Abstract
The Us9 gene is highly conserved among the alphaherpesviruses sequenced to date, yet its function remains unknown. In this report, we demonstrate that the pseudorabies virus (PRV) Us9 protein is present in infected cell lysates as several phosphorylated polypeptides ranging from 17 to 20 kDa. Synthesis is first detected at 6 h postinfection and is sensitive to the DNA synthesis inhibitor phosphonoacetic acid. Unlike the herpes simplex virus type 1 Us9 homolog, which was reported to be associated with nucleocapsids in the nuclei of infected cells (M. C. Frame, D. J. McGeoch, F. J. Rixon, A. C. Orr, and H. S. Marsden, Virology 150:321–332, 1986), PRV Us9 localizes to the secretory pathway (predominately to the Golgi apparatus) and not to the nucleus. By fusing the enhanced green fluorescent protein (EGFP) reporter molecule to the carboxy terminus of Us9, we demonstrated that Us9 not only is capable of targeting a Us9-EGFP fusion protein to the Golgi compartment but also is able to direct efficient incorporation of such chimeric molecules into infectious viral particles. Moreover, through protease digestion experiments with Us9-EGFP-containing viral particles, we demonstrated that the Us9 protein is inserted into the viral envelope as a type II, tail-anchored membrane protein.
Despite conservation of the Us9 gene in many alphaherpesvirus genomes, the Us9 gene product has not been assigned a function in vitro or in vivo (8, 10, 13, 14, 23, 31, 43, 46, 60, 62, 68). Its preliminary assignment as a phosphorylated tegument protein in herpes simplex virus type 1 (HSV-1) (14) has been widely adapted for all of the Us9 homologs, although only the HSV-1 protein has been studied. Several observations made with pseudorabies virus (PRV) (a swine alphaherpesvirus) prompted us to reassess the structure and function of PRV Us9. First, some attenuated strains of PRV with spontaneous mutations conferring reduced virulence have been isolated, and a subset of these strains have a deletion in the unique short (Us) region of the viral genome that removes the Us9 coding sequence. For instance, the vaccine strain Bartha (Ba) has a large Us deletion encompassing gI, gE, Us9 (the old nomenclature was 11K), and Us2 (the old nomenclature was 28K) (35, 42, 45). The attenuated strain Norden has a similar Us deletion including gI, gE, and Us9 (35, 41, 42). While it is generally accepted that the lack of the gI and gE genes accounts for most of the reduced virulence and tropism defects in these attenuated strains (1, 4–6, 19, 24, 25, 28, 36, 41, 42, 55, 66), it is not clear why Us9 or Us2 should be deleted in these natural isolates. Another observation concerning Us9 was made by Pol et al. (48, 49), who reported that mutations in both Us9 and gE affected viral egress in PRV-infected porcine nasal turbinate cultures, but the contribution of individual genes to this phenotype was not elucidated.
Us9 homologs have been identified in the human pathogens HSV-1 (14, 38) and varicella-zoster virus (10) as well as in many animal herpesviruses, including PRV (46, 62), bovine herpesvirus type 1 (31), equine herpesvirus type 1 (8, 13, 60), feline herpesvirus type 1 (68), and simian herpes B virus (23). In PRV, as well as in many other alphaherpesviruses, the Us9 gene is located in the Us region of the viral genome adjacent to a cluster of genes coding for well-characterized membrane proteins (gG, gD, gI, and gE). The DNA sequences of all of the Us9 gene homologs encode proteins with several common motifs, including potential tyrosine and casein kinase I and II phosphorylation sites. The homologous proteins in PRV, feline herpesvirus type 1, and equine herpesvirus type 1 all have a conserved N-glycosylation sequence (NXS/T). Moreover, all of the Us9 homologs contain a 25- to 30-residue hydrophobic stretch near the carboxy terminus preceded by several clusters of charged residues (Fig. 1) (8, 10, 13, 14, 23, 31, 38, 46, 60, 62, 68). In the case of the HSV-1 Us9 homolog, the string of six arginines just preceding the hydrophobic stretch has been suggested to be a nuclear localization sequence (14). McGeoch et al. classified the Us9 gene as one of several HSV-1 genes that may be associated with membranes, because its product has a carboxy-terminal hydrophobic domain that may function to span membranes (37). McGeoch et al. (37) also noted the lack of a canonical N-terminal signal sequence in the Us9-coding sequence.
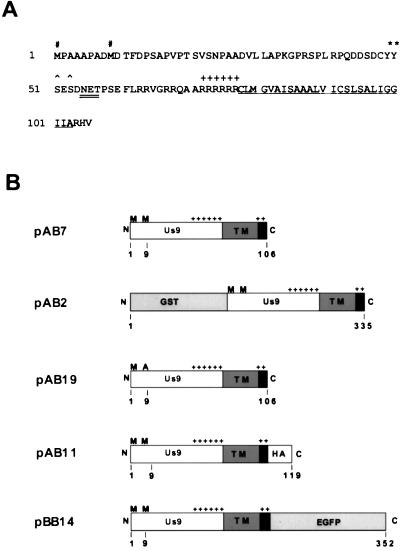
FIG. 1.
Us9 amino acid sequence and gene constructs. (A) Amino acid sequence of the Us9 open reading frame. The two in-frame methionine residues (#), the potential tyrosine kinase phosphorylation sites (∗), and the potential casein kinase I and II sites (∧) are indicated. The potential N-linked glycosylation (NXS/T) sequence is double underlined. The putative transmembrane domain is underlined, and the surrounding basic residues are indicated (+). (B) Diagrams of all the Us9 constructs used in this study. The two in-frame methionine residues (M), the putative transmembrane domain (TM), and the clusters of basic charges (+) are marked. The Us9 protein fusions with GST, the influenza A virus HA epitope, and EGFP are indicated. All constructs depicted are under the transcriptional control of the CMV immediate-early promoter.
The protein product of the HSV-1 Us9 gene was identified by Frame et al. (14) by using an antiserum made against a synthetic oligopeptide whose sequence was deduced from the DNA sequence. These workers provided evidence that the HSV-1 Us9 protein was a virion structural protein localized to the tegument. Moreover, they demonstrated that the protein was phosphorylated and, using immunogold electron microscopy, found it to be associated with nucleocapsids in the nuclei of infected cells. The HSV-1 Us9 protein was predicted to have a molecular mass of 10 kDa, yet 12 distinct polypeptides ranging from 12 to 20 kDa were observed and characterized as phosphoforms of Us9 (14). The HSV-1 Us9 gene is not required for virus growth in tissue culture, and no obvious phenotype for HSV-1 Us9 null mutants has been observed in animal studies (43).
We expressed the PRV Us9 open reading frame in bacteria as a glutathione S-transferase (GST) fusion protein and prepared a Us9-specific antiserum in rabbits to characterize the protein in infected tissue culture cells. We first found that PRV Us9 was present in PRV Becker (Be)-infected cells as multiple phosphorylated polypeptides. As we proceeded, we discovered that the PRV Us9 gene product did not follow the paradigm established for HSV-1 Us9 (14). Using standard immunohistochemistry and fusions to enhanced green fluorescent protein (EGFP), we established that PRV Us9 is a tail-anchored type II membrane protein, a class of membrane proteins previously uncharacterized in alphaherpesvirus envelopes. The protein appears to be targeted to the Golgi compartment in both infected and transfected cells. This may have important ramifications for models of PRV envelopment.
MATERIALS AND METHODS
Virus strains and cells.
The PRV Be and Ba strains used in this study were propagated on PK15 (pig kidney epithelial) cells as described previously (66). PRV Ba harbors a deletion removing the entire Us9-coding sequence (35, 42) and was used as a negative control in our experiments.
PK15-BB14 cells, which express a Us9-EGFP fusion protein described below (Fig. 1), were constructed as follows. A 40% confluent monolayer of PK15 cells growing in a 100-mm-diameter dish was transfected with 10 μg of pBB14. Forty-eight hours after tranfection, cells were split at 1:20, replated in Dulbecco modified Eagle medium with 10% fetal calf serum, and allowed to attach for 24 h before being placed under selection with Dulbecco modified Eagle medium–10% fetal calf serum containing 1 mg of G418 per ml (Gibco/BRL). G418-resistant cells were pooled and grown to confluence. A uniform population of Us9-EGFP cells expressing intermediate levels of the fusion protein was separated from nonexpressing, low-expressing, and high-expressing cells by using a FACScan cell sorter (Becton Dickinson). These cells were designated PK15-BB14, expanded, and frozen for use in subsequent experiments.
Cloning of PRV Us9.
