Abstract
Objectives:
The aim of this study was to establish an automatic classification model for chronic inflammation of the choledoch wall using deep learning with CT images in patients with pancreaticobiliary maljunction (PBM).
Methods:
CT images were obtained from 76 PBM patients, including 61 cases assigned to the training set and 15 cases assigned to the testing set. The region of interest (ROI) containing the choledochal lesion was extracted and segmented using the UNet++ network. The degree of severity of inflammation in the choledochal wall was initially classified using the ResNeSt network. The final classification result was determined per decision rules. Grad-CAM was used to explain the association between the classification basis of the network and clinical diagnosis.
Results:
Segmentation of the lesion on the common bile duct wall was roughly obtained with the UNet++ segmentation model and the average value of Dice coefficient of the segmentation model in the testing set was 0.839 ± 0.150, which was verified through fivefold cross-validation. Inflammation was initially classified with ResNeSt18, which resulted in accuracy = 0.756, sensitivity = 0.611, specificity = 0.852, precision = 0.733, and area under curve (AUC) = 0.711. The final classification sensitivity was 0.8. Grad-CAM revealed similar distribution of inflammation of the choledochal wall and verified the inflammation classification.
Conclusions:
By combining the UNet++ network and the ResNeSt network, we achieved automatic classification of chronic inflammation of the choledoch in PBM patients and verified the robustness through cross-validation performed five times. This study provided an important basis for classification of inflammation severity of the choledoch in PBM patients.
Advances in knowledge:
We combined the UNet++ network and the ResNeSt network to achieve automatic classification of chronic inflammation of the choledoch in PBM. These results provided an important basis for classification of choledochal inflammation in PBM and for surgical therapy.
Introduction
Pancreaticobiliary maljunction (PBM) is a rare congenital malformation with a long common channel between the pancreatic duct and the bile duct, resulting in two-way reflux of bile and pancreatic juice through the common channel. 1 Due to frequent reflux of pancreatic juice into the biliary tract, PBM can result in multiple types of serious complications, including chronic choledochal inflammation, which is the major reason for cholangitis and carcinogenesis of the bile duct. PBM is also commonly associated with a choledochal cyst, while early detection of recurrent cholangitis is vital for the long-term effect and precise complete resection of the cyst and Roux-en-Y hepaticojejunostomy in PBM. 2–4
Currently, magnetic resonance cholangiopancreatography (MRCP), endoscopic retrograde cholangiopancreatography (ERCP), and endoscopic ultrasound scan (EUS), in combination with clinical symptoms and primary pathogenesis, are effective diagnostic strategies for cholangitis, mainly with an irregular thickened biliary wall (>0.8 mm). 5–8 However, there is no image or comparative pathological study related to PBM-induced chronic choledochal inflammation; the quantitative study and deep learning study of chronic inflammation of the biliary wall are also lacking in recent years. A lack of these studies causes a potential risk of safety for surgical treatment of PBM and related choledochal cyst. Therefore, establishing the classification of chronic choledochal inflammation through deep learning, which has been used for classification of inflammation, 9 may be helpful for accurate assessment of the safety of Roux-en-Y surgery and treatment of PBM-related complications.
Currently, deep learning has achieved great success in image classification and segmentation in all fields. 10–12 Although there have been no reports documenting the use of deep learning to study choledochal cysts, deep learning has played a very important role in the computer-aided diagnosis of other cystic diseases. Monaco et al studied autosomal dominant polycystic kidney disease (ADPKD) and used UNet network to segment cysts to assist doctors in evaluating the effects of treatment for ADPKD to find more effective treatments. 13 Kurita et al used a deep learning method to diagnose malignant pancreatic cystic lesions. The classification is far more sensitive than that of traditional methods. Deep learning can improve the diagnostic ability of pancreatic cystic lesions as a support system. 14 Narmatha et al conducted a classification study of seven types of ovarian cysts, using a new deep Q-Network and Harris Hawks Optimization classifier for cyst classification after feature extraction from ultrasound images using CNN. 15
In the deep learning field, the U-Net and the ResNet have shown excellent performance in segmentation and classification, respectively, of biomedical images of pediatric patients. 16–19 However, there is no segmentation and classification network frame used for chronic choledochal inflammation. Because of the thin wall and inflammation of the choledochal wall in PBM patients, the surrounding structure cannot be displayed clearly; therefore, the need for segmentation of the tissue lesion in the choledoch in PBM is higher. UNet++ is one of the deep learning methods with the most promising prospective in the segmentation tasks, 20 and ResNeSt is a recently developed classification method of deep learning based on ResNet. 21
To our knowledge, no previous work has used deep learning to assess the choledochal inflammation in PBM patients. In this prospective study, we segmented and classified of choledochal inflammation in PBM patients by combining the UNet++ network and the ResNeSt network.
