Abstract
Objective:
The purpose is to establish and validate a machine-learning-derived radiomics approach to determine the existence of atrial fibrillation (AF) by analyzing epicardial adipose tissue (EAT) in CT images.
Methods:
Patients with AF based on electrocardiographic tracing who underwent contrast-enhanced (n = 200) or non-enhanced (n = 300) chest CT scans were analyzed retrospectively. After EAT segmentation and radiomics feature extraction, the segmented EAT yielded 1691 radiomics features. The most contributive features to AF were selected by the Boruta algorithm and machine-learning-based random forest algorithm, and combined to construct a radiomics signature (EAT-score). Multivariate logistic regression was used to build clinical factor and nested models.
Results:
In the test cohort of contrast-enhanced scanning (n = 60/200), the AUC of EAT-score for identifying patients with AF was 0.92 (95%CI: 0.84–1.00), higher than 0.71 (0.58–0.85) of the clinical factor model (total cholesterol and body mass index) (DeLong’s p = 0.01), and higher than 0.73 (0.61–0.86) of the EAT volume model (p = 0.01). In the test cohort of non-enhanced scanning (n = 100/300), the AUC of EAT-score was 0.85 (0.77–0.92), higher than that of the CT attenuation model (p < 0.001). The two nested models (EAT-score+volume and EAT-score+volume+clinical factors) for contrast-enhanced scan and one (EAT-score+CT attenuation) for non-enhanced scan showed similar AUCs with that of EAT-score (all p > 0.05).
Conclusion:
EAT-score generated by machine-learning-based radiomics achieved high performance in identifying patients with AF.
Advances in knowledge:
A radiomics analysis based on machine learning allows for the identification of AF on the EAT in contrast-enhanced and non-enhanced chest CT.
Introduction
Atrial fibrillation (AF) is the most common arrhythmia with clinical significance. 1 The prevalence of AF in adults is between 2 and 4%. 2 The European Society of Cardiology predicted that 18 million people in the European Union will suffer from AF by 2060. 3 The clinical symptoms of patients with AF are often non-specific, such as fatigue and palpitations. The diagnosis of AF requires rhythm documentation based on electrocardiographic (ECG) tracing. But AF is usually asymptomatic, and paroxysmal AF occurs in 20–30% of diagnosed AF cases. 4 A single standard ECG test cannot rule it out. 5 Because AF is often underestimated or missed, it increases the risk of stroke, systemic thromboembolism, and heart failure. 6
Epicardial adipose tissue (EAT) is located between the visceral pericardium and the myocardium, which secretes proinflammatory factors or cytokines. 7 EAT promotes the occurrence and progression of AF through increased inflammation, adipose infiltration, autonomic nervous system dysfunction, fibrosis, and structural remodeling. 8 Al Chekakie et al demonstrated that patients with AF had more EAT compared with those with sinus rhythm, independent of factors such as left atrial enlargement and body mass index (BMI). 9
With its high-spatial resolution, CT provides a convenient and reproducible way to quantify EAT. 10 Militello et al proposed a semi-automatic method to segment and quantify epicardial fat volume on cardiac CT scans. 11 According to the Centers for Medicare and Medicaid Services, the overall utilization of non-ECG-triggered chest CT scans has risen abruptly to 114 examinations per 1000 individuals per year, which is much higher than ECG-triggered cardiac CT. 12,13 Taking advantage of the extensive availability of chest CT is conducive to screening research.
Given that AF is sometimes asymptomatic and paroxysmal, and sometimes its symptoms are atypical, it can be easily missed. The early detection and treatment of AF are vital to prevent its complications. 3 The existing biological evidence strongly supports the hypothesis that the occurrence and progression of AF are closely associated with EAT. Data mining of EAT images may be a breakthrough in identifying AF. We hypothesized that chest CT, while completing the radiological diagnostic task, would also discriminate AF patients even if there was no AF attack at the time of examination. Once identified as AF based on CT image features, patients can visit a cardiologist for diagnosis to avoid complications caused by delayed diagnosis. This study aimed to establish and validate a machine-learning-derived radiomics approach to determine the existence of AF by analyzing EAT in CT images.
Methods and materials
Study population
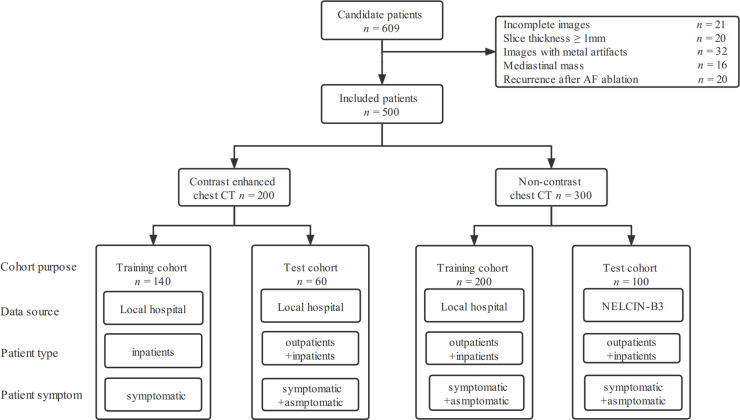
The patients from January 2013 to September 2020 were retrospectively included. The inclusion criteria were: (1) patients underwent standard 12-lead ECG recording or single-lead ECG tracing for ≥30 s; (2) followed by contrast-enhanced or non-enhanced chest CT scanning within 2 weeks; (3) two cardiologists with more than 10 years of experience diagnosed AF or normal sinus rhythm (NSR) according to the European Society of Cardiology guideline 2020. 2 Figure 1 displays a flow diagram of patient selection and cohort details. This study complied with the Declaration of Helsinki. The local Institutional Review Board approved this retrospective study and waived the need for written informed consent.
Figure 1.
Workflow diagram of patient selection and cohort composition. AF, atrial fibrillation; NELCIN-B3, Netherlands–China Big-3; NSR, normal sinus rhythm.
