Abstract
Genetic and biologic observations suggest that pigs may serve as “mixing vessels” for the generation of human-avian influenza A virus reassortants, similar to those responsible for the 1957 and 1968 pandemics. Here we demonstrate a structural basis for this hypothesis. Cell surface receptors for both human and avian influenza viruses were identified in the pig trachea, providing a milieu conducive to viral replication and genetic reassortment. Surprisingly, with continued replication, some avian-like swine viruses acquired the ability to recognize human virus receptors, raising the possibility of their direct transmission to human populations. These findings help to explain the emergence of pandemic influenza viruses and support the need for continued surveillance of swine for viruses carrying avian virus genes.
This century has seen the emergence of four pandemic strains of human influenza virus: the Spanish influenza virus of 1918, the Asian influenza virus of 1957, the Hong Kong influenza virus of 1968, and the Russian influenza virus of 1977. The pandemic viruses isolated in 1957 and 1968 were reassortants possessing genes of both human and avian viruses. Avian virus genes encoding the hemagglutinin (HA), neuraminidase, and PB1 proteins were introduced into the 1957 strain, while the 1968 strain acquired only the avian virus HA and PB1 genes (20, 27, 37).
Avian influenza viruses replicate less efficiently in humans (1) and in other primates (more than 100-fold) (29). Although immunity to currently circulating human H1 and H3 viruses may explain the poor replication of these subtypes of avian viruses in human hosts, it does not account for the limited replication of avian viruses with other subtypes not found in humans (i.e., H4, H6, H9, and H10) (1). Similarly, human viruses do not replicate efficiently in waterfowl when introduced by natural routes (18). Therefore, although avian influenza viruses can be directly transmitted to humans as indicated by a recent incident in Hong Kong (9, 39), the probability of these viruses becoming established in human populations is low, thereby limiting opportunities for the generation of human-avian reassortant viruses in these host animals. How, then, does one explain the apparent breach of this host range restriction during the generation of pandemic influenza viruses? Scholtissek et al. (36) proposed that pigs may serve as “mixing vessels” for the production of reassortant influenza viruses. Indeed, a variety of avian and human influenza viruses replicate efficiently in pigs upon experimental infection (17, 23). Results of phylogenetic and epidemiologic analyses indicate that avian and human viruses have also been transmitted to pigs in nature (12, 38) and that they have reassorted in pigs (7) and been transmitted to humans (8). Despite this considerable body of circumstantial evidence, the molecular basis for human-avian viral gene reassortment in pigs remains unknown.
Although influenza A viruses uniformly recognize cell surface oligosaccharides with a terminal sialic acid, their receptor specificity varies. Most avian influenza viruses preferentially bind to the N-acetylneuraminic acid-α2,3-galactose (NeuAcα2,3Gal) linkage on sialyloligosaccharides, while human influenza viruses prefer the NeuAcα2,6Gal linkage (32, 33). However, very little is known about the sialyloligosaccharide structures at viral replication sites in pigs or in other common hosts. Since the presence or absence of viral species-specific receptors on host cells would have a key role in generating viruses with pandemic potential, we sought to identify the types of sialyloligosaccharides at the replication sites of influenza viruses in ducks and pigs and to test their suitability for binding various avian and swine viruses. The results we report help to clarify how avian influenza A viruses could overcome normal host range barriers and infect human populations.
MATERIALS AND METHODS
Viruses.
The influenza A viruses used in this study (Table 1) were maintained in repositories at St. Jude Children’s Research Hospital in Memphis, Tenn., the Istituto Superiore di Sanita in Rome, Italy, and the Graduate School of Veterinary Medicine of Hokkaido University in Sapporo, Japan.
TABLE 1.
