Abstract
Background
Nutrient availability is among the most widespread means by which environmental variability affects developmental outcomes. Because almost all cells within an individual organism share the same genome, structure-specific growth responses must result from changes in gene regulation. Earlier work suggested that histone deacetylases (HDACs) may serve as epigenetic regulators linking nutritional conditions to trait-specific development. Here we expand on this work by assessing the function of diverse HDACs in the structure-specific growth of both sex-shared and sex-specific traits including evolutionarily novel structures in the horned dung beetle Onthophagus taurus.
Results
We identified five HDAC members whose downregulation yielded highly variable mortality depending on which HDAC member was targeted. We then show that HDAC1, 3, and 4 operate in both a gene- and trait-specific manner in the regulation of nutrition-responsiveness of appendage size and shape. Specifically, HDAC 1, 3, or 4 knockdown diminished wing size similarly while leg development was differentially affected by RNAi targeting HDAC3 and HDAC4. In addition, depletion of HDAC3 transcript resulted in a more rounded shape of genitalia at the pupal stage and decreased the length of adult aedeagus across all body sizes. Most importantly, we find that HDAC3 and HDAC4 pattern the morphology and regulate the scaling of evolutionarily novel head and thoracic horns as a function of nutritional variation.
Conclusion
Collectively, our results suggest that both functional overlap and division of labor among HDAC members contribute to morphological diversification of both conventional and recently evolved appendages. More generally, our work raises the possibility that HDAC-mediated scaling relationships and their evolution may underpin morphological diversification within and across insect species broadly.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13227-024-00223-5.
Keywords: Onthophagus, Chromatin, Plasticity, Nutrition
Background
Phenotypic plasticity is the ability of an organism to change its phenotype in response to environmental stimuli [1, 2], a universal phenomenon in the living world. Diverse abiotic (e.g., temperature, photoperiod) and biotic (e.g., conspecific density) factors may influence growth and differentiation [3–5]. Among those, nutrition is one of the most widespread means by which environmental variability affects developmental outcomes. Lack of essential nutrients can slow or arrest development, and sometimes trigger alternative developmental programs, such as the diapause of insects and worms [6]. Nutrition also serves as a major determinant of animal size and shape [7, 8], with poor nutrition generally yielding reduced growth and final adult body size in animals with determinate growth such as insects and mammals. However, different body parts within an individual typically differ in their response to nutrient availability. For instance, brain size in mammals and male genital size in arthropods are relatively nutrition-insensitive [9, 10], whereas secondary sexual traits such as male horns of dung and rhinoceros beetles [11, 12], or mandibles of stag and broad-horned flour beetles [13, 14] are exquisitely sensitive to nutritional variation during development. Such trait-specific scaling relationships therefore contribute in important ways to shape morphological diversity within and among taxa.
Because all cells within an individual organism essentially share the same genome, organ- or structure-specific growth must result from changes in gene regulation. Much recent work has described changes in transcription profiles in response to environmental modifications such as nutrient availability, and has begun to identify key regulators of condition-responsive growth (e.g., insulin/IIS [15–17], doublesex [18], and hedgehog [19]). However, how such variation in gene expression is achieved in the first place, and then subsequently transduced into organ-specific growth is much less well understood. Here, we investigate the role of epigenetic modifications in enabling organ-specific growth, with a particular emphasis on histone modifications.
Histone acetylation/deacetylation is crucial in the organization of euchromatin (which enables transcription) and heterochromatin (which inhibits transcription), thereby mediating changes in gene expression. Histone deacetylases (HDACs) are members of an ancient enzyme family that reverses the acetylation of protein substrates. HDAC-mediated removal of acetylation from histone tail lysines generally correlates with gene silencing by decreasing the ability of transcription factors to access DNA [20]. In insects, studies have confirmed that HDACs play a role in various developmental processes, such as growth [21], metamorphosis [22–24], long-term memory [25], reproduction [26], longevity [27], caste differentiation [28, 29], diapause [30], and immunity [31]. Furthermore, in the broad-horned flour beetle Gnatocerus cornutus, HDAC1 and HDAC3 differentially participate in the nutrition-dependent growth of wings and male-exaggerated mandibles, suggesting that HDACs may serve as epigenetic regulators linking nutritional conditions to trait-specific development [21]. Here, we expand on this work by assessing the function of diverse HDACs in the structure-specific growth of both sex-shared and sex-specific traits including evolutionarily novel structures in a horned dung beetle.
Horned dung beetles (genus Onthophagus) have emerged as promising model systems to investigate the development and diversification of scaling relationships. Here we employ one such model, the bull-headed dung beetle O. taurus, to investigate the function of five different HDAC members in the development of four different morphological structures. We selected hind legs as examples of traits that exhibit moderate nutrition-responsiveness and therefore scale roughly isometrically with body size. We also investigated male genitalia because of their relatively muted nutrition response and corresponding hypoallometric scaling. Finally, we assessed thoracic horns and head horns because of their highly sex-specific growth and scaling relationships [32]. Thoracic horns are observed only in the pupal stage of both sexes where they function as molting devices in the shedding of the larval head capsule during the larval–pupal molt, and exhibit exaggerated growth in males [33, 34]. In partial contrast, head horns are found only in male pupae as well as adults and exhibit extreme nutrition-responsive growth resulting in hyper-allometric scaling with body size. Head horns function as weapons in competition between adult males over reproductive access to females. While thoracic horns have recently been identified as partial wing serial homologs [35], head horns lack any obvious homology with other structures and are thus considered evolutionary novelties even by the strictest of definitions [36, 37]. Below we detail our results and discuss them in the light of the developmental regulation of growth and plasticity in horned beetles in particular and insects broadly.