Image data sets
We analyzed the data sets of contrast-enhanced and non-enhanced CT, separately. The contrast-enhanced CT data set from Shanghai General Hospital (n = 200) was divided into the training cohort of 70% (from January 2013 to June 2019) and the test cohort of 30% (from July 2019 to September 2020). The non-enhanced CT dataset (n = 300) included the training cohort (n = 200) from Shanghai General Hospital and the external test cohort (n = 100) from the NELCIN-B3 trial (Netherlands–China Big-3 disease screening: lung cancer, coronary atherosclerosis, and chronic obstructive pulmonary disease). 14
CT image acquisition
A total of four CT scanners were used in this study, namely SOMATOM Definition Flash (Siemens Healthineers, Erlangen, Germany), uCT760 (United-imaging, Shanghai, China), Revolution CT (GE Healthcare, Milwaukee, US), and HD 750 (GE Healthcare, Milwaukee, US). All subjects underwent non-ECG-triggered chest CT. Patients with enhanced scans were injected with 60–80 ml contrast media (Iopamiro 300, Bracco, Milan, Italy) at a rate of 3–4 ml s−1 through the antecubital vein. Supplementary Table 1 shows the details of the scanning protocol.
EAT segmentation and radiomics feature extraction
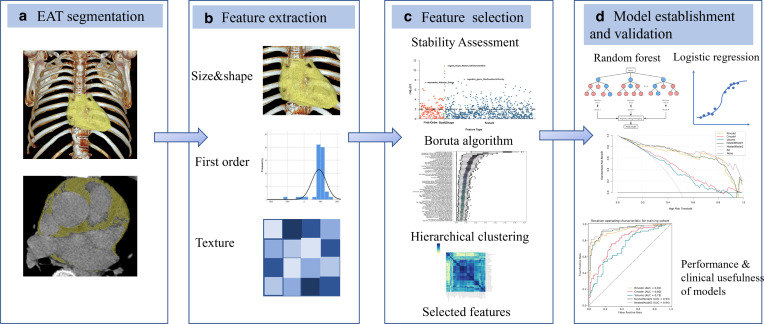
Figure 2 depicts the radiomics analysis approach and Figure 3 demonstrates several representative EAT segmentation cases. The process of EAT segmentation and radiomics feature extraction was the same in contrast-enhanced and non-enhanced CT scans. The Supplementary Methods describe the detailed segmentation procedures. Dedicated analysis software (Radiomics 1.0.9a, Siemens Healthineers, Erlangen, Germany) extracts radiomics features based on the Pyradiomics library (Pyradiomics 3.0, https://pyradiomics.readthedocs.io/en/latest/) in conformance with the Image Biomarker Standardization Initiative. 15 Finally, 1691 features, including three major categories: 18 first-order features, 75 texture features, and 17 size and shape features, were extracted from each package-segmented EAT volume. Figure 1 and Supplementary Table 3 to S9 list the composition and interpretation of features.
Figure 2.
Process of establishing a machine-learning-derived radiomics approach. (A) Epicardial adipose tissue on chest CT images was semi-automatically segmented. (B) Automatic extraction of 1691 radiomics features, including size and shape features, first-order features, and texture features. (C) Stability assessment, Boruta algorithm, and hierarchical clustering were used to select the contributive features to atrial fibrillation. Finally, the most contributive features were selected to create an EAT-score by random forest algorithm. (D) Random forest algorithm and multivariate logistic regression were used to identify patients with atrial fibrillation. EAT, Epicardial adipose tissue.
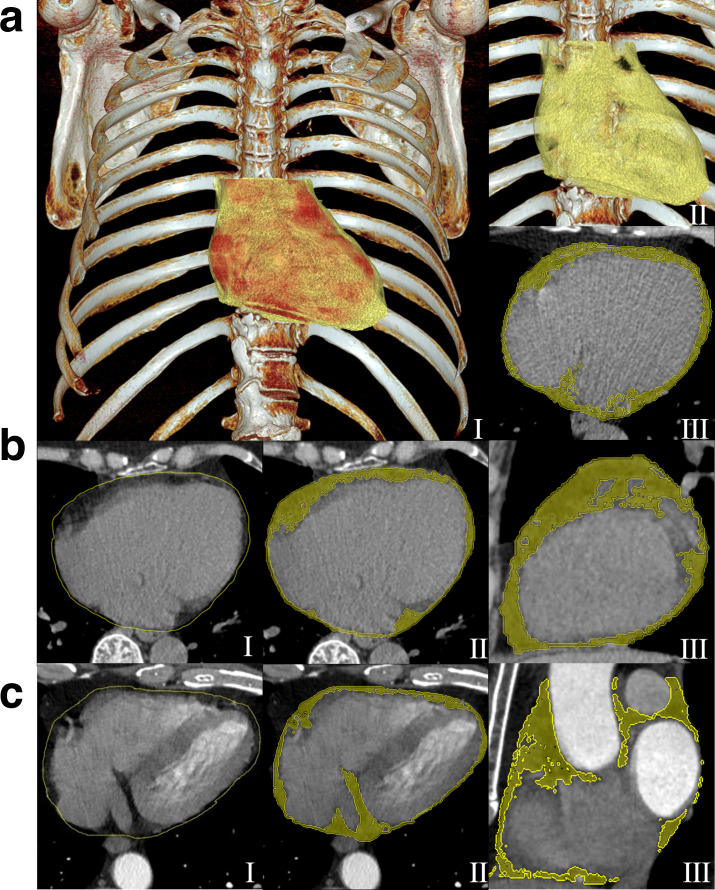
Figure 3.
Schematic diagram for EAT segmentation. (A) Two 3D volume rendering images (I and II) and an axial CT image (III) show the heart (red color) and EAT (yellow). (B, C) The manually delineated pericardial contours (yellow) (I). A dedicated radiomics analysis package overlays the pericardial contours into chest CT images to segment EAT volume, which is defined by a CT value range for fatty tissues within the pericardial contours (II-axial and III-sagittal). (B) The images of non-enhanced scan of a 70-year-old female in the validation cohort. The radiomics model generated an EAT-score of 0.17 and considered her as normal sinus rhythm, that was finally conformed by 12-lead ECG tracing. (C) The images of contrast-enhanced chest CT of a 52-year-old male in the test cohort. The radiomics model generated an EAT-score of 0.97 and considered him as atrial fibrillation, that was finally proved by 12-lead ECG tracing. EAT, epicardial adipose tissue; ECG, electrocardiogram.
