Summary
Chemical modifications in mRNAs, such as pseudouridine (psi), can control gene expression. Yet, we know little about how they are regulated, especially in neurons. We applied nanopore direct RNA sequencing to investigate psi dynamics in SH-SY5Y cells in response to two perturbations that model a natural and unnatural cellular state: retinoic-acid-mediated differentiation (healthy) and exposure to the neurotoxicant, lead (unhealthy). We discovered that the expression of some psi writers change significantly in response to physiological conditions. We also found that globally, lead-treated cells have more psi sites but lower relative occupancy than untreated cells and differentiated cells. Interestingly, examples of highly plastic sites were accompanied by constant expression for psi writers, suggesting trans-regulation. Many positions were static throughout all three cellular states, suggestive of a “housekeeping” function. This study enables investigations into mechanisms that control psi modifications in neurons and its possible protective effects in response to cellular stress.
Keywords: Pseudouridine, RNA sequencing, mRNA modification, nanopore DRS, plasticity, epitranscriptome, differentiation, neurotoxicant
Introduction
RNA modifications are enzyme-mediated chemical changes to the canonical structure of RNA nucleotides. Over 170 types of RNA modifications have been discovered in all classes of RNAs1 and play roles in diverse biological processes such as RNA metabolism2,3, translational control4-6, gene expression3,7, splicing3,8 RNA-protein interactions7, and immune response9. Of all uridines (U) in mammalian mRNA, 0.2% - 0.6% are pseudouridine (psi)10,11, a similar frequency as the abundance of m6A with respect to all adenosines12,13. Psi is an isomer of uridine14 in which a new hydrogen bond is available to base-pair primarily with adenosine but also other nucleobases when in an RNA duplex for a stabilizing effect15, 3,16,17. This duplex stabilization is thought to modulate cellular interactions with proteins and other biomolecules3,8.
Various next-generation sequencing methods have been utilized for psi mapping in mRNAs6,10,18-21; however, these methods all require chemical mediators (i.e., CMC labeling and bisulfite conversion) combined with reverse transcription to cDNA before amplification and sequencing. We and others have recently developed algorithms to classify psi sites from nanopore direct RNA sequencing (DRS)22-25. Our method, Mod-p ID25, compares the frequency of systematic basecalling errors at the modification site to an in vitro transcribed (IVT) unmodified transcriptome26. Mod-p ID accounts for the sequence context surrounding individual psi modifications and coverage at a given site to determine a statistical probability of a modification and provides a lower-limit occupancy value. A major benefit to this method is that it can provide a relative occupancy when the same position within the transcriptome is compared across different conditions. However, a caveat of this and other nanopore-based methods for psi-calling is that the methods alone are insufficient to validate psi modifications, necessitating exhaustive orthogonal validation approaches such as synthetic controls27-29 or biochemical assays30. Another suitable route for transcriptome-wide validation is to utilize knockdown/knockout of psi synthases (PUS) and measure changes in psi occupancies8,21 at the sites matching the motif for respective PUS.
In this work, we aim to understand how psi occupancies in mRNAs of neurons respond to changes in cellular state. Dysregulation of genes encoding PUS enzymes is associated with neuronal impairment.31,32 However, the plasticity of specific psi sites in neurons remains unknown. We used SH-SY5Y cells as a neuron-like system to model changes in cellular state and assess whether psi distribution and occupancy levels adjust in response to the environment. SH-SY5Y cells continuously express markers similar to immature catecholaminergic neurons while maintaining the ability to divide33. To understand the plasticity of psi, transcriptome-wide, we applied two different perturbations: one that models a healthy change in cellular state and one that models an unhealthy change in cellular state. The rationale for employing two distinct perturbations to investigate plasticity is that their mechanistic divergence would facilitate the identification of universally changing sites, stable sites, and those that may be condition dependent.