The PRV Us9 open reading frame was amplified by PCR with Taq DNA polymerase (Gibco/BRL) and the plasmid pALM94 (5), which contains the BamHI 7 fragment of PRV Be, as a template. The forward PCR primer corresponded to the 5′ end of the Us9 gene, beginning 24 nucleotides upstream and including the first ATG. The forward PCR primer also introduced an EcoRI site upstream of the first ATG. The reverse primer corresponded to the 3′ end of the Us9 gene, including the stop codon and 12 downstream nucleotides. This 360-bp PCR product was cloned directly into pBKS+ (Stratagene) which had been digested with EcoRV and T tailed with ddTTP and terminal deoxynucleotidyl transferase as described previously (17), to yield plasmid pAB1. The PCR-amplified portion of this construct was sequenced to confirm that no mutations had been introduced into the cloned Us9 gene.
Us9 constructs.
Figure 1 shows the predicted amino acid sequences of Us9 and the constructs used in this study. The nucleotide sequence of the Us9 gene in PRV Be is identical to that reported by Petrovskis and Post (46) for the PRV Rice strain except for a 1-bp change that results in a threonine (ACC)-to-alanine (GCC) amino acid change at residue 3 (18). The nucleotide sequences of all Us9 constructs were verified by DNA sequencing.
Plasmid pAB7 carries the Us9 gene under the control of the cytomegalovirus (CMV) immediate-early promoter. pAB7 was constructed by ligating the EcoRI fragment of pAB1 containing the Us9 gene into the EcoRI site of the eukaryotic expression vector pcDNA1/Amp (Invitrogen).
Plasmid pAB2 carries a Us9-GST gene fusion that was used to produce antiserum specific for Us9. To create pAB2, a 369-bp EcoRI fragment containing the Us9 gene was isolated from pAB1 and ligated into the EcoRI site of pGEX1, a GST fusion expression vector (Pharmacia Biotech).
Plasmid pAB19 carries a mutant Us9 gene in which the second methionine residue at position 9 was mutated to an alanine residue by PCR mutagenesis. PCR was performed with Taq DNA polymerase and with pAB1 as the template. The forward primer corresponded to the 5′ end of the Us9 gene, beginning 24 nucleotides upstream and including the first in-frame ATG. This PCR primer mutated the methionine residue at amino acid 9 (ATG) to an alanine residue (GCG). The reverse primer corresponded to the 3′ end of the Us9 gene, including the stop codon and 12 downstream nucleotides. The 351-bp Us9 M9A PCR fragment was ligated to the T-cloning vector pT7Blue (Novagen) before being transferred into the expression vector pcDNA1/Amp.
pAB11 carries a hybrid Us9 gene encoding a product in which a 9-amino-acid epitope from the influenza A virus hemagglutinin (HA) gene was fused to the carboxy terminus of Us9. Plasmid pAB11 was created by PCR amplification with Taq DNA polymerase and the plasmid pAB1 as template DNA. The forward PCR primer (see above) introduced an EcoRI site prior to the first Us9 ATG, while the reverse primer placed the HA epitope sequence (YPYDVPDYA) immediately downstream of amino acid 106. The reverse primer also introduced a 4-amino-acid alanine linker between the Us9 sequence and the HA epitope and an amber codon (TAG) after the last HA epitope amino acid. This 420-bp PCR product was ligated to the T-cloning vector, pT7Blue, and subsequently cloned into pcDNA1/Amp.
Plasmid pBB14 carries a hybrid gene in which the EGFP open reading frame was fused to sequences encoding the carboxy terminus of Us9. To construct pBB14, PCR mutagenesis was used to introduce an EcoRI site upstream of the first methionine of Us9 and to replace the Us9 stop codon (TAG, codon 107) with the codon for an arginine residue (CGG). To allow an in-frame fusion of Us9 with EGFP, a BamHI site was also introduced after the mutated stop codon. PCR was performed with pAB1 as template DNA, and the 358-bp amplified product was cloned directly into pT7Blue. This construct was then digested with EcoRI and BamHI, and the Us9 fragment was ligated into pEGFP-N1 (Clontech). This plasmid was designated pBB14 and contains the Us9-EGFP gene fusion under the control of the CMV immediate-early promoter.
PRV Us9 polyclonal antiserum.
Expression of the GST-Us9 fusion protein was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a culture of DH5α cells transformed with pAB2. The fusion protein was isolated as inclusion bodies essentially as described by Strebel et al. (58), dialyzed against phosphate-buffered saline (PBS), and injected (250 μg) into two New Zealand White rabbits. Complete Freund’s adjuvant was used in the preparation of antigen for the first injection. Incomplete Freund’s adjuvant was used in all subsequent injections, which were performed every 2 weeks for 8 weeks. Serum was collected at 2-week intervals beginning 1 month after the initial injection and tested by Western blot analysis for reactivity against Us9.
The polyvalent goat PRV gC (Ab 282) and gB (Ab 284) antisera have been described previously (51, 54). A mouse monoclonal antibody directed against GFP was purchased from Clontech.
Isolation of virions.
Three confluent 150-mm-diameter dishes of PK15-BB14 cells were infected with PRV Be or Ba (multiplicity of infection [MOI] = 10). At 16 h postinfection, the medium was collected and centrifuged at 3,000 rpm (Sorvall H1000B) to remove cellular debris. The clarified supernatant was then layered onto a 7-ml 30% sucrose cushion (in PBS) and centrifuged in an SW28 rotor at 23,000 rpm for 3 h. The sucrose cushion was removed, and the virion pellet was resuspended in 0.5 ml of medium by brief sonication. The virions were centrifuged through a 1-ml 30% sucrose cushion at 28,000 rpm for 90 min in an SW50.1 rotor. The pelleted virions were resuspended in PBS. Virions prepared in this manner appeared to be free of intracellular membrane contamination as assessed by the lack of PRV glycoprotein precursors after Western blot analysis.
For preparation of radiolabeled virions, two 100-mm-diameter dishes of PK15 cells infected with either PRV Be or Ba (MOI = 10) were incubated overnight with [35S]cysteine-methionine (50 μCi/ml; New England Nuclear) starting at 5 h postinfection. After 16 h of infection, the medium was collected and virions were prepared as described above.
Protease treatment of virions.
Isolated virions were treated with 10 μg of proteinase K (Sigma) per ml in either the presence or absence of 1% Nonidet P-40 (NP-40). After incubation for 60 min at room temperature, phenylmethylsulfonyl fluoride was added to each sample to a final concentration of 2 mM to inhibit further proteolysis. Samples were then immediately loaded onto a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel and electroblotted onto nitrocellulose, and Us9 or GFP was detected with specific antiserum.
Immunoprecipitations.
Monolayers of PK15 cells were infected with PRV Be or PRV Ba at an MOI of 10. After 5 h of infection, the cells were labeled with [35S]cysteine-methionine (170 μCi/ml; New England Nuclear) as described previously (66). For phosphate labeling of viral proteins, [33P]orthophosphate (100 μCi/ml; Amersham) was added to PRV Be- and Ba-infected monolayers at 5 h postinfection. At 16 h postinfection, the cells were washed with PBS (or Tris-buffered saline [pH 7.4] for [33P]orthophosphate-labeled cells) and harvested in 0.5 ml of lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.8], 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS). Labeled cell extracts (100 μl) were immunoprecipitated as described previously (66). The immunoprecipitated samples were suspended in sample buffer, heated to 90°C for 2.5 min, and loaded onto an SDS–12.5% polyacrylamide gel.
Western blot analysis.
Cellular extracts were electrophoresed on an SDS–12.5% polyacrylamide gel and transferred to nitrocellulose membranes (Amersham) by using a semidry protein transfer apparatus. Proteins were visualized by using polyclonal or monoclonal primary antibodies and enhanced chemiluminescence detection as recommended by the manufacturer (SuperSignal; Pierce).
In vitro translation.
Us9 (pAB7), Us9 M9A (pAB19), and Us9-HA (pAB11) proteins were translated in vitro by using a rabbit reticulocyte lysate coupled transcription-translation system (Promega) in the presence of [35S]methionine (Amersham). Three microliters of the translation reaction mixture was precipitated with 4 volumes of 100% acetone for 1 h at −20°C prior to visualization by SDS-polyacrylamide gel electrophoresis and autoradiography. This step was necessary to remove unincorporated label and labeled charged tRNAs.
N-Glycosidase F treatment.
Monolayers of PK15 cells in 100-mm-diameter dishes were infected with PRV Be (MOI = 10). At 16 h postinfection, the cells were washed with PBS, harvested, and lysed in 1 ml of lysis buffer without SDS (lysis buffer is described above). A 15-μl aliquot of the cellular extract was incubated overnight at 37°C with 0.2 U of N-glycosidase F in 1× buffer (50 mM Tris-HCl [pH 6.5], 1% SDS, 1% β-mercaptoethanol, 4% NP-40, 100 mM sodium citrate [pH 5.5]). The reaction mixture was electrophoresed on an SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and reacted with antiserum specific for Us9 or gC.