Receptor specificities of H1N1 influenza A virusesa
| Virus | Hemagglutination titer by use of erythrocytes containing:b
|
||
|---|---|---|---|
| Native linkage | NeuAcα2,3Gal | NeuAcα2,6Gal | |
| Human | |||
| A/USSR/77 | 512 | 0 | 512 |
| A/Lackland/78 | 128 | 0 | 64 |
| A/Brazil/78 | 256 | 0 | 16 |
| A/Baylor/3724/81 | 64 | 0 | 64 |
| A/Chile/83 | 128 | 0 | 64 |
| A/Taiwan/1/86 | 64 | 0 | 64 |
| A/Singapore/6/86 | 256 | 0 | 128 |
| Duck | |||
| A/duck/Alberta/35/76 | 1,024 | 2,048 | 0 |
| A/duck/Bavaria/1/77 | 32 | 32 | 0 |
| A/mallard/Alberta/740/80 | 128 | 128 | 128 |
| A/mallard/Tennessee/11464/85 | 16 | 16 | 0 |
| Swine | |||
| Classic | |||
| A/swine/Iowa/15/30 | 16 | 0 | 8 |
| A/swine/Wisconsin/1/61 | 128 | 0 | 64 |
| A/swine/Hokkaido/2/81 | 64 | 0 | 32 |
| Avian-like | |||
| A/swine/Belgium/1/79 | 128 | 128 | 128 |
| A/swine/Finland/55/80 | 256 | 128 | 128 |
| A/swine/Netherlands/3/80 | 256 | 64 | 32 |
| A/swine/Belgium/1/83 | 1,024 | 512 | 512 |
| A/swine/Italy-Vir/383/84 | 8 | 4 | 8 |
| A/swine/Italy-Vir/492/85 | 32 | 0 | 64 |
| A/swine/Italy-Vir/547/85 | 64 | 0 | 64 |
| A/swine/Netherlands/12/85 | 32 | 0 | 2 |
| A/swine/Italy-Vir/671/87 | 16 | 0 | 8 |
Erythrocyte preparations included unmodified cells (native linkage) and sialidase-treated cells which were enzymatically resialylated, as described earlier (32, 33), so that they contained the terminal NeuAcα2,3Gal or the NeuAcα2,6Gal linkage on cell surface oligosaccharides. Native (unmodified) and asialo (sialidase-treated) cells were used as controls. Although the amounts of incorporated sialic acid varied among the erythrocyte preparations used, they remained within the previously reported ranges: NeuAcα2,3Gal, 98 to 164 nmol/ml; NeuAcα2,6Gal, 20 to 50 nmol/ml (10, 32, 33).
Hemagglutination titers are expressed as the reciprocal of the maximal dilution required to give complete agglutination (0 = ≤2). All virus samples presented hemagglutination titers of ≤2 when asialo cells were used (data not shown).
Nucleotide sequencing.
Molecular cloning, nucleotide sequencing, and nucleotide analysis were performed as previously described (20).
Receptor specificity analysis.
The receptor specificity of viruses was determined by hemagglutination tests, using derivatized erythrocytes at room temperature (32). In this assay, the titration of end points was performed after 30 min and 1 h, with essentially no differences in results.
Immunologic detection of NeuAcα2,3Gal and NeuAcα2,6Gal linkages.
The colons of 4-week-old F1 hybrid ducks (Pekin, Anas platyrhynchos domesticos, and mallard, Anas platyrhynchos platyrhyncos) and the tracheas of 70-day-old F1 hybrid pigs (Landrace and Durock lines) were collected immediately after exsanguination by cardiac puncture, rinsed with phosphate-buffered saline (PBS; pH 7.2), and cut into cubes (3 mm3 each). The tissue blocks were then embedded in OCT compound (Miles Inc., Indianapolis, Ind.) and frozen in liquid nitrogen. Tissue sections (6 μm each) were cut from the blocks with a microtome cryostat, air dried, and fixed for 15 min with cold acetone before immunostaining. The presence or absence of NeuAcα2,3Gal and NeuAcα2,6Gal linkages in these organs was determined with linkage-specific lectins (glycan differentiation kit; Boehringer Mannheim Biochemicals). Briefly, each section was incubated with 50 μl of digoxigenin (DIG)-labeled Sambucus nigra (1 μg/ml; specific for NeuAcα2,6Gal) or Maackia amurensis (5 μg/ml; specific for NeuAcα2,3Gal) agglutinin for 1 h at room temperature. After three washes with cold PBS, the S. nigra and M. amurensis agglutinin-exposed sections were incubated with fluorescein- and rhodamine-conjugated anti-DIG antibody (Boehringer Mannheim Biochemicals), respectively, for 1 h at room temperature. After three washes with cold PBS, the tissue sections were mounted on slides in buffered glycerol (pH 9.0) for observation with a fluorescence microscope (BH-RFL; Olympus Optics, Tokyo, Japan) equipped with a dark-field condenser and UV excitation capability. Control slides were incubated with PBS instead of lectin.