Feature selection and EAT-score construction
The same methods analyzed the data sets of contrast-enhanced and non-enhanced scans. The selection for usable radiomics features depends on feature stability. The radiologist with 4 years of experience in thoracic imaging randomly selected 25 patients from the training cohort to perform EAT segmentation and feature extraction, and repeated them 4 weeks later. The radiomics features with an intraclass correlation coefficient >0.8 were selected as stable features. 16
Subsequently, highly correlated features with AF were selected based on the Boruta algorithm, a wrapper around random forest algorithm, 17 which iteratively removes the features and eventually selects all relevant features for AF recognition. Once the features were selected, hierarchical clustering will converge to similar features. In each cluster, according to the Boruta algorithm, the feature with the highest average importance was the candidate feature. Finally, the random forest algorithm, a supervised machine-learning algorithm composed of a few decision tree classifiers, 18 identified the features which contributed the most to identifying AF.
A radiomics signature was constructed by a random forest algorithm. To avoid overfitting, parameter estimation was performed by grid search with 10-fold cross-validation in the training cohort. Gini impurity decreases overall decision trees were calculated to evaluate the feature importance. Gini impurity measures how often a randomly chosen element from the set would be mislabeled if it were randomly labeled according to the distribution of labels in the subset. 19 Thereafter, a radiomics signature, namely EAT-score ranging from 0 to 1, was created to represent the probability of AF in each patient. With the increase of EAT-score, AF is more likely to occur.
Model establishment
For contrast-enhanced CT, six models were established to identify AF based on EAT-score, EAT volume, CT attenuation, clinical factors, and two nested models (EAT-score+volume and EAT-score+volume + clinical factors). The volume model and the CT attenuation model were established via univariate logistic regression analysis and the model with a p < 0.05 could be used to build the nested models. CT attenuation refers to the average CT attenuation of EAT. In constructing the clinical factor model, the factors with a p < 0.05 of univariable logistic regression analysis were included in multivariate logistic regression (MLR) to evaluate its discrimination ability. The independent factors that yielded a p < 0.05 in MLR were included to generate the clinical factor and nested models.
For non-enhanced CT, four models were established to identify AF based on EAT-score, EAT volume, CT attenuation, and a nested model (EAT-score+CT attenuation) in the same way as contrast-enhanced CT. Among the 300 patients with non-enhanced scans, 109 (36.3%) were outpatients without clinical data, so the clinical factor model could not be established.
Statistical analysis
Continuous variables are expressed as median (interquartile range) according to the data normality by the Kolmogorov–Smirnov test, and categorical variables are represented as frequencies (percentages). Categorical variables were tested with the χ2 or Fisher’s exact tests. The Wilcoxon rank-sum test compared continuous variables between cohorts. The receiver operating characteristic (ROC) curve, area under ROC curve (AUC), accuracy, sensitivity, and specificity reflected the discrimination ability of the models. Youden’s index determined the optimal cut-off value of each model. The DeLong test compared the AUCs between models.
A two-sided p < 0.05 was considered statistically significant. Statistical analysis was performed using open-source packages (R v. 3.6.0, http://www.Rproject.org; Python v. 3.7 with Scikit-survival library v. 0.13.2, https://scikit-survival.readthedocs.io/en/latest/). Supplementary Table 2 lists the details of software packages and functions. The Supplementary Materials provides the methods and results of calibration curves and decision curve analysis.
Results
Patient characteristics
500 patients (292 men, median age: 67 years, interquartile range: 59–74) were eligible for this retrospective study, including 250 patients with AF. 200 (120 men, 65 years, 59–70) who underwent contrast-enhanced chest CT were included. Another 300 patients (172 men, 69 years, 59–76) who underwent non-enhanced chest CT were enrolled, including 200 and 100 as the training and test cohorts, respectively. Table 1 summarizes the patient characteristics.
Table 1.
Characteristics of patients who underwent contrast-enhanced or non-enhanced chest CT scans
| Patients with contrast-enhanced chest CT scan | |||
|---|---|---|---|
| Training cohort (n = 140) | Test cohort (n = 60) | p-value | |
| Age, year | 65 (58–71) | 65.0 (60–70) | 0.90 |
| Sex (males, n [%]) | 85 (60.7%) | 35 (58.3%) | 0.88 |
| Total cholesterol, mmol/l | 4.46 (3.80–5.22) | 4.44 (3.94–4.87) | 0.31 |
| HDL, mmol/l | 1.18 (0.99–1.41) | 1.18 (1.01–1.25) | 0.23 |
| BMI, kg/m² | 23.9 (21.8–26.0) | 24.0 (21.7–26.2) | 0.92 |
| Hypertension therapy, n (%) | 64 (45.7%) | 22 (36.7%) | 0.30 |
| Diabetes, n (%) | 27 (19.3%) | 4 (6.67%) | 0.04 |
| Smoking, n (%) | 21 (15.0%) | 10 (16.7%) | 0.93 |
| Drinking, n (%) | 10 (7.1%) | 5 (8.3%) | 0.77 |
| Heart failure, n (%) | 7 (5.0%) | 3 (5.0%) | >0.99 |
| Hyperthyroidism, n (%) | 1 (0.7%) | 1 (1.7%) | 0.51 |
| OSAHS, n (%) | 0 (0) | 0 (0) | N/A |
| |
Patients with non-enhanced chest CT scan | ||
| Training cohort (n = 200) | Test cohort (n = 100) | p-value | |
| Age, year | 71 (62–78) | 65 (55–71) | 0.01 |
| Sex (males, n [%]) | 129 (64.5%) | 44 (44.0%) | <0.001 |
BMI, body mass index; HDL, high density lipoprotein cholesterol; N/A, not available; OSAHS, obstructive sleep apnea hypopnea syndrome.
p-value refers to the significance between training and internal test cohorts. Continuous variables are expressed as median (interquartile range).
