Abstract
Over the past decade, information for precision disease medicine has accumulated in the form of textual data. To effectively utilize this expanding medical text, we proposed a multi-task learning-based framework based on hard parameter sharing for knowledge graph construction (MKG), and then used it to automatically extract gastric cancer (GC)-related biomedical knowledge from the literature and identify GC drug candidates. In MKG, we designed three separate modules, MT-BGIPN, MT-SGTF and MT-ScBERT, for entity recognition, entity normalization, and relation classification, respectively. To address the challenges posed by the long and irregular naming of medical entities, the MT-BGIPN utilized bidirectional gated recurrent unit and interactive pointer network techniques, significantly improving entity recognition accuracy to an average F1 value of 84.5% across datasets. In MT-SGTF, we employed the term frequency-inverse document frequency and the gated attention unit. These combine both semantic and characteristic features of entities, resulting in an average Hits@ 1 score of 94.5% across five datasets. The MT-ScBERT integrated cross-text, entity, and context features, yielding an average F1 value of 86.9% across 11 relation classification datasets. Based on the MKG, we then developed a specific knowledge graph for GC (MKG-GC), which encompasses a total of 9129 entities and 88,482 triplets. Lastly, the MKG-GC was used to predict potential GC drugs using a pre-trained language model called BioKGE-BERT and a drug-disease discriminant model based on CNN-BiLSTM. Remarkably, nine out of the top ten predicted drugs have been previously reported as effective for gastric cancer treatment. Finally, an online platform was created for exploration and visualization of MKG-GC at https://www.yanglab-mi.org.cn/MKG-GC/.
Keywords: Drug discovery, Entity recognition, Entity normalization, Gastric cancer, Relation classification
Graphical Abstract
Highlights
-
•
MKG is a framework that performs knowledge extraction using a multi-task learning approach with hard parameter sharing.
-
•
With MKG, we built a KG for GC and predicted candidate GC drugs, confirming the clinical value of the MKG.
-
•
An online platform was developed for MKG-GC exploration and visualization.
1. Introduction
Gastric cancer (GC) is one of the most deadly and malignant diseases that human beings are confronted with. According to stomach cancer cases statistics in 2020, stomach cancer causes about 800,000 deaths and is the fourth leading cause of death among cancers [1]. There is a large amount of research material related to gastric cancer. For example, the number of relevant medical publications retrieved from PubMed using "(gastric cancer [Title/Abstract]) OR (stomach cancer [Title/Abstract])" exceeds 80,000, and the number of publications is rapidly increasing.
Knowledge graphs (KG) are increasingly used to effectively organize and manage diverse medical research data, enabling their comprehensive use in various medical applications, including drug discovery, clinical diagnosis, and medical data analysis systems [2]. Recently, a number of KGs have been developed for cancer research based on literature, EMRs, or databases. Examples of such KGs include KGHC [3], DSTKG [4], and TBKG [5]. However, KGs have rarely been employed in the field of gastric cancer. There is an urgent need for an infrastructure to perform simple, quick, and routine annotations of textual data related to gastric cancer. Moreover, the features of biomedical texts are not fully utilized in most knowledge extraction models, and entity normalization is seldom performed.
To address these issues, we proposed a knowledge graph construction framework (MKG) using a hard parameter sharing-based multi-task learning approach to extract knowledge from medical literature and then applied it to gastric cancer knowledge graph construction and drug discovery. It is freely accessible and available at https://www.yanglab-mi.org.cn/MKG-GC/.
2. Methods and materials
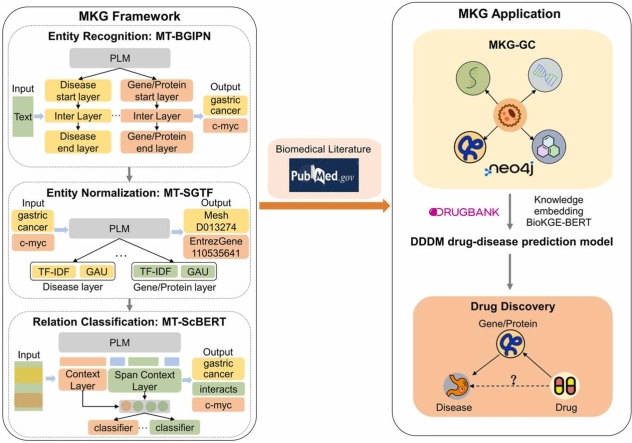
2.1. MKG framework
In the MKG framework, we designed three separate modules, MT-BGIPN, MT-SGTF and MT-ScBERT, for entity recognition, entity normalization, and relation classification, which employ multi-task learning with hard parameter-sharing approach.
2.2. Pre-trained language models
In our multi-task knowledge extraction framework, we used a pre-trained language model (PLM) as an embedding layer. BioBERT[6], which is a deep neural network model built on a transformer to extract potential semantic features from text through a multi-headed attention mechanism, was used as a PLM for the entity recognition model and the relation classification model. In addition, BioBERT leverages a massive dataset from PubMed and PMC (biomedical literature databases) for unsupervised pre-training based on the original BERT architecture, making it is exquisitely tailored for biomedical applications, including those explored in our study. The entity normalization used SapBERT [7] as a PLM.
2.3. Entity recognition
In MKG, entity recognition was considered as a sequence labeling task, and the BIO format was used to annotate the dataset, marking the first word of the entity as B-type, the rest of the words in the entity as I-type, and other words as O-type.
2.3.1. Dataset
Five public medical entity recognition datasets, including BC2GM [8], BC4CHEMD [9], JNLPBA [10], NCBI-disease [11], and LINNAEUS [12], were collected to train and evaluate models; their statistics are shown in Table S1. To train and evaluate the multi-task entity recognition model, we merged the five datasets described above to form a new one, Dataset-5, which includes eight entity types: Gene/Protein, Chemical/Drug, Disease, DNA, RNA, Cell type, Cell line, and Species. Table S2 provides the statistics for Dataset-5.