Methods and materials
Subjects
In total, we collected the data of choledochal cysts from 76 pediatric patients (≤14 years of age) who were admitted to the Children’s Hospital of Soochow University from March 2015 to October 2019, including 64 females and 12 males. The general information of the subjects is listed in Table 1. According to the situation of hyperemia, edema, inflammatory infiltration, exfoliation of the mucous epithelium, and proliferation of fibrous tissue by pathological examination, chronic inflammation of the choledochal cystic wall was classified into different grades based on a previous study. 22 The classification was also based on the pathological grading criteria of chronic gastritis and intestinal mucosal damage, the surgical record, and the prognosis of these cases. The grading of the lesion region and inflammation were provided by a radiologist of the Children’s Hospital of Soochow University.
Table 1.
Demographic and baseline characteristics of the patients
| N (76) | Lesion size (mm2):7806.93 ± 11073.51 | |
|---|---|---|
| Gender | ||
| Male | 12 | 6735.74 ± 4382.30 |
| Female | 64 | 8007.77 ± 11906.22 |
| Age | ||
| 0–2 | 43 | 7850.22 ± 9316.36 |
| 3–10 | 29 | 6306.44 ± 11712.52 |
| >10 | 4 | 18220.01 ± 16656.44 |
| Inflammation grade score | ||
| Mild (≤4) | 47 | 8346.05 ± 12387.08 |
| Severity (>4) | 29 | 6933.18 ± 8452.40 |
The study protocol was approved by the Institutional Review Board of the Children’s Hospital of Soochow University (No. 20160606013). Written informed consent was provided by the parents or legal guardians of the children.
Imaging acquisition and training environment
Equipment used in the present study was helical CT scanning set (GE Optima) with 5 mm slice thickness, 1 mm slice distance, and 512 × 512 matrix. The patients were put in a spine position after anal administration of 10% chloral hydrate (0.5 ml/kg). Enhanced CT scanning was performed with non-ionic contrast Omnipaque (General Motors Electric Pharmaceutical Industry (Shanghai) Limited Company, China). The contrast (1.5–2 mg/kg) was diluted with saline at a certain ratio and injected within 15–20 s at a flow rate of 1–1.5 ml s−1. The images were processed in the AW462 workstation with AW Volume Share five sdc. The training environment used in the present study was as follows: Inter Core i9-9900k, internal memory Kingston DDR4 2400 MHz 32 GB, graphics card Nvidia GeForce RTX 2080Ti (11 GB), and Windows 10 Pro.
Method
The algorithm of inflammation classification of the choledochal cyst mainly included the following two major components: (1) automatic extraction of ROI of the choledochal cyst and (2) prediction of inflammation grading based on images (Figure 1). The present study extracted the data of CT images containing the cystic lesion of the choledoch, and performed network training for two-dimensional segmentation and classification.
Figure 1.
The flowchart of the algorithm. ROI, region of interest.
Automatic extraction of ROI containing the cystic lesion of the choledochal wall
After obtaining the slices containing the lesion, the grayscale value of image data was adjusted to 0–255 with the standard of window width and window level (40 of window level and 250 of window width) to extract the ROI, referring to the observation parameters of clinical imaging examination of choledochal cysts. The rules for ROI extraction were as follows: (1) to determine the upper and bilateral boundaries of the cystic wall after localizing the chest wall in CT images, (2) to extract the ROI with the ¾ width of the bilateral boundary of the cystic wall from the basis of upper and left boundaries, and, finally, (3) to resample the ROI with a resolution of 256 × 256 as the input of segmental network for model training. Within the 76 samples, we randomly selected 61 cases as the training set and 15 cases as the testing set.