Results
We sought to characterize the presence and function of HDACs during the development of the bull-headed dung beetle O. taurus. We identified five HDACs in the annotated genome, which, based on phylogenetic analysis, could be classified into three classes: class I (HDAC1 and HDAC3), class II (HDAC4 and HDAC6), and class IV (HDAC11) (Additional file 1: Fig. S1). It is worth noting that two HDAC proteins that were predicted as HDAC-Rpd3 (reference number: XP_022902140.1) and HDAC5 (reference number: XP_022905538.1) for O. taurus in the NCBI database were found to be nested within the cluster containing HDAC1 and HDAC4 proteins (Additional file 1: Fig. S1). As a result, we have re-annotated these two proteins as Ot-HDAC1 and Ot-HDAC4, respectively. We then performed RNAi experiments by injecting double-stranded RNA (dsRNA) corresponding to each of the five HDACs into newly molted final-instar O. taurus larvae and assessed their influence on pupal and adult morphologies. Bioinformatic analyses indicated that the maximum number of identical sequences in loci other than the targeted genes did not exceed 11mers for HDAC1 dsRNA, 8mers for HDAC3 dsRNA, 14mers for HDAC4 dsRNA, 16mers for HDAC6 dsRNA, and 7mers for HDAC11 dsRNA, respectively, suggesting that off-target effects are an unlikely explanation for the phenotypes documented below though we cannot completely rule out the possibility of off-target effects. Wildtype morphology for each focal phenotype is shown in Fig. 1.
Fig. 1.
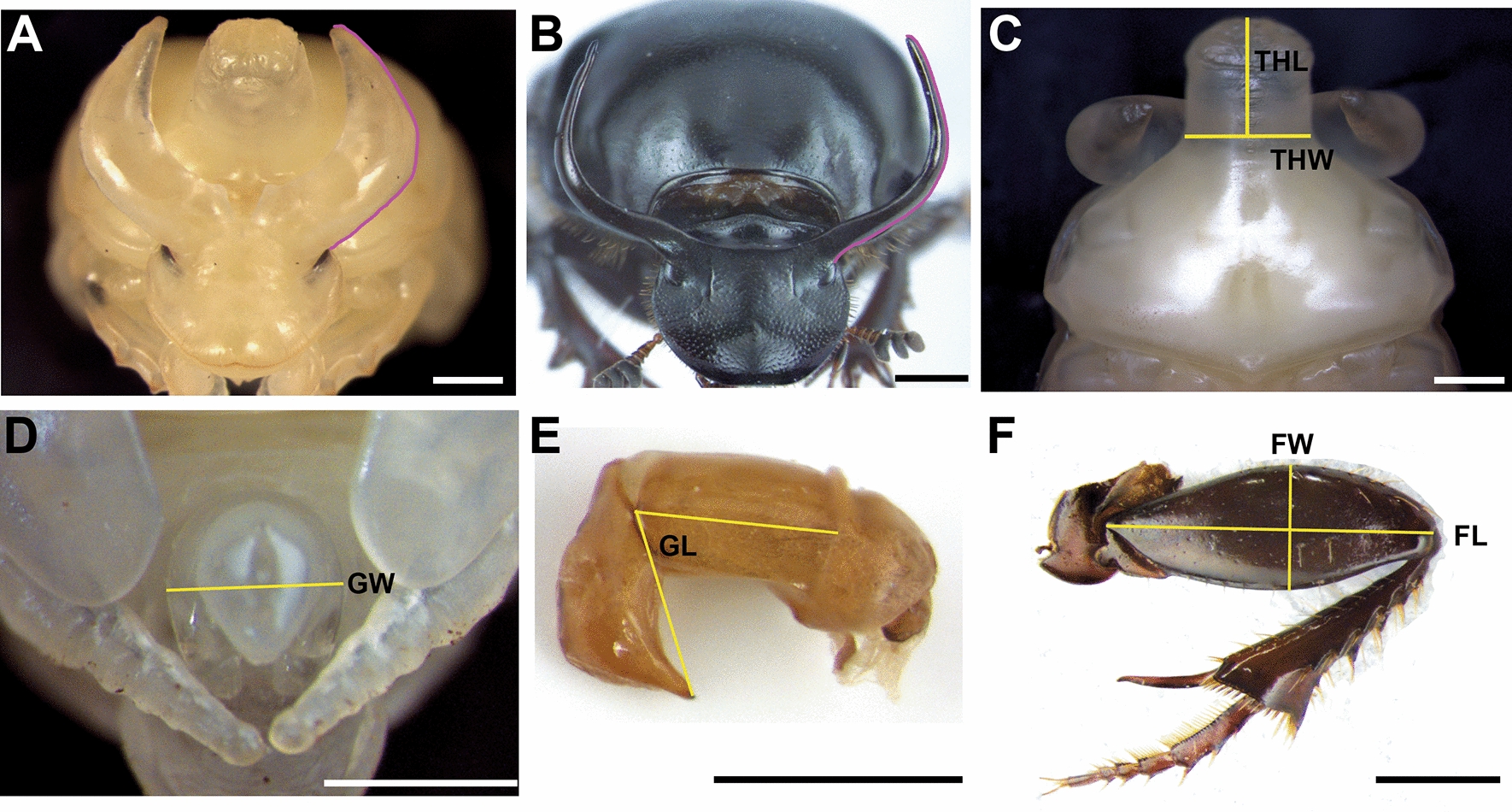
Wildtype morphology and morphometric landmarks used for morphological quantification. A–F Morphology of the male pupal head (A), male adult head (B), pupal pronotum from dorsal view (C), pupal reproductive organ (D), adult aedeagus (E), and hind leg (F), respectively, and the morphometric landmarks used for measurements (purple and yellow line). THW thoracic horn width, THL thoracic horn length, GW genital width, GL genital length, FL femur length, FW femur width. Scale bars: 1 mm
HDAC-RNAi resulted in highly variable mortality depending on the target HDAC
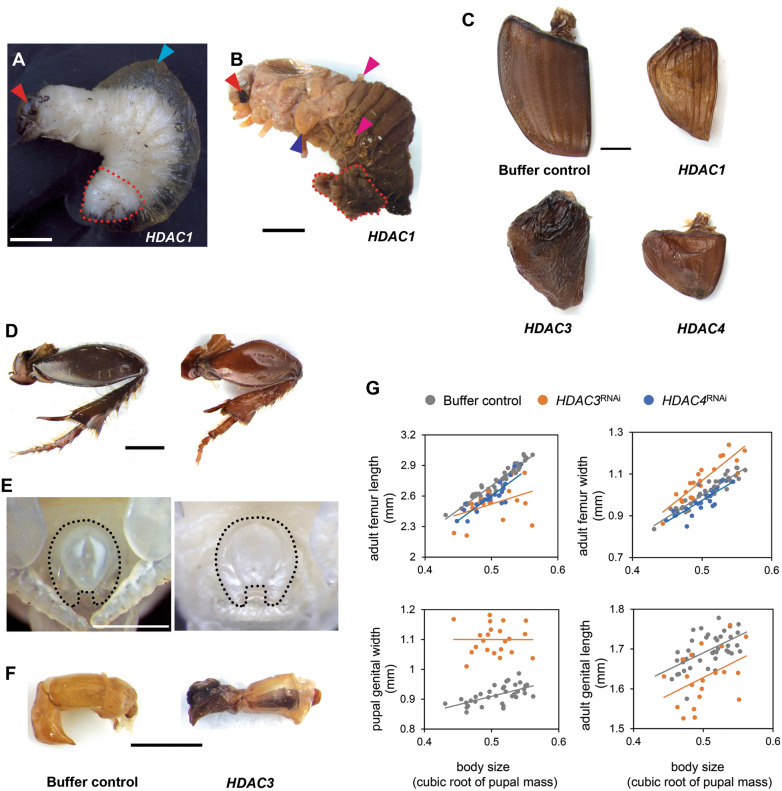
RNAi-mediated knockdown of HDAC1 resulted in 100% larval mortality at the initial 1 μg/μl dsRNA injection dosage, as well as subsequent dosages as low as 0.25 μg/μl (Additional file 1: Table S1). Most individuals exhibited molting defects at the prepupal stages and eventually died with pupal traits, such as compound eyes, observed underneath the larval integument (Fig. 2A). Pupa-specific features, such as wings and pupal support structures, became visible when the larval cuticle was carefully removed (Fig. 2B). A mass of fat body accumulated underneath the posterior region of the developing pupal abdomen, resulting in a cavity filled with hemolymph between the larval integument and newly formed pupal cuticle (Fig. 2A). When dsRNA concentration was decreased to 0.01 μg/μl, very few individuals succeeded to develop to pupal (3/44) and adult (2/44) stages (Additional file 1: Table S1) amenable to phenotyping. In marked contrast, RNAi targeting HDAC 3, 4, 6, or 11 resulted in mortalities ranging from 16.7% to 86.7% depending on dosage and permitted more nuanced and quantitative analysis of phenotypic effects (Additional file 1: Table S1). Because no observable phenotypes were found following HDAC6RNAi or HDAC11RNAi even at high dsRNA dosage, we focus on HDAC 1, 3, and 4 in the remainder.