RESULTS
Pig trachea contains receptors for both avian and human viruses.
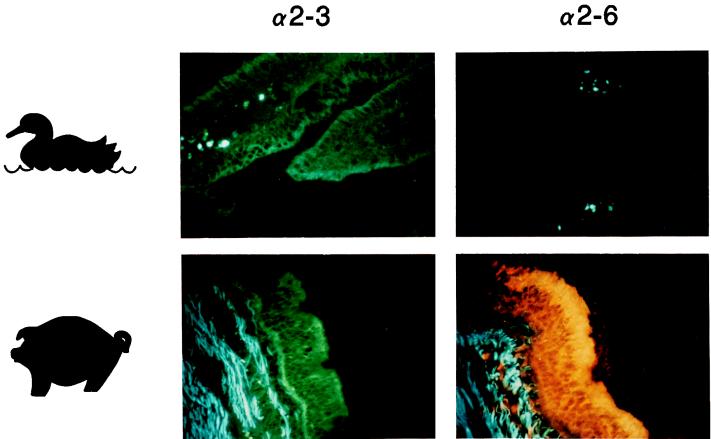
Because avian and human influenza viruses show different receptor preferences, NeuAcα2,3Gal versus NeuAcα2,6Gal (32, 33), and epithelial cells lining human trachea possess mainly the latter sialyloligosaccharide (11), we considered that one of the determinants of the replicative potential of such viruses in different hosts might be the sialic acid-galactose linkage present on sialyloligosaccharides. Using linkage-specific lectins, we examined the epithelial cells of duck intestine and pig trachea for NeuAcα2,3Gal and NeuAcα2,6Gal. Duck intestine was chosen for these experiments because avian influenza viruses replicate well in this site and only marginally in duck trachea (24). Duck intestine showed a strong reaction with M. amurensis lectin (specific for NeuAcα2,3Gal) but not with S. nigra lectin (specific for NeuAcα2,6Gal), whereas pig trachea reacted with both linkages (Fig. 1). These results, together with the findings of Couceiro et al. (11), using the same lectin method, that ciliated epithelial cells in human trachea contain NeuAcα2,6Gal but not NeuAcα2,3Gal, suggest that the lack of a suitable receptor accounts for the inefficient replication of human viruses in duck intestine and of avian viruses in humans. Moreover, detection of both linkages in pig trachea provides a molecular basis for efficient replication of human and avian influenza viruses in this host (17, 23).
FIG. 1.
Comparison of lectin staining in duck intestine (colon) and pig trachea. The M. amurensis lectin specific for NeuAcα2,3Gal (designated α2,3; detected with fluorescein isothiocyanate-labeled anti-DIG antibody) bound to both duck intestinal epithelium and pig tracheal epithelium, whereas S. nigra lectin specific for NeuAcα2,6Gal (designated α2,6; detected with rhodamine-labeled anti-DIG antibody) bound only to the latter. Blue staining in the connective tissue represents autofluorescence. Magnification, ×300.
Changes in receptor specificity during the adaptation of avian viruses to pigs.
Three types of influenza viruses are circulating in pigs: classic H1N1, maintained in this species for more than 60 years; human-like H3N2, present in pigs since 1969 (26); and avian-like H1N1, introduced into European pigs in 1979 (35). Since pig trachea contains both the NeuAcα2,6Gal and the NeuAcα2,3Gal linkages (Fig. 1), we were uncertain whether the avian-like H1N1 virus had retained its preference for NeuAcα2,3Gal after the parental avian strain was introduced into the pig population. A receptor specificity analysis indicated that all of the human and classic swine viruses preferentially recognize NeuAcα2,6Gal, whereas most avian viruses prefer NeuAcα2,3Gal (Table 1), as previously reported (32, 33). One avian virus, A/mallard/Alberta/740/80, recognized both NeuAcα2,3Gal and NeuAcα2,6Gal. Other avian viruses with dual receptor specificities have been reported (14, 32). Surprisingly, the avian-like swine viruses showed a shift in receptor specificity over time. Viruses isolated from European pigs up to 1984 recognized both sialic acid-galactose linkages, whereas those isolated after 1985 recognized only NeuA cα2,6Gal (Table 1). A/Swine/Netherlands/12/85 recognized NeuAcα2,6Gal-containing erythrocytes appreciably less efficiently than those with native linkages, for unknown reasons. Nonetheless, the shift in receptor specificity suggests a mechanism that would allow avian viruses to replicate in humans efficiently.