Radiomics feature selection
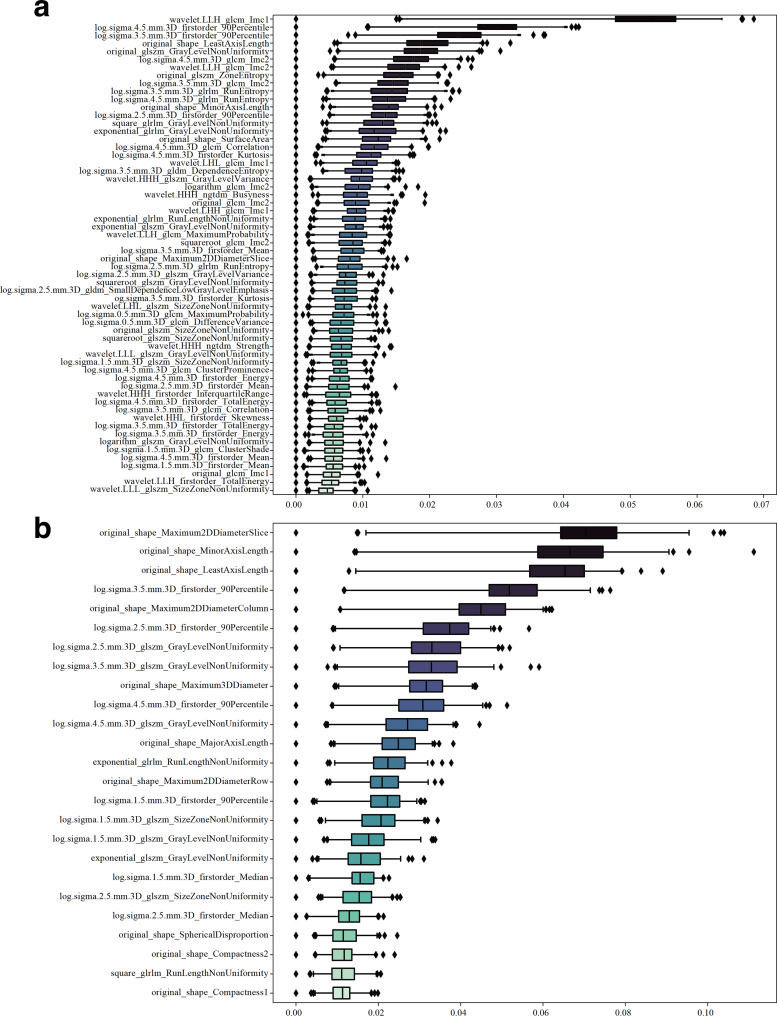
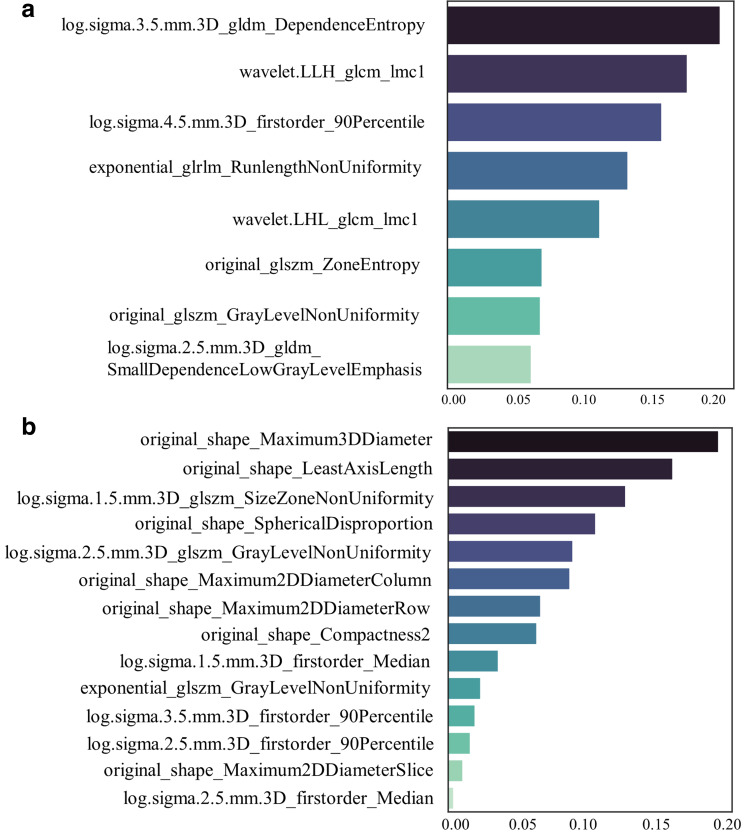
Among the features extracted in the contrast-enhanced and non-enhanced scans, 1186/1691 (70%) and 1280/1691 (76%) had good stability (intraclass correlation coefficient>0.8), respectively, and thus were selected for further analysis. The Boruta algorithm identified 60 and 25 features related to the occurrence of AF (Figure 4), and hierarchical clustering yielded 10 and 16 clusters, respectively (Figures 2 and 3). Based on the 10 and 16 features, the random forest algorithm identified 8 and 14 that were closely associated with the presence of AF. These features were ranked according to their importance in generating an EAT-score (Figure 5). The swarm plots (Figures 4 and 5) illustrate the distribution of features between the AF and NSR groups in the training cohorts. The correlation between the selected features is shown in Supplementary Figure 6 and 7.
Figure 4.
Importance ranking for feature selection using Boruta algorithm. (A) For the dataset of contrast-enhanced chest CT scans, 60 features were selected by Boruta algorithm; (B) For the data set of non-enhanced scans, 25 features were selected. GLCM, gray level co-occurrence matrix; GLDM, gray level dependence matrix; GLRLM, gray level run length matrix; GLSZM, gray level size zone matrix.
Figure 5.
Importance ranking of the most-contributive radiomics features to identify atrial fibrillation. (A) for contrast-enhanced chest CT scans and (B) for non-enhanced scans. The darker the color is, the higher the importance. The sum of the importance index of features is one. GLCM, gray level co-occurrence matrix; GLDM, gray level dependence matrix; GLRLM, gray level run length matrix; GLSZM, gray level size zone matrix.