2.3.2. Model architecture
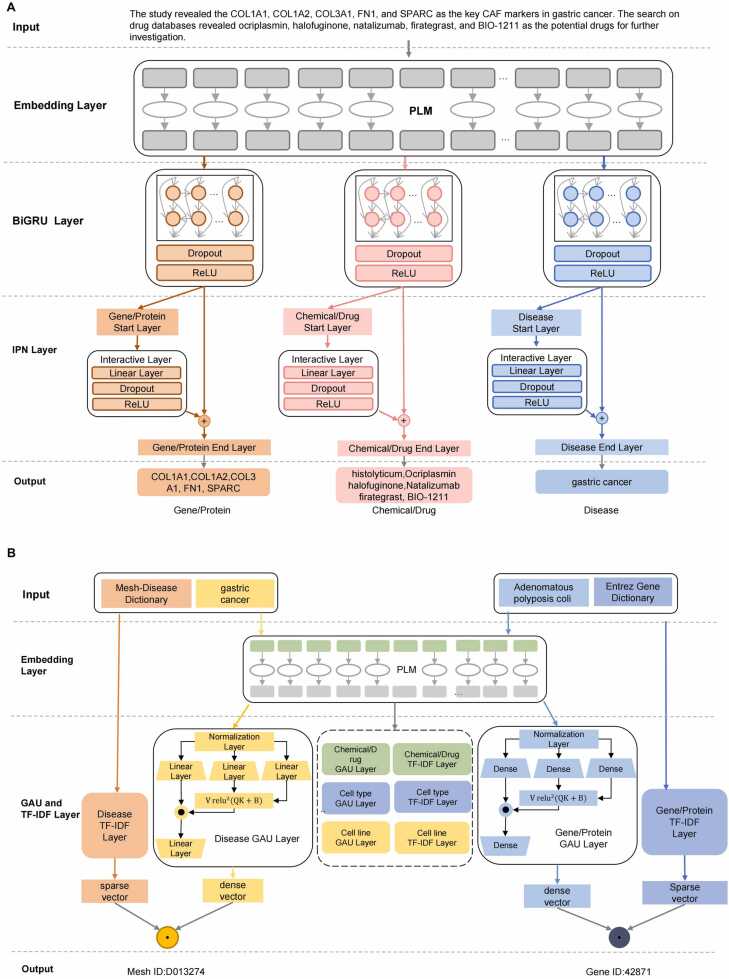
The MT-BGIPN consists of a shared PLM and separate task-specific layers for each entity type, as shown in Fig. 1A. The MT-BGIPN recognizes over eight entity types, as mentioned above.
Fig. 1.
Architecture of entity extraction modules. A. MT-BGIPN. B. MT-SGTF.
2.3.2.1. Input
The entity recognition model needs only medical text as input and does not need additional introduced feature information. "[CLS]" and "[SEP]" are special tokens required as input to the PLM.
2.3.2.2. Embedding layer
BioBERT was used as the embedding layer in MT-BGIPN to extract symbol-level semantic feature vectors from text and share semantic feature information between subtasks.
2.3.2.3. BiGRU layer
The second layer of MT-BGIPN is the Bi-directional Gated Recurrent Network (BiGRU), which is used to extract sentence-level semantic feature information oriented to a specific subtask and consists of a dropout layer, and the ReLU activation function. The gated recurrent unit consists of three parts: a reset gate, an update gate, and a memory unit, and is implemented to retain historical state information.
2.3.2.4. IPN layer
The third layer is the Interactive Pointer Network (IPN) decoder layer, which decodes according to the extracted feature information to accurately recognize medical entities. The IPN layer is composed of a start layer, an interactive layer, and an end layer, among which the start and end layers are fully connected and predict the start and end boundaries of the entity, respectively. The interactive layer is composed of a fully connected layer, a dropout layer, and a ReLU activation function. The feature information of the predicted entity start boundary is used to predict the entity end boundary, enabling two independent pieces of feature information to interact and thereby identifying the entity boundary more accurately.
The text is input to BioBERT and BiGRU to obtain the global feature vector and then input to the start layer to obtain the entity start boundary feature vector . The calculation is shown in Eq. (1), where denotes the feature dimension and denotes the entity type numbers:
| (1) |
The entity start boundary feature vector is fed into the Interactive Layer, and the output is obtained as the interaction feature vector . Then the interaction feature vector is summed with the global feature vector , and the result is input to the End Layer to obtain the entity end boundary feature vector . The calculation is shown in (2), (3):
| (2) |
| (3) |
2.3.2.5. Output
Finally, the entity start boundary vector and the entity end boundary vector are decoded to obtain the medical entities in the text.
2.4. Entity normalization
2.4.1. Dataset
For entity normalization, each dataset contains a medical dictionary for mapping entities to the dictionary. The model was trained and evaluated on three public datasets, as shown in Table S3 (BC5CDR-Chemical [13], BC5CDR-Disease [13] and NCBI-Disease [11]), as well as five new datasets introduced in this paper. Because the public entity normalization dataset contains only two entity types, five medical dictionaries and five entity normalization datasets were constructed based on four medical databases to enable the model to normalize all entity types (Table S4).
2.4.2. Model architecture
The MT-SGTF architecture is shown in Fig. 1B and consists of two parts: a shared SapBERT and a multiple gated attention unit (GAU) + term frequency-inverse document frequency (TF-IDF).
2.4.2.1. Input
The input is entities and medical dictionaries.
2.4.2.2. Embedding layer
The first layer of MT-SGTF uses SapBERT as the embedding layer, which is used to extract semantic and synonym features from medical entities and serves as a shared layer to share effective feature information among various steps.