In the steps of model training, we used the UNet++ network model, 20 which is based on the classical U-Net network and achieves the enhancement of network features by improving the skip connection between the encoding pathway and the decoding pathway. The architecture is shown in Figure 2. During network training, each iteration shuffled the training data, which were then processed with a batch size of 4. The network parameters were initialized with Xavier and adaptive learning rate, and the initial learning rate was set at 1e-5. When the network feature was not enhanced by more than 30 iterations, the learning rate was decreased to 1/10 of the initial learning rate. The network parameters were optimized with Adam optimizer and Dice loss function. To achieve guaranteed convergence, the iteration time was set at 150. The ADAM optimizer combines the advantages of the AdaGrad and RMSProp optimization algorithms, comprehensively considers the first-order moment estimation and second-order moment estimation of the gradient, calculates the update step size, and optimizes the network parameters. The Dice loss function is shown in the formula, where X is the ground truth and Y is the predicted results of segmentation.
Figure 2.
Architecture of the UNet++.
To assess the performance of the Unet++ network, the UNet 23 and the R2U-Net 24 networks were used to train the segmentation model using the same training parameters and training data. The Dice coefficient (DC) was used to evaluate the segmentation performance of the models. After obtaining segmentation with UNet++, three representative slices with the maximal area of the cystic lesion were selected. When extracting the image ROI, in order to ensure the integrity of the lesion information, such as the adhesion between the lesion and the surrounding tissue, the lesion area and the surrounding tissue within six pixels was defined as the ROI, which was resampled with a sampling rate of 64 × 64 and set as the input of the second step. The flowchart is shown in Figure 3.
Figure 3.
Flowchart of extraction of the ROI in the lesion area
Grading prediction of the inflammatory pathology based on images
In the data of 76 cases, there were 47 cases of mild inflammation and 29 cases of severe inflammation. The training set comprised 61 cases, including randomly selected 38 cases of mild inflammation and 23 cases of severe inflammation. The other 15 cases were used for the testing set. After statistical analysis of each patient’s data, we found that the number of slices containing lesions in the CT data of different patients was always at least 3. In order to ensure the consistency of the preprocessing steps, the first three slices with the larger cross-sectional area of the lesion were selected per case. In the training set, the number of cases with mild inflammation and severe inflammation was enhanced with rotation operation and shear transformation by six- and ninefold, respectively, reaching 228 cases and 207 cases.
During network training, each iteration randomly shuffled the training data, which were then processed with a batch size of 8. The initial learning rate was set at 1e-5. When the network feature was not enhanced by more than 30 iterations, the learning rate was decreased to 1/10 of the initial learning rate. The network parameters were optimized with the ADAM optimizer and BCELoss function. To achieve guaranteed convergence, the iteration time was set at 300. BCELoss is the cross-entropy loss function used for two classification problems. The calculation formula is shown in the formula, where N represents the sample’s identification number, represents the score of predicting the nth sample as a positive example, represents the label of the nth sample, and represents the sigmoid function.
To assess the performance of the ResNeSt network, the ResNeXt 25 and the SENet 26 were used to train the classification model using the same training parameters and training data. The accuracy, sensitivity, specificity, precision, and receiver operating characteristic (ROC) were used to evaluate the classification performance of the models. The Grad-CAM 27 was used to visualize the area activated by the network. When the classified model was tested, three slices obtained from the same case were individually tested and the grading of chronic inflammation of the choledochal cystic wall was classified using the one-vote veto, which means that if at least one of the three slices is classified as severe inflammation, the patient is considered to have severe inflammation. The flowchart is shown in Figure 4.
Figure 4.
Flowchart of inflammation classification of the cystic wall.
Assessment parameters
The DC was used as the assessment parameter for the ROI of extraction of the choledochal cystic lesion:
① .
For the inflammation classification of the choledochal cystic wall, severe inflammation and mild inflammation was assigned a value of 1 and 0, respectively. Accuracy, sensitivity, specificity, precision, and area under curve (ROC) were used as the assessment parameters, and they were calculated as follows:
| , |
| , |
| , |
| , |
where TP, FP, TN, and FN were true positive, false positive, true negative, and false negative, respectively.
Results
In total, we collected the CT image data of 76 cases and classified them through pathological comparison as 47 cases of mild inflammation and 29 cases of severe inflammation.
