RNA extraction. RNAs on the spotted membrane were crosslinked in a UV254 nm Stratalinker 2400. Then, membranes were blocked and incubated with an anti‐ac4C antibody (1:200, Abcam, ab252215) at 4°C overnight. The membranes were washed with phosphate‐buffered saline with Tween (PBST), incubated with 0.02% methylene blue solution, and scanned as an internal reference.
Acetylated RNA immunoprecipitation and sequencing (acRIP‐seq) and qPCR
ac4C‐RIP was performed using an RNA immunoprecipitation kit (Sigma‐Aldrich, 17 700) following the manufacturer's instructions. Briefly, cells were lysed with 1 mL RIP lysis buffer. The anti‐ac4C antibody (Abcam, ab252215) or normal rabbit IgG (CST, 2729S) was allowed to bind to the beads overnight and then incubated with the RNA samples at 4°C for 2 h. After washing the beads with buffer, the RNA was extracted from the beads and used for both sequencing and q‐PCR analyses.
Statistical analysis
The data are presented as the mean ± standard deviation, and the experiments were replicated thrice. Differences between groups were determined using paired two‐tailed student's t‐test or two‐way analysis of variance (ANOVA). Within the series of IHC assays performed on paraffin‐embedded tissue samples, the Pearson χ2 test was used to determine the correlation between NAT10 and MDR1/BRCP. All the statistical analyses were conducted using GraphPad Prism 9.0. A significance threshold of p‐value less than 0.05 was considered statistically significant when compared against the controls.
RESULTS
NAT10 is highly expressed in breast cancer
In this study, we investigated the expression of NAT10 in breast cancer. To achieve this, we performed a comprehensive analysis using the data from the Cancer Genome Atlas (TCGA) database. Our study yielded a substantial upregulation of NAT10 in breast cancer patients compared to healthy controls (Figure 1a). Furthermore, we examined NAT10 expression in tissue samples from breast cancer patients. Our analyses consistently demonstrated a marked elevation of NAT10 expression in tissues from breast cancer patients relative to adjacent nonmalignant tissues (Figure 1b). Moreover, our investigation extended to assessing the potential clinical significance of NAT10 expression in breast cancer. Interestingly, higher expression of NAT10 predicted an adverse overall survival (OS) outcome among breast cancer patients (Figure 1c). To gain additional insight, we screened the NAT10 expression across a diverse array of cancer cell lines. This was performed utilizing the data from the Cancer Cell Line Encyclopedia (CCLE) cell bank, encompassing 81 distinct types of cancer cell lines. NAT10 expression exhibited notable prominence, especially in notable breast cancer cell lines, including MCF‐7 and MDA‐MB‐231(Figure 1d). By using qPCR and western blotting assays, we also validated that the expression of NAT10 in MCF‐7, MDA‐MB‐231, and MDA‐MB‐468 cells was significantly higher than normal breast cell lines, such as MCF10A and MCF12A (Figure 1e, f). These results suggest that NAT10 expression is dysregulated in breast cancer tissues, and aberrant NAT10 expression is correlated with the disease progression.
FIGURE 1.

High N‐acetyltransferase 10 (NAT10) expression is correlated with breast cancer. (a) Analysis of NAT10 expression levels in breast cancer patients using the Cancer Genome Atlas (TCGA) database (tumor = 1099, normal = 292, https://shiny.hiplot.cn/ucsc‐xena‐shiny/). (b) Detection of NAT10 expression by immunohistochemistry (IHC) in breast cancer patient tissues and normal controls at protein level. (c) Effect of NAT10 expression level on breast cancer patient overall survival according to the Kaplan–Meier Plotter database (NAT10 low patients = 168, NAT10 high patients = 172, https://kmplot.com/analysis/). (d) Analysis of NAT10 levels across different breast cancer cell lines (81 variants) using the Cancer Cell Line Encyclopedia (CCLE) cell bank, including MCF‐7 and MDA‐MB‐231 cells. NAT10 expression at mRNA level (e) and protein level (f) in breast cancer cell lines (MCF‐7, MDA‐MB‐231, MDA‐MB‐468). Two‐tailed p‐values were calculated by two‐way analysis of variance (ANOVA) test. Data are presented as mean ± s.d. The data shown is representative of three (n = 3) biologically independent experiments unless stated otherwise. A p‐value less than 0.05 was considered statistically significant when compared to the controls.