2.4.2.3. GAU and TF-IDF layers
The GAU is a new attention mechanism combining an attention mechanism with a Gated Linear Unit, which has a faster computation speed and stronger feature extraction ability than the Transformer. The formula for calculating the GAU is shown in (4), (5).
| (4) |
| (5) |
where A is the attention calculation result; is the parameter; and are affine transformations; is the bias value; is the normalization factor to eliminate the length effect; is an activation function; is another activation function; and and denote feature dimensions.
The second layer of the model is oriented towards specific subtasks and consists of GAU and TF-IDF. Each subtask is used to normalize a specific class of medical entities. TF-IDF is used to extract character feature vectors of entities, whereas GAU is used to extract semantic feature vectors of entities. The medical entity is input to SapBERT to obtain the feature vector H. The sparse vector and the dense vector are calculated as shown in (6), (7), where denotes the number of inputs:
| (6) |
| (7) |
Then the sparse vector and the dense vector are weighted and summed to obtain the entity feature vector , as shown in Eq.(8), where W is a learnable weight parameter and ⨁ denotes vector summation:
| (8) |
Next, the feature vector of the input entity and the feature vector of all entities in the dictionary are computed separately, and the similarity scores of the input entity and of all entities in the dictionary are computed. The entity with the highest similarity score is the synonym of the input entity in the dictionary, thus realizing the unique identification of the input entity with ID, as shown in Eq.(9), where ⨂ represents the inner product operation and represents the similarity scores between the candidate entity and all entities in the dictionary:
| (9) |
2.4.2.4. Output
In the example shown in Fig. 1B, the model maps “Gastric Cancer” to the Disease dictionary, identified by “D013274″, and “Adenomatous polyposis coli 2″ to the EGene dictionary, identified by “42871″.
2.5. Relation classification
In our investigation, we have approached relation extraction as a form of relation classification, similar to previous research conducted in the biomedical and clinical domains [14], [15]. This approach involves identifying whether a potential pair of entities possesses a semantic relation within a given text sequence, defining the nature of that relation, and finally forming triplets.
There are two rules for generating candidate entity pairs: (a) limit entity type; (b) limit distance between entity pairs. The MT-ScBERT identifies relationship categories as Gene/Protein-Gene/Protein Interaction (PPI), Chemical/Drug-Chemical/Drug Interaction (DDI), Chemical/Drug-Disease Interaction (CDI), Gene/Protein-Disease Interaction (GDI), and Chemical/Drug-Gene/Protein Interaction (CPI). It relies on context, entity, and span context to predict the semantic relationship between entity pairs. The distance between the statements in which the entity pairs are located is limited to less than or equal to two due to the limitation on the input length of the model. When the distance between pairs of entities is larger, the possibility of semantic relationship decreases.
2.5.1. Dataset
Eleven biomedical relation classification datasets, including BC5CDR [13], EU-ADR [16], DDI2013 [17], BC6ChemProt [14], BC7DrugProt [18], GAD [19], LLL [20], IEPA [21], HPRD50 [21], BioInfer [21], and AIMed [21], were collected for relation classification model training and evaluation (Table S5). To train and evaluate the multi-task model, the above eleven relation classification datasets were merged into a new dataset, Dataset-11, which contains five semantic relationship categories: PPI, DDI, CDI, GDI, and CPI (Table S6).
2.5.2. Model Architecture
2.5.2.1. Input
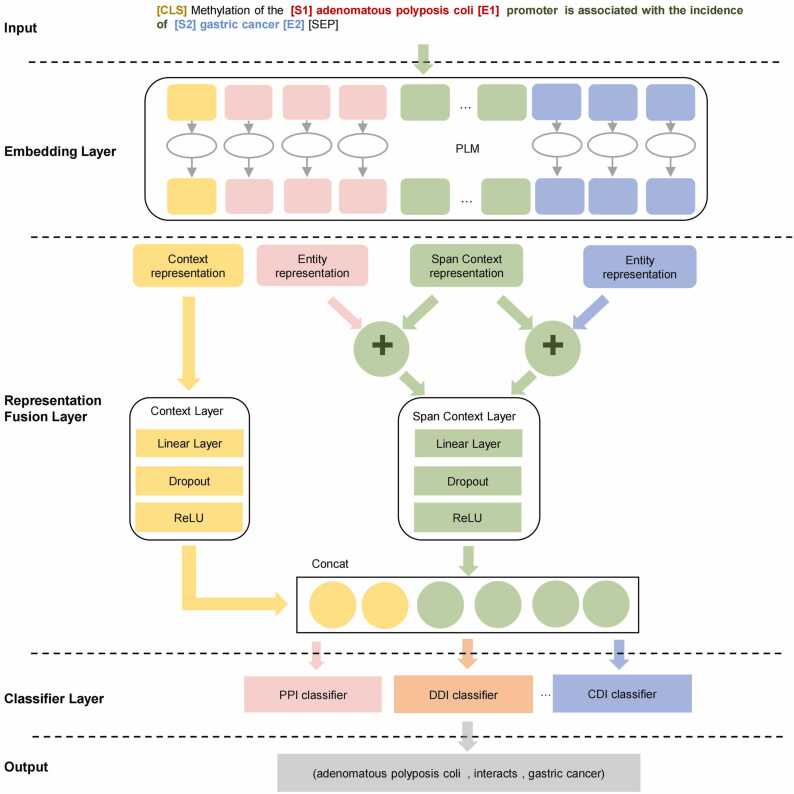
Text must be transformed before entering it into the model. "[CLS]" is a special token required for PLM inputs. “[S1][E1]” and “[S2][E2]” are used to mark the positions of candidate entity pairs in the medical text. An example of the input format is shown in Fig. 2.
Fig. 2.
Architecture of MT-ScBERT relation classification model based on representation fusion.