PAA treatment.
PK15 monolayers were treated with phosphonoacetic acid (PAA) (Sigma) at concentrations ranging from 200 to 800 μg/ml 60 min prior to and during infection with PRV Be (MOI = 10). Cellular extracts were prepared at 10 h postinfection as described above and analyzed for Us9, gC, and gB expression by Western blot analysis.
Transient transfections.
PK15 cells were grown on glass coverslips to approximately 40% confluency and transfected with plasmid pAB7, pBB14, or pEGFP-N1 (Clontech) by a calcium phosphate coprecipitation method (15). The cells were analyzed either within 24 to 48 h for EGFP expression (pBB14 and pEGFP-N1) by fluorescence microscopy or at 72 h posttransfection for Us9 expression (pAB7) by indirect immunofluorescence microscopy.
Indirect immunofluorescence.
PK15 cells grown to approximately 70% confluency on glass coverslips were infected with virus at an MOI of 10. At various times postinfection, the cells were fixed and analyzed with Us9, gB, or gC polyvalent antiserum as described previously (61). For Brefeldin A (BFA) treatment of virus-infected cells, BFA (Epicentre Technologies) was added (2.5 μg/ml) to the medium of Us9-EGFP-transfected monolayers at 48 h posttransfection. After 1 h, the coverslips were fixed with 4% paraformaldehyde and examined by confocal microscopy. Alternatively, cells were allowed to recover from BFA treatment by washing three times in fresh medium and incubating for 60 min at 37°C.
RESULTS
Identification of the Us9 protein in PRV Be-infected cell lysates and virions.
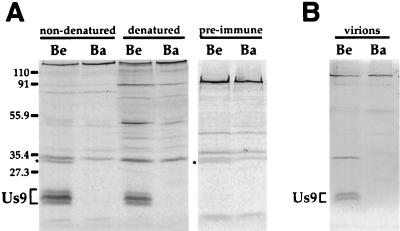
The Us9 antiserum precipitated three polypeptides from [35S]cysteine-methionine-labeled PRV Be-infected cell lysates that were not present in Ba-infected cell lysates or in Be lysates precipitated with preimmune serum (Fig. 2A). The Us9 immunoreactive polypeptides were precipitated with Us9 antiserum under both denaturing and nondenaturing conditions, indicating that the antiserum recognized both native and denatured conformations of the proteins. The apparent molecular masses of these polypeptides were estimated to be approximately 20, 18, and 17 kDa. This calculation was based on the relative migrations of nonstained molecular weight markers electrophoresed on an SDS–12.5% polyacrylamide gel. Although in this experiment the middle form (18 kDa) appeared to be the more abundant species immunoprecipitated from PRV Be-infected cell lysates, the apparent concentrations of these three forms of Us9 varied depending on the time after infection and the labeling protocol. As predicted, the Us9-specific serum reacted with polypeptides only in PRV Be-infected, and not PRV Ba-infected, cells, as the Us9 open reading frame is missing in PRV Ba.
FIG. 2.
Us9 protein in PRV Be- and Ba-infected cell lysates and virions. Monolayers of PK15 cells were infected with PRV Be or Ba virus at an MOI of 10. PRV-infected monolayers were radiolabeled for 11 h beginning at 5 h postinfection. (A) Cell lysates were either denatured with SDS and dithiothreitol or left untreated prior to immunoprecipitation with Us9 antiserum or preimmune serum. The identity of the polypeptide in nondenatured Be lysates immunoprecipitated with either Us9 antiserum or preimmune serum (indicated by an asterisk) is currently unknown. (B) Radiolabeled PRV virions were collected at 16 h postinfection by centrifugation through a 30% sucrose cushion and subjected to immunoprecipitation with Us9 antiserum. Positions of molecular mass markers (kilodaltons) are indicated on the left.
Under nondenaturing conditions, Us9 antiserum also precipitated an additional protein of approximately 31 kDa that was present in PRV Be-infected cell lysates (Fig. 2A). This polypeptide was also precipitated in nondenatured PRV Be-infected cell lysates with preimmune serum (Fig. 2A) or with gE and gB antisera (data not shown). However, the unidentified polypeptide was not detected in PRV Ba- or PRV BaBe (PRV Be with the Ba Us deletion) (5)-infected cell lysates immunoprecipitated with any of the antisera tested (data not shown). The identity of this molecule was not pursued.
We next identified proteins in PRV virions that reacted with the Us9 antiserum. Virions were isolated as described in Materials and Methods, and immunoreactive Us9 proteins were identified by immunoprecipitation. As shown in Fig. 2B, two Us9-specific polypeptides were recognized by the antiserum in virions isolated from PRV Be- but not Ba-infected cells. Although it appeared that only two of the three Us9 polypeptides detected in PRV Be-infected cell lysates were incorporated into viral particles, because of the inherent variability of labeling of these peptides, we cannot conclude which Us9 polypeptide, if any, is excluded from virions. An additional polypeptide was also observed in the Be virions that was absent from the Ba virions, which migrated at approximately 30 kDa. The identity of this polypeptide is not known. The precise nature of the Us9-specific polypeptides present in PRV Be virions is currently under investigation.
Time course of Us9 expression in PK15 cells.
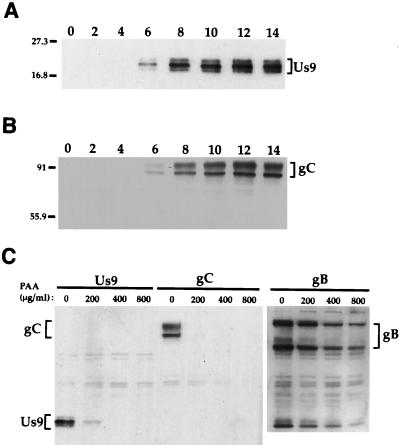
Us9 protein was first detected in infected cell lysates by Western blot analysis at approximately 6 h postinfection (Fig. 3A). This was also approximately the same time that the PRV glycoprotein gC was detected in PRV Be-infected cell lysates under these conditions (Fig. 3B). All three species of Us9 were detected by 6 h postinfection. As observed with the immunoprecipitated Us9 polypeptides (Fig. 2A), the middle form appeared to be the predominant species (Fig. 3A). Although the upper and lower species (Fig. 3A) appeared to be present in approximately equal amounts, this was not always the case (Fig. 3C).
FIG. 3.
Analysis of Us9 protein kinetics in PRV Be-infected cell lysates. (A and B) Monolayers of PK15 cells were infected with PRV Be (MOI = 10), and cellular extracts were prepared at 0, 2, 4, 6, 8, 10, 12, and 14 h after infection. Ten micrograms of total cell lysate per lane was fractionated on an SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and Western blotted with either Us9 (A) or gC (B) antiserum. (C) For PAA treatment, 0, 200, 400, or 800 μg of PAA per ml was added to PK15 cells 1 h prior to and during viral infection. The cells were harvested and lysed at 10 h postinfection, and the cell lysate (10 μg) was analyzed by Western blotting with Us9, gC, or gB antiserum. The migration of molecular mass markers is indicated on the left in kilodaltons.
Determination of the kinetic class of the Us9 gene.
Herpesvirus genes are grouped into three kinetic classes: immediate early, early, or late. The criteria are that immediate-early genes are transcribed before any new viral gene products are synthesized, early genes are expressed prior to viral DNA replication, and late genes are dependent on viral DNA replication for transcription and translation (reviewed in reference 52). To test whether the Us9 gene was expressed as an early or late gene, we tested its sensitivity to the DNA synthesis inhibitor PAA as described in Materials and Methods. Us9 expression was sensitive to PAA, as shown in Fig. 3C. In this experiment, PK15 cells were infected with PRV Be in the presence of 0, 200, 400, or 800 μg of PAA per ml. The expression of both Us9 and gC (a known late protein) was inhibited in the presence of 200 μg of PAA per ml, whereas expression of gB, an early-late PRV protein, was only slightly inhibited with PAA concentrations of up to 800 μg/ml. These results show that the protein product of the Us9 gene is not synthesized when viral DNA replication is inhibited. Although these findings suggest that Us9, like gC, is a PRV late protein, we cannot exclude the possibility that Us9 translation is delayed under these conditions.
The primary translation products of the Us9 gene.
The amino acid sequence of Us9 predicts that the protein has a molecular mass of 11.3 kDa, yet Us9 antiserum recognized three distinct polypeptides with molecular masses ranging between 17 and 20 kDa in infected cells. The unusual electrophoretic mobility implies that one or more of the following affects the Us9 protein: it migrates aberrantly in gel electrophoresis due to its amino acid composition; it has multiple primary translation products; or it is modified after translation by covalent addition of sugars, phosphates, or other small molecules.