Segmentation of the lesion area
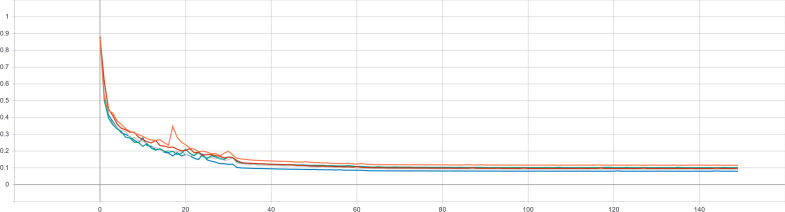
In the present study, we first extracted the ROI of the choledochal cystic lesion through the UNet++ segmentation model. The loss curve of the training set during the training procedure is shown in Figure 5, and the DC curve of the test set during the training procedure is shown in Figure 6, indicating a small loss of accuracy. The DC in the testing set was 0.839 ± 0.150, which was verified by cross-validation performed five times. The DC of the UNet in the testing set was 0.809 ± 0.016, and the DC of the R2U-Net in the testing set was 0.826 ± 0.021. The comparisons of the DC are shown in Figure 7. They showed that the UNet++ network has the best segmentation effect under our data set. Figure 8 shows a qualitative comparison between the segmentation results of U-Net, R2U-Net, UNet++, and the ground truth. In this section, the main purpose of the segmentation of the lesion area is to locate the lesion. The segmentation result of the UNet++ network can completely cover the lesion area to provide a basis for the extraction of ROI in the following work.
Figure 5.
Loss curve of the training set during the training procedure.
Figure 6.
DC curve of the testing set with cross-validation performed five times. DC, Dice coefficient.
Figure 7.
Dice coefficient comparison among U-Net, R2U-Net, and UNet++.
Figure 8.
Comparison of the segmentation results of U-Net, R2U-Net, and UNet++.
Results of the pathological classification network
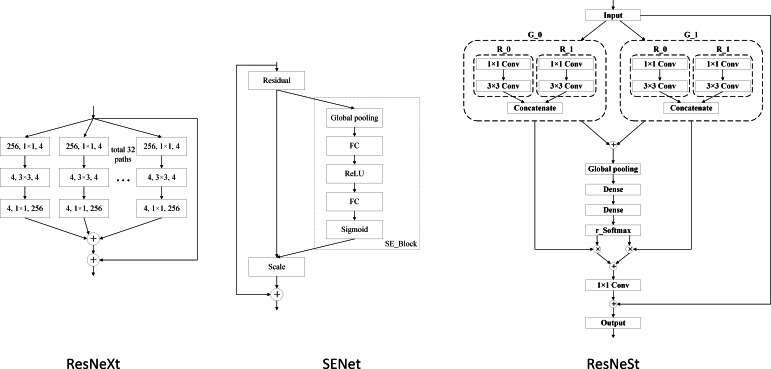
When selecting the classification model, we tested several improved networks of ResNet, including ResNeXt-50, SENet-50, and ResNeSt-50, The architecture of the network as shown in Figure 9. The classification results were assessed with the same training method (Table 2).
Figure 9.
Architecture of the ResNeXt, SENet, and ResNeSt.
Table 2.
Assessment of classification networks
| Accuracy | Specificity | False positive | ROC (Area under curve) | |
|---|---|---|---|---|
| ResNeXt-50 | 0.644 | 0.815 | 11 | 0.592 |
| SENet-50 | 0.711 | 0.704 | 5 | 0.677 |
| ResNeSt-50 | 0.756 | 0.852 | 7 | 0.703 |
ROC, receiver operating characteristic.
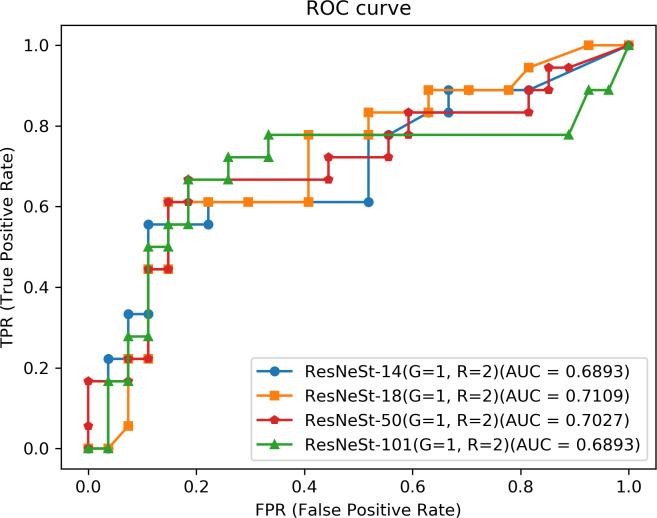
After comparison, we finally used the ResNeSt network for training of the classification network. By selecting the input of the classification network based on real label of lesions and extracting the ROI, we investigated the classification effect of the ResNeSt network under the combination of different network depths, number of group convolutions in the ResNeSt module (G), and split-attention (R). The results of inflammation classification are listed in Table 3 and Figure 10.