Amino acid residues responsible for the shift in receptor specificity among avian-like swine viruses.
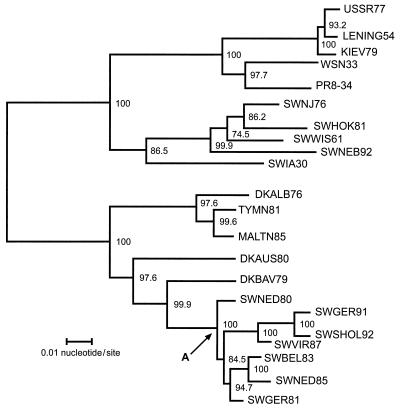
To understand the receptor specificity conversion that occurred in avian-like swine viruses at some point between 1984 and 1985, we first determined the phylogenetic relationships among the H1 HAs (Fig. 2). The analysis established that the HA of avian-like swine viruses was introduced from birds once and then evolved thereafter, as did other genes of these viruses (6). Comparison of the amino acid sequences of the HA molecules (Fig. 3 and Table 2) showed that an amino acid change at residue 142 (145 in the H3 numbering system) was the only substitution that occurred between 1983 and 1985 and was associated with loss of NeuAcα2,3Gal recognition. Avian-like swine viruses isolated in 1985 or later (A/swine/Netherlands/12/85, A/swine/Italy- Vir/671/87, A/swine/Germany/3/91, and A/swine/Schleswig- Holstein/1/92) contained Leu at this position, whereas those isolated earlier had different amino acids: A/swine/Arnsberg/79 and A/swine/Netherlands/80, Ser; A/swine/Germany/2/81, His; and A/swine/Belgium/83, Arg. Residue 142 (145 in the H3 numbering system) is located on the loop of the HA near the receptor-binding pocket (Fig. 4), supporting our contention that a mutation at this position may have contributed to a shift in receptor specificity.
FIG. 2.
Phylogenetic tree of influenza A virus H1 HA genes. Nucleotide residues 1 to 1731 of each H1 HA were analyzed by the neighbor-joining method (34). Horizontal distances are proportional to the minimum number of nucleotide differences required to join nodes and H1 HA sequences. Vertical lines are for spacing branches and labels. Bootstrap values (1,000 replications) are presented for each node. The node shown by the arrow at A indicates the hypothetical introductory virus in pigs that originated from birds. The HA nucleotide sequences represent USSR77 (A/USSR/90/77), KIEV79 (A/Kiev/59/79), SWHOK81 (A/swine/Hokkaido/2/81), SWWIS61 (A/swine/Wisconsin/1/61), SWIW30 (A/swine/Iowa/15/30), DKALB (A/duck/Alberta/35/76), TYMN81 (A/turkey/Minnesota/1661/81), MALTN85 (A/mallard/Tennessee/11464/85), DKAUS80 (A/duck/Australia/749/80), DKBAV (A/duck/Bavaria/1/77), SWNED80 (A/swine/Netherlands/3/80), SWGER91 (A/swine/Germany/8533/91), SWSHOL (A/swine/Schleswig-Holstein/1/92), SWVIR (A/swine/Italy-Vir/671/87), SWBEL83 (A/swine/Belgium/1/83), SWNED85 (A/swine/Netherlands/12/85), and SWGER81 (A/swine/Germany/2/81) from this study; LENING54 (A/Leningrad/1/54), reported by Beklemishev et al. (2); WSN33 (A/WSN/33), reported by Hiti et al. (19); PR8-34 (A/Puerto Rico/8/34), reported by Winter et al. (41); SWNJ76 (A/swine/New Jersey/11/76), reported by Both et al. (5); and SWNEB92 (A/swine/Nebraska/1/92) reported by Olsen et al. (30).