The DNA sequence indicates that Us9 could have two translation initiation sites. The predicted first methionine codon is followed by a second in-frame methionine eight codons downstream (Fig. 1). Accordingly, some of the multiple Us9 infected-cell polypeptides may arise by alternative initiation of translation. This possibility was also noted by Petrovskis and Post, who first sequenced PRV Us9 (46). To investigate this, we determined the apparent molecular mass of the primary Us9 translation product after in vitro translation in the presence of [35S]methionine. If both methionines were used to initiate translation, two polypeptides should be observed, one 8 amino acids shorter than the other. We also mutated the second methionine to an alanine (Us9 M9A, encoded by pAB19) and predicted that this mutation would give rise to a single unique polypeptide that corresponded to the larger Us9 species. To ensure that we could detect the predicted Us9 polypeptides, we also constructed a control Us9 hybrid protein with a fusion of the product of 13 codons of the influenza virus HA gene to the carboxy terminus of Us9.
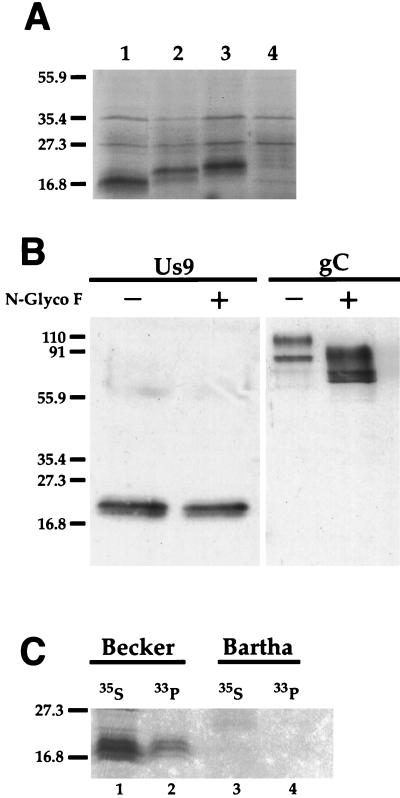
The results of this experiment are shown in Fig. 4A. To our surprise, mutagenesis of the second methionine to an alanine residue (pAB19) (lane 2) resulted in a primary translation product with an apparent molecular mass greater than that of the wild-type Us9 primary translation product (pAB7) (lane 1) yet lower than that of the Us9-HA primary translation product (pAB11) (lane 3). Lane 4 shows the results of a control reaction showing the nonspecific translation products from the pcDNA1/Amp vector alone. We interpret this finding to mean that the Us9 primary translation product is initiated not at the first methionine residue but rather at methionine residue 9. Changing this methionine residue to an alanine residue resulted in translation initiation at the first methionine residue. This novel Us9 primary translation product is predicted to be 106 amino acids with a molecular mass between those of the wild-type Us9 (98 amino acids) (Fig. 4A, lane 1) and the Us9-HA (111 amino acids) (lane 3) primary translation products. Indeed, the Us9 M9A primary translation product (lane 2) migrated to a position closer to the primary translation product of Us9-HA (lane 3) than that of the wild-type Us9 (lane 1). These results are also consistent with the nucleotide sequence surrounding the methionine residue at codon 9 being closer to the Kozak consensus sequence (26, 27) than the methionine at codon 1. These data also indicate that the Us9 polypeptide has an intrinsic anomalous electrophoretic mobility.
FIG. 4.
Us9 posttranslational modifications. (A) In vitro translation of Us9, Us9 M9A, and Us9-HA-tagged constructs. Plasmids pAB7 (lane 1), pAB19 (lane 2), and pAB11 (lane 3) were transcribed and translated in vitro in the presence of [35S]methionine by a rabbit reticulocyte lysate procedure. The bands migrating at approximately 27 and 35 kDa represent nonspecific translation products, as they are also detected during in vitro translation of the parental vector alone (pcDNA1/Amp) (lane 4). (B) N-glycosidase F treatment of Us9. PRV Be-infected cell lysates were incubated in either the presence (+) or absence (−) of N-glycosidase F (N-Glyco F). The treated lysates were separated on an SDS–12.5% polyacrylamide gel, electroblotted onto nitrocellulose, and analyzed by Western blotting with Us9 or gC antiserum. (C) Analysis of Us9 phosphoforms. PRV Be (lanes 1 and 2)- or Ba (lanes 3 and 4)-infected PK15 cells were radiolabeled overnight in the presence of either [35S]methionine-cysteine (lanes 1 and 3) or [33P]orthophosphate (lanes 2 and 4). Cellular extracts were prepared at 16 h postinfection and subjected to immunoprecipitation with Us9 antiserum. All of the immunoprecipitated products were analyzed by electrophoresis on an SDS–12.5% polyacrylamide gel followed by autoradiography. Positions of molecular mass markers (kilodaltons) are indicated on the left.
Us9 posttranslational modifications.
The predicted Us9 amino acid sequence contains a single NXT N-linked glycosylation motif at position 55. To determine if the Us9 protein was N glycosylated, PRV Be-infected cell lysates were treated with N-glycosidase F, which removes both simple and complex N-linked oligosaccharides from the peptide backbone. The treated lysates were then analyzed by Western blot analysis with Us9 and gC antisera. If Us9 was glycosylated at position 55, N-glycosidase F treatment would reduce the apparent molecular mass by approximately 1.5 to 3.0 kDa. The electrophoretic migration pattern of Us9 protein was not altered by treatment with N-glycosidase F, whereas the migration pattern of gC, which contains eight N-linked glycosylation sites, was altered significantly following treatment (Fig. 4B). These results indicated that Us9 was not modified by N-linked glycosylation. In this particular experiment, the uppermost Us9 species was not readily detectable even in the untreated lysates. As noted above, the intensity of the multiple Us9 species is variable from experiment to experiment.
We also tested whether Us9 was posttranslationaly modified by the addition of O-linked oligosaccharides to one of its several serine or threonine residues (Fig. 1). Treatment of PRV Be-infected cell lysates with O-glycosidase did not alter the migration pattern of Us9 (data not shown). However, we cannot conclude from these findings that the Us9 protein does not contain any O-linked oligosaccharides, as O-glycosidase is unable to cleave sugars modified with sialic acid, fucose, or N-acetylglucosamine.
The HSV-1 Us9 homolog has been shown to be a phosphorylated protein; it has a predicted molecular mass of 10 kDa, yet it migrates as 12 distinct polypeptides ranging from 12 to 20 kDa, presumably due to various degrees of phosphorylation (14). Accordingly, we determined if PRV Us9 was modified by phosphorylation by labeling PRV Be-infected cells overnight with [33P]orthophosphate beginning at 5 h postinfection (see Materials and Methods). At 16 h postinfection, the cells were harvested and lysed, and the extracts were immunoprecipitated with Us9 antiserum. The results are shown in Fig. 4C. Lane 1 contains Us9-specific proteins precipitated from extracts labeled with [35S]cysteine-methionine to show the three Us9 immunoreactive species present in PRV Be-infected cell extracts. Lane 2 shows the immunoprecipitation of [33P]orthophosphate-labeled cellular extracts with Us9 antiserum. At least three Us9-related polypeptides were labeled. This is consistent with the presence of four potential phosphorylation consensus sites in the Us9 protein (Fig. 1). As a control for this experiment, no Us9 immunoreactive polypeptides were precipitated from PRV Ba-infected cell lysates labeled with either [35S]cysteine-methionine (Fig. 4C, lane 3) or [33P]orthophosphate (lane 4).
In PRV-infected cells, Us9 localizes rapidly and predominantly to the Golgi apparatus.
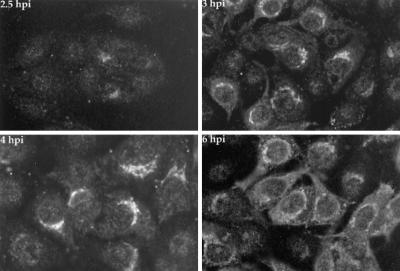
We used immunofluorescence microscopy to determine the localization of Us9 in PRV Be-infected cells at various times after viral infection. PRV Be-infected cells were fixed, permeabilized, and stained for Us9 as described in Materials and Methods at 2.5, 3, 4, and 6 h postinfection (Fig. 5). This sensitive technique enabled us to detect Us9 in infected cells as early as 2.5 h postinfection. In striking contrast to the results reported for HSV-1 (14), the PRV Us9 protein was not localized to the nucleus. In fact, Us9 was first detected in a staining pattern reminiscent of the Golgi apparatus. This localization of Us9 to the Golgi apparatus was most apparent at 3 and 4 h postinfection. The labeling of small cytoplasmic vesicles, which became prevalent by 6 h postinfection, was also frequently observed. As shown below, localization to the Golgi apparatus was verified by using drugs that disrupt the Golgi compartment (see Fig. 7), by colocalization with Golgi-specific dyes (data not shown), or by colocalization with the Golgi resident protein p115 (data not shown) (63). The salient observation was that no detectable nuclear staining was visible at any time.