Fig. 2.
HDACRNAi effects on molting and appendage development. A and B Larval–pupal intermediate induced by HDAC1 knockdown. The fat body accumulated outside of newly formed pupa is outlined (red dotted line) before (A) and after (B) peeling away the larval cuticle, respectively. Hemolymph in the cavity between larval and newly formed pupal cuticle, pupal compound eyes, wings, and pupal support structures are indicated by cyan, red, blue, and magenta arrowheads, respectively. C Representative wing phenotypes are shown as follows: buffer injection, HDAC1RNAi, HDAC3RNAi, and HDAC4RNAi. D-F Morphology of the hind leg (D), pupal genitalia (E), and adult aedeagus (F) compared to buffer-injected (left column) and HDAC3RNAi (right column) individuals, respectively. G Knockdown of HDAC3 (orange dots) or HDAC4 (blue dots) on femur length and width compared to buffer-injected control (gray dots). Effects of HDAC3RNAi on the male pupal and adult reproductive organ (bottom row in G). The RNAi phenotypes and their corresponding negative controls are shown at the same magnification. Scale bars: 1 mm
HDACs function during appendage development
To determine affected traits, we measured the scaling relationship of each trait to the cubic root of pupal mass as a proxy of body size since the more commonly used measure of thorax width was affected by HDAC3 knockdown (treatment: P < 0.001, Additional file 1: Table S2). HDAC1RNAi resulted in curtailment of both forewings (i.e., elytra) and hindwings at the pupal stage, which was retained into the adult (Fig. 2C). Similarly, both HDAC3 or HDAC4 knockdown diminished wing size (Fig. 2C). Thus, HDAC 1, 3, and 4 appear to regulate wing development in similar ways. RNAi targeting HDAC3 and HDAC4 also affected leg development, but specific effects diverged. To assess leg phenotypes quantitatively we selected the femur, which is especially amenable to width and length measurement, for morphometric analyses. HDAC4RNAi led to a reduction in femur length (treatment: P < 0.001, Additional file 1: Table S2) while the slope of the body size/femur length allometry was decreased in HDAC3RNAi animals (treatment: P = 0.163; treatment × body size: P = 0.004, Fig. 2D and G, and Additional file 1: Table S2). In contrast, whereas HDAC3RNAi increased femur width compared to control individuals (treatment: P < 0.001, Additional file 1: Table S2), the same measure was reduced in HDAC4 knockdown animals (treatment: P = 0.001, Fig. 2D and G, and Additional file 1: Table S2). Lastly, we found that knockdown of HDAC3 also affected the development of the male reproductive organ, the aedeagus, itself composed of the more proximal phallobase and the more distal parameres. Specifically, genital width increased and attained a more rounded shape at the pupal stage (treatment: P < 0.001, Fig. 2E and G, and Additional file 1: Table S2), while adult genitalia exhibited a deformation and overall shortening of both parameres and phallobase, which combined yielded a shortening of overall aedeagus length across all body sizes (treatment: P < 0.001, Fig. 2F and G, and Additional file 1: Table S2).