NAT10 promotes malignant phenotypic traits of breast cancer cells
To elucidate the involvement of NAT10 in shaping the attributes of breast cancer cells, we utilized lentivirus‐carrying RNA interference or overexpressing constructs to manipulate the NAT10 expression. Knockdown of NAT10 in MCF‐7 and MDA‐MB‐231 cells (Figure 2a) resulted in significant suppression of cellular proliferation of breast cancer cells (Figure 2b, c). This cell growth reduction was accompanied by a markedly elevated apoptotic rate (Figure 2d). Meanwhile, we observed that suppression of NAT10 in breast cancer cells inhibited colony‐forming ability compared to the control (Figure 2e–g). Conversely, cell proliferation markedly accelerated (Figure 2i, j) when successfully overexpressing NAT10 in MCF‐7 and MDA‐MB‐231 cells (Figure 2h). In addition, the induction of cell apoptosis by capecitabine was significantly inhibited compared to the vector controls (Figure 2k, l). Collectively, these findings conclusively demonstrate the critical role of NAT10 in orchestrating the regulation of malignant phenotypes in breast cancer cells.
FIGURE 2.

N‐acetyltransferase 10 (NAT10) promotes breast cancer cell proliferation in vitro. (a) Representative western blotting, depicting the knockdown effects in MCF‐7 and MDA‐MB‐231 cells infected with lentivirus carrying NAT10 shRNAs, in comparison to the nontarget control (NT Ctrl). (b, c) Assessment of cell viability of MCF‐7 and MDA‐MB‐231 cells of NAT10 knockdown using cell counting kit‐8 (CCK‐8) assay. (d) Flow cytometry analysis of cell apoptosis in MCF‐7 and MDA‐MB‐231 cells infected with lentivirus carrying NAT10 shRNAs, in comparison to the NT Ctrl. The cells were treated with capecitabine. (e) Execution of colony formation assays on MCF‐7 and MDA‐MB‐231 cells infected with nontarget control or NAT10 shRNA. (f, g) Corresponding quantitative from the assays in (e). (h) Representative western blotting presenting the overexpressed NAT10 in MCF‐7 and MDA‐MB‐231 cells. (i, j) Utilization of CCK‐8 assay to assess cell viability in MCF‐7 and MDA‐MB‐231 cells following NAT10 overexpression. (k, l) Flow cytometric analysis of cell apoptosis in MCF‐7 and MDA‐MB‐231 cells infected with lentivirus carrying NAT10 plasmid in comparison with the vector controls. The cells were treated with capecitabine. Two‐tailed p‐values were determined using a two‐way analysis of variance (ANOVA) test. The data are presented as mean ± s.d. The results are representative of three (n = 3) biologically independent experiments unless stated otherwise. A p‐value less than 0.05 was considered statistically significant when compared to the controls.