2.5.2.2. Embedding layer
BioBERT is used as a shared embedding layer of MT-ScBERT to extract contextual, entity, and span context representations from the input text, that is, the feature vector corresponding to “[CLS]” (the context representation vector ), the feature vectors corresponding to “[S1]entity[E1]” and “[S2]entity[E2]” (the entity representation feature vectors and ), and the feature vector between two entity pairs, which is used as the Span Context representation feature vector .
2.5.2.3. Representation fusion layer
The representation fusion layer consists of the context layer and the entity layer. It is used to fuse the three extracted features just described. First, is fed to the Context Layer for semantic feature extraction and dimensionality reduction, as shown in Eq. (10), where d denotes the feature dimension:
| (10) |
and are then added to , and fed to the span context layer for feature extraction and dimensionality reduction to obtain new entity feature vectors and , as shown in (11), (12), where represents element-wise addition of eigenvectors:
| (11) |
| (12) |
These three features are then spliced to obtain the fusion feature vector as shown in Eq. (13), where ⨁ represents concatenation of features:
| (13) |
2.5.2.4. Classifier layer
The third layer of the model is the classification layer. According to the input entity pair category, the fusion feature vector is input to the corresponding binary classification layer. The output is the semantic relationship category, as shown in Eq.(14), where c denotes the relationship category number:
| (14) |
2.5.2.5. Output
In the example in Fig. 2, the model extracted the triplet (adenomatous polyposis coli, interacts, Gastric Cancer) from the text.
2.6. Evaluation Metric for Knowledge Extraction Models
The precision (P), recall (R), accuracy (ACC) and F1-score (F1) were used as the evaluation metrics, as shown in (15), (16), (17), (18):
| (15) |
| (16) |
| (17) |
| (18) |
As described in previous papers [7], [22], Hits@k was used as an evaluation metric, which means the proportion of correct entities in the top-k, and k equals 1 is regarded as the accuracy rate, as shown in Eq. (19):
| (19) |
where indicates the number of predicted samples, and the function indicates that the condition returns 1 if it holds, otherwise it returns 0. indicates the ranking of the i-th sample according to the predicted probability, from highest to lowest.
2.7. Knowledge source and preprocessing for MKG-GC
The knowledge sources for constructing the MKG-GC were 3791 biomedical literature abstracts, which were related to gastric cancer from PubMed through manual screening and keyword retrieval. The retrieval keywords used in PubMed were (("gastric can*"[Title/Abstract] OR "stomach can*"[Title/Abstract]) AND "drug"[Title/Abstract]) NOT "review"[Publication Type]) AND (2000:2022[pdat]). The research described in the literature deals mainly with the relationships between gastric cancer and genes, or, between genes, and the therapeutic effects of drugs on gastric cancer. Biomedical literature abstracts were preprocessed using NLTK [23], including word and sentence segmentation.
2.8. Gastric cancer drug candidates prediction based on MKG-GC
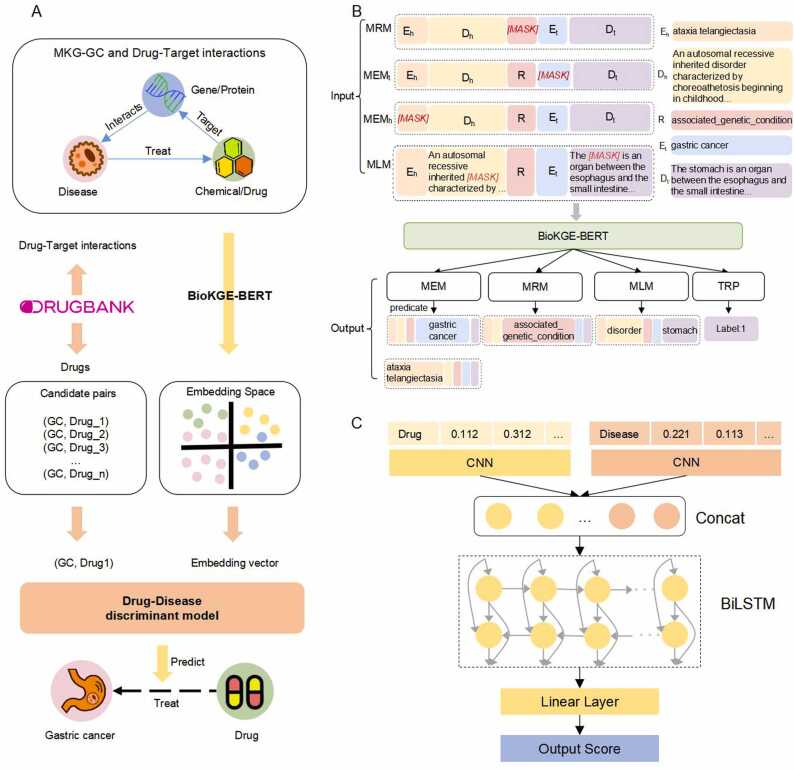
2.8.1. A BERT-based PLM for biomedical knowledge embedding
A PLM BioKGE-BERT for knowledge embedding was constructed for the characteristics of biomedical triplets, and a neural network model was used to extract the semantic feature information of entities and relationships in the triplets. This information was used to transform the MKG-GC into a knowledge embedding vector. Details of BioKGE-BERT construction are shown in the Supplementary Materials.
2.8.2. Drug-disease discriminant model based on CNN and BiLSTM
We constructed a drug-disease discrimination model (DDDM) based on CNN-BiLSTM to predict the potential of a drug in treating a specific disease (Fig. 3C). The inputs of the DDDM are candidate drugs and diseases; the corresponding drug embedding and disease embedding vectors are found in the knowledge embedding vector table. Then local feature information is extracted by inputting these vectors into two CNN models; the two output vectors are concatenated and inputted into BiLSTM to extract global logical feature information. Finally, the prediction scores are output using the fully-connected layer, where a higher score represents a higher probability of the drug being a candidate to treat the disease.