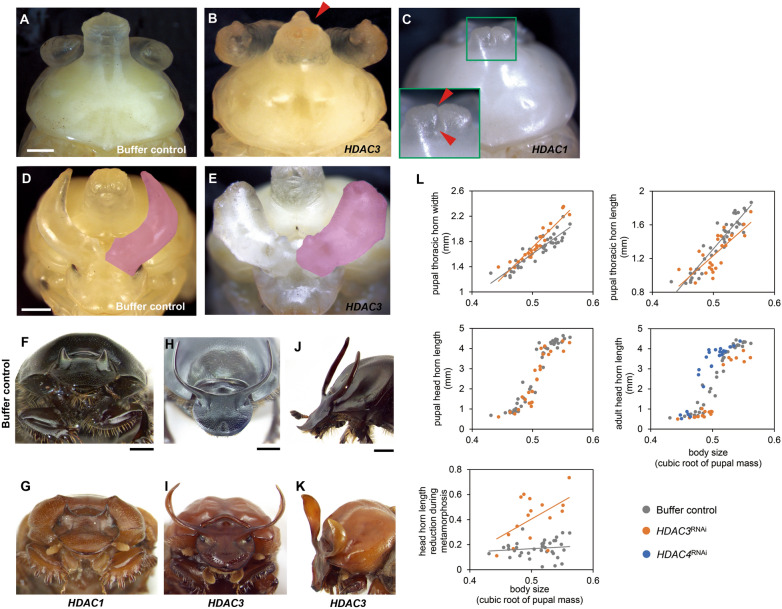
HDACs function during thoracic and head horn formation
Head and thoracic horns are textbook examples of evolutionary novelties, and we sought to determine whether HDAC function may have been co-opted during the evolution of one or both horn types. HDAC1RNAi resulted in a reduction in thoracic horn length and a split tip at the pupal stage (Fig. 3A and C), whereas the corresponding area in the adult exhibited a broad indentation (compared to the smoothly convex outline observed in wildtype or buffer control-injected individuals) and small bilateral projections at the respective edge of the indentation (Fig. 3F and G). Effects on head horns could not be quantified with certainty due to the high degree of natural variability of the trait and the very low number of surviving males, which in addition were too small to develop fully formed head horns. However, HDAC3 knockdown caused measurable shape and scaling changes in thoracic horns. Specifically, HDAC3RNAi increased pupal thoracic horn width (treatment: P < 0.001, Fig. 3A, B, L, and Additional file 1: Table S3), but decreased pupal thoracic horn length (treatment: P < 0.001, Fig. 3L and Additional file 1: Table S3). In addition, we found a bilateral indentation to the distal region of the thoracic horn, causing the thoracic horns of large HDAC3RNAi pupae to attain a more conical shape (Fig. 3B). In contrast to thoracic horn phenotypes, HDAC3RNAi yielded drastically enlarged head horns, in particular concerning head horn width across the entire range of male body sizes (Fig. 3D, E, H–K, and Additional file 1: Fig. S2). However, due to the highly varied nature of these phenotypes, we were unable to arrive at reliable landmarks for quantitative measure, hence this observation could only be made qualitatively. To determine whether head horn length was also affected, we further measured the scaling relationship of head horn length to body size. HDAC3RNAi steepened the slope of the head horn length allometry at both pupal (treatment: P = 0.025, Fig. 3L and Additional file 1: Table S3) and adult stages (treatment: P = 0.012, Fig. 3L and Additional file 1: Table S3). Notably, HDAC3RNAi also reduced the maximum asymptotic horn length in adults (treatment: P < 0.001, Fig. 3L and Additional file 1: Table S3), which was not detected at the pupal stage (treatment: P = 0.107, Fig. 3L and Additional file 1: Table S3). The quantification of head horn length reduction from pupa to adult further confirmed this observation (treatment: P < 0.05, Additional file 1: Table S2), and this effect was enhanced with increasing body size (treatment × body size effect: P = 0.010, Fig. 3L and Additional file 1: Table S2). These results suggest that HDAC3RNAi affects head horn width during the horn growth phase (which takes place during the larval-to-pupal transition, thereby resulting in visible pupal phenotypes), but affects horn length during the pupal remodeling phase of ontogeny (thus becoming apparent in adults only). HDAC4RNAi in turn not only decreased maximum asymptotic horn length in adults (treatment: P = 0.027, Additional file 1: Table S3), but also altered the body size threshold of sigmoidal allometry separating small hornless from large and fully horned males, causing relatively small males which would normally remain hornless to develop relatively large head horns instead (treatment: P < 0.001, Fig. 3L and Additional file 1: Table S3). In contrast, we did not find abnormal phenotypes with respect to thoracic horns in HDAC4RNAi individuals.
Fig. 3.
HDACRNAi effects on thoracic and head horn formation. A–C The pronotum in buffer-injected (A), HDAC3RNAi (B), and HDAC1RNAi (C) individuals. Inset in C shows the prothoracic horn. The furrow between paired horn vestiges is indicated by red arrowheads. D and E Representative head horn of negative control (D) and HDAC3RNAi (E) individuals, respectively. The right head horn is colored magenta. F–K The morphology of the adult pronotum in front view (F and G), front view of head horns (H and I), as well as head horn viewed from lateral (J and K) in negative control (third row) individuals and following HDAC RNAi (bottom row), respectively. L Changes in thoracic and head horns resulting from HDAC3RNAi and HDAC4RNAi. Reduction in head horn length during the transition from pupa to adult is increased by HDAC3RNAi compared to buffer-injections (measured as the difference between pupal head horn length and adult head horn length in the same individual). RNAi phenotypes and their corresponding negative controls are shown at the same magnification. Scale bars: 1 mm
Discussion
The significance of HDACs in horned beetle development and evolution
Earlier work on the broad-horned flour beetle G. cornutus was the first to document the role of HDACs in the regulation of nutrition-responsive plasticity in insects [21]. Large males in this species develop conspicuous mandibular projections (called mandibular horns) which were reduced following HDAC1RNAi, whereas HDAC3RNAi led to hypertrophy. Opposite effects were observed with respect to wing size, yet none in genitalia [21]. These results were the first to suggest that HDACs operate in a trait-specific manner, and in particular contribute to the plastic and sex-specific expression of exaggerated male mandibles. Our results presented here further support a role of HDACs in the regulation of trait-specific plasticity, as well as add important new aspects to our understanding of HDAC function in insect development.
First, similar to the horns of rhinoceros beetle Trypoxylus dichotomus, Onthophagus horns frequently exhibit pronounced nutrition-dependent plasticity, in contrast to the more isometric growth typical of wings and legs, or the hypoallometric growth of genitalia [15, 32]. Several mechanisms have been proposed as possibly underlying module-specific conditional growth (e.g., insulin/insulin-like growth factor [14, 15], FOXO [16, 38, 39], HDAC [21]). Ozawa et al. [21], in particular, proposed a mechanistic explanation of epigenetic flexibility in which developmentally plastic organs (e.g., mandibular horns in G. cornutus) are more susceptible to epigenetic (i.e., HDAC) perturbation, whereas developmentally robust organs (e.g., genitalia) are non-responsive to HDAC perturbation. However, results presented here are at odds with this model. Specifically, even though head and thoracic horn development in O. taurus exhibit exaggerated nutritional plasticity, the effects of HDAC3RNAi were considerably more pronounced in genitalia and wings. Genitalia in particular exhibited a considerable reduction of size relative to body size across all body sizes following HDAC3RNAi, thereby highlighting a previously unexpected role of HDAC in regulating the development of traits generally assumed to be robust to nutritional variation. Intriguingly, a similar outcome was observed following insulin receptor (InR1/2) transcript depletion in O. taurus [38]. Hence, the epigenetic flexibility hypothesis proposed in Gnatocerus beetles is unlikely to explain the findings seen here in Onthophagus, consistent with divergences in HDAC function across the Coleoptera, again similar to what has recently been reported for the insulin signaling pathway [40]. This in turn raises the possibility that, in addition to its primary function in regulating epigenetic status, HDAC3 may also function in aspects of trait morphogenesis not related to nutritional status and developmental plasticity.