FIG. 5.
Immunofluorescence analysis of Us9 in PRV Be-infected cells. PK15 cells grown on glass coverslips to approximately 70% confluency were infected with PRV Be at an MOI of 10. At 2.5, 3, 4, and 6 h postinfection (hpi), the cells were fixed with formaldehyde and processed to detect Us9 protein by indirect immunofluorescence and confocal microscopy. A fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobin G secondary antibody was used to visualize the Us9 antigen-antibody complexes.
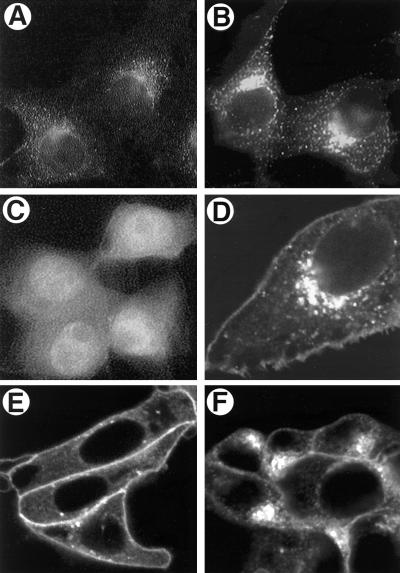
FIG. 7.
Transfection of Us9 constructs. PK15 cells were grown on glass coverslips and transfected by the calcium phosphate method with pAB7 (A), pBB14 (B, D, E, and F), or EGFP (C). At 72 h posttransfection, Us9 was detected by indirect immunofluorescence microscopy with Us9 antiserum (A) as described in the legend to Fig. 5. The localization of pBB14 (B, D, E, and F) and EGFP (C) was determined at 48 h posttransfection by confocal microscopy under UV illumination. Panel D is a higher magnification of the Us9-EGFP-transfected cells shown in panel B, clearly demonstrating expression of the fusion protein in the Golgi compartment and plasma membrane. (E and F) Sensitivity of Us9-EGFP (pBB14) to BFA treatment. PK15 cells transfected with pBB14 were treated with 2.5 μg of BFA per ml for 60 minutes (E). Some BFA-treated cells were then washed three times with fresh medium and allowed to recover for 1 h (F).
Us9 partially colocalizes with gB and gC in infected cells.
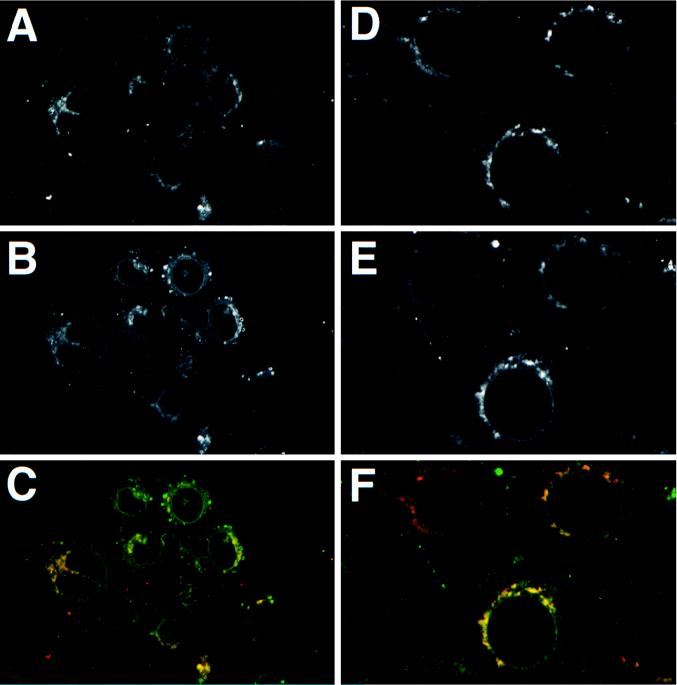
Localization of Us9 to vesicles and to the Golgi apparatus suggested that this protein was associated with membranes of the secretory pathway. The PRV gB and gC envelope proteins are typical type I membrane proteins and are found in all aspects of the secretory pathway in infected cells (Fig. 6B and E). We determined if Us9 (Fig. 6A and D) colocalized with gB (Fig. 6B) and gC (Fig. 6E) during PRV infection. While there was colocalization of Us9 with gB (Fig. 6C) and with Us9 and gC (Fig. 6F) in the Golgi apparatus and some vesicles (yellow signal), Us9 protein could not be detected in the nuclear membrane or the plasma membrane of infected cells. These results suggested that Us9 is concentrated in the Golgi apparatus relative to the other membranes populated by gB and gC in PRV-infected cells.
FIG. 6.
Colocalization of Us9 with envelope proteins gB and gC. PK15 cells grown on glass coverslips were infected with PRV Be at an MOI of 10. The cells were fixed and stained for Us9 and gB (A, B, and C) at 6 h postinfection and for Us9 and gC (D, E, and F) at 4 h post-infection. Us9 (A and D) was detected with an indocarbocyanine-conjugated (Cy3) donkey anti-rabbit secondary antibody. gB (B) and gC (E) were visualized with a fluorescein isothiocyanate-conjugated donkey anti-goat secondary antibody. The Us9 (red) and gB (green) colocalization is shown in panel C. The Us9 (red) and gC (green) colocalization is shown in panel F.
Localization of Us9 protein in transfected cells.
The concentration of Us9 to the Golgi apparatus may be affected by other proteins in the PRV-infected cell. To test this idea, the staining pattern of Us9 in transiently transfected cells was examined. Plasmid pAB7, containing the Us9 gene under the control of the CMV immediate-early promoter, was transfected into PK15 cells. At 72 h posttransfection, Us9 was detected by indirect immunofluorescence microscopy (Fig. 7A). The staining pattern of Us9 resembled that observed in PRV Be-infected cells at early times after infection, suggesting that Us9 localization to the Golgi apparatus was independent of viral infection (compare Fig. 5 and 7A). The staining of Us9 was predominantly restricted to the Golgi apparatus even in transfected cells which expressed high levels of the Us9 protein (data not shown).
A Us9-EGFP fusion protein localizes to the Golgi apparatus.
It was of interest to determine if Us9 could direct a cytoplasmic reporter protein to the Golgi compartment. This would aid in identifying putative targeting signals found in the Us9 protein. To this end, EGFP was fused to the carboxy terminus of Us9. pBB14, carrying a Us9-EGFP gene fusion, was transfected into PK15 cells as described above, and its localization was determined by fluorescence microscopy (Fig. 7B). The localization of Us9-EGFP in transfected cells was indistinguishable from that of transfected wild-type Us9 in that the Golgi compartment was heavily labeled. As a control, transfection of DNA encoding unfused EGFP into PK15 cells revealed that the reporter protein alone (Fig. 7C) filled every compartment in the cell, including the nucleus, producing a pattern of fluorescence that was clearly distinguishable from the specific localization of the Us9-EGFP protein (compare Fig. 7B and C). We conclude that the Us9 component of the Us9-EGFP fusion targets the fusion protein to the Golgi apparatus.
Localization of the Us9-EGFP fusion protein is sensitive to BFA treatment.
While it was clear that Us9 and the Us9-EGFP fusion protein localized to a region reminiscent of the Golgi in infected and transfected cells, we were uncertain if the Golgi apparatus per se was directly involved. An indication of functional Golgi involvement is the sensitivity of protein localization to the drug BFA. BFA is a fungal metabolite that reversibly inhibits transport of vesicles from the endoplasmic reticulum (ER) to the Golgi while maintaining Golgi-to-ER transport (33, 34). The net result of BFA treatment is dissolution of the Golgi and redistribution of Golgi proteins to the ER. Us9-EGFP-transfected PK15 cells were treated with 2.5 μg of BFA per ml for 60 min, and the localization of Us9 was determined by immunofluorescence and confocal microscopy (Fig. 7E). BFA effects are reversible in that Golgi structure and function can be restored by removal of the drug (Fig. 7F).