Fig. 3.
Knowledge representation and application of MKG-GC in drug discovery. A. The drug discovery process. B. The model architecture of the BioKGE-BERT. C. The model architecture of DDDM.
A total of 7774 drugs were collected from DrugBank, and all the drugs and GC were combined into 7774 (Drug, GC) pairs. The drug-disease discrimination model predicted a score for each (Drug, GC), indicating the likelihood that the drug was helpful in gastric cancer treatment.
2.8.3. Data storage and application
The MKG-GC webserver was built by Django (https://www.djangoproject.com/), and the web interface was developed using ECharts (https://echarts.apache.org/) for visualization. Neo4j was used for KG data storage and management.
3. Data availability
Source data and code for MKG-GC construction and application are available at the GitHub repository (https://github.com/KeDaCoYa/MKG-GC).
4. Results
4.1. MT-BGIPN entity recognition module in MKG
Due to the irregularity and complexity of biomedical entity naming and the length of entity names, the entity recognition task in the biomedical domain is more challenging than in the general domain. A multi-task entity recognition model, MT-BGIPN, was constructed based on BiGRU and IPN to improve model performance by focusing on the entity boundary information (Fig. 1A). Details on the technologies used in the model are available in Methods section.
The IPN layer facilitates interaction between features and improves model performance by using the feature information of the entity’s start boundary to predict the entity’s end boundary. To verify the effectiveness of our model using IPN as the decoder layer, it was compared with the Pointer Network, the Conditional Random Field, and the Multi-Layer Perceptron. As shown in Table S7, our model achieved an average F1-score of 84.5% on the eight datasets, which was 0.5%, 0.8%, and 1.3% higher than PN, CRF, and MLP, respectively.
Then we compared the performance of MT-BGIPN with single-task learning approach ST-BGIPN on a new dataset Dataset-5 to verify the effectiveness of the multi-task learning approach (Table 1). The average F1 of MT-BGIPN was 83.9%, which is 0.8% higher than ST-BGIPN. The MT-BGIPN model was also compared with the previously published BERN2 [24], MTM-CW [25], and PTC [26] multi-task models. The average F1-score of MT-BBIPN was 83.9%, which represented the highest performance.
Table 1.
Performance comparison of MT-BGIPN with other models on Dataset-5.
| Type | MTM-CW | PTC | BERN2 | ST-BGIPN | MT-BGIPN |
|---|---|---|---|---|---|
| Gene/Protein | 0.808 | 0.867 | 0.835 | 0.831 | 0.845 |
| Chemical/Drug | 0.894 | 0.895 | 0.904 | 0.901 | 0.913 |
| Disease | 0.865 | 0.837 | 0.894 | 0.888 | 0.908 |
| Species | 0.889 | 0.854 | 0.889 | 0.881 | 0.881 |
| Cell line | - | - | 0.777 | 0.754 | 0.779 |
| Cell type | - | - | 0.793 | 0.788 | 0.791 |
| DNA | - | - | 0.769 | 0.767 | 0.764 |
| RNA | - | - | 0.750 | 0.832 | 0.832 |
| Average | - | - | 0.827 | 0.831 | 0.839 |
Note: (1) All reported scores are best micro F1-score; (2) the best micro F1-score was highlighted as bold.
4.2. MT-SGTF entity normalization module in MKG
The name length and complexity of medical entities are challenging for entity normalization. A multi-task biomedical entity normalization model (MT-SGTF) was constructed based on a shared SapBERT as PLM and the GAU + term frequency-inverse document frequency (TF-IDF) to extract the semantic and character features of entities and to improve model performance by fusing the two kinds of features. The MT-SGTF architecture is shown in Fig. 1B.
We first compared the performance of MT-SGTF with two single-task learning approach, BioSYN and SapBERT, on three public entity normalization datasets (Table 2). The results show that MT-SGTF achieved the best performance, with an average Hits@ 1 of 94.4% and an average Hits@ 5 of 96.9%. To further validate the effectiveness of the multi-task model, MT-SGTF was trained and evaluated on the five entity normalization datasets that were constructed in this paper. Again, MT-SGTF performed significantly better than SapBERT, with an average Hits@ 1 value of 95.0%, which was 0.9% higher than SapBERT (Table 3). MT-SGTF has a total of five subtasks, which enabled the shared model to fully learn the feature information of the facilitated subtasks, thus improving model performance. This result indicates that the multi-task model can achieve higher performance when the number of subtasks increase.
Table 2.
Performance comparison of MT-SGTF with other models on three public datasets.
| Dataset | Metric | BioSYN | SapBERT | MT-SGTF |
|---|---|---|---|---|
| BC5CDR-Disease | Hits@ 1 | 0.911 | 0.936 | 0.937 |
| Hits@ 5 | 0.939 | 0.962 | 0.966 | |
| BC5CDR-Chemical | Hits@ 1 | 0.966 | 0.968 | 0.969 |
| Hits@ 5 | 0.972 | 0.984 | 0.982 | |
| NCBI-Disease | Hits@ 1 | 0.932 | 0.925 | 0.926 |
| Hits@ 5 | 0.960 | 0.962 | 0.960 | |
| Average | Hits@ 1 | 0.936 | 0.943 | 0.944 |
| Hits@ 5 | 0.957 | 0.969 | 0.969 |
Table 3.