NAT10 facilitates ac4C‐modification of MDR1 and BCRP genes
Given that NAT10 operates as a lysine acetyltransferase involved in mRNA acetylation processes, we investigated the overall level of mRNA acetylation MCF‐7 cells subjected to NAT10 knockdown (NAT10‐KD). We discovered that the ac4C level of mRNAs was markedly suppressed in response to the NAT10 knockdown (Figure 3a). We further performed immunoprecipitation and acetylated RNA sequencing (acRIP‐seq) experiments in both the NAT10‐KD MCF‐7 cells and the parental control cells to screen ac4C‐modified genes, which promotes breast cancer progression. The analysis of the ac4C modification distribution across different mRNA regions revealed distinct patterns in the NAT10‐KD cells and control cells. Specifically, the ac4C peaks were distributed on 3′‐UTR (53.6% vs.57.1%), coding sequence (CDS) (26.4% vs. 22.4%), 5′‐UTR (5.2% vs. 7.9%) and stop codon (14.8% vs. 12.6%) regions in both the NAT10‐KD cells and the controls respectively, indicating that ac4c‐modification mainly affects the transcription initiation and termination (Figure 3b). Volcano plot visualization showed that 577 mRNAs displayed heightened ac4C‐modification and 551 genes exhibited decrease ac4C‐modification in the NAT10‐KD cells compared to the control cells (Figure 3c). Further analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) elucidated the functional relevance of these differentially modified genes. KEGG analysis revealed that these differently modified genes were correlated with positive regulations of cell migration, apoptotic processes, responses to pharmaceutical agents, and aging (Figure 3d). Evidently, a cluster of ATP binding cassette (ABC) transporters assumes an active role in fundamental cellular mechanism including cell proliferation, cellular migration, the sustenance of stemness in cancer cells, and the genesis of multidrug resistance. 11 Remarkably, our observations underlined that a pair of constituents within this family, MDR1 and BCRP, manifested the most significantly decreased ac4C modification level within cells subjected to NAT10‐KD cells (Figure 3e). To further validate our acRIP‐seq findings, we performed acRIP‐qPCR assays. The results from these assays confirmed a substantial reduction in ac4C enrichment of MDR1 and BCRP gene upon NAT10 knockdown in MCF‐7 cells (Figure 3f). In summary, these findings indicated that MDR1 and BCRP are ac4C‐modified mRNA targets regulated by NAT10 in breast cancer.
FIGURE 3.
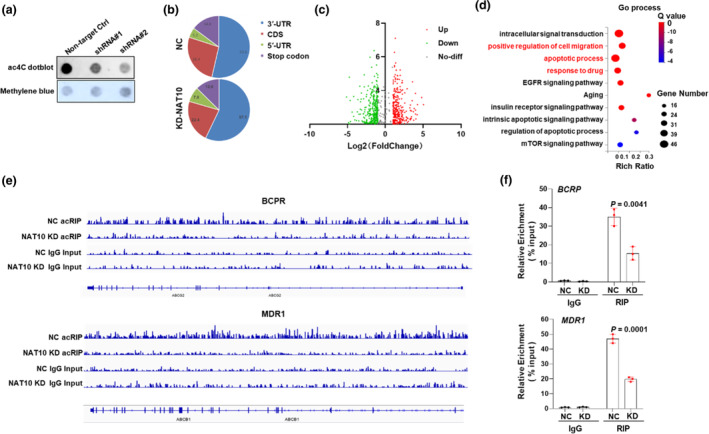
N‐acetyltransferase 10 (NAT10)‐mediated ac4C‐modification of multidrug resistance protein 1 (MDR1) and breast cancer resistance protein (BCRP). (a) Dot blot assay detecting mRNA acetylation level in MCF‐7 cells with or without NAT10 knockdown. (b) acRIP‐seq analysis and corresponding pie chart depicting the distribution of ac4C peaks in the indicated regions, including 3′‐ UTR, CDS, 5′‐UTR and stop codons in control and NAT10 knockdown cells. (c) Volcano plots showing 577 genes with upregulated ac4C modification and 551 genes with downregulated ac4C modification in NAT10‐knockdown cells compared to control cells. (d) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealing the correlation of mRNA acetylated genes with positive regulation of cell migration, apoptotic process, response to drugs, aging, and more. (e) Integrative genomics viewer (IGV) tracks displaying read distributions across MDR1 and BCRP in acRIP‐seq data with NAT10‐KD and NC cells. (f) Quantification of ac4C‐modified mRNA enrichment in MDR1 and BCRP genes with or without knockdown of NAT10 using acRIP quantitative polymerase chain reaction (qPCR). Two‐tailed p‐values were determined by the two‐way analysis of variance (ANOVA) test. The results are presented as mean ± s.d. Data are representative of three (n = 3) biologically independent experiments unless stated otherwise. A p‐value less than 0.05 was considered statistically significant compared to controls.