Model construction in contrast-enhanced CT scans
The EAT-scores for the AF (median: 0.69, interquartile range: 0.61–0.97) and NSR groups (0.28, 0.17–0.36) were significantly different in the training cohort (p < 0.001). The optimal threshold of EAT-score determined by ROC analysis was 0.57, so patients with an EAT-score>0.57 were considered to have AF. In the training cohort, the EAT-score achieved an excellent AUC of 0.92 [95% confidence interval (CI): 0.88–0.96], the high specificity of 97% (89–100%), and an accuracy of 87% (80–92%). The high specificity indicates a low false-positive rate, which may help avoid unnecessary delays when screening patients with NSR. The MLR analysis showed that total cholesterol [odds ratio: 0.47 (95%CI: 0.31–0.69), p < 0.001] and BMI [1.36 (1.19–1.58), p < 0.001] were significantly associated with the presence of AF, so they were included in the clinical factor model. The CT attenuation model was not significantly associated with AF (p = 0.74), so CT attenuation was not incorporated into the nested model.
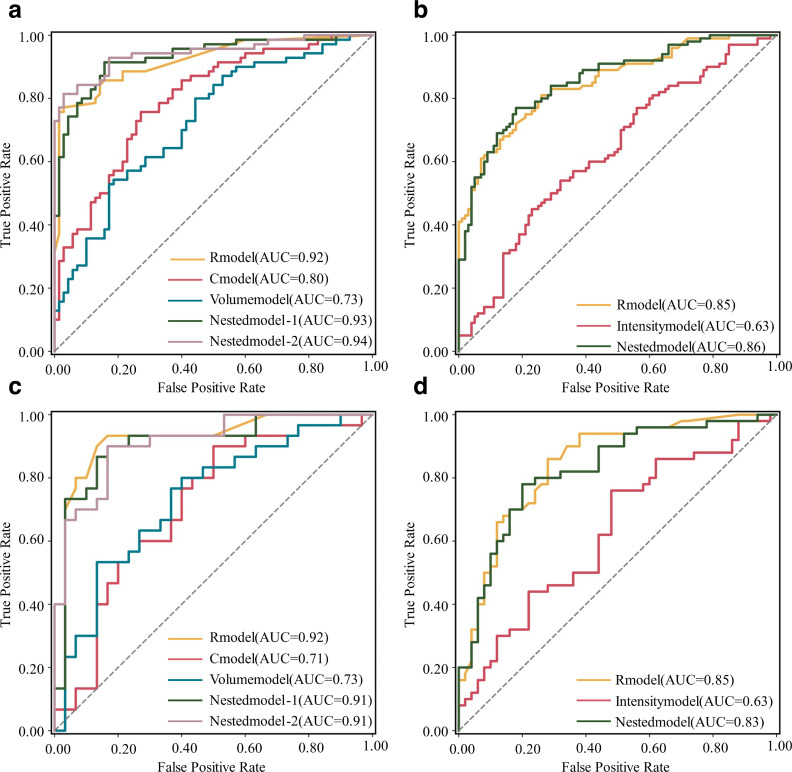
In the training cohort, the nested model (EAT-score+volume) performed excellently with an AUC of 0.93 (0.89–0.97). Meanwhile, the other nested model (EAT-score+volume + clinical factors) also achieved a high AUC of 0.94 (0.90–0.98). However, these two nested models did not significantly improve the AUCs compared with EAT-score (DeLong’s p = 0.25 and 0.46, respectively). The AUC of the clinical factor model was 0.80 (0.72–0.87), significantly lower than EAT-score (p = 0.01). The AUC of the volume model was 0.73 (0.64–0.81), also significantly lower than EAT-score (p < 0.001). Figure 6A and C demonstrate the ROCs. Table 2 lists the diagnostic performance metrics.
Figure 6.
ROC curves of all models for the datasets of contrast-enhanced and non-enhanced CT scans. (A, C) are ROC curves in the training and test cohorts for the dataset of contrast-enhanced scans. (B, D) are ROC curves in the training and test cohorts for the dataset of non-enhanced scans. AUC, area under the ROC curve; Cmodel, clinical factor model; ROC, receiver operating characteristic; Rmodel, radiomics model.
Table 2.
Performance metrics of all models of contrast-enhanced chest CT scans
| Model | AUC | p-value | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Training cohort | |||||
| Radiomics model | 0.92 [0.88–0.96] | N/A | 87% (122/140) [80–92] | 77% (54/70) [65–86] | 97% (68/70) [89–100] |
| Clinical factor model | 0.80 [0.72–0.87] | 0.01 | 74% (104/140) [66–81] | 76% (53/70) [64–85] | 73% (51/70) [61–82] |
| Volume model | 0.73 [0.64–0.81] | <0.001 | 68% (95/140) [59–75] | 53% (37/70) [41–65] | 83% (58/70) [72–90] |
| Nested model-1 (EAT- score+volume) | 0.93 [0.89–0.97] | 0.25 | 88% (123/140) [81–93] | 91% (64/70) [82–96] | 84% (59/70) [73–92] |
| Nested model-2 (EAT-score+volume + clinical factors) | 0.94 [0.90–0.98] | 0.46 | 89% (125/140) [83–94] | 81% (57/70) [70–89] | 97% (68/70) [89–100] |
| Test cohort | |||||
| Radiomics model | 0.92 [0.84–1.00] | N/A | 80% (48/60) [68–89] | 63% (19/30) [44–80] | 97% (29/30) [81–100] |
| Clinical factor model | 0.71 [0.58–0.85] | 0.01 | 67% (40/60) [53–78] | 77% (23/30) [57–89] | 57% (17/30) [38–74] |
| Volume model | 0.73 [0.61–0.86] | 0.01 | 68% (41/60) [55–80] | 57% (17/30) [38–74] | 80% (24/30) [61–92] |
| Nested model-1 (EAT- score+volume) | 0.91 [0.83–0.99] | 0.46 | 82% (49/60) [70–90] | 93% (28/30) [76–99] | 70% (21/30) [50–85] |
| Nested model-2 (EAT-score+volume + clinical factors) | 0.91 [0.84–1.00] | 0.90 | 80% (48/60) [68–89] | 73% (22/30) [54–87] | 87% (26/30) [68–96] |
AUC, area under the receiver operating characteristic curve.