Second, we found that HDAC1RNAi induced developmental arrest at the prepupal stages in line with previous studies in Tribolium castaneum [22], which suggests that HDAC1 expression is required for suppressing the expression of genes involved in juvenile hormone (JH) action. In Tribolium, HDAC1 knockdown prevents the larval-to-pupal transition via derepressing the expression of JH-response genes, thereby influencing JH actions and thus halting metamorphosis [22]. Similarly, severe HDAC1 knockdown caused developmental failures during the pupal stage in Gnatocerus, indicating a possibly conserved role in basic developmental process mediating metamorphosis. However, despite this putative conservation of HDAC1 function across the Coleoptera assessed to date, we also found that the precise nature and direction of HDACRNAi effects on appendage formation diverged between Gnatocerus and Onthophagus beetles even beyond those already noted above: for example, in Onthophagus downregulation of HDAC1, 3, and 4 all appears to affect wing size similarly, whereas in Gnatocerus HDAC1RNAi and HDAC3RNAi yield opposite effects. Likewise, in Onthophagus, HDAC3RNAi increased femur width, but HDAC4RNAi decreased it, whereas leg morphology was generally unaffected in Gnatocerus beetle. Lastly, our results document the recruitment of HDAC function into the formation of an evolutionarily novel structure—head and thoracic horns—suggesting that HDAC function is not just evolutionarily labile among conserved insect traits but also contributed to the comparatively recent evolution of Onthophagus weaponry, including the regulation of size, shape, and key components of scaling.
Development and evolution of pupal remodeling
The horns of adult beetles are the product of developmental processes operating at at least two distinct stages of development, a rapid growth phase approximately 48 h immediately prior to the larval-to-pupal molt and a remodeling phase during the pupal stage [33]. While generally given less attention, pupal remodeling can be quite extensive and fully formed pupal horns may be subject to considerable reduction and even complete resorption in many species. Thus, the morphological diversity of adult horns is not only influenced by the differential regulation of growth during the prepupal stage, but also by the developmental processes underlying the differential resorption of horn tissue during the pupal stage [33, 41, 42]. Previous work identified that differential programmed cell death facilitates species, sex, and body-region specific resorption of horn primordia [43]. However, the mechanisms regulating horn resorption during the pupal stage remain largely unknown. Our results implicate HDAC3 as a regulator of both prepupal growth and pupal remodeling of horn primordia. Specifically, we show that HDAC3RNAi increased head horn width, that this phenotype was already prominently visible at the pupal stage, and must therefore have resulted from modifications to the prepupal growth phase of horn formation (Fig. 3). In addition, however, we also find that HDAC3RNAi altered horn length in a manner not evident at the pupal stage but clearly discernible in the resulting adults, and thus a consequence of HDAC3RNAi effects on the pupal remodeling phase of horn formation. As such, HDAC3 is one of relatively few genes identified to date to be involved in horn remodeling during the pupal stage of horned beetles [42, 44].
Chromatin modifications and developmental plasticity
This work is the first to implicate chromatin modifications in the regulation of development and plasticity in horned dung beetles. While earlier work documented the existence of the complete methylation machinery in the O. taurus genome alongside sex- and nutrition-dependent differences in methylation signatures, the functional significance of chromatin modifications, if any, had remained unknown [45, 46]. We now show that downregulation of HDAC3 and HDAC4 affect critical aspects of horn formation including size, shape, and the location of the inflection point separating alternate male morphs. Future work will need to explore if and how HDAC functions may be contributing to the genome-wide remodeling patterns, and more generally the roles of cis-regulatory elements in the development and evolution of plasticity in insects.
Methods
Insects
Adult O. taurus were collected, courtesy of John Allen, from Paterson Farm near Ravenswood, Western Australia. A laboratory population was maintained at 25 ℃ in a sand/soil mixture and fed cow manure twice a week. Larvae used for injection were collected and prepared as described previously [47].
Identification of O. taurus orthologs for HDACs
The Onthophagus orthologs of HDAC genes were identified via reciprocal BLAST to T. castaneum, G. cornutus, Drosophila melanogaster, Bombyx mori, Apis mellifera, and Homo sapiens in NCBI databases. Amino acid sequences of HDAC genes and sirtuin-1, a NAD-dependent protein deacetylase used as outgroup to the above species were aligned with MUSCLE algorithm implemented in MEGA X [48]. Neighbor-Joining tree (bootstrap replicates 1000) was constructed using MEGA X.
Gene clone, double-stranded RNA (dsRNA) synthesis, and injection
To exclude potential off-target effect, we executed a bioinformatic search of selected gene regions for dsRNA design against the whole genome of O. taurus using the BlastN algorithm in NCBI, which enables sequence identity searches of a word-size down to seven bases, to ensure that no more than 20mers of identical sequences in loci other than the targeted genes existed within the genome. Total RNA was extracted with RNeasy Mini Plus Kit (QIAGEN) and reverse transcribed with iScript cDNA Synthesis kit (Bio-Rad). Partial fragments of each genes were amplified with PCR by using gene-specific primers (Additional file 1: Table S4) and cloned into pCR4-TOPO TA vector (Invitrogen, Thermo Fisher Scientific). After the sequences of the inserted gene fragment were confirmed by sequencing (Eurofins Genomics), DNA templates for in vitro transcription were produced with PCR by using TOPO RNAi primers (Additional file 1: Table S4) [49]. PCR products were purified and concentrated using the QIAquick PCR Purification Kit (QIAGEN) and subjected to in vitro transcription (MEGAscript T7 Transcription Kit, Thermo Fisher Scientific) and dsRNA purification (MEGAclear Transcription Clean-Up Kit, Thermo Fisher Scientific), according to the manufacturer’s protocol. DsRNA was quantified and stored at – 80 ℃ until use. Each individual was injected with 3 μl dsRNA at the early stage of the last larval instar (i.e., the third larval instar, L3). Past work showed that neither control injections with non-specific dsRNA derived from exogenous vectors nor buffer solution alone affect morphological trait formation including scaling in Onthophagus [18, 19, 50–53], and we therefore selected injections using buffer solution as a negative control treatment in this study. Control animals were injected with the same volume of injection buffer (1.4 mM NaCl, 0.07 mM Na2HPO4, 0.03 mM KH2PO4, and 4 mM KCl) and kept at the same condition as dsRNA injected animals (see Additional file 1: Table S1 for detailed information of injection).