MDR1 and BCRP are associated with adverse impact on the prognosis of breast cancer
The aforementioned findings suggest that MDR1 and BCRP may play a role in the downstream function of NAT10, highlighting the potential involvement of MDR1 and BCRP in the prognostic implications of breast cancer. Therefore, we further investigated the protein levels of MDR1 and BCRP in response to NAT10 knockdown or overexpression. In the NAT10‐KD breast cancer cells, both MDR1 and BCRP proteins exhibit a remarkable reduction expression (Figure 4a). On the contrary, the artificial expression of NAT10 considerably augmented of the protein levels of MDR1 and BCRP (Figure 4b). Notably, although mRNA levels of MDR1 and BCRP exhibited no statistically significant correlation in breast cancer (Figure 4c), an evident positive correlation was observed between MDR1 and BCRP protein levels and NAT10 within the identical tissue samples extracted from the same patient (Figure 4d, e). Importantly, upon reintroducing elevated levels of MDR1 and BCRP expressions in the NAT10‐KD cells, the apoptosis induced by capecitabine in MCF‐7 cells was noticeably alleviated (Figure 4f–h). Clinically, follow‐up analysis revealed a negative correlation between the expression of MDR1 and BCRP, and overall survival (OS) of breast cancer patients (Figure 4i, j). Collectively, these findings demonstrated that MDR1 and BCRP function as downstream targets of NAT10, governing breast cancer progression.
FIGURE 4.

Multidrug resistance protein 1 (MDR1) and breast cancer resistance protein (BCRP) pose adverse implications for breast cancer prognosis. (a) Assessment of protein levels of MDR1 and BCRP in the N‐acetyltransferase 10 (NAT10) knockdown cells. (b) Representative Western blot (WB) showing the protein levels of MDR1 and BCRP in NAT10 overexpression cells. (c) The correlation analysis between NAT10 and MDR1/BCRP mRNA levels in breast cancer according to the GEPIA2 database (http://gepia2.cancer‐pku.cn/). (d) Immunohistochemical (IHC) detection of NAT10, MDR1, and BCRP expression in a tissue sample from a breast cancer patient. (e) Correlation between NAT10 and MDR1/BCRP protein levels in breast cancer patients. (f) Representative WB showing the protein levels of MDR1 and BCRP upon overexpressing of MDR1 and BCRP in the NAT10‐KD cells. (g, h) Flow cytometry analysis of cell apoptosis in NAT10‐KD MCF‐7 cells infected with lentivirus‐carrying plasmids for MDR1 and BCRP overexpression. (i, j) Effect of MDR1 and BCRP expressions on the overall survival (OS) of breast cancer patients according to the GEPIA2 database (BCRP low patients = 531, BCRP high patients = 535, MDR1 low patients = 534, MDR1 high patients = 534, http://gepia2.cancer‐pku.cn/). Two‐tailed p‐values were determined by the two‐way analysis of variance (ANOVA) test. The results are presented as mean ± s.d. Data are representative of three (n = 3) biologically independent experiments unless stated otherwise. A p‐value less than 0.05 was considered statistically significant compared to controls.
NAT10 inhibition sensitizes capecitabine‐resistant breast cancer cells to chemotherapy in vitro and in vivo
To explore the translational potential of targeting NAT10 in circumventing chemoresistance in breast cancer cells, we established capecitabine‐resistant MCF‐7 cells, with a marked elevation in their IC50 values for capecitabine (Figure 5a). Concomitantly, we observed an upregulation in the expressions of MDR1 and BCRP (Figure 5b). Upon subjecting the capecitabine‐resistant cells to treatment with remodelin, we observed a reversal in the IC50 values for capecitabine treatment (Figure 5c), accompanied by a simultaneous downregulation of MDR1 and BCRP expressions (Figure 5d). Meanwhile, suppression of cell proliferation was evidently noted in both in MCF‐7 and MDA‐MB‐231 capecitabine‐resistant cells upon exposure to remodelin (Figure 5e). This effect was coupled with distinct augmentation in cell apoptosis triggered by capecitabine (Figure 5f).
FIGURE 5.