First, we exposed SH-SY5Y cells to retinoic acid (RA) to achieve differentiation, which models a healthy change in cellular state 34. Upon RA-mediated differentiation, SH-SY5Y cells become morphologically similar to primary neurons; their proliferation rate is decreased (similar to mature neurons), and the activity of enzymes specific to neuronal tissues is increased35. Second, we exposed them to lead (Pb2+), a systemic toxicant affecting virtually every organ system, but primarily the central nervous system36. Pb2+ is an environmental toxin that has been shown to adversely affect neuron functionality, especially during the developmental stages of the human brain.37,38,39
We performed DRS and Mod-p ID analysis to identify psi sites across conditions. To validate the identified sites as psi and assign them to specific psi synthases, we performed a siRNA knock-down of two key PUS enzymes, TRUB1 (motif GUUCN)40 and PUS7 (motif UNUAR)41. In addition to knockdown experiments, we performed rigorous validation of the query sites using five orthogonal methods: CeU-Seq10, BID-seq21, PRAISE18, CLAP30, and RBS-Seq20. These methods include RNA bisulfite sequencing, CMC-based sequencing, and biochemical CMC analysis by gel. Identifying a site as knocked down and through multiple orthogonal methods provides confidence that these sites are indeed psi.
We performed differential analysis, i.e., comparing psi occupancy at psi sites in untreated vs RA-differentiated cells, and untreated vs Pb2+-treated SH-SY5Y cells. Comparing three very different states can reveal the relative plasticity of psi sites at steady state (i.e., untreated SH-SY5Y cells) and in response to environmental cues for both differentiation and Pb2+ exposure. Such plasticity has been observed with m6A, for which modification levels increase significantly throughout brain development, which is suggested as a mechanism to achieve higher-order brain function42.
Results
Retinoic acid-induced differentiation of SH-SY5Y cells leads to a change in cell state.
SH-SY5Y cells were differentiated into neuron-like cells by supplementing them with retinoic acid43, according to Kovalevich et al.44 (Fig. 1a). To confirm differentiation, we compared the cellular morphology between conditions and observed the characteristic elongation and branching of neurite-like processes from the differentiated cells (Fig. 1b). Poly-A selected RNA was extracted from differentiated and untreated SH-SY5Y cells (n = 3 biological replicates for each group) and sequenced on a MinION device using R9 flow cells. We observed differential mRNA expression when comparing the two groups, supporting a change in cell state (Fig 1d). Importantly, we observed the expected differential mRNA expression of known differentiation markers34,45,46. CRABP2, RARB, RGS2, RET, and DKK2 exhibited upregulation, and ISOC1, MYC, SPRY2, and ASCL1 displayed decreased RNA expression in differentiated SH-SY5Y cells compared to the untreated counterparts (Supplementary Fig. 1). To assess the effects of retinoic-acid-mediated differentiation on psi machinery, we evaluated expression levels for 13 different PUS enzymes from untreated and differentiated libraries (Fig. 1c). While most PUS enzymes remained constant between conditions, we observed a significant difference in the RNA expression levels for RPUSD3 and PUS7L in response to differentiation (p < 0.05). Rpusd3 is a pseudouridine synthase that is thought to act on mitochondrial RNA and ribosomal RNAs47. While the exact function of Pus7L is unknown, it may be regulated in response to physiological conditions as evidenced by tissue specificity of PUS7L in different brain tissues, as compared to the homolog PUS7 which is ubiquitously expressed48.
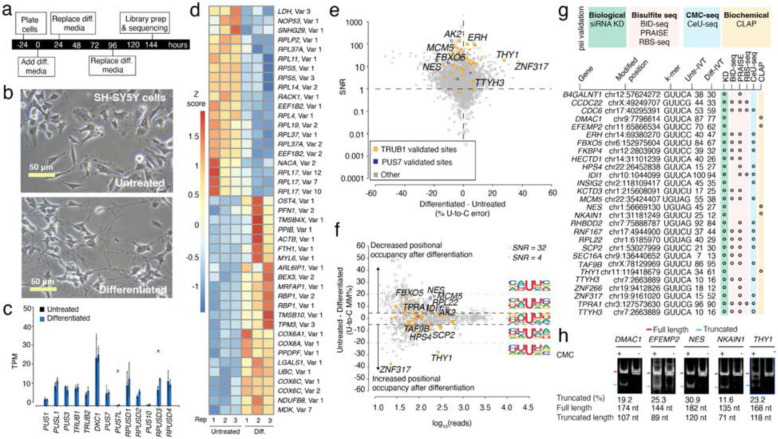
Figure 1. Effects of RA-mediated differentiation on mRNA psi modification and machinery in SH-SY5Y cells.