Addition of BFA to the Us9-EGFP-transfected cells resulted in the redistribution of the fusion protein from a defined structure to a diffuse staining pattern, as predicted if Us9 was a Golgi-associated protein and BFA treatment dispersed it to the ER (compare Fig. 7B and E). Note that the Us9-EGFP fusion protein present at the plasma membrane is insensitive to BFA treatment. When BFA was removed from the medium and the cells were washed and allowed to recover, Us9-EGFP returned to a defined structure characteristic of the Golgi (Fig. 7F). A higher-magnification image of Us9-EGFP fluorescence in transfected PK15 cells is shown in Fig. 7D. This image reveals the fusion protein in the plasma membrane, something that had been difficult to discern for the unfused Us9 protein by indirect immunofluorescence. This may reflect the use of detergents to permeabilize the cells transfected with wild-type Us9 (pAB7), resulting in extraction of the molecule from the membrane.
While the BFA experiment of Fig. 7 was performed with the Us9-EGFP construct, we obtained similar results with the wild-type Us9 protein in BFA-treated infected cells (data not shown). As the behaviors of the Us9 and Us9-EGFP proteins were indistinguishable, we conclude that the presence of EGFP on the carboxy terminus of the Us9 molecule does not interfere with targeting to the Golgi apparatus.
Topology of the Us9 and Us9-EGFP proteins.
As noted by McGeoch et al. (37), the predicted protein sequences of all Us9 homologs sequenced to date have no obvious signal sequence motif characteristic of type I membrane proteins, but they do contain a 25- to 30-amino acid stretch of hydrophobic amino acids near the carboxy terminus that may function as a membrane-spanning domain. This hydrophobic sequence is preceded by several positively charged arginine residues and is followed by a single positively charged residue in most Us9 homologs. This arrangement, with the N-terminal side of the transmembrane domain having a highly positive charge relative to the C-terminal side, is typical of type II membrane proteins, in which the amino terminus is found on the cytosolic face of the membrane.
If PRV Us9 is indeed a type II membrane protein, it would be predicted to have only three amino-terminal amino acids on the outside face of the plasma membrane or the virus envelope, 26 amino acids spanning the membrane, and 68 amino acids found on the cytoplasmic side of the plasma membrane or in the tegument of the virion. To test this hypothesis, we made use of the hybrid Us9-EGFP gene described above. If this chimeric protein attains the type II membrane protein conformation and is incorporated into virus particles, the EGFP moiety will be exposed on the surface of the virus and will therefore be accessible to protease digestion.
The outline of these experiments is as follows. First, we constructed a stable cell line expressing the Us9-EGFP fusion protein as described in Materials and Methods. This cell line was then infected with PRV Be or PRV Ba to determine if the fusion protein could be incorporated into virions. Finally, the topology of the fusion protein in purified virions was tested directly.
The PK15 EGFP-Us9 cell line (PK15-BB14) was infected with either PRV Be or PRV Ba at an MOI of 10. At 16 h postinfection, the medium was collected and the virions were purified by centrifugation through sucrose as described in Materials and Methods. The yield of infectious virus per cell was indistinguishable from that with the PK15 parental cell line used as a control (data not shown).
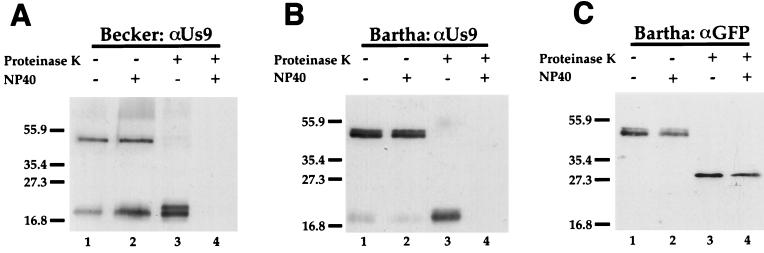
As shown in Fig. 8, the EGFP-Us9 chimeric protein was incorporated into purified PRV Be or PRV Ba virions as efficiently as wild-type Us9 protein when these viruses were grown on the PK15-BB14 cell line. The incorporation was efficient, because in PRV Be, but not Ba, virions the parental isoforms of Us9 were detected in approximately equimolar quantities with Us9-EGFP. The Us9-EGFP chimeric protein was resolved as a doublet in the PRV Ba virion preparation, but the doublet was not as distinct in the PRV Be virions.
FIG. 8.
Us9 is a type II membrane protein. Us9-EGFP-containing virions were isolated from the medium of Us9-EGFP-expressing cells infected with PRV Be or Ba (MOI = 10) as described in Materials and Methods. The purified PRV Be virions (A) or Ba virions (B and C) were treated with (lanes 3 and 4) or without (lanes 1 and 2) proteinase K in either the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 1% NP-40. The samples were then fractionated on an SDS–12.5% polyacrylamide gel and analyzed by Western blotting with either Us9 (A and B) or GFP (C) antiserum. Positions of molecular mass markers (kilodaltons) are indicated on the left.
We next determined the topology of the Us9-EGFP protein in the virions by treatment with proteinase K. Treated and untreated virions were analyzed by Western blot analysis with antiserum specific for Us9 or for GFP. As a control, virions were treated with or without 1% NP-40 in the presence or absence of protease to verify that the proteins were sensitive to the protease and that the detergent alone had no effect on protein migration or stability.
The PRV Be Us9-EGFP virion controls show that both the Us9-EGFP molecule (approximately 47 kDa) and the wild-type Us9 protein (17 to 20 kDa) were efficiently incorporated into virions (Fig. 8A, lanes 1 and 2). The Us9-EGFP fusion protein was sensitive to proteinase K (Fig. 8A), while the wild-type Us9 protein was protected from degradation (lane 3). It is important to note that a protease-resistant Us9 immunoreactive product migrating slightly slower than wild-type Us9 remained after protease treatment (Fig. 8A, lane 3). This band, representing wild-type Us9 with a small portion of the EGFP molecule attached, was protected from digestion by the viral envelope, thereby arguing against the possibility of cellular contamination. When detergent was added in addition to protease, not only the EGFP portion but also the Us9 moiety of the chimeric protein was digested (Fig. 8A, lane 4). The same results were obtained when the experiment was conducted with virions containing Us9-EGFP from PRV Ba-infected cells (Fig. 8B). The PRV Ba virions do not contain wild-type Us9 protein, and the only Us9 immunoreactive polypeptide was the Us9-EGFP fusion protein itself (Fig. 8B, lanes 1 and 2). The faint bands detected in PRV Ba virions (Fig. 8B, lanes 1 and 2) migrating at approximately the same position as wild-type Us9 (Fig. 8A, lanes 1 and 2) are thought to be due to a low level of degradation of the Us9-EGFP fusion protein during isolation and manipulation of the viral particles. When the Us9-EGFP Ba virions shown in Fig. 8B were analyzed by using antiserum specific for GFP (Fig. 8C), it was clear that the protease digested the Us9-EGFP fusion protein into an EGFP immunoreactive protease-resistant fragment (approximately 27 kDa) in both the presence and absence of NP-40 (Fig. 8C).
When PRV Be virions without the Us9-EGFP fusion protein were digested with proteinase K, Us9 was resistant to digestion, and it became completely sensitive only when detergent plus protease was added (data not shown). These experiments demonstrate that the Us9-EGFP fusion protein is oriented as a type II membrane protein: the carboxy-terminal EGFP moiety is accessible to protease, while the Us9 moiety is not. Accordingly, given that in virions Us9 protein itself is resistant to external protease, we conclude that the Us9 protein is also a type II membrane protein.
DISCUSSION
The Us9 protein is highly conserved among the various alphaherpesviruses. This observation and the fact that its absence causes no obvious phenotype in standard tissue culture cell lines imply a conserved function in natural animal infections with a positive selective advantage. The fact that some naturally attenuated PRV strains have deletions of Us9 provides a hint that the protein affects viral pathogenesis. Consequently, in this study, we produced a Us9-specific polyclonal antiserum to identify and characterize the protein product of the PRV Us9 gene. Prior to this study, the Us9-homologous proteins had been classified as structural proteins residing in the tegument, based on a seminal study performed by Frame and colleagues on the HSV-1 Us9 homolog (14).
Us9 contains posttranslational modifications.
The deduced Us9 amino acid sequence predicts a protein of 98 amino acids and 10.6 kDa. Using immunoprecipitation and Western blot analysis with Us9 polyclonal antiserum and PRV Be-infected cell lysates, we found several Us9 polypeptides with relative molecular masses of between 17 and 20 kDa. Even after in vitro translation, multiple species of Us9 protein could be readily identified. We first thought that these polypeptides resulted from two translation starts at one of two in-frame methionines found at the beginning of the predicted open reading frame. However, the data from site-directed mutagenesis experiments suggested that the multiple forms were not due to alternative translation starts. Moreover, these data suggested that the second methionine is the major site of translation initiation. Our experiments support the notion that the multiple forms of Us9 are due primarily to phosphorylation. We have shown by direct phosphate labeling that there are at least three phosphoforms of Us9 protein in PRV Be-infected cell lysates. Furthermore, treatment of Us9 immunoprecipitates with phosphatases also shifts the electrophoretic mobility of the proteins, consistent with direct Us9 phosphorylation (unpublished data). Previous reports demonstrated that the HSV-1 Us9 protein is also phosphorylated and ubiquitinated (2, 14).