Table 3.
The results of the classification network with different parameters
| Classification network | G | R | Accuracy | Specificity | False positive | ROC (Area under curve) |
|---|---|---|---|---|---|---|
| ResNeSt14 | 1 | 2 | 0.733 | 0.852 | 8 | 0.689 |
| 2 | 2 | 0.667 | 0.704 | 7 | 0.600 | |
| 1 | 4 | 0.733 | 0.741 | 5 | 0.659 | |
| ResNeSt18 | 1 | 2 | 0.756 | 0.852 | 7 | 0.711 |
| 2 | 2 | 0.711 | 0.778 | 7 | 0.644 | |
| 1 | 4 | 0.733 | 0.889 | 9 | 0.687 | |
| ResNeSt50 | 1 | 2 | 0.756 | 0.852 | 7 | 0.703 |
| 2 | 2 | 0.733 | 0.815 | 7 | 0.699 | |
| 1 | 4 | 0.711 | 0.815 | 8 | 0.653 | |
| ResNeSt101 | 1 | 2 | 0.756 | 0.815 | 6 | 0.689 |
| 2 | 2 | 0.711 | 0.815 | 8 | 0.521 | |
| 1 | 4 | 0.711 | 0.704 | 5 | 0.660 |
ROC, receiver operating characteristic.
Figure 10.
ROC curve of inflammation classification. ROC, receiver operating characteristic.
Using the classification model of ResNeSt training, we obtained the following confusion matrix of the testing set (Table 4). The confusion matrix produced accuracy = 0.756, sensitivity = 0.611, specificity = 0.852, precision = 0.733, and AUC = 0.711.
Table 4.
Confusion matrix of the testing set
| Diagnosed mild inflammation | Diagnosed severe inflammation |
|---|---|
| Mild inflammation | |
| 23 (TP) | 4 (FN) |
| Severe inflammation | |
| 7 (FP) | 11 (TN) |
FN, false negative; FP, false positive; TN, true negative; TP, true positive.
After classification, the recognition sensitivity of the testing set was 12/15 = 80%, according to the criteria that the inflammation would be classified as severe if one of the three slices were severe.
Visualization of heatmap features of the classification network
After verification with cross-validation performed five times, we selected the training model with the best segmental effect to extract the ROI of the lesion area as the training data for the classification network. After calculation, the cover-rate of the ROI against the lesion area with real label was 100%.
To verify the clinical reliability of the inflammation classification model obtained with ResNeSt, we used a gradient-based classification activation map (Grad-CAM) to interpret the model and visualized the features obtained from the classification model. The class activation map was shown superimposed on the original map, as shown in Figure 11. In the figure, the first row is the original image, and the second row is the class activation map. Grad-CAM correctly revealed the lesion area and produced similar distribution of inflammation classification of the choledochal cyst. According to the ground truth of the region of the choledochal cyst, we used the red curve to outline the wall of choledochal cyst in the figure. The red regions correspond to high scores for class.
Figure 11.
Visualization of the inflammation classification model with Grad-CAM. Grad-CAM, gradient-based classification activation map.
Discussion
Our previous study indicated that the choledochal wall in PBM is usually combined with different extents of inflammation. 8 Chronic inflammation of the choledochal wall has an important effect on the surgical safety and prognosis of patients with PBM, and the degree of inflammation severity is also one of the important bases for selection of the surgery method. Damage to the bile duct during Roux-en-Y surgery is mainly related to fragility of the choledochal wall due to inflammatory edema and increased rupture risk of the cystic wall due to difficulty in isolating the cystic wall that is adhered to the surrounding tissues. For such cases with severe inflammation of the choledochal wall, previous studies proposed inside resection to avoid intraoperative bleeding. 2,4,8
Currently, the diagnosis of bile duct inflammation is mainly dependent on image examination, including ultrasound, MR, and endoscope, 5–8 CT is more widely used in clinical practice. An enhanced CT can clearly display the structure of the choledochal wall and the situation of its adhesion with the surrounding tissue, and therefore, it is used as one of the common examination methods. Recently, deep learning has been used for analysis of CT images to guide clinical practice, including hepatocellular carcinoma 28 and classification of lung disease, 29 and it has shown valuable feasibility and advantages. However, there is no related report about the classification of inflammation of the bile duct wall in PBM.