Capecitabine combined with NAT10 inhibitors synergistically hinders breast cancer cell proliferation in vitro. (a) Shift in IC50 values upon capecitabine treatment in the MCF‐7 and MDA‐MB‐231 cells. (b) Elevation of multidrug resistance protein 1 (MDR1) and breast cancer resistance protein (BCRP) in capecitabine‐resistant breast cancer MCF‐7 and MDA‐MB‐231 cells by Western blot (WB) analysis. (c) Alteration of IC50 values in capecitabine‐resistant cells with treatment of remodelin in the MCF‐7 and MDA‐MB‐231 cells. (d) WB detection of MDR1 and BCRP in the capecitabine‐resistant MCF‐7 and MDA‐MB‐231 cells treated with remodelin. (e) Assessing the viability of the capecitabine‐resistant MCF‐7 and MDA‐MB‐231 cells treated with remodelin by cell counting kit‐8 (CCK‐8) assay. (f) Flow cytometry analysis depicting apoptosis induction in capecitabine‐resistant MCF‐7 and MDA‐MB‐231 cells by remodelin treatment. Two‐tailed p‐values were determined by the two‐way analysis of variance (ANOVA) test. The results are presented as mean ± s.d. Data represent three (n = 3) biologically independent experiments unless stated otherwise. A p‐value less than 0.05 was considered statistically significant compared to controls.
To elucidate the in vivo effect of NAT10 in targeting breast cancer, we established a subcutaneous xenograft tumor model in immunodeficient nude mice using the capecitabine‐resistant MCF‐7 cells. The mice were then subjected to a combination treatment involving capecitabine and remodelin (Figure 6a). Notably, when compared to the administration of capecitabine alone, the combined therapy of capecitabine and remodelin resulted in a remarkable suppression of tumor growth, leading to a significant reduction in tumor volume (Figure 6b, c) along with the prolonged survival rate for the mice (Figure 6d). Moreover, upon subjecting tumors tissues to TUNEL staining for apoptosis assessment, we observed that the combination treatment induced a notably higher count of apoptotic cells in comparison to capecitabine treatment alone (Figure 6e, f). Collectively, these findings strongly indicate that targeting NAT10 holds promise for effectively overcoming chemoresistance in breast cancer in vivo.
FIGURE 6.

Synergistic inhibition of breast cancer cell tumorigenesis in vivo by the combination of capecitabine and N‐acetyltransferase 10 (NAT10) inhibitor treatment. (a) Establishment of a subcutaneous transplantation tumor model in nude mice and treatment with capecitabine and remodelin collaboratively. 1 × 106 MCF‐7 cells were subcutaneously injected into the left flank of mice. Treatment consisting of 0.55 mmol/kg/day capecitabine with or without 100 mg/kg remodelin was started on day 7, twice a week. Tumor volumes were monitored every other day (n = 12/group). (b) Tumor volumes in the groups treated with remodelin, or remodelin in combination with capecitabine (brown). Relative ratios of tumor volume are shown in (c). (d) Assessment of overall survival (OS) in mice using Kaplan–Meier survival analysis. (e) Representative images of immunofluorescent TUNEL staining of tumor tissues, TUNEL (green), nuclei (DAPI, blue) at day 28. (f) Quantification assessment of TUNEL‐positive cells. Two‐tailed p‐values were determined by the two‐way analysis of variance (ANOVA) test. The results are presented as mean ± s.d. Data are representative of three (n = 3) biologically independent experiments unless stated otherwise. A p‐value less than 0.05 was considered statistically significant compared to controls.
DISCUSSION
In the current study, we unveiled the association between NAT10 expression and breast cancer, both in breast cancer patients and cell lines. Remarkably, higher NAT10 expression was observed in breast cancer, and intriguingly, this elevation was negatively correlated with clinical outcomes. We also deciphered a novel mechanism involving ac4C‐modification of MDR1 and BCRP mRNA orchestrated by NAT10, which influences the progression of breast cancer. Furthermore, we performed translational assessments, evaluating the therapeutic potential of targeting NAT10 to impede the progression of breast cancer, both in vitro and in vivo.