It is thought that the degree of phosphorylation of a polypeptide in virions may differ from that in the infected cell (40). For example, the HSV-1 tegument protein VP22 has been recently shown by Elliott et al. (11) to be phosphorylated in the infected cell, yet only the nonphosphorylated polypeptide is packaged in the viral particle. It is conceivable that phosphorylation of the Us9 molecule modulates its ability to be incorporated into viral particles, location in the cell, or interaction with other viral or cellular proteins.
Us9 is a tail-anchored, type II membrane protein.
Although the PRV Us9 polypeptide was classified as a viral tegument protein based on homology to the HSV-1 Us9 homolog, our data indicate that it is a type II membrane protein. As such, PRV Us9 would have a 3-amino-acid carboxy-terminal extension on the outside of the viral particle, 26 amino acids spanning the lipid bilayer, and 68 amino acids in the tegument region. It is easy to see, based on this model, how Us9 may have been characterized as a tegument protein, as the majority of the protein does in fact reside in the tegument and the three carboxy-terminal amino acids on the outside of the viral particle will most likely not be recognized either by antibodies or by protease digestion, two important criteria for classification of a tegument protein. To aid in the identification of Us9 as a type II membrane protein, we fused the EGFP reporter molecule to the carboxy terminus of the Us9 protein. This fusion protein provided independent verification of the unique localization and topology of Us9.
Three additional lines of evidence support the type II topology of Us9. First, the Us9 protein has a single N-linked glycosylation consensus sequence, at residue 55 preceding the putative transmembrane domain, which is not utilized. If Us9 is a type II membrane protein, the N-linked glycosylation site would be in the cytosol (and not the lumen of the secretory system) and consequently not available for modification. Second, as a membrane protein with type II topology, the tyrosine and casein kinase I and II phosphorylation sites would be exposed to the cytosol and available for modification by either cellular or viral protein kinases. If Us9 was oriented as a type I molecule, these sites would be inaccessible to these common protein kinases. Finally, preliminary results have indicated that a truncated Us9 mutant protein lacking the hydrophobic domain predicted to span the membrane was found dispersed in the cytoplasm and not in the lumen of the secretory system (data not shown). This result is consistent only with a membrane protein of type II and not type I topology.
Type II membrane proteins have been identified in several virus families. For example, the influenza A virus neuraminidase protein (12), the respiratory syncytial virus G protein (56, 64), and the vaccinia virus A36R (44) and A33R (53) proteins are all type II membrane proteins. As discussed below, the Us9 protein may be distinct from these proteins in the mechanism of insertion into membranes. To the extent of our knowledge, the only type II membrane proteins identified in the Herpesviridae prior to this report are the bcl-2 homolog (BHFR1) of Epstein-Barr virus, a gammaherpesvirus (47), and the UL45 gene product of HSV-2 (7).
We suggest that the PRV Us9 protein falls into a unique class of type II membrane proteins called tail-anchored proteins (29, 30, 32, 67). Tail-anchored proteins have no obvious signal sequence, an amino terminus exposed to the cytoplasm, and a hydrophobic membrane-spanning sequence near the carboxy terminus. Other examples of proteins in this class include cytochrome b5, Bcl-2, syntaxin, and several proteins involved in vesicular transport (BOS1 and SLY2 of Saccharomyces cerevisiae and eukaryotic synaptobrevins) (29). In contrast to canonical type I or type II molecules with an active signal sequence promoting cotranslational membrane insertion, the tail-anchored proteins are predicted to be posttranslationally inserted into membranes, as most of the membrane anchor will be buried within the translating ribosome when the termination codon is reached (29, 30, 32, 67). This is particularly true of Us9, since the hydrophobic domain is essentially at the carboxy terminus of the protein. Although most tail-anchored proteins are posttranslationally inserted into the ER before transport to their final destination, it is believed that some tail-anchored proteins may be incorporated directly into the target organelle (29, 30). In support of this concept, synaptobrevin (30), cytochrome b5 (9, 59), and Bcl-2 (20) have all been demonstrated to be inserted into a variety of membranes in vitro.
Implications of a type II tail-anchored membrane protein in the PRV envelope.
Because we find Us9 localized primarily to the Golgi apparatus at early times in infection and in transfected cells, we speculate that Us9 may be inserted into the secretory system after translation in the cytoplasm and may possibly be inserted directly into vesicles of the Golgi apparatus. Our inability to detect obvious nuclear or ER staining by indirect immunofluorescence microscopy or by EGFP fluorescence supports this speculation. Obviously, a detailed examination using genetics and biochemistry to determine the viral protein composition of the ER and inner nuclear membrane must be conducted, but the rapid and early localization of Us9 to the Golgi apparatus and cytoplasmic vesicles is striking. If the Us9 protein is inserted posttranslationally into late compartments of the secretory system and not the inner nuclear membrane, the mechanism of its incorporation into the viral envelope may challenge some of the current models for herpesvirus envelopment (3, 16, 21, 22, 50, 57, 65, 69). Indeed, Pol et al. have observed that infection of swine nasal turbinate cells with a PRV gE/Us9 double mutant revealed abnormal envelopment (48, 49). We are currently creating defined point mutations and internal deletions in the Us9 molecule in an attempt to reveal potential targeting defects and protein-protein interactions. Infection of various animal models with these mutant viruses may provide insight into function.
Efficient incorporation of Us9-EGFP into viral particles.
A Us9-EGFP fusion protein was efficiently incorporated into either PRV Be or Ba virions after infection of a Us9-EGFP-expressing cell line. These particles offer several unique advantages to study PRV biology. First, it appears that the Us9 protein can target foreign proteins to viral membranes and present them on the surface of virions. If so, we will be able to identify specific envelopment and assembly targeting signals. Alternatively, it is possible that the EGFP protein is not specifically targeted to virion envelopes by Us9 and instead is incorporated only passively by being in the appropriate compartment at high concentrations. At this time, we do not believe incorporation is passive, as we find that the endogenous Us9 and the Us9-EGFP fusion protein are incorporated into PRV Be virions with similar efficiencies. This may suggest that the amount of Us9 protein incorporated into viral particles is controlled by the virus, as was observed for the HSV-1 tegument protein UL37 (39). Experiments necessary to test this idea are in progress.
We are now studying these novel Us9-EGFP viral particles to ensure that no defects in viral replication result from the incorporation of the fusion protein. Initial examination of the Us9-EGFP viral particles indicated that the virions were capable of undergoing a productive infection and plated on indicator cells with the same efficiency as wild-type virus (data not shown). Our unpublished results indicate that PRV Be and Ba virions containing the Us9-EGFP fusion protein may serve as useful tools for visualizing the processes of herpesvirus entry and egress in real time. Moreover, as Us9 is nonessential for viral replication in tissue culture and is able to target a chimeric fusion protein into viral particles, this molecule may be useful in altering the tropism of alphaherpesviruses for potential use in gene therapy applications.
ACKNOWLEDGMENTS
We gratefully acknowledge the expert technical assistance of J. Goodhouse and D. Hasara. Thanks also go to G. M. Waters and the members of the Enquist laboratory for advice and helpful comments.
A.D.B. is supported by NIH training grant 5T32GM07312. B.W.B. is supported by a postdoctoral fellowship from the Medical Research Council of Canada. This work was supported by NINDS grant 1RO133506 to L.W.E.