FIG. 3.
The HA1 sequences of avian and European avian-like swine influenza viruses. The HA1 portions, which form the receptor-binding sites, are compared, using the SWARN79 sequence as a baseline. Abbreviations are given in the legend to Fig. 2.
TABLE 2.
HA variability among European avian-like swine influenza viruses
| Strain | Amino acid position in the HA1a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 96 | 121 | 132 | 138 | 142 | 207 | 267 | 311 | |
| Sw/Arnsberg/6554/79b | I | N | V | Y | S | N | T | K |
| Sw/Netherlands/3/80c | I | N | V | Y | S | N | T | K |
| Sw/Germany/2/81b | I | N | V | Y | H | N | T | K |
| Sw/Belgium/1/83c | I | N | I | Y | R | N | T | K |
| Sw/Netherlands/12/85c | I | N | I | Y | L | N | T | K |
| Sw/Italy-Vir/671/87c | A | T | A | H | L | Y | M | Q |
| Sw/Germany/3/91b | A | T | V | H | L | Y | M | Q |
| Sw/Schleswig-Holstein/1/92b | A | T | A | H | L | Y | M | Q |
Only those positions in the HA1 at which more than three strains contain an amino acid change are shown.
Based on nucleotide sequence obtained from GenBank.
Determined as described in Materials and Methods.
FIG. 4.
Globular head of the influenza virus HA molecule (ribbon, cyan), illustrating the location of Ser145 (space-filled, purple) relative to that of bound sialic acid (ball and stick, red). Inset shows the entire molecule. This figure is based on the H3 HA structure of A/Aichi/2/68 (H3N2) complexed with sialic acid as determined by Weis et al. (40) and is not intended to represent the actual three-dimensional structure of the H1 molecule.
DISCUSSION
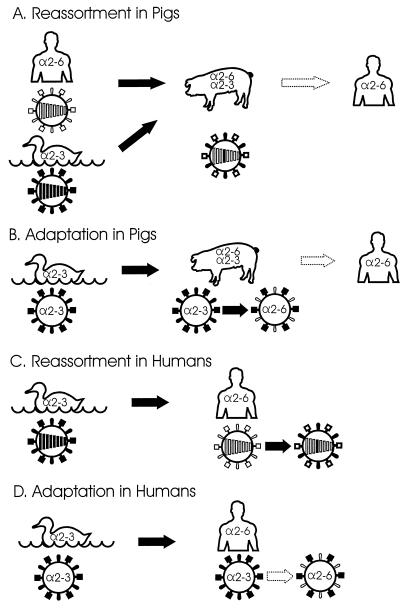
We have demonstrated that pig trachea contains receptors for both human and avian influenza viruses, providing a milieu conducive to the replication, and hence the genetic reassortment, of these viruses. Moreover, with continued replication in pigs, the avian viruses appear to undergo a shift in their receptor specificity to NeuAcα2,6Gal linkages exclusively. These observations suggest at least two mechanisms, both dependent on HA-receptor interactions, that would permit pigs to serve as intermediate hosts for the generation of pandemic influenza viruses (Fig. 5). In one, avian and human viruses would reassort in classical fashion, giving rise to a hybrid strain with pandemic potential (Fig. 5A). In the other, an avian virus would acquire the ability to bind efficiently to human cell surface receptors so that it could be readily transmitted to a human host without a requirement for genetic recombination (Fig. 5B). These models may not be mutually exclusive. Quite possibly, an avian virus could combine with a human virus before or after becoming adapted to the NeuAcα2,6Gal linkage, resulting in a reassortant with enhanced proliferative capacity.
FIG. 5.