Data in the brackets are 95% confidence interval and data in the parentheses are the numerator and denominator for percentages. p values represent the comparison of AUC between radiomics model and other models.
Model validation in contrast-enhanced CT scans
In the test cohort, the EAT-scores of the AF (median: 0.66, interquartile range: 0.55–0.78) and NSR groups (0.32, 0.19–0.47) were significantly different (p < 0.001). The EAT-score reached an AUC of 0.92 (95%CI: 0.84–1.00), a specificity of 97% (89–100%), and an accuracy of 80% (68–89%). In the test cohort, the nested models performed as well as EAT-score. The two nested models yielded AUCs of 0.91 (0.83–0.99, DeLong’s p = 0.46 compared with EAT-score) and 0.91 (0.84–0.98, p = 0.90), respectively. Similar to the training cohort, the AUC of the volume model was 0.73 (0.61–0.86), significantly lower than that of EAT-score (p = 0.01). The AUC of the clinical factor model was 0.71 (0.58–0.85), also significantly lower than that of EAT-score (p = 0.01). Table 2 provides the results of model performance.
Model construction in non-enhanced CT scans
The EAT-score of the AF (median: 0.69, interquartile range: 0.47–0.86) and NSR groups (0.29, 0.18–0.42) were significantly different in the training cohort (p < 0.001). The ROC analysis showed that the patients with an EAT-score>0.40 had AF. In the training cohort, the AUC of EAT-score was 0.85 (0.80–0.90), higher than 0.63 (0.55–0.71) of the CT attenuation model (DeLong’s p < 0.001). The EAT volume cannot be included to build a nested model, based on univariable logistic regression analysis (p > 0.05). The AUC of the nested model (EAT-score+CT attenuation) was 0.86 (0.77–0.92), which was not significantly better than that of only EAT-score (p = 0.41). Table 3 lists diagnostic performance metrics.
Table 3.
Performance metrics of all models of non-contrast chest CT scans
| Model | AUC | Accuracy (%) | Sensitivity (%) | Specificity (%) | p-value |
|---|---|---|---|---|---|
| Training cohort | |||||
| Radiomics model | 0.85 [0.80–0.90] | 78% (155/200) [71–83] | 81% (81/100) [72–88] | 74% (74/100) [64–82] | N/A |
| CT attenuation model | 0.63 [0.55–0.71] | 61% (122/200) [54–68] | 54% (54/100) [44–64] | 68% (68/100) [58–77] | <0.001 |
| Nested model (EAT-score+CT attenuation) | 0.86 [0.81–0.91] | 80% (159/200) [73–85] | 77% (77/100) [67–85] | 82% (82/100) [73–89] | 0.41 |
| Test cohort | |||||
| Radiomics model | 0.85 [0.77–0.92] | 77% (77/100) [68–85] | 70% (35/50) [55–82] | 84% (42/50) [70–92] | N/A |
| CT attenuation model | 0.63 [0.52–0.74] | 51% (51/100) [41–61] | 88% (44/50) [75–95] | 14% (7/50) 6–27 | <0.001 |
| Nested model (EAT-score+CT attenuation) | 0.83 [0.74–0.91] | 76% (76/100) [66–84] | 68% (34/50) [53–80] | 84% (42/50) [70–92] | 0.10 |
AUC, area under the receiver operating characteristic curve; EAT, epicardial adipose tissue.
Data in the brackets are 95% confidence interval and data in the parentheses are the numerator and denominator for percentages. The p-value represents significance of AUC between the radiomics model and each of other models.
Validation of models in non-enhanced CT scans
In the test cohort, the EAT-scores for the AF (median: 0.50, interquartile range: 0.34–0.65) and NSR groups (0.21, 0.12–0.32) were significantly different (p < 0.001). The EAT-score proved excellent performance in identifying AF. The EAT-score reached an AUC of 0.85 (95%CI: 0.77–0.92) and a specificity of 84% (70–92%). The AUC of the nested model was 0.83 (0.74–0.91), which was not significantly different from that of EAT-score (DeLong’s p = 0.100). Both the EAT-score and nested models outperformed the CT attenuation model (p < 0.001), whose AUC was only 0.63 (0.52–0.74). Figure 6B and D demonstrate the ROCs. Table 3 provides the results of model performance.
Discussion
Regardless of whether the patients were subjected to contrast-enhanced or non-enhanced scans, the radiomics model, or called EAT-score, can distinguish the patients with AF from those with NSR. The EAT-score yielded an excellent diagnostic performance of identifying AF with a high AUC of 0.92 and 0.85 in the test cohorts of contrast-enhanced and non-enhanced scans, respectively. The nested models showed diagnostic values similar to those of the EAT-scores. The EAT-score and nested model exhibited higher performance than that of the traditional predictors, such as clinical factors (total cholesterol and BMI, AUC: 0.71), EAT volume (0.73), and CT attenuation (0.63).
CT attenuation is often used to evaluate adipose tissue, but in this study, the CT attenuation model of contrast-enhanced CT was insignificant to determine AF. It might be due to the influence of contrast-media enhancement of EAT, which interferes with the CT values between AF and NSR groups. For EAT volume, the volume model was significant to determine AF. The EAT volume of patients with AF was higher than those with NSR in contrast-enhanced CT (96 vs 86 ml), and in non-enhanced scans (133 vs 90 ml). These findings are consistent with the meta-analysis of studies that reported higher EAT volume in patients with AF. 20 The morphological changes of EAT in patients with AF suggest that the extraction of high-throughput EAT image features may be more effective and reliable to identify patients with AF.
Radiomics is a thriving field that can mine useful quantitative information from various imaging modalities. The potential of radiomics in cardiac imaging has gradually been recognized. Recently, researchers developed several models for differential diagnosis of cardiac diseases, such as myocardial infarction and vulnerable coronary plaques. 21,22 Kolossváry et al proved that radiomics features outperformed conventional CT signs (AUC of 0.72 vs 0.59), i.e. positive remodeling, spotty calcification, and napkin-ring, to identify vulnerable plaques in coronary CTA. 21 Our study proposed using the radiomics-based EAT-score to determine patients with AF, which is an innovative research direction in cardiac imaging.