REFERENCES
- 1.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 2.Brandimarti R, Roizman B. Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteosomes. Proc Natl Acad Sci USA. 1997;94:13973–13978. doi: 10.1073/pnas.94.25.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Card J P, Rinaman L, Schwaber J S, Miselis R R, Whealy M E, Robbins A K, Enquist L W. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropsim and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Card J P, Whealy M, Robbins A, Moore R, Enquist L. Two alphaherpesvirus strains are transported differentially in the rodent visual system. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 7.Cockerell A S, Muggeridge M I. Herpes simplex virus type 2 UL45 is a type II membrane protein. J Virol. 1998;72:4430–4433. doi: 10.1128/jvi.72.5.4430-4433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullinane A A, Rixon F J, Davison A J. Characterization of the genome of equine herpesvirus 1 subtype 2. J Gen Virol. 1988;69:1575–1590. doi: 10.1099/0022-1317-69-7-1575. [DOI] [PubMed] [Google Scholar]
- 9.Daily H A, Strittmatter P. Structural and functional properties of the membrane binding segment of cytochrome b5. J Biol Chem. 1978;253:8203–8209. [PubMed] [Google Scholar]
- 10.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 11.Elliott G, O’Reilly D, O’Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 12.Fields S, Winter G, Brownlee G G. Structure of the neuraminidase gene in human influenza virus A/PR/8/34. Nature. 1981;290:213–217. doi: 10.1038/290213a0. [DOI] [PubMed] [Google Scholar]
- 13.Flowers C C, O’Callaghan D J. The equine herpesvirus type 1 (EHV-1) homolog of herpes simplex virus type 1 Us9 and the nature of a major deletion within the unique short segment of the EHV-1 KyA strain genome. Virology. 1992;190:307–315. doi: 10.1016/0042-6822(92)91217-i. [DOI] [PubMed] [Google Scholar]
- 14.Frame M C, McGeoch D J, Rixon F J, Orr A C, Marsden H S. The 10K virion phosphoprotein encoded by gene Us9 from herpes simplex virus type I. Virology. 1986;150:321–332. doi: 10.1016/0042-6822(86)90297-7. [DOI] [PubMed] [Google Scholar]
- 15.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 16.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holton T A, Graham M W. A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res. 1990;19:1156. doi: 10.1093/nar/19.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husak, P. J., and L. W. Enquist. Unpublished data.
- 19.Jacobs L, Rziha H J, Kimman T G, Gielkens A L J, van Oirschot J T. Deleting valine-125 and cysteine-126 in glycoprotein gI of pseudorabies virus strain NIA-3 decreases plaque size and reduces virulence in mice. Arch Virol. 1993;131:251–264. doi: 10.1007/BF01378630. [DOI] [PubMed] [Google Scholar]
- 20.Janiak F, Leber B, Andrews D W. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J Biol Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 21.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones R, Grose C. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J Virol. 1988;62:2701–2711. doi: 10.1128/jvi.62.8.2701-2711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killeen A M, Harrington L, Wall L V M, Kelly D C. Nucleotide sequence analysis of a homologue of herpes simplex virus type I gene Us9 found in the genome of simian herpes B virus. J Gen Virol. 1992;73:195–199. doi: 10.1099/0022-1317-73-1-195. [DOI] [PubMed] [Google Scholar]
- 24.Kimman T G, de Wind N, Oei-Lie N, Pol J M A, Berns A J M, Gielkens A L J. Contribution of single genes within the unique short region of Aujeszky’s disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 25.Knapp A C, Enquist L W. Pseudorabies virus recombinants expressing functional virulence determinants gE and gI from bovine herpesvirus 1.1. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. The scanning model for translation: an update. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type and gC-, gE- and gI-deleted mutants of Aujeszky’s disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 29.Kutay U, Hartmann E, Rapoport T A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 30.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport T A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung-Tack P, Audonnet J, Riviere M. The complete DNA sequence and the genetic organization of the short unique region (Us) of the bovine herpesvirus type 1 (ST strain) Virology. 1994;199:409–421. doi: 10.1006/viro.1994.1139. [DOI] [PubMed] [Google Scholar]
- 32.Linstedt A D, Foguet M, Renz M, Seelig H P, Glick B S, Hauri H P. A C-terminally-anchored Golgi protein is inserted into the endoplasmic reticulum and then transported to the Golig apparatus. Proc Natl Acad Sci USA. 1995;92:5102–5105. doi: 10.1073/pnas.92.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lippincott-Schwartz J, Donaldson J, Schweizer A, Berger E, Hauri H P, Yuan L, Klausner R. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 34.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomniczi B, Blankenship M L, Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the gene. J Virol. 1984;49:970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomniczi B, Watanabe S, Ben-Porat T, Kaplan A. Genetic basis of the neurovirulence of pseudorabies virus. J Virol. 1984;52:198–206. doi: 10.1128/jvi.52.1.198-205.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the unique long region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 38.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type I. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 39.McLauchlan J. The abundance of the herpes simplex virus type I UL37 tegument protein in virus particles is closely controlled. J Gen Virol. 1997;78:189–194. doi: 10.1099/0022-1317-78-1-189. [DOI] [PubMed] [Google Scholar]
- 40.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter T C, Lomniczi B, Sugg N, Schreurs C, Ben-Porat T. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J Virol. 1988;62:12–19. doi: 10.1128/jvi.62.1.12-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mettenleiter T C, Lukacs N, Rziha H J. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J Virol. 1985;56:307–311. doi: 10.1128/jvi.56.1.307-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiyama Y, Kurachi R, Daikoku T, Umene K. The Us 9, 10, 11, and 12 genes of herpes simplex virus type I are of no importance for its neurovirulence and latency in mice. Virology. 1993;194:419–423. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- 44.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr 43-50K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 45.Petrovskis E A, Timmins J, Gierman T, Post L. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrovskis E A, Post L E. A small open reading frame in pseudorabies virus and implications for evolutionary relationships between herpesviruses. Virology. 1987;159:193–195. doi: 10.1016/0042-6822(87)90368-0. [DOI] [PubMed] [Google Scholar]
- 47.Pfitzner A J, Tsai E C, Strominger J L, Speck S H. Isolation and characterization of cDNA clones corresponding to transcripts from the BamHI H and F regions of the Epstein-Barr virus genome. J Virol. 1987;61:2902–2909. doi: 10.1128/jvi.61.9.2902-2909.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pol J M A, Wagenaar F, Gielkens A. Morphogenesis of three pseudorabies virus strains in porcine nasal mucosa. Intervirology. 1991;32:327–337. doi: 10.1159/000150217. [DOI] [PubMed] [Google Scholar]
- 49.Pol J M A, Quint W G V, Kok G L, Broekhuysen-Davies J M. Pseudorabies virus infections in explants of porcine nasal mucosa. Res Vet Sci. 1991;50:45–53. doi: 10.1016/0034-5288(91)90052-p. [DOI] [PubMed] [Google Scholar]
- 50.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hubinger A, Reschke M. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol. 1996;141:557–572. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 51.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J C, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1043–1107. [Google Scholar]
- 53.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan J P, Whealy M E, Robbins A K, Enquist L W. Analysis of pseudorabies virus glycoprotein gIII localization and modification by using novel infectious viral mutants carrying unique EcoRI sites. J Virol. 1987;61:2251–2257. doi: 10.1128/jvi.61.10.2962-2972.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rziha H J, Mettenleiter T C, Ohlinger V, Wittmann G. Herpesvirus (pseudorabies virus) latency in swine: occurrence and physical state of viral DNA in neural tissues. Virology. 1986;155:600–613. doi: 10.1016/0042-6822(86)90220-5. [DOI] [PubMed] [Google Scholar]
- 56.Satake M, Coligan J E, Elango N, Norrby E, Venkatesan V. Respiratory syncytial virus envelope protein (G) has a novel structure. Nucleic Acids Res. 1985;13:7795–7812. doi: 10.1093/nar/13.21.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stackpole C W. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J Virol. 1969;4:75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strebel K, Beck E, Strohmaier K, Schaller H. Characterization of foot-and-mouth disease virus gene products with antisera against bacterially synthesized fusion proteins. J Virol. 1986;57:983–991. doi: 10.1128/jvi.57.3.983-991.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takagaki Y, Radhakrishnan R, Wirtz K W, Khorana H G. The membrane-embedded segment of cytochrome b5 as studied by cross-linking with photoactivatable phospholipids. II. The nontransferable form. J Biol Chem. 1983;258:9136–9142. [PubMed] [Google Scholar]
- 60.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 61.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Zijl M, van der Gulden H, de Wind N, Gielkens A, Berns A. Identification of two genes in the unique short region of pseudorabies virus: comparison with herpes simplex virus and varicella-zoster virus. J Gen Virol. 1990;71:1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]
- 63.Waters G M, Clary D O, Rothman J E. A novel 115-kd peripheral membrane protein is required for intercisternal transport in the golgi stack. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wertz G W, Collins P L, Huang Y, Gruber C, Levine S, Ball L A. Nucleotide sequence of the G protein of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci USA. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitley P, Grahn E, Kutay U, Rapoport T A, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 68.Willemse M J, Strijdveen I G L, van Schooneveld S H B, van Den Berg M C, Sondermeijer P J A. Transcriptional analysis of the short segment of the feline herpesvirus type 1 genome and insertional mutagenesis of a unique reading frame. Virology. 1995;208:704–711. doi: 10.1006/viro.1995.1202. [DOI] [PubMed] [Google Scholar]
- 69.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]