Models for the generation of pandemic influenza virus strains in pigs. (A) In the classical genetic reassortment model, avian and human viruses bind, respectively, to NeuAcα2,3Gal and NeuAcα2,6Gal (α2,3 and α2,6) linkages in pig trachea, setting the stage for the emergence of a reassortant that infects a large fraction of the human population. The segments in the center of each particle represent the viral genome. The reassortant HA gene (black) is derived from an avian virus. (B) In this adaptation model, avian viruses acquire the ability to replicate efficiently in humans during adaptation to the NeuAcα2,6Gal linkage in pigs. This change is mediated by a mutation in the HA gene. (C) Alternatively, an avian influenza virus is transmitted directly to humans where it reassorts with a human virus, or (D) it acquires the ability to recognize the NeuAcα2,6Gal linkage after direct introduction from birds, leading to efficient replication in humans.
Circumstantial evidence (7, 8, 36) favors the involvement of pigs in genetic reassortment; however, direct transmission of avian viruses to humans does occur, as exemplified by the recent incident in Hong Kong (9, 39), so that reassortment in humans (Fig. 5C) or adaptation to recognize receptors in humans (Fig. 5D) cannot be discounted. With the rarity of human influenza pandemics, it is difficult to predict which of the models in Fig. 5 is more likely to generate a potentially hazardous virus. In addition to the Hong Kong H5N1 outbreak, avian viruses have been transmitted directly from birds to a variety of mammals, including seals, mink, and horses (13, 16, 25).
What types of selective pressure drive the shift in receptor specificity of avian-like swine influenza viruses from both NeuAcα2,3Gal and NeuAcα2,6Gal to NeuAcα2,6Gal exclusively? One possibility is that the NeuAcα2,6Gal linkage is more abundant than NeuAcα2,3Gal on the epithelial cells of pig trachea, thus providing a replicative advantage for viruses that recognize the former receptor. Alternatively, glycoproteins containing NeuAcα2,3Gal may exist at viral replication sites in pigs, serving as receptor analog inhibitors. Detection of the NeuAcα2,3Gal linkage in mucin from human trachea (11) supports this suggestion.
One avian virus in our study, A/mallard/Alberta/740/80, recognized both NeuAcα2,3Gal and NeuAcα2,6Gal, consistent with reports by others (10, 31, 32). In a natural setting, would avian viruses with this dual binding capacity be the ones transmitted to pigs? Although a number of avian viruses have been shown to replicate in pigs in experimental infection (17, 23), their receptor specificities are largely unknown. Of the avian viruses we have tested, most recognize only the NeuAcα2,3Gal linkage and those tested for replicative activity in pigs have multiplied efficiently, suggesting that a lack of affinity for the NeuAcα2,6Gal linkage would not preclude natural transmission of an avian virus to pigs.
A single substitution at position 145 (H3 numbering), located on the loop near the receptor-binding site of the HA molecule, appears responsible for the inability of avian-like swine viruses isolated after 1984 to bind to NeuAcα2,3Gal linkages. A mutation at the same position was also identified in variant human influenza A viruses during their adaptation in eggs (28); however, the receptor specificity change in the avian-like swine viruses described here is not likely an artifact of egg adaptation, as classic swine viruses such as A/swine/Iowa/15/30 (H1N1) have been passaged many times in eggs without loss of NeuAcα2,6Gal specificity (Table 1). Also, swine viruses directly isolated in Madin-Darby canine kidney (MDCK) cells or in eggs show identical receptor specificities (21a). The precise mechanism by which this mutation causes a shift in receptor specificity is unknown and will likely remain so until the crystal structure of the mutant HA is determined.
The molecular features of the HA that influence the virulence of avian influenza viruses in birds are well characterized (3, 4, 21, 22) and have been used successfully to predict whether an emerging influenza virus poses a threat to large concentrations of poultry. Progress in identifying viruses with pandemic potential in human populations has been less impressive. None of the avian-human reassortants identified since 1968 (7), or any of the avian-like swine viruses, has produced a global outbreak of influenza in humans. This suggests that the generation of pandemic strains of influenza A virus is a complex process, requiring critical combinations of avian and human viral genes or mutations beyond those affecting the HA molecule. Indeed, multiple genes affect the interspecies transmission of influenza viruses (18, 23, 36). One could also argue that immunity to H1 HAs in human populations would protect against infection by the H1N1 avian-like swine viruses. However, since human H1 viruses and avian-like swine viruses show appreciable differences in antigenicity (15), it will be necessary to evaluate the latter in primates before making conclusions as to their replicative capacity in humans.