In the present study, we collected the CT image data of 76 pediatric patients with PBM and classified them through pathological comparison into 47 cases of mild inflammation and 29 cases of severe inflammation. In the segmentation network, through the comparison experiment, the UNet++ network performed better segmentation than UNet and the R2U-Net using our data set. In the classification network, ResNeSt outperformed SENet and ResNeXt. Futhermore, the ResNeSt network with 18 convolutional layers and the hyperparameters of G = 1, R = 2 showed the best classification performance.
UNet++ has most promising prospective in the segmentation task of any deep learning method. 20 The ResNeSt network is based on the ResNeSt module, which is a version of ResNet module improved by combining group convolution and split-attention module. 21 By combining the UNet++ network and the ResNeSt network, we segmented the cystic lesion of the choledochal wall to extract the ROI and obtained a DC of 0.839 ± 0.150 in the testing set of the segmentation model. Segmentation was verified with cross-validation performed five times and it produced an accuracy of 0.80 in the classification of inflammation. Thus, we achieved a high feature in classifying inflammation of the choledochal wall by using the ResNeSt18 training classification model.
In addition, we used Grad-CAM to interpret the model and visualized the features extracted from the classification model, which improved the explanation of the inflammation classification of the choledochal wall and also verified the reliability of the classification model. In addition, visualization could show a smaller structure like the peripheral bile duct, which is advantageous for the choice of surgical methods and better prognosis. 27 Taken together, the present study introduced a network model to establish an automatic recognition model to classify choledochal inflammation at the pixel level, which plays an important role in precisely recognizing inflammation of the common bile duct in PBM. However, the clinical significance of this should be further verified.
Limitation
The present study was a single-center study with a small sample size and the thickness of contrast-enhanced CT scanning was only 5 mm without any comparative study in scanning thickness. To assess choledochal inflammation in PBM, future study should be performed at multiple centers and with larger sample sizes and different scanning thicknesses.
Conclusion
In the present study, we combined the UNet++ network and the ResNeSt network to achieve automatic classification of chronic inflammation of the choledoch in PBM. With cross-validation performed five times, the robustness of the algorithm was verified and the clinical reliability of the algorithm was verified further with analysis of the feature heatmap of the classification network. These results provided an important basis for classification of choledochal inflammation in PBM and for surgical therapy.
Footnotes
Acknowledgments: This work was supported partially by National Natural Science Foundation (No. 81971685), Science and Technology Project of Suzhou (No. SS201868, SS202072), Suzhou Health Science & Technology Project (GWZX201904), Quancheng 5150 Project, Medical Research Project of Jiangsu Provincial Health and Family Planning Commission (No. M2020068), and Youth Innovation Promotion Association CAS (No. 2021324).
Conflict of interest :We have no conflicts of interest to declare in relation to this study
Contributor Information
Wan-liang Guo, Email: gwlsuzhou@163.com, Department of Radiology, Children’s Hospital of Soochow University, Suzhou, China .
An-kang Geng, Email: 2214512963@qq.com, School of Biomedical Engineering (Suzhou), Division of Life Sciences and Medicine, University of Science and Technology of China, 88 Keling Road, Suzhou, China ; Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, 88 Keling Road, Suzhou, China .
Chen Geng, Email: gengc@sibet.ac.cn, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, 88 Keling Road, Suzhou, China .
Jian Wang, Email: wj196312@vip.163.com, Pediatric Surgery, Children’s Hospital of Soochow University, Suzhou, China .
Ya-kang Dai, Email: daiyk@sibet.ac.cn, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, 88 Keling Road, Suzhou, China ; Jinan Guoke Medical Engineering Technology Development Co. LTD, Jinan, China .