Performance comparison of MT- SGTF with other models on the five datasets.
| Dataset | Metric | SapBERT | MT-SGTF |
|---|---|---|---|
| MeSH-DIS | Hits@ 1 | 0.963 | 0.965 |
| Hits@ 5 | 0.982 | 0.982 | |
| MeSH-CD | Hits@ 1 | 0.951 | 0.955 |
| Hits@ 5 | 0.974 | 0.973 | |
| EGene | Hits@ 1 | 0.929 | 0.942 |
| Hits@ 5 | 0.961 | 0.971 | |
| CO-CT | Hits@ 1 | 0.904 | 0.934 |
| Hits@ 5 | 0.986 | 0.985 | |
| Cellosaurus-CL | Hits@ 1 | 0.956 | 0.956 |
| Hits@ 5 | 0.977 | 0.975 | |
| Average | Hits@ 1 | 0.941 | 0.950 |
| Hits@ 5 | 0.976 | 0.977 |
Moreover, we also verified the effectiveness of GAU and the combination of semantic and character features in MT-SGTF. The average Hits@ 1 value of GAU on the five datasets was 0.5% higher than Transformer (Table S8). For feature combination, MT-SGTF achieved the highest performance by combining semantic and character features, with Hits@ 1 values increased by 1% and 2.8% compared to character features alone and semantic features alone, respectively (Table S9).
4.3. MT-ScBERT relation classification module in MKG
A multi-task relationship classification model based on span context information (MT-ScBERT) was proposed to capture the semantic relationships that exist between entity pairs, and forms triplets. The model contains five subtasks: PPI, DDI, CDI, GDI, and CPI. The structure of MT-ScBERT is shown in Fig. 2.
First, we verified the effectiveness of the cross-text features that ScBERT uses and compared it with Baseline [6], MTB [27], and RBERT [28] on 11 public datasets. As shown in Table S10, the performance of ScBERT was significantly better than the other three models. The average F1-score of ScBERT on the 11 datasets was 86.9%, which was 0.5%, 1.8%, and 2.8% higher than RBERT, MTB, and the Baseline model, respectively. This indicates the effectiveness of span context representation for the relation classification task. To assess the effectiveness of the multi-task learning approach, we then compared the performance of the multi-task model with that of the single-task model on Dataset-11, which was created by combining the 11 datasets mentioned earlier. The results showed that the performance of the multi-task model relational classification outperforms that of the single-task model (Table 4) with average F1-score of 92%, which was 0.6% higher than that of ST-ScBERT.
Table 4.
Performance comparison of MT-ScBERT with other models on Dataset-11.
| Relationship Category | ST-RBERT | ST-ScBERT | MT-RBERT | MT-ScBERT |
|---|---|---|---|---|
| PPI | 0.888 | 0.888 | 0.889 | 0.890 |
| DDI | 0.968 | 0.967 | 0.964 | 0.966 |
| CPI | 0.983 | 0.984 | 0.984 | 0.987 |
| CDI | 0.915 | 0.912 | 0.853 | 0.866 |
| GDI | 0.814 | 0.820 | 0.890 | 0.893 |
| Average | 0.913 | 0.914 | 0.916 | 0.920 |
Note: (1) All reported scores are best micro F1-score; (2) the best micro F1-score was highlighted as bold.
Moreover, we also compared the multi-task and single-task whole learning extraction models in terms of number of parameters and computation speed. As shown in Fig. S1, the multi-task model exhibited significantly faster computation speed and fewer parameters compared to the single-task model. The average time required by the multi-task model to process a literature abstract was 0.875 s, which is 4.7 times faster than that of the single-task model.
4.4. Construction of gastric cancer knowledge graph based on MKG
Based on the MKG proposed above, we successfully extracted entities and triplets from 3791 GC-related medical literature abstracts. In total, we identified 137,698 medical entities and 195,238 triplets. Knowledge fusion was then performed on the results to reduce redundancy and ambiguity. The final MKG-GC included a total of 9129 entities and 88,482 triplets (Table S11). A user-friendly web interface was also provided for exploration and visualization of MKG-GC (Fig. S2 and S3).
4.5. Identification of gastric cancer candidate drug based on MKG-GC
The current cost of drug development is still exorbitant, and the aim of drug discovery is to minimize this cost by repurposing existing drugs for treating other diseases. In this study, we explored the application of MKG-GC for drug discovery to provide new possibilities for GC treatment. The knowledge representation and application of MKG-GC in drug discovery is shown in Fig. 3A.
We first developed a BERT-based pre-trained language model known as BioKGE-BERT, which was used to convert the extracted relations from MKG-GC and the drug-target triplets from DrugBank into a low-dimensional vector space (Fig. 3B). This method was compared with six knowledge embedding methods: TransE [22], HoLE [29], DistMult [30], KG-BERT [31], StAR [32], and LpBERT [33] (Table S12). BioKGE-BERT achieved the highest performance, with a Hits@ 1 value of 0.431.
Next, we constructed a drug-disease neural network discriminant model (DDDM) based on CNN-BiLSTM to predict the suitability of each candidate drug for gastric cancer treatment. To verify the effectiveness of the CNN-BiLSTM hybrid model DDDM, the CNN-based model alone and BiLSTM-based model alone were constructed for performance comparison (Table S13). Our DDDM achieved the highest performance, with an F1 score of 0.856 and an accuracy of 0.816. The results indicate that the combination of the CNN extracting local feature information and the BiLSTM extracting global feature information effectively improved model performance.
The candidate drugs were ranked according to the DDDM prediction scores from highest to lowest, and the descriptions of the top 30 candidates are presented in Table S14. Nine out of the top ten predicted drugs have been previously reported as effective for GC treatment. For example, Tegafur-uracil was used as an adjuvant therapy for GC as early as 1997 [34]. Amlodipine was found to have the potential as a targeted therapy for GC in 2021 because it can significantly reduce the number of tumor spheres [35]. The anticancer effect of Chloroquine may be due to its inhibitory effect on autophagy, which enhances the efficacy of anticancer drugs in treating tumors [36].
5. Discussion
With the rapid development of life science and medicine, the scientific literature began to grow exponentially, which provided masses of information and offered unprecedented opportunities for gastric cancer diagnosis and treatment. However, the complexity and diversity of scientific texts pose challenges for automated information extraction and knowledge representation.