Many new viral pathogens have emerged in humans (e.g., human immunodeficiency virus, influenza A viruses, Ebola virus, hantavirus, monkeypox virus, and Borna disease virus). Learning the precise molecular changes that allow these agents to cross host species barriers is essential to developing an effective means of prevention. The evidence we present supports the role of pigs as a source of potentially hazardous influenza A viruses, arising through classical genetic reassortment or a novel adaptation to human virus receptors or perhaps through both mechanisms. Thus, continued intensive monitoring of swine populations for avian-like influenza viruses should be an integral part of global health planning.
ACKNOWLEDGMENTS
We thank Krisna Wells for excellent technical assistance, Clayton Naeve and the St. Jude Children’s Research Hospital Molecular Resource Center for preparation of oligonucleotides, Patricia Eddy and the Molecular Biology Computer Facility for computer support, Yuko Kawaoka for illustration, and John Gilbert for scientific editing.
This work was supported by Public Health Service research grants AI-29680 and AI-33898 from the National Institute of Allergy and Infectious Diseases, Cancer Center Support (CORE) grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 2.Beklemishev A B, Blinov V M, Vasilenko S K, Golovin S I, Gutorov V V. Sintez polnorazmernoi DNK-kopii gena gemaggliutinina virusa grippa A H1N1-podtipa, ee klonirovanie i opredelenie pervichnoi struktury. Bioorg Khim. 1984;10:1535–1543. [PubMed] [Google Scholar]
- 3.Bosch F X, Garten W, Klenk H D, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 4.Bosch F X, Orlich M, Klenk H D, Rott R. The structure of the hemagglutinin, a determinant for the pathogenicity of influenza viruses. Virology. 1979;95:197–207. doi: 10.1016/0042-6822(79)90414-8. [DOI] [PubMed] [Google Scholar]
- 5.Both G W, Shi C H, Kilbourne E D. Hemagglutinin of swine influenza virus: a single amino acid change pleiotropically affects viral antigenicity and replication. Proc Natl Acad Sci USA. 1983;80:6996–7000. doi: 10.1073/pnas.80.22.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown I H, Ludwig S, Olsen C W, Hannoun C, Scholtissek C, Hinshaw V S, Harris P A, McCauley J W, Strong I, Alexander D J. Antigenic and genetic analyses of H1N1 influenza A viruses from European pigs. J Gen Virol. 1997;78:553–562. doi: 10.1099/0022-1317-78-3-553. [DOI] [PubMed] [Google Scholar]
- 7.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 8.Claas E C J, Kawaoka Y, De Jong J C, Masurel N, Webster R G. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology. 1994;204:453–457. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]
- 9.Claas E C J, Osterhaus A D M E, Van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 10.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 11.Couceiro J N S S, Paulson J C, Baum L G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 12.Gorman O T, Bean W J, Kawaoka Y, Donatelli I, Guo Y, Webster R G. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991;65:3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Wang M, Kawaoka Y, Gorman O, Ito T, Saito Y, Webster R G. Characterization of a new avian-like influenza A virus from horses in China. Virology. 1992;188:245–255. doi: 10.1016/0042-6822(92)90754-d. [DOI] [PubMed] [Google Scholar]
- 14.Higa H H, Rogers G N, Paulson J C. Influenza virus hemagglutinins differentiate between receptor determinants bearing N-acetyl-, N-glycollyl-, and N,O-diacetylneuraminic acids. Virology. 1985;144:279–282. doi: 10.1016/0042-6822(85)90325-3. [DOI] [PubMed] [Google Scholar]
- 15.Hinshaw V S, Alexander D J, Aymard M, Bachmann P A, Easterday B C, Hannoun C, Kida H, Lipkind M, Mackenzie J S, Nerome K, Schild G C, Scholtissek C, Senne D A, Shortridge K F, Skehel J J, Webster R G. Antigenic comparisons of swine-influenza-like H1N1 isolates from pigs, birds, and humans: an international collaborative study. Bull W H O. 1984;62:871–878. [PMC free article] [PubMed] [Google Scholar]
- 16.Hinshaw V S, Bean W J, Webster R G, Rehg J E, Fiorelli P, Early G, Geraci J R, St. Aubin D J. Are seals frequently infected with avian influenza viruses? J Virol. 1984;51:863–865. doi: 10.1128/jvi.51.3.863-865.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinshaw V S, Webster R G, Easterday B C, Bean W J. Replication of avian influenza A viruses in mammals. Infect Immun. 1981;34:354–361. doi: 10.1128/iai.34.2.354-361.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinshaw V S, Webster R G, Naeve C W, Murphy B R. Altered tissue tropism of human-avian reassortant influenza viruses. Virology. 1983;128:260–263. doi: 10.1016/0042-6822(83)90337-9. [DOI] [PubMed] [Google Scholar]
- 19.Hiti A, Davis A R, Nayak D P. Complete sequence analysis shows that the hemagglutinins of the H0 and H2 subtypes of human influenza are closely related. Virology. 1981;111:113–124. doi: 10.1016/0042-6822(81)90658-9. [DOI] [PubMed] [Google Scholar]
- 20.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaoka Y, Naeve C W, Webster R G. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology. 1984;139:303–316. doi: 10.1016/0042-6822(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 21a.Kawaoka, Y., and Y. Suzuki. Unpublished data.