Effect of HDAC3- and HDAC4-RNAi on the scaling relationships between several morphological traits and body size
Since the usual measure of Onthophagus body size—thorax width (e.g., [38, 54])—was clearly affected by our RNAi treatments (Fig. 3B and Additional file 1: Table S2), we measured the cube root of pupal mass as a proxy for individual body size [55, 56]. We used t-tests to compare body sizes between each RNAi-treated and control group.
Consistent with previous studies, we analyzed nonlinear horn allometries using untransformed data (e.g., [38, 54]). We used the package drc [57] in R 3.5.2 [58] to fit the body size/head horn length distribution a four‐parameter log-logistic (Hill) function in the form:
With x = body size, y = head horn length, a = body size at the point of inflection of the sigmoid curve, b = slope of the curve, c = minimum and d = maximum asymptotic horn lengths [54]. We then inferred whether a complex model including a sigmoidal regression per treatment (i.e., control and RNAi treatments) fitted our data better than a simpler model with one sigmoidal regression including the whole sample by means of the Akaike Information Criterion (AIC) [59]. The AIC measures relative model fit—the lower its value, the better the model fits to the experimental data [60]. Upon finding the complex model more fitting, we used Welch’s t-tests (with Holm–Bonferroni sequential correction where applicable) to compare parameter means (a, b, c, d) between control-injected and RNAi treatment groups [38, 61]. We compared control individuals to HDAC3RNAi and HDAC4RNAi individuals in the case of the adult horn allometry. As for pupal horn allometry, we compared HDAC3RNAi to control individuals.
To inspect the effect of RNAi manipulations on the linear allometries of all the other morphological traits considered, we used the ANCOVA procedure implemented in SPSS Statistics 25 [62] to model trait size as a function of body size, treatment (HDAC3RNAi or HDAC4RNAi vs control-injected) and their interaction. Interactions were removed if non-significant. Data were log-transformed prior to analyses [63]. The reduction in horn size during metamorphosis (defined as pupal horn length—adult horn length) was analyzed similarly, using untransformed data as in other analyses of horn morphology. Statistics for morphometric analyses are provided in Additional file 1: Tables S2 and S3.
Image processing
All images were captured with a digital camera (Scion) mounted to a dissecting microscope (Leica MZ16, Germany). Brightness and contrast of images were adjusted across the entire image with Adobe Photoshop CC 2017 (Adobe, USA).
Supplementary Information
Additional file 1: Fig. S1. Phylogenetic analysis of HDAC orthologues. Fig. S2. HDAC3RNAi head horn phenotypes across the range of male body sizes. Table S1. Injection information for this study. Table S2. Effects of HDAC3 and HDAC4 knockdowns on trait sizes. Table S3. Effects of HDAC3 and HDAC4 knockdowns on pupal and adult head horns. Table S4. The primer information used in this study.
Acknowledgements
We would like to thank Kayla Copper and Levi Burdine for help with beetle care, and Patrick Rohner for discussion of statistical analyses.
Author contributions
Y.H. conceived the initial idea, Y.H. and A.P.M. designed the research, Y.H. and J.R.C. performed the experiments, Y.H. and A.L.M.M. analyzed the data, Y.H., A.P.M., and A.L.M.M. wrote the manuscript with contribution from J.R.C.
Funding
Support for this study was provided by awards from the National Science Foundation (IOS1256689) and John Templeton Foundation (61369) to APM. Additional support was provided by the Natural Science Foundation of Chongqing for Distinguished Young Scholars award to YH (CSTB2022NSCQ-JQX0025). The opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the National Science Foundation, the John Templeton Foundation, or the Natural Science Foundation of Chongqing.
Availability of data and materials
All data are available in the main text and Additional file 1.
Declarations
Ethical approval and consent to participate.
Not applicable.