The strategic modification of biological molecules through specific chemical alterations has proven to be a highly effective avenue for regulating molecular function within intricate biological systems. 12 The mRNA modifications and their associated regulators have emerged as a critical driver in tumorigenesis and the progression of various cancers. 13 In this spectrum, NAT10 stands out as the only acetyltransferase that catalyzes the ac4C modification, significantly associated with the prognosis of various malignant tumors and potentially promoting tumor progression. Indeed, high expression of NAT10 has been linked to poor prognosis in patients with liver and gastric cancer. 14 Similarly, a study centered on head and neck squamous cell carcinoma showed that inhibiting NAT10 with siRNA led to reduced proliferation, migration, and invasive potential of head and neck squamous cell carcinoma cell lines. 13 While the precise role of NAT10 in breast cancer proliferation and tumorigenesis remains to be elucidated, we identified the higher expression level of NAT10 in breast cancer patients, correlating with reduced survival. Additionally, we revealed elevated levels of NAT10 in well‐established breast cancer cell lines, including MCF‐7 and MDA‐MB‐231. Interestingly, the depletion of NAT10 compromised the proliferation capacity of breast cancer cells.
Our investigation of molecular mechanisms revealed that NAT10 knockdown exerts a significant impact on the ac4C enrichment of two ATP binding cassette (ABC) transporters, MDR1 and BCRP, substantiated by ac4C RNA immunoprecipitation (ac4c‐RIP) and acRIP‐qPCR assays. Of significance, the expression of these two target genes was correlated with NAT10 expression and unfavorable outcomes of breast cancer prognosis. This suggests that NAT10 expression is accountable for ac4C modification of target transcripts, including MDR1 and BCRP. Previous studies have identified ABC transporters as causal factors in the genesis of multidrug resistance, 15 with MDR1 in particular, encoded by ABC subfamily B member 1 (ABCB1), being recognized for conferring resistance to cytotoxic and targeted chemotherapy. 16 A previous study reported that blocking BCRP‐mediated active efflux could hold therapeutic promise for cancer treatment. 17 In various solid tumors, ABC transporters are highly expressed in less differentiated subtypes and metastases. In breast cancer, ABCA1, MDR1, and BCRP are critical players in the development of chemoresistance, and suppressing their activity holds substantial potential in reversing this phenomenon. 18 Our study provides novel insights into the molecular mechanisms underlying the role of ABC transporters in breast cancer progression, focusing on the perspective of mRNA ac4c modification.
In the domain of cancer treatment, conventional chemotherapy has long been used albeit accompanied by systemic side effects such as hair loss and gastrointestinal toxicity. Therefore, a strategy involving combined and synergistic chemotherapy has gained traction, offering improved therapeutic efficacy while mitigating systemic toxicity. 19 , 20 , 21 Our subsequent studies have confirmed that NAT10 inhibitors effectively modulate MDR1 and BCRP expression, leading to the attenuation of breast cancer cell proliferation in vitro. Moreover, the in vivo subcutaneous transplantation tumor model demonstrated that remodelin significantly inhibited tumor proliferation and enhanced the therapeutic efficacy of capecitabine in breast cancer. All these findings demonstrated that NAT10 serves as a viable prognostic indicator. Moreover, the utilization of remodelin as a means to target NAT10 displays the potential to enhance the efficacy of conventional chemotherapy agents in restraining breast cancer progression. This not only presents a prospective avenue for prognosis assessment but also introduces an encouraging focal point for therapeutic exploration within the realm of breast cancer treatment.
In conclusion, our study illuminates the essential role of NAT10‐mediated ac4c‐modification in breast cancer proliferation and tumorigenesis. Additionally, we present novel insights into the dynamic of MDR1 and BCRP mRNA modification via NAT10‐dependent acetylation. Our study highlights the therapeutic potential of the combination of capecitabine and remodelin, which effectively modulates the MDR1 and BCRP expression, attenuating breast cancer cell proliferation. These findings extend to the in vivo scenario, providing a promising avenue for combating breast cancer progression through innovative therapeutic strategies.
AUTHOR CONTRIBUTIONS
Cui‐Cui Zhao and Xuan Sun performed the experiments, statistical analyses and conducted the animal studies. Cui‐Cui Zhao, Jing Chen and Bill D. Geng contributed to writing the manuscript. Cui‐Cui Zhao and Jing Chen provided the patient samples with clinical data, statistical analyses and also contributed to the study design.
CONFLICT OF INTEREST STATEMENT
The authors have no potential conflicts of interests.
ACKNOWLEDGMENTS
This work was supported by the Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK‐009A).