In this study, contrast- and non-enhanced CT scans differed in the imaging feature types that contributed to establishing the EAT score. For contrast-enhanced CT scans, 88% (7/8) of the most relevant features were texture features. For non-enhanced scans, 50% (7/14) were size and shape features. Towards the difference of the selected features, we considered that contrast-enhanced scanning can capture the histological changes induced by inflammation (such as fibrosis and neoangiogenesis), which is one main mechanism of AF. 23,24 Texture features depict the spatial relationship and voxel distribution, represent the heterogeneity of voxels, 25 and reflect the phenotype of histological changes in adipose tissue. Oikonomou et al found that radiomics texture features can reflect the histological heterogeneity of perivascular fat (affected by fibrosis and microvascular remodeling). 26 Thereafter, the radiomics features in contrast-enhanced CT of adipose tissue may help elucidate the basis of EAT-induced AF from the computing perspective of inflammation.
At present, there are multiple methods available to detect AF. Insertable cardiac monitors are helpful to screen AF, but they are invasive and expensive. Smartphone apps and wearable activity monitors have rapidly evolved to discriminate between AF and sinus rhythm. 27 These devices still formulate diagnoses based on ECGs converted from the information acquired by sensors that cannot avoid the problems of potentially undetectable AF in the absence of an attack. The application of deep learning to the analysis of ECG signals can compensate for some disadvantages of ECG. Attia et al created an artificial intelligence ECG with an AUC of 0.90 with convolutional neural networks to identify AF even when the ECG exhibited NSR. 28
This study has limitations. First, this is a retrospective analysis of a non-ethnically diverse population. The conclusions of this study can be expanded and generalized if conducting a prospective study in another region. Second, in the NSR subjects, there may be false-negative patients on a single ECG if they did not have a history of AF. However, our results should not be significantly influenced by this limitation as paroxysmal AF generally involves only a few NSR subjects. The prevalence of AF in adults was estimated to be 2–4%, 2 of which paroxysmal AF accounted for 20–30%. 4 Third, this study focused on identifying patients with AF and NSR based on the image feature analysis of EAT. It is essential to investigate whether machine-learning-derived analysis of EAT can identify other types of arrhythmias.
Conclusion
This proof-of-concept study established diagnostic models based on machine-learning-derived radiomics to identify patients with AF using EAT images in contrast-enhanced and non-enhanced chest CT. These models exhibited high diagnostic performance in identifying AF. As a new biological insight on AF from a computational perspective, machine-learning provides an innovative research direction for diagnosing patients with AF. Considering the high prevalence of AF and the extensive use of chest CT, the machine-learning-derived radiomics model is feasible to screen patients with AF who are asymptomatic, paroxysmal, or with atypical symptoms, but have not been diagnosed before.
Supplementary Material
Footnotes
Acknowledgments: Zhihan Xu is an employee of Siemens Healthineers Ltd., the CT manufacturer and software provider in this study, who has no control over any data and information that may cause conflicts of interest.
Funding: This work was supported by the Ministry of Science and Technology of China (2016YFE0103000), the National Natural Science Foundation of China (project no. 81971612 and 81471662), and Shanghai Jiao Tong University (ZH2018ZDB10).
Contributor Information
Lu Zhang, Email: zhangluuuu@hotmail.com, Department of Radiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China .
Zhihan Xu, Email: zhihan.xu@siemens-healthineers.com, Siemens Healthineers Ltd., Zhouzhu Rd.278, Shanghai, China .
Beibei Jiang, Email: jennifer.chiang@hotmail.com, Department of Radiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China .
Yaping Zhang, Email: zhangyaping.ok@163.com, Department of Radiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China .
Lingyun Wang, Email: wanglingyun0514@163.com, Department of Radiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China .
Geertruida H de Bock, Email: g.h.de.bock@umcg.nl, Department of Epidemiology, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands .
Rozemarijn Vliegenthart, Email: r.vliegenthart@umcg.nl, Department of Radiology, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands .
Xueqian Xie, Email: xiexueqian@hotmail.com, Department of Radiology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China .
REFERENCES
- 1. Camm AJ, Lip GYH, De Caterina R, Savelieva I, Atar D, Hohnloser SH, et al. . 2012 focused update of the ESC guidelines for the management of atrial fibrillation: an update of the 2010 ESC guidelines for the management of atrial fibrillation. developed with the special contribution of the european heart rhythm association . Eur Heart J 2012. ; 33: 2719 – 47 . doi: 10.1093/eurheartj/ehs253 [DOI] [PubMed] [Google Scholar]
- 2. Hindricks G, Potpara T, Dagres N, Arbelo E, Bax JJ, Blomström-Lundqvist C, et al. . 2020 ESC guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the european association for cardio-thoracic surgery (EACTS): the task force for the diagnosis and management of atrial fibrillation of the european society of cardiology (ESC) developed with the special contribution of the european heart rhythm association (ehra) of the ESC . Eur Heart J 2021. ; 42: 373 – 498 . doi: 10.1093/eurheartj/ehaa612 [DOI] [PubMed] [Google Scholar]
- 3. Rahman F, Kwan GF, Benjamin EJ . Global epidemiology of atrial fibrillation . Nat Rev Cardiol 2014. ; 11: 639 – 54 . doi: 10.1038/nrcardio.2014.118 [DOI] [PubMed] [Google Scholar]