Competing interests
All authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schlichting C, Pigliucci M. 1998. Phenotypic evolution: a reaction norm perspective. Sinauer
- 2.West-Eberhard MJ. Developmental plasticity and evolution. Oxford: Oxford University Press; 2003. [Google Scholar]
- 3.Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity. 2006;97:192–9. [DOI] [PubMed]
- 4.Prudic KL, Jeon C, Cao H, Monteiro A. Developmental plasticity in sexual roles of butterfly species drives mutual sexual ornamentation. Science. 2011;331:73–5. [DOI] [PubMed]
- 5.Régnière J, Powell J, Bentz B, Nealis V. Effects of temperature on development survival and reproduction of insects: Experimental design data analysis and modeling. J Insect Physiol. 2012;58:634–47. [DOI] [PubMed]
- 6.Lafuente E, Beldade P. Genomics of developmental plasticity in animals. Front Genet. 2019;10:720. doi: 10.3389/fgene.2019.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen DS, Colombani J, Léopold P. Coordination of organ growth: principles and outstanding questions from the world of insects. Trends Cell Biol. 2013;23:336–344. doi: 10.1016/j.tcb.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Rohner PT, Hu Y, Moczek AP. Developmental bias in the evolution and plasticity of beetle horn shape. Proc R Soc B Biol Sci. 2022;289:20221441. doi: 10.1098/rspb.2022.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberhard WG. Static allometry and animal genitalia. Evolution. 2009;63:48–66. doi: 10.1111/j.1558-5646.2008.00528.x. [DOI] [PubMed] [Google Scholar]
- 10.Koh I, Lee MS, Lee NJ, Park KW, Kim KH, Kim H, Rhyu IJ. Body size effect on brain volume in Korean youth. NeuroReport. 2005;16:2029–2032. doi: 10.1097/00001756-200512190-00012. [DOI] [PubMed] [Google Scholar]
- 11.Ito Y, Harigai A, Nakata M, Hosoya T, Araya K, Oba Y, Ito A, Ohde T, Yaginuma T, Niimi T. The role of doublesex in the evolution of exaggerated horns in the Japanese rhinoceros beetle. EMBO Rep. 2013;14:561–567. doi: 10.1038/embor.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moczek AP. Horn polyphenism in the beetle Onthophagus taurus: larval diet quality and plasticity in parental investment determine adult body size and male horn morphology. Behav Ecol. 1998;9:636–641. doi: 10.1093/beheco/9.6.636. [DOI] [Google Scholar]
- 13.Gotoh H, Miyakawa H, Ishikawa A, Ishikawa Y, Sugime Y, Emlen DJ, Lavine LC, Miura T. Developmental Link between sex and nutrition; doublesex regulates sex-specific mandible growth via juvenile hormone signaling in stag beetles. PLOS Genet. 2014;10:e1004098. doi: 10.1371/journal.pgen.1004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y, Katsuki M, Okamoto N, Fujioka H, Okada K. A specific type of insulin-like peptide regulates the conditional growth of a beetle weapon. PLOS Biol. 2019;17:e3000541. doi: 10.1371/journal.pbio.3000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science. 2012;337:860–864. doi: 10.1126/science.1224286. [DOI] [PubMed] [Google Scholar]
- 16.Snell-Rood EC, Moczek AP. Insulin signaling as a mechanism underlying developmental plasticity: the role of FOXO in a nutritional polyphenism. PLoS ONE. 2012;7:e34857. doi: 10.1371/journal.pone.0034857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H-J, Xue J, Lu B, Zhang X-C, Zhuo J-C, He S-F, Ma X-F, Jiang Y-Q, Fan H-W, Xu J-Y, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015;519:464–467. doi: 10.1038/nature14286. [DOI] [PubMed] [Google Scholar]
- 18.Kijimoto T, Moczek AP, Andrews J. Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proc Natl Acad Sci. 2012;109:20526–20531. doi: 10.1073/pnas.1118589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kijimoto T, Moczek AP. Hedgehog signaling enables nutrition-responsive inhibition of an alternative morph in a polyphenic beetle. Proc Natl Acad Sci. 2016;113:5982–5987. doi: 10.1073/pnas.1601505113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog. 2015;20:35–47. doi: 10.1615/CritRevOncog.2015012997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozawa T, Mizuhara T, Arata M, Shimada M, Niimi T, Okada K, Okada Y, Ohta K. Histone deacetylases control module-specific phenotypic plasticity in beetle weapons. Proc Natl Acad Sci U S A. 2016;113:15042–15047. doi: 10.1073/pnas.1615688114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.George S, Gaddelapati SC, Palli SR. Histone deacetylase 1 suppresses Krüppel homolog 1 gene expression and influences juvenile hormone action in Tribolium castaneum. Proc Natl Acad Sci. 2019;116:17759–17764. doi: 10.1073/pnas.1909554116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George S, Palli SR. Histone deacetylase 3 is required for development and metamorphosis in the red flour beetle Tribolium castaneum. BMC Genom. 2020;21:420. doi: 10.1186/s12864-020-06840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George S, Palli SR. Histone deacetylase 11 knockdown blocks larval development and metamorphosis in the red flour beetle Tribolium castaneum. Front Genet. 2020;11:683. doi: 10.3389/fgene.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzsimons HL, Schwartz S, Given FM, Scott MJ. The histone deacetylase HDAC4 regulates long-term memory in Drosophila. PLoS ONE. 2013;8:e83903. doi: 10.1371/journal.pone.0083903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J-L, Yuan X-B, Chen S-J, Chen H-H, Xu N, Xue W-H, Fu S-J, Zhang C-X, Xu H-J. The histone deacetylase NlHDAC1 regulates both female and male fertility in the brown planthopper Nilaparvata lugens. Open Biol. 2018;8:180158. doi: 10.1098/rsob.180158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel S, Woods J, Ziafazeli T, Rogina B. RPD3 histone deacetylase and nutrition have distinct but interacting effects on Drosophila longevity. Aging. 2015;7:1112–1129. doi: 10.18632/aging.100856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spannhoff A, Kim YK, Raynal NJ-M, Gharibyan V, Su M-B, Zhou Y-Y, Li J, Castellano S, Sbardella G, Issa J-PJ, et al. Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Rep. 2011;12:238–243. doi: 10.1038/embor.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki R, Yaguchi H, Maekawa K. Histone modifying genes are involved in the molting period during soldier differentiation in Zootermopsis nevadensis. J Insect Physiol. 2019;117:103892. doi: 10.1016/j.jinsphys.2019.103892. [DOI] [PubMed] [Google Scholar]
- 30.Cui J, Lin K, Xu L, Yue F, Yu L, Zhang Q. Transcriptome analysis of beet webworm shows that histone deacetylase may affect diapause by regulating juvenile hormone. Insects. 2022;13:835. doi: 10.3390/insects13090835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee K, Fischer R, Vilcinskas A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front Zool. 2012;9:25. doi: 10.1186/1742-9994-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casasa S, Schwab DB, Moczek AP. Developmental regulation and evolution of scaling: novel insights through the study of Onthophagus beetles. Curr Opin Insect Sci. 2017;19:52–60. doi: 10.1016/j.cois.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Moczek AP. Pupal Remodeling and the development and evolution of sexual dimorphism in horned beetles. Am Nat. 2006;168:711–729. doi: 10.1086/509051. [DOI] [PubMed] [Google Scholar]