To address these challenges, this study first employed a multi-task learning framework for KG construction with hard parameter sharing, which consists of three modules, including MT-BGIPN, MT-SGTF, and MT-ScBERT. By sharing feature information among subtasks, performance has been effectively improved (MT-BBIPN: F1 score = 0.854, MT-SGTF: Hits@5 = 0.969, MT-ScBERT: F1 score = 0.920) with fewer parameters and in less time. The MKG framework was then applied in GC knowledge graph MKG-GC construction and drug discovery. The MKG-GC encompasses a total of 9129 entities and 88,482 triples. In drug discovery, a biomedical knowledge embedding pre-trained language model named BioKGE-BERT was built to map triples in MKG-GC to a low-dimensional vector space to obtain embedding vectors. A drug-disease discrimination model DDDM based on CNN-BiLSTM was then used to predict candidate gastric cancer drugs. Among the top ten predicted drugs, nine have been validated by existing literature to have a beneficial effect on gastric cancer treatment, confirming the clinical value of the MKG-GC. Finally, an online platform was developed for exploration and visualization. It is accessible free for academic research purposes at https://www.yanglab-mi.org.cn/MKG-GC/.
There are, however, some potential limitations to the current MKG framework. First, in the entity recognition task, the proposed model achieves accurate identification of long complex medical entities by focusing on the boundary information of the entities. However, recognizing nested entities from text remains a challenge. In the future, new decoding layers should be built to better identify nested medical entities. Second, there is conflicting information among different literature papers, and certain knowledge is only valid in specific contexts, which requires the assistance of experts to establish rules or perform multi-level verification for resolution. Finally, the current version of the MKG-GC mainly relies on biomedical literature, which has a single source and limited data. In the future, updated versions of the MKG-GC will have to be complemented by incorporating other information sources, such as medical databases and clinical electronic medical records, to expand the MKG-GC into a larger and more comprehensive medical knowledge base for GC.
In conclusion, the MKG can be described as an open and robust framework for KG construction and personalized application to GC. It will enhance the reasoning ability of the KG, assist in diagnosis and treatment, and promote medical research on GC.
CRediT authorship contribution statement
Fuliang Qian: Funding acquisition, Supervision. Wenying Yan: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Yang Yang: Methodology, Software, Visualization, Writing – original draft, Writing – review & editing, Project administration, Validation. Yuwei Lu: Methodology, Software, Visualization, Writing – original draft, Formal analysis, Validation. Zixuan Zhen: Data curation, Investigation, Resources, Software. Hao Wu: Data curation, Investigation, Resources. Yuxin Lin: Data curation, Investigation.
Declaration of Competing Interest
None.
Acknowledgements
This work was supported by the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJA520010); The Medical and Health Science and Technology Innovation Project of Suzhou (SKY2022010, SKYD2022097); The National Natural Science Foundation of China (82272439, 32200533); Foundation of Suzhou Medical College of Soochow University (MP13405423, MX13401423); Collaborative Innovation Center of Novel Software Technology and Industrialization at Soochow University; Priority Academic Program Development of Jiangsu Higher Education Institutions. We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.03.021.
Contributor Information
Fuliang Qian, Email: flqian@suda.edu.cn.
Wenying Yan, Email: wyyan@suda.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y., Lu Y., Yan W. A comprehensive review on knowledge graphs for complex diseases. Brief Bioinforma. 2022;24 doi: 10.1093/bib/bbac543. [DOI] [PubMed] [Google Scholar]
- 3.Li N., Yang Z., Luo L., Wang L., Zhang Y., Lin H., et al. KGHC: a knowledge graph for hepatocellular carcinoma. BMC Med Inf Decis Mak. 2020;20:135. doi: 10.1186/s12911-020-1112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiu X., Qian Q., Wu S. Construction of a digestive system tumor knowledge graph based on chinese electronic medical records: development and usability study. JMIR Med Inform. 2020;8 doi: 10.2196/18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., Ma X., Si J., Tang H., Wang H., Li T., et al. Adverse drug reaction discovery using a tumor-biomarker knowledge graph. Front Genet. 2020;11 doi: 10.3389/fgene.2020.625659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019;36:1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F., Shareghi E., Meng Z., Basaldella M., Collier N. Association for Computational Linguistics; 2021. Self-Alignment Pretraining for Biomedical Entity Representations; pp. 4228–4238. (Online) (Online) [Google Scholar]
- 8.Smith L., Tanabe L.K., Ando R.J. nee, Kuo C.-J., Chung I.-F., Hsu C.-N., et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;9:1–19. doi: 10.1186/gb-2008-9-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krallinger M., Rabal O., Leitner F., Vazquez M., Salgado D., Lu Z., et al. The CHEMDNER corpus of chemicals and drugs and its annotation principles. J Chemin- 2015;7 doi: 10.1186/1758-2946-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.N. Collier, J.-D. Kim, Introduction to the Bio-entity Recognition Task at JNLPBA, International Joint Workshop on Natural Language Processing in Biomedicine and its Applications COLING, Geneva, Switzerland, 2004, pp. 73–78.