a. Timeline illustrating the stages and duration of the RA treatment applied to SH-SY5Y cells.
b. A representative photomicrograph of untreated and differentiated SH-SY5Y cells is shown.
c. TPM of various PUS enzymes in untreated and differentiated SH-SY5Y cells determined by DRS. Individual colored bars represent each experimental condition, with error bars describing the standard error of the mean (SEM) across downsampled replicates. Individual replicates are shown as black dots. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
d. We used Deseq2 to identify the transcripts with the highest fold change between the untreated and differentiated samples. Three biological replicates for each condition were used. The color scale shows a Z-score based on the relative fold-change.
e. SNR vs. the difference in U-to-C error % between the untreated and differentiated samples. Orange dots represent uridine positions that are validated TRUB1 substrates, and blue dots represent uridine positions that are validated PUS7 substrates.
f. Putative psi-positions determined by Mod-p ID are plotted according to the difference in U-to-C basecalling error in the untreated and differentiated samples against the reads for each position. Dotted line at the +5% and −5% marks indicate the cutoff for a position to be changed in response to perturbation. Inset shows the sequencing logo for positions within the TRUB1 motif, gray points above the threshold line, and total points above the threshold line.
g. Annotation of genes containing a psi modification validated by PUS7 or TRUB1 KD (Figure 2) and orthogonal methods.
h. CLAP gel result of psi incorporation in mRNAs for the sites confirmed by Nanopore DRS and KD data, but not by any other orthogonal method. See Supplementary Fig. 2 for original gel source data.
Transcriptome-wide mapping of psi-modifications before and after differentiation
Psi positions were identified in differentiated and untreated SH-SY5Y cells by enriching our paired in vitro transcribed (IVT) dataset with additional human IVT data26 (see Methods), allowing us to achieve sufficient coverage for an additional 878 putative psi sites. We then used Mod-p ID25 to identify putative psi sites based on significant differences in U-to-C basecalling error (p < 0.001) in the untreated and differentiated libraries compared to the IVT control library.
Sites with > 40% U-to-C error (i.e., hypermodification type I25) may be considered “high-occupancy” and have the potential for stronger phenotypes. These sites were assessed across samples (Supplementary Table 1). It should be noted that the percentage here is only a qualitative metric of occupancy. From the untreated sample, 131 hypermodified sites were detected, and 132 sites were detected from the differentiated library. (Supplementary Fig. 2a, b). Of these sites, 83 were hypermodified in both conditions (Supplementary Fig. 2d). Most of these sites fall within mRNAs that encode proteins of various functions. Notably, for the sites that are unique to differentiated samples, CAMKV, KIF2A, DBNL, and CRKL are involved in neuronal and synaptic function.
The occupancy levels of sites common to both libraries were compared to identify differences in psi profiles due to the differentiation process beyond just the hypermodified sites. First, we selected positions present in both the untreated and differentiated libraries and defined a signal-to-noise ratio (SNR) for each site (see Methods). This filtering step was introduced to guarantee a strong DRS signal level as compared to the IVT while considering coverage and size of error signatures (Fig 1e). We selected sites with SNR ≥ 1 and read coverage of ≥ 10 reads in both libraries (Fig. 1f). We observed that 1,786 sites exceeded this threshold, indicating that the signal-to-noise ratio was sufficient for analysis (Supplementary Table 2).
Next, we performed a differential analysis comparing the relative occupancy of identified sites in the untreated and differentiated libraries. We found a mean difference in U-to-C mismatch of 2.46% with a 95% CI = [2.02, 2.91], indicating a statistically significant change in modification occupancy between the differentiated and untreated SH-SY5Y cells. This result suggests that the relative modification occupancy is lower in differentiated SH-SY5Y cells compared to the untreated cells for conserved targets. We focused on sites with a differential U-to-C mismatch level outside the CI. To make our differential analysis more conservative, we defined sites as “changed” if samples had observed a ≥ 5% difference in U-to-C error in both directions (Fig. 1f).