REFERENCES
- 1. Kamisawa T, Kaneko K, Itoi T, Ando H . Pancreaticobiliary maljunction and congenital biliary dilatation . Lancet Gastroenterol Hepatol 2017. ; 2: 610 – 18 . doi: 10.1016/S2468-1253(17)30002-X [DOI] [PubMed] [Google Scholar]
- 2. Ishibashi H, Shimada M, Kamisawa T, Fujii H, Hamada Y, Kubota M, et al. . Japanese study group on congenital biliary dilatation (JSCBD) . Japanese Clinical Practice Guidelines for Congenital Biliary Dilatation J Hepatobiliary Pancreat Sci 2017. ; 24: 1 – 16 . [DOI] [PubMed] [Google Scholar]
- 3. Qiao G, Li L, Li S, Tang S, Wang B, Xi H, et al. . Laparoscopic cyst excision and roux-Y hepaticojejunostomy for children with choledochal cysts in china: a multicenter study . Surg Endosc 2015. ; 29: 140 – 44 . doi: 10.1007/s00464-014-3667-7 [DOI] [PubMed] [Google Scholar]
- 4. Guo W-L, Zhan Y, Fang F, Huang S-G, Deng Y-B, Zhao J-G, et al. . Factors affecting the operating time for complete cyst excision and roux-en-Y hepaticojejunostomy in paediatric cases of congenital choledochal malformation: a retrospective case study in southeast china . BMJ Open 2018. ; 8( 5 . doi: 10.1136/bmjopen-2018-022162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naitoh I, Nakazawa T, Ohara H, Ando T, Hayashi K, Tanaka H, et al. . Endoscopic transpapillary intraductal ultrasonography and biopsy in the diagnosis of igg4-related sclerosing cholangitis . J Gastroenterol 2009. ; 44: 1147 – 55 . doi: 10.1007/s00535-009-0108-9 [DOI] [PubMed] [Google Scholar]
- 6. Kobori I, Suda T, Nakamoto A, Saito H, Okawa O, Sudo R, et al. . Two cases of immunoglobulin G4-related sclerosing cholangitis in which transabdominal ultrasonography was useful in diagnosis and follow-up observation . J Med Ultrason (2001) 2016. ; 43: 271 – 77 . doi: 10.1007/s10396-015-0676-7 [DOI] [PubMed] [Google Scholar]
- 7. Mohammad Alizadeh AH . Cholangitis: diagnosis, treatment and prognosis . J Clin Transl Hepatol 2017. ; 5: 404 – 13 . doi: 10.14218/JCTH.2017.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo W-L, Wang J . Preoperative one-stop magnetic resonance imaging evaluation of the pancreaticobiliary junction and hepatic arteries in children with pancreaticobiliary maljunction: a prospective cohort study . Surg Today 2021. ; 51: 79 – 85 . doi: 10.1007/s00595-020-02077-5 [DOI] [PubMed] [Google Scholar]
- 9. Lin B-S, Chen J-L, Tu Y-H, Shih Y-X, Lin Y-C, Chi W-L, et al. . Using deep learning in ultrasound imaging of bicipital peritendinous effusion to grade inflammation severity . IEEE J Biomed Health Inform 2020. ; 24: 1037 – 45 . doi: 10.1109/JBHI.2020.2968815 [DOI] [PubMed] [Google Scholar]
- 10. Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng PA . H-denseunet: hybrid densely connected unet for liver and tumor segmentation from CT volumes . IEEE Trans Med Imaging 2018. ; 37: 2663 – 74 . doi: 10.1109/TMI.2018.2845918 [DOI] [PubMed] [Google Scholar]
- 11. Li J, Lin X, Che H, Li H, Qian X . Probability map guided bi-directional recurrent unet for pancreas segmentation . 2019. . [DOI] [PubMed]
- 12. Pan T, Shu H, Coatrieux J-L, Yang G, Wang C, Lu Z, et al. . ( n.d .). A multi-task convolutional neural network for renal tumor segmentation and classification using multi-phasic CT images . International Conference on Image Processing ; 809 – 13 . [Google Scholar]
- 13. Monaco S, Bussola N, Butto S, Sona D, Apiletti D, Jurman G, et al. . Cyst segmentation on kidney tubules by means of U-Net deep-learning models . In : Jurman G ed . 2021 IEEE International Conference on Big Data (Big Data) ; Orlando, FL, USA . ; 15 December 2021. . doi: 10.1109/BigData52589.2021.9671669 [DOI] [Google Scholar]
- 14. Kurita Y, Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, et al. . Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions . Sci Rep 2019. ; 9( 1 . doi: 10.1038/s41598-019-43314-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Narmatha C, Manimegalai P, Krishnadass J, Valsalan P, Manimurugan S . Ovarian cysts classification using novel deep Q-learning with harris hawks optimization method . 2021. .