Zhao C‐C, Sun X, Chen J, Geng BD. NAT10‐mediated mRNA N4‐acetylcytidine modification of MDR1 and BCRP promotes breast cancer progression. Thorac Cancer. 2024;15(10):820–829. 10.1111/1759-7714.15262
Cui‐Cui Zhao, Xuan Sun and Jing Chen contributed equally to this work and are cofirst authors.
DATA AVAILABILITY STATEMENT
The data generated in this study are available upon request from the corresponding author.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA‐Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- 2. Harbeck N, Penault‐Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5(1):66. [DOI] [PubMed] [Google Scholar]
- 3. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624. [DOI] [PubMed] [Google Scholar]
- 5. Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175(7):1872–1886 e1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng Z, Li K, Qin K, Liang J, Shi M, Ma Y, et al. The LINC00623/NAT10 signaling axis promotes pancreatic cancer progression by remodeling ac4C modification of mRNA. J Hematol Oncol. 2022;15(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Jin C, Wang T, Zhang D, Yang P, Zhang C, Peng W, et al. Acetyltransferase NAT10 regulates the Wnt/beta‐catenin signaling pathway to promote colorectal cancer progression via ac(4)C acetylation of KIF23 mRNA. J Exp Clin Cancer Res. 2022;41(1):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin J, Xiang Y, Huang J, Zeng H, Zeng Y, Liu J, et al. NAT10 maintains OGA mRNA stability through ac4C modification in regulating oocyte maturation. Front Endocrinol. 2022;13:907286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin G, Xu M, Zou M, Duan S. The processing, gene regulation, biological functions, and clinical relevance of N4‐Acetylcytidine on RNA: a systematic review. Mol Ther Nucleic Acids. 2020;20:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y, Jing Y, Wang Y, Tang J, Zhu X, Jin WL, et al. NAT10 promotes gastric cancer metastasis via N4‐acetylated COL5A1. Signal Transduct Target Ther. 2021;6(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang JQ, Wu ZX, Yang Y, Teng QX, Li YD, Lei ZN, et al. ATP‐binding cassette (ABC) transporters in cancer: a review of recent updates. J Evid Based Med. 2021;14(3):232–256. [DOI] [PubMed] [Google Scholar]
- 12. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20(6):303–322. [DOI] [PubMed] [Google Scholar]
- 13. Tao W, Tian G, Xu S, Li J, Zhang Z, Li J. NAT10 as a potential prognostic biomarker and therapeutic target for HNSCC. Cancer Cell Int. 2021;21(1):413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang X, Chen J, Jiang S, He S, Bai Y, Zhu L, et al. N‐acetyltransferase 10 enhances doxorubicin resistance in human hepatocellular carcinoma cell lines by promoting the epithelial‐to‐mesenchymal transition. Oxid Med Cell Longev. 2019;2019:7561879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug‐resistant cancer. Nat Rev Cancer. 2018;18(7):452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pote MS, Gacche RN. ATP‐binding cassette efflux transporters and MDR in cancer. Drug Discovery Today. 2023;28(5):103537. [DOI] [PubMed] [Google Scholar]
- 17. Nakanishi T, Ross DD. Breast cancer resistance protein (BCRP/ABCG2): its role in multidrug resistance and regulation of its gene expression. Chin J Cancer. 2012;31(2):73–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Modi A, Roy D, Sharma S, Vishnoi JR, Pareek P, Elhence P, et al. ABC transporters in breast cancer: their roles in multidrug resistance and beyond. J Drug Target. 2022;30(9):927–947. [DOI] [PubMed] [Google Scholar]
- 19. Li B, Shao H, Gao L, Li H, Sheng H, Zhu L. Nano‐drug co‐delivery system of natural active ingredients and chemotherapy drugs for cancer treatment: a review. Drug Delivery. 2022;29(1):2130–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Z, Ren Y, Weng S, Xu H, Li L, Han X. A new trend in cancer treatment: the combination of epigenetics and immunotherapy. Front Immunol. 2022;13:809761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Najafi M, Majidpoor J, Toolee H, Mortezaee K. The current knowledge concerning solid cancer and therapy. J Biochem Mol Toxicol. 2021;35(11):e22900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.