To explore whether a specific sequence motif is changed, we exported the sequencing logo for positions categorized into three groups: positions with higher relative occupancy following differentiation, positions that remain unchanged, and positions with lower relative occupancy following differentiation (Fig. 1f). For the positions in which the positional occupancy decreased after differentiation, the +1-nucleotide neighboring the psi position was frequently uridine while the unchanged and higher occupancy sites equally represented all nucleotides in this position.
Among the sites with significant differences between samples (i.e., sites that fell outside of the CI), 26 positions were assigned to a specific PUS enzyme by siRNA mediated knockdown (KD) of PUS7 and TRUB1 (Supplementary Table 3). 86% of these positions were additionally validated as psi using our siRNA KD with at least 1 out of these four orthogonal methods: BID-Seq21, PRAISE18, RBS-seq20, and CeU-Seq10. We found that five positions (DMAC1 chr9:7796614, EFEMP2 chr11:65866534, NES chr1:56669130, NKAIN1 chr1:31181249, THY1 chr11:119418679) were validated by our knockdowns but have not been discovered by these four orthogonal methods (Fig. 1g) so we proceeded to validate these sites with CLAP (CMC-RT and ligation assisted PCR analysis of psi modification)30. CLAP confirmed all these sites identified that were only by Mod-p ID as psi serving as orthogonal validation (Fig. 1g-h, Supplementary Fig. 3).
The most substantial changes were for ZNF317 (chr19:9161020), which increased from a 15% U-to-C error in the untreated sample to 52% in the differentiated sample. The next most substantial change was for THY1 (chr11:119418679), which increased from a 34% U-to-C error in the untreated sample to a 61% U-to-C error in the differentiated sample. The modified position for ZNF317 is within the CDS and the encoded protein is part of the zinc finger protein family and is involved in transcriptional regulation and cellular adaptation to changes49. The modified position for THY1 is in the 3’UTR, and the encoded protein is a cell surface glycoprotein involved in cell adhesion processes and modulates neurite outgrowth50. An example of a reduced psi occupancy upon differentiation (25% to 12%) is found for NKAIN1, which encodes a protein involved in sodium-potassium transport in the brain51.
Next, we explored differences in mRNA expression for transcripts that harbor a psi position. Interestingly, the mRNA levels remained unchanged, with differences in psi occupancy. (Supplementary Fig. 4a) We found that only IDI1 (chr10:1044099; 15.3 TPM in the untreated sample and 58.9 TPM in the differentiated sample) showed a significant difference in mRNA expression between the two conditions (p < 0.001). Other methods have also reported that this site is a substrate for Trub118. IDI1 encodes an enzyme that is involved in the synthesis of cholesterol metabolites which are essential for the proper differentiation and function of neurons. Next, we examined the protein expression levels in cellular compartments for the two dominant PUS enzymes for humans, Pus7 and Trub1, using immunofluorescence in untreated and differentiated SH-SY5Y cells. Our analysis revealed no significant differences in the subcellular distribution of these two PUS enzymes (Supplementary Fig. 5).
Absolute quantification of psi occupancy in response to differentiation
While our differential analysis highlights sites with significant relative changes in occupancy, we were interested in assigning absolute occupancy to some of these positions. Sequence-specific synthetic controls were generated to match the mRNA sequences bearing psi in the undifferentiated and differentiated samples. One library with uridine in the modification site and one with psi in the modification site was generated and sequenced using DRS. Using these sequence-specific synthetic controls, we trained 10 supervised machine-learning models (ML) to quantify the occupancy of 10 psi-sites (see Supplementary Table 4) in the native RA-differentiated and untreated transcripts (see Methods) using ModQuant29. Each ML model had a classification accuracy of > 90%. PSMB2 (chr1:35603333) is an example of a site with similar occupancies in the untreated (62% ML-predicted level, 61% U-to-C mismatch) and differentiated library (61% ML-predicted level, 63% U-to-C mismatch). An opposite example is PRPSAP1 (chr17:76311411), which changes both in the untreated (67% ML-predicted level, 47% U-to-C mismatch) and differentiated sample (62% ML-predicted level, 33% U-to-C mismatch). This highlights the importance of generating synthetic controls for absolute quantifications.
When applied to the DRS libraries, the lowest level of occupancy predicted by the ML models is 16.9% (ML model for MRPS14). These stoichiometry levels are expected as the sites we tested with synthetic standards have at least 20% U-to-C mismatch in the native datasets. However, this approach can reach single-read classification accuracy of 95%, as shown by the model error calculated on each synthetic standard (Supplementary Fig. 6).