- 16. Sriram SA, Paul A, Zhu Y, Sandfort V, Pickhardt PJ, Summers R, et al. . Multilevel UNet for pancreas segmentation from non-contrast CT scans through domain adaptation . Computer-Aided Diagnosis ; Houston, United States . ; 19 March 2020. . doi: 10.1117/12.2551093 [DOI] [Google Scholar]
- 17. Yi X, Adams S, Babyn P, Elnajmi A . Automatic catheter and tube detection in pediatric x-ray images using a scale-recurrent network and synthetic data . J Digit Imaging 2020. ; 33: 181 – 90 . doi: 10.1007/s10278-019-00201-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim TK, Yi PH, Wei J, Shin JW, Hager G, Hui FK, et al. . Deep learning method for automated classification of anteroposterior and posteroanterior chest radiographs . J Digit Imaging 2019. ; 32: 925 – 30 . doi: 10.1007/s10278-019-00208-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Somasundaram E, Dillman JR, Crotty EJ, Trout AT, Towbin AJ, Anton CG, et al. . Automatic detection of inadequate pediatric lateral neck radiographs of the airway and soft tissues using deep learning . Radiol Artif Intell 2020. ; 2. doi: 10.1148/ryai.2020190226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Z, Siddiquee MR, Tajbakhsh N, Liang J . UNet++: a nested U-net architecture for medical image segmentation . 2018. ; 3 – 11 . [DOI] [PMC free article] [PubMed]
- 21. Zhang H, Wu C, Zhang Z, Zhu Y, Zhang Z, Lin H, et al. . ResNeSt: split-attention networks . 2020. .
- 22. Chiu C-J, McArdle AH, Brown R, Scott HJ, Gurd FN . Intestinal mucosal lesion in low-flow states. I. A morphological, hemodynamic, and metabolic reappraisal . Arch Surg 1970. ; 101: 478 – 83 . doi: 10.1001/archsurg.1970.01340280030009 [DOI] [PubMed] [Google Scholar]
- 23. Ronneberger O, Fischer P, Brox T . U-net: convolutional networks for biomedical image segmentation . Med Image Comput Comput Assist Interv 2015. ; 234 – 41 . [Google Scholar]
- 24. Alom MZ, Yakopcic C, Taha TM, Asari VK . Nuclei segmentation with recurrent residual convolutional neural networks based U-net (R2U-Net) . NAECON 2018 - IEEE National Aerospace and Electronics Conference ; Dayton, OH, USA . ; July 2018. . doi: 10.1109/NAECON.2018.8556686 [DOI] [Google Scholar]
- 25. Xie S, Girshick R, Dollar P, Tu Z, He K . Aggregated Residual Transformations for Deep Neural Networks . 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) ; Honolulu, HI . ; July 2017. . pp . 5987 – 95 . doi: 10.1109/CVPR.2017.634 [DOI] [Google Scholar]
- 26. Hu J, Shen L, Sun G . Squeeze-and-excitation networks . Computer Vision and Pattern Recognition 2018. ; 7132 – 41 . [Google Scholar]
- 27. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D . Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization . In : Batra D ed . 2017 IEEE International Conference on Computer Vision (ICCV) ; Venice . ; October 2017. . doi: 10.1109/ICCV.2017.74 [DOI] [Google Scholar]
- 28. Shi W, Kuang S, Cao S, Hu B, Xie S, Chen S, et al. . Deep learning assisted differentiation of hepatocellular carcinoma from focal liver lesions: choice of four-phase and three-phase CT imaging protocol . Abdom Radiol (NY) 2020. ; 45: 2688 – 97 . doi: 10.1007/s00261-020-02485-8 [DOI] [PubMed] [Google Scholar]
- 29. Huang S, Lee F, Miao R, Si Q, Lu C, Chen Q . A deep convolutional neural network architecture for interstitial lung disease pattern classification . Med Biol Eng Comput 2020. ; 58: 725 – 37 . doi: 10.1007/s11517-019-02111-w [DOI] [PubMed] [Google Scholar]