- 11.Dogan R.I., Leaman R., Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inf. 2014;47:1–10. doi: 10.1016/j.jbi.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerner M., Nenadic G., Bergman C.M. LINNAEUS: a species name identification system for biomedical literature. Bmc Bioinforma. 2010;11:85. doi: 10.1186/1471-2105-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. Li, Y. Sun, R.J. Johnson, D. Sciaky, C.-H. Wei, R. Leaman, A.P. Davis, C.J. Mattingly, T.C. Wiegers, Z. Lu, BioCreative V CDR task corpus: a resource for chemical disease relation extraction, Database, 2016 (2016). [DOI] [PMC free article] [PubMed]
- 14.Krallinger M., Rabal O., Akhondi S.A., Pérez M.P., Santamaría J., Rodríguez G.P., et al. Overview of the BioCreative VI chemical-protein interaction Track. Proc Sixth BioCreative Chall Eval Workshop. 2017:141–146. [Google Scholar]
- 15.Henry S., Buchan K., Filannino M., Stubbs A., Uzuner O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inf Assoc. 2020;27:3–12. doi: 10.1093/jamia/ocz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Mulligen E.M., Fourrier-Reglat A., Gurwitz D., Molokhia M., Nieto A., Trifiro G., et al. The EU-ADR corpus: annotated drugs, diseases, targets, and their relationships. J Biomed Inf. 2012;45:879–884. doi: 10.1016/j.jbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Herrero-Zazo M., Segura-Bedmar I., Martinez P., Declerck T. The DDI corpus: an annotated corpus with pharmacological substances and drug-drug interactions. J Biomed Inf. 2013;46:914–920. doi: 10.1016/j.jbi.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 18.A. Miranda, F. Mehryary, J. Luoma, S. Pyysalo, A. Valencia, M. Krallinger, Overview of DrugProt BioCreative VII track: quality evaluation and large scale text mining of drug-gene/protein relations, BioCreative VII challenge and workshopCecilia Arighi, USA, 2021.
- 19.Bravo A., Pinero J., Queralt-Rosinach N., Rautschka M., Furlong L.I. Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC Bioinforma. 2015;16:55. doi: 10.1186/s12859-015-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nédellec C. ACM-Association for Computing Machinery,; 2005. Learning language in logic-genic interaction extraction challenge, 4. Learning language in logic workshop (LLL05) [Google Scholar]
- 21.Pyysalo S., Airola A., Heimonen J., Björne J., Ginter F., Salakoski T. Comparative analysis of five protein-protein interaction corpora. BMC Bioinforma. 2008;9 doi: 10.1186/1471-2105-9-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordes A., Usunier N., Garcia-Duran A., Weston J., Yakhnenko O. Neural Information Processing Systems (NIPS); South Lake Tahoe, United States: 2013. Translating Embeddings for Modeling Multi-relational Data; pp. 1–9. [Google Scholar]
- 23.E. Loper, S. Bird, NLTK: the Natural Language Toolkit, Proceedings of the ACL-02 Workshop on Effective tools and methodologies for teaching natural language processing and computational linguistics - Volume 1, Association for Computational Linguistics, Philadelphia, Pennsylvania, 2002, pp. 63–70.
- 24.Sung M., Jeong M., Choi Y., Kim D., Lee J., Kang J. BERN2: an advanced neural biomedical named entity recognition and normalization tool. Bioinformatics. 2022;38:4837–4839. doi: 10.1093/bioinformatics/btac598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Zhang Y., Ren X., Zhang Y., Zitnik M., Shang J., et al. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics. 2018;35:1745–1752. doi: 10.1093/bioinformatics/bty869. [DOI] [PubMed] [Google Scholar]
- 26.Wei C.-H., Allot A., Leaman R., Lu Z. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 2019;47:W587–W593. doi: 10.1093/nar/gkz389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L. Baldini Soares, N. FitzGerald, J. Ling, T. Kwiatkowski, Matching the Blanks: Distributional Similarity for Relation Learning, Association for Computational Linguistics, Florence, Italy, 2019, pp. 2895–2905.
- 28.S. Wu, Y. He, Enriching Pre-trained Language Model with Entity Information for Relation Classification, Proceedings of the 28th ACM International Conference on Information and Knowledge Management, Association for Computing Machinery, 2019, pp. 2361–2364.
- 29.Nickel M., Rosasco L., Poggio T. Holographic embeddings of knowledge graphs. Proc AAAI Conf Artif Intell. 2016;30 [Google Scholar]
- 30.B. Yang, W.-t Yih, X. He, J. Gao, L. Deng, Embedding Entities and Relations for Learning and Inference in Knowledge Bases, International Conference on Learning Representations 2014, pp. arXiv:1412.6575.
- 31.L. Yao, C. Mao, Y. Luo, KG-BERT: BERT for Knowledge Graph Completion, (2019) arXiv:1909.03193.
- 32.B. Wang, T. Shen, G. Long, T. Zhou, Y. Wang, Y. Chang, Structure-Augmented Text Representation Learning for Efficient Knowledge Graph Completion, Proceedings of the Web Conference 2021, Association for Computing Machinery, Ljubljana, Slovenia, 2021, pp. 1737–1748.
- 33.D. Li, S. Yang, K. Xu, M. Yi, Y. He, H. Wang, Multi-task Pre-training Language Model for Semantic Network Completion, (2022) arXiv:2201.04843.
- 34.Yen H.H., Chen C.N., Yeh C.C., Lai I.R. Adjuvant tegafur-uracil (UFT) or S-1 monotherapy for advanced gastric cancer: a single center experience. World J Surg Oncol. 2021;19:124. doi: 10.1186/s12957-021-02233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiozaki A., Katsurahara K., Kudou M., Shimizu H., Kosuga T., Ito H., et al. Amlodipine and verapamil, voltage-gated Ca(2+) channel inhibitors, suppressed the growth of gastric cancer stem cells. Ann Surg Oncol. 2021;28:5400–5411. doi: 10.1245/s10434-021-09645-0. [DOI] [PubMed] [Google Scholar]
- 36.Ke X., Qin Q., Deng T., Liao Y., Gao S.J. Heterogeneous responses of gastric cancer cell lines to tenovin-6 and synergistic effect with chloroquine. Cancers (Basel) 2020;12 doi: 10.3390/cancers12020365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Source data and code for MKG-GC construction and application are available at the GitHub repository (https://github.com/KeDaCoYa/MKG-GC).