- 22.Kawaoka Y, Webster R G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc Natl Acad Sci USA. 1988;85:324–328. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge K F, Kawaoka Y, Webster R G. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 24.Kida H, Yanagawa R, Matsuoka Y. Duck influenza lacking evidence of disease signs and immune response. Infect Immun. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klingeborn B, Englund L, Rott R, Juntti N, Rockborn G. An avian influenza A virus killing a mammalian species—the mink. Arch Virol. 1985;86:347–351. doi: 10.1007/BF01309839. [DOI] [PubMed] [Google Scholar]
- 26.Kundin W D. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature (London) 1970;228:857. doi: 10.1038/228857a0. [DOI] [PubMed] [Google Scholar]
- 27.Laver W G, Webster R G. Studies on the origin of pandemic influenza. III. Evidence implicating duck and equine influenza viruses as possible progenitors of the Hong Kong strain of human influenza. Virology. 1973;51:383–391. doi: 10.1016/0042-6822(73)90437-6. [DOI] [PubMed] [Google Scholar]
- 28.Meyer W J, Wood J M, Major D, Robertson J S, Webster R G, Katz J M. Influence of host cell-mediated variation on the international surveillance of influenza A (H3N2) viruses. Virology. 1993;196:130–137. doi: 10.1006/viro.1993.1461. [DOI] [PubMed] [Google Scholar]
- 29.Murphy B R, Sly D L, Tierney E L, Hosier N T, Massicot J G, London W T, Chanock R M, Webster R G, Hinshaw V S. Reassortant virus derived from avian and human influenza A viruses is attenuated and immunogenic in monkeys. Science. 1982;218:1330–1332. doi: 10.1126/science.6183749. [DOI] [PubMed] [Google Scholar]
- 30.Olsen C W, McGregor M W, Cooley A J, Schantz B, Hotze B, Hinshaw V S. Antigenic and genetic analysis of a recently isolated H1N1 swine influenza virus. Am J Vet Res. 1993;54:1630–1636. [PubMed] [Google Scholar]
- 31.Rogers G N, D’Souza B L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 32.Rogers G N, Paulson J C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 33.Rogers G N, Pritchett T J, Lane J L, Paulson J C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983;131:394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. Polymorphism and evolution of influenza A virus genes. Mol Biol Evol. 1986;3:57–74. doi: 10.1093/oxfordjournals.molbev.a040381. [DOI] [PubMed] [Google Scholar]
- 35.Scholtissek C, Burger H, Bachmann P A, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983;129:521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 36.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 37.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtype H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 38.Schultz U, Fitch W M, Ludwig S, Mandler J, Scholtissek C. Evolution of pig influenza viruses. Virology. 1991;183:61–73. doi: 10.1016/0042-6822(91)90118-u. [DOI] [PubMed] [Google Scholar]
- 39.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 40.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature (London) 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 41.Winter G, Fields S, Brownlee G G. Nucleotide sequence of the hemagglutinin gene of a human influenza virus H1 subtype. Nature (London) 1981;292:72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]