- 34.Moczek AP, Cruickshank TE, Shelby A. When ontogeny reveals what phylogeny hides: gain and loss of horns during development and evolution of horned beetles. Evolution. 2006;60:2329–2341. doi: 10.1111/j.0014-3820.2006.tb01868.x. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, Linz DM, Moczek AP. Beetle horns evolved from wing serial homologs. Science. 2019;366:1004–1007. doi: 10.1126/science.aaw2980. [DOI] [PubMed] [Google Scholar]
- 36.Moczek AP. The evolution and development of novel traits, or how beetles got their horns. Bioscience. 2005;55:937–951. doi: 10.1641/0006-3568(2005)055[0937:TEADON]2.0.CO;2. [DOI] [Google Scholar]
- 37.Müller GB, Wagner GP. Novelty in evolution: restructuring the concept. Annu Rev Ecol Syst. 1991;22:229–256. doi: 10.1146/annurev.es.22.110191.001305. [DOI] [Google Scholar]
- 38.Casasa S, Moczek AP. Insulin signalling’s role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc R Soc B Biol Sci. 2018;285:20181631. doi: 10.1098/rspb.2018.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang HY, Smith-Caldas MSB, Driscoll MV, Salhadar S, Shingleton AW. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 2011;7:e1002373. [DOI] [PMC free article] [PubMed]
- 40.Rohner PT, Casasa S, Moczek AP. Assessing the evolutionary lability of insulin signalling in the regulation of nutritional plasticity across traits and species of horned dung beetles. J Evol Biol. 2023;36:1641–1648. doi: 10.1111/jeb.14240. [DOI] [PubMed] [Google Scholar]
- 41.Moczek AP. Pupal remodeling and the evolution and development of alternative male morphologies in horned beetles. BMC Evol Biol. 2007;7:151. doi: 10.1186/1471-2148-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita S, Ando T, Maeno A, Mizutani T, Mase M, Shigenobu S, Niimi T. Precise staging of beetle horn formation in Trypoxylus dichotomus reveals the pleiotropic roles of doublesex depending on the spatiotemporal developmental contexts. PLOS Genet. 2019;15:e1008063. doi: 10.1371/journal.pgen.1008063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kijimoto T, Andrews J, Moczek AP. Programed cell death shapes the expression of horns within and between species of horned beetles. Evol Dev. 2010;12:449–458. doi: 10.1111/j.1525-142X.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda K, Adachi H, Gotoh H, Inoue Y, Kondo S. Adhesion and shrinkage transform the rounded pupal horn into an angular adult horn in Japanese rhinoceros beetle. Development. 2024 doi: 10.1242/dev.202082. [DOI] [PubMed] [Google Scholar]
- 45.Choi J-H, Kijimoto T, Snell-Rood E, Tae H, Yang Y, Moczek AP, Andrews J. Gene discovery in the horned beetle Onthophagus taurus. BMC Genom. 2010;11:703. doi: 10.1186/1471-2164-11-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snell-Rood EC, Moczek AP. Horns and the role of development in the evolution of beetle contests. In: Hardy ICW, Briffa M, editors. Animal Contests. Cambridge: Cambridge University Press; 2013. pp. 178–198. [Google Scholar]
- 47.Shafiei M, Moczek AP, Nijhout HF. Food availability controls the onset of metamorphosis in the dung beetle Onthophagus taurus (Coleoptera: Scarabaeidae) Physiol Entomol. 2001;26:173–180. doi: 10.1046/j.1365-3032.2001.00231.x. [DOI] [Google Scholar]
- 48.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philip BN, Tomoyasu Y. Gene knockdown analysis by double-stranded RNA injection. In: Orgogozo V, Rockman MV, editors. Molecular methods for evolutionary genetics. Methods in molecular biology. Totowa: Humana Press; 2011. pp. 471–497. [DOI] [PubMed] [Google Scholar]
- 50.Hu Y, Moczek AP. Wing serial homologues and the diversification of insect outgrowths: insights from the pupae of scarab beetles. Proc R Soc B Biol Sci. 2021;288:20202828. doi: 10.1098/rspb.2020.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linz DM, Hu Y, Moczek AP. The origins of novelty from within the confines of homology: the developmental evolution of the digging tibia of dung beetles. Proc R Soc B Biol Sci. 2019;286:20182427. doi: 10.1098/rspb.2018.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moczek AP, Rose DJ. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc Natl Acad Sci. 2009;106:8992–8997. doi: 10.1073/pnas.0809668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohner PT, Linz DM, Moczek AP. Doublesex mediates species-, sex-, environment- and trait-specific exaggeration of size and shape. Proc R Soc B Biol Sci. 2021;288:20210241. doi: 10.1098/rspb.2021.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macagno ALM, Edgerton TJ, Moczek AP. Incipient hybrid inferiority between recently introduced, diverging dung beetle populations. Biol J Linn Soc. 2021;132:931–944. doi: 10.1093/biolinnean/blaa228. [DOI] [Google Scholar]
- 55.Fleming MJ, Carter AW, Sheldon KS. Dung beetles show metabolic plasticity as pupae and smaller adult body size in response to increased temperature mean and variance. J Insect Physiol. 2021;131:104215. doi: 10.1016/j.jinsphys.2021.104215. [DOI] [PubMed] [Google Scholar]
- 56.Marchini M, Sparrow LM, Cosman MN, Dowhanik A, Krueger CB, Hallgrimsson B, Rolian C. Impacts of genetic correlation on the independent evolution of body mass and skeletal size in mammals. BMC Evol Biol. 2014;14:258. doi: 10.1186/s12862-014-0258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-response analysis using R. PloS One. 2015;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team . R: A language and environment for statistical computing. Vienna: R Core Team; 2018. [Google Scholar]
- 59.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 60.Burnham KP, Anderson DR, Huyvaert KP. AIC model selection and multimodel inference in behavioral ecology: some background, observations, and comparisons. Behav Ecol Sociobiol. 2011;65:23–35. doi: 10.1007/s00265-010-1029-6. [DOI] [Google Scholar]
- 61.Moczek A, Hunt J, Emlen D, Simmons L. Threshold evolution in exotic populations of a polyphenic beetle. Evol Ecol Res. 2002;4:587–601. [Google Scholar]
- 62.IBM Corp . IBM SPSS statistics for windows, version 25.0. Armonk, NY: IBM Corp; 2017. [Google Scholar]
- 63.Huxley JS. Constant differential growth-ratios and their significance. Nature. 1924;114:895–896. doi: 10.1038/114895a0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Phylogenetic analysis of HDAC orthologues. Fig. S2. HDAC3RNAi head horn phenotypes across the range of male body sizes. Table S1. Injection information for this study. Table S2. Effects of HDAC3 and HDAC4 knockdowns on trait sizes. Table S3. Effects of HDAC3 and HDAC4 knockdowns on pupal and adult head horns. Table S4. The primer information used in this study.
Data Availability Statement
All data are available in the main text and Additional file 1.