- 4. Zoni-Berisso M, Lercari F, Carazza T, Domenicucci S . Epidemiology of atrial fibrillation: european perspective . Clin Epidemiol 2014. ; 6: 213 – 20 . doi: 10.2147/CLEP.S47385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zimetbaum P . Atrial fibrillation . Ann Intern Med 2017. ; 166: ITC33-ITC48 . doi: 10.7326/AITC201703070 [DOI] [PubMed] [Google Scholar]
- 6. Bisson A, Bodin A, Fauchier L . Why and how to screen for atrial fibrillation . Heart 2018. ; 104: 1474 – 75 . doi: 10.1136/heartjnl-2018-313085 [DOI] [PubMed] [Google Scholar]
- 7. Sacks HS, Fain JN . Human epicardial adipose tissue: A review . Am Heart J 2007. ; 153: 907 – 17 . doi: 10.1016/j.ahj.2007.03.019 [DOI] [PubMed] [Google Scholar]
- 8. Wong CX, Ganesan AN, Selvanayagam JB . Epicardial fat and atrial fibrillation: current evidence, potential mechanisms, clinical implications, and future directions . Eur Heart J 2017. ; 38: 1294 – 1302 . doi: 10.1093/eurheartj/ehw045 [DOI] [PubMed] [Google Scholar]
- 9. Al Chekakie MO, Welles CC, Metoyer R, Ibrahim A, Shapira AR, Cytron J, et al. . Pericardial fat is independently associated with human atrial fibrillation . J Am Coll Cardiol 2010. ; 56: 784 – 88 . doi: 10.1016/j.jacc.2010.03.071 [DOI] [PubMed] [Google Scholar]
- 10. Davidovich D, Gastaldelli A, Sicari R . Imaging cardiac fat . Eur Heart J Cardiovasc Imaging 2013. ; 14: 625 – 30 . doi: 10.1093/ehjci/jet045 [DOI] [PubMed] [Google Scholar]
- 11. Militello C, Rundo L, Toia P, Conti V, Russo G, Filorizzo C, et al. . A semi-automatic approach for epicardial adipose tissue segmentation and quantification on cardiac CT scans . Comput Biol Med 2019. ; 114: 103424 . doi: 10.1016/j.compbiomed.2019.103424 [DOI] [PubMed] [Google Scholar]
- 12. Kamel SI, Levin DC, Parker L, Rao VM . Utilization trends in noncardiac thoracic imaging, 2002-2014 . J Am Coll Radiol 2017. ; 14: 337 – 42 . doi: 10.1016/j.jacr.2016.09.039 [DOI] [PubMed] [Google Scholar]
- 13. Goldfarb JW, Weber J . Trends in cardiovascular MRI and CT in the U.S. medicare population from 2012 to 2017 . Radiol Cardiothorac Imaging 2021. ; 3: e200112 . doi: 10.1148/ryct.2021200112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Du Y, Li Q, Sidorenkov G, Vonder M, Cai J, de Bock GH, et al. . Computed tomography screening for early lung cancer, COPD and cardiovascular disease in shanghai: rationale and design of a population-based comparative study . Acad Radiol 2021. ; 28: 36 – 45 . doi: 10.1016/j.acra.2020.01.020 [DOI] [PubMed] [Google Scholar]
- 15. Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. . The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping . Radiology 2020. ; 295: 328 – 38 . doi: 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liljequist D, Elfving B, Skavberg Roaldsen K . Intraclass correlation - A discussion and demonstration of basic features . PLoS One 2019. ; 14( 7 ): e0219854 . doi: 10.1371/journal.pone.0219854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kursa M, Rudnicki W . Feature selection with boruta package . J Stat Softw 2010. ; 36: 1 – 13 . doi: 10.18637/jss.v036.i11 [DOI] [Google Scholar]
- 18. Tin Kam H . The random subspace method for constructing decision forests . IEEE Trans Pattern Anal Machine Intell 1998. ; 20: 832 – 44 . doi: 10.1109/34.709601 [DOI] [Google Scholar]
- 19. Zhi T, Luo H, Liu Y . A gini impurity-based interest flooding attack defence mechanism in NDN . IEEE Commun Lett 2018. ; 22: 538 – 41 . doi: 10.1109/LCOMM.2018.2789896 [DOI] [Google Scholar]
- 20. Gaeta M, Bandera F, Tassinari F, Capasso L, Cargnelutti M, Pelissero G, et al. . Is epicardial fat depot associated with atrial fibrillation? A systematic review and meta-analysis . Europace 2017. ; 19: 747 – 52 . doi: 10.1093/europace/euw398 [DOI] [PubMed] [Google Scholar]
- 21. Kolossváry M, Park J, Bang J-I, Zhang J, Lee JM, Paeng JC, et al. . Identification of invasive and radionuclide imaging markers of coronary plaque vulnerability using radiomic analysis of coronary computed tomography angiography . Eur Heart J Cardiovasc Imaging 2019. ; 20: 1250 – 58 . doi: 10.1093/ehjci/jez033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin A, Kolossváry M, Yuvaraj J, Cadet S, McElhinney PA, Jiang C, et al. . Myocardial infarction associates with a distinct pericoronary adipose tissue radiomic phenotype: a prospective case-control study . JACC Cardiovasc Imaging 2020. ; 13: 2371 – 83 . doi: 10.1016/j.jcmg.2020.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crewe C, An YA, Scherer PE . The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis . J Clin Invest 2017. ; 127: 74 – 82 : 88883 . doi: 10.1172/JCI88883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oikonomou EK, Antoniades C . The role of adipose tissue in cardiovascular health and disease . Nat Rev Cardiol 2019. ; 16: 83 – 99 . doi: 10.1038/s41569-018-0097-6 [DOI] [PubMed] [Google Scholar]
- 25. Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ . CT texture analysis: definitions, applications, biologic correlates, and challenges . Radiographics 2017. ; 37: 1483 – 1503 . doi: 10.1148/rg.2017170056 [DOI] [PubMed] [Google Scholar]
- 26. Oikonomou EK, Williams MC, Kotanidis CP, Desai MY, Marwan M, Antonopoulos AS, et al. . A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography . Eur Heart J 2019. ; 40: 3529 – 43 . doi: 10.1093/eurheartj/ehz592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wasserlauf J, You C, Patel R, Valys A, Albert D, Passman R . Smartwatch performance for the detection and quantification of atrial fibrillation . Circ Arrhythm Electrophysiol 2019. ; 12( 6 ): e006834 . doi: 10.1161/CIRCEP.118.006834 [DOI] [PubMed] [Google Scholar]
- 28. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, et al. . An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction . Lancet 2019. ; 394: 861 – 67 . doi: 10.1016/S0140-6736(19)31721-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.