SH-SY5Y cell state changes in response to lead (Pb2+) exposure
Next, we introduced the neurotoxicant Pb2+ to SH-SY5Y cells for six days as an alternative, “unhealthy” change in cellular state to contrast with differentiation as a “healthy” change (Fig. 2a). Previous studies have shown that 5 μM is close to the Pb2+ levels in human blood that can cause encephalopathy in children53; however, we chose to use a higher Pb2+ concentration (50 μM) because the in vitro tolerance for cytotoxicity is higher than in vivo53 while maintaining cell viability and typical cellular morphology following exposure (Fig. 2b).
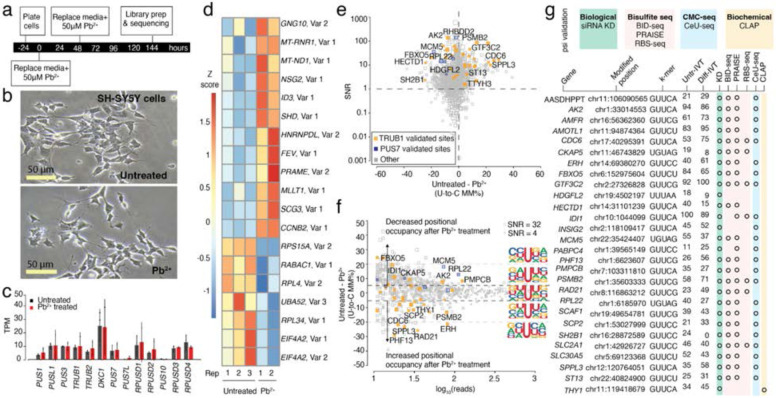
Figure 2. Effects of Pb2+ treatment on mRNA psi modification and machinery in SH-SY5Y cells.
a. Timeline illustrating the stages and duration of the Pb2+ treatment applied to SH-SY5Y cells.
b. A representative photomicrograph of untreated and Pb2+ treated SH-SY5Y cells is shown.
c. DRS determined the TPM of various PUS enzymes in untreated and Pb2+-treated SH-SY5Y cells. Individual colored bars represent each experimental condition, with error bars describing the standard error of the mean (SEM) across downsampled replicates. Individual replicates are shown as black dots. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
d. We used Deseq2 to identify the transcripts with the highest fold change between the untreated and Pb2+ treated samples. Three biological replicates of each condition were used. Color scale shows a Z-score based on the relative fold-change.
e. SNR vs. the difference in U-to-C error % between untreated and Pb2+ treated samples. Orange dots represent uridine positions that are validated TRUB1 substrates, and blue dots represent uridine positions that are validated PUS7 substrates.
f. Putative psi-positions determined by Mod-p ID are plotted according to the difference in U-to-C basecalling error in the untreated and Pb2+ treated samples against the reads for each position. A dotted line at the +5% and −5% marks indicate the cutoff for a position to be changed in response to perturbation. The inlet shows the sequencing logo for positions within the TRUB1 motif, gray points above the threshold line, and total points above the threshold line.
g. Annotation of genes containing a psi modification that changed in response to perturbation and validated by PUS7 or TRUB1 KD (Figure 2) and orthogonal methods.
Poly-A selected RNA was extracted from untreated and Pb2+-exposed SH-SY5Y cells (n = 2 biological replicates) and sequenced on a MinION device using R9 flow cells. Global gene expression profiling of untreated and Pb2+-exposed libraries support a change in cellular state (Fig. 2d). To assess the effects of Pb2+ exposure on psi machinery, we evaluated mRNA expression levels for 13 different psi-synthases from Pb2+ treated and untreated SH-SY5Y cell libraries, and we found no significant differences in expression levels (Fig. 2c).
Transcriptome-wide mapping of psi-modifications following Pb2+ exposure.
To explore changes in psi modification between untreated and Pb2+ treated cells, we generated a paired, unmodified transcriptome for untreated SH-SY5Y cells to identify putative psi positions. We enriched the paired IVT (see Methods), recovering 333 sites that would have been discarded because of insufficient coverage in the paired IVT26. We then applied Mod-p ID25 to identify putative psi sites based on significant differences in U-to-C basecalling error (p < 0.001 in at least two biological replicates, see Methods).
First, the frequency of sites with > 40% U-to-C error (i.e., hypermodification type I54) was assessed. We detected 74 hypermodified sites from the Pb2+-treated library, of which 48 were shared with the untreated library (Supplementary Fig. 2c, d). We selected positions found in both the untreated and Pb2+ treated libraries and calculated the SNR for each site as described above (Fig. 2e), obtaining 946 sites that match our filtration criteria. From these sites, we found a mean difference in U-to-C mismatch error of 2.72% and a 95% CI = [2.14,3.29], indicating a statistically significant change in modification occupancy between the Pb2+-treated and untreated SH-SY5Y cells. This result indicates that the global modification occupancy is lower in Pb2+-treated cells compared to the untreated cells for conserved mRNA targets. Many sites unique to the Pb2+ treated sample fall within mRNAs encoding proteins involved in oxidative stress response, including MRPS14, DHCR7, RTN4, SIGMAR1, and SAE1.
Sites were defined as changed in response to Pb2+ treatment using the same CI-based criteria applied to RA-differentiated SH-SY5Y cells explained above (Fig. 2f, Supplementary Table 5). We exported the sequencing logo for positions categorized into three groups: positions with higher relative occupancy following Pb2+ treatment, positions that remained unchanged, and positions with lower relative occupancy following Pb2+ treatment (Fig. 2f). We found that psi sites with increased positional occupancy following Pb2+ treatment tend to be flanked by two uridines, while those with decreased positional occupancy following Pb2+ treatment have a uridine in the n+1 position.
Among the sites with significant differences between samples, we found 28 positions that were assigned to a specific PUS enzyme using our KD experiments (Supplementary Table 7). In addition to our siRNA KD validation, we confirmed these sites using the same orthogonal methods that were used for differentiated cells (Fig. 2g). Among these sites, 26 (93%) were validated with at least one additional orthogonal method. Two positions, HDGFL2 (chr19:4502197) and THY1 (chrl 1:119418679) transcripts, were not validated by previously annotated datasets so we validated using the CLAP method. The site within HDGFL2 was not possible to test with this method because it is only 10 nucleotides away from the end of the 3′ UTR, leaving insufficient space to design CLAP primers. We were able to apply the method to THY1, which was confirmed by CLAP as a psi-modified site (Fig. 1h, Supplementary Fig. 3). In the comparison of psi sites in untreated and Pb2+ treated samples, the biggest change was for PHF13 (chr1:6623607), which increased from 26% U-to-C error in the untreated library to 56% U-to-C error following Pb2+ treatment. Phf13 modulates chromatin structure and DNA damage response.
We calculated TPMs for each Trub1 and Pus7 mRNA substrate with differences in psi levels to explore differences in mRNA expression for transcripts that harbor a psi. We found that only ERH (chr14:69380270; 158 TPM in the untreated sample and 207 TPM in the Pb2+ treated sample) showed a significant difference in mRNA expression between the two conditions (p < 0.05; Supplementary Fig. 4b). We then examined the protein expression levels in cellular compartments for the two dominant PUS enzymes for humans, Pus7 and Trub1, using immunofluorescence in untreated and Pb2+ treated SH-SY5Y cells and found no significant differences in the subcellular distribution of these two PUS enzymes (Supplementary Fig. 7).
Absolute quantification of psi occupancy in response to Pb2+ exposure
Following the same approach described for RA-differentiation stoichiometry assessment, we used ModQuant29 and trained 9 supervised ML models to quantify the occupancy of 9 psi-sites (see Supplementary Table 4) in the transcripts from the Pb2+-treated and untreated libraries (see Methods). As in the differentiated library, PSMB2 (chr1:35603333) is an example of a site with similar occupancies in both untreated (62% ML-predicted level, 61% U-to-C mismatch) and lead-treated library (78% ML-predicted level, 74% U-to-C mismatch). Alternatively, U-to-C mismatch and ML-predicted occupancy were different for SLC2A1 (chr1:42926727) in both untreated (67% ML-predicted level, 46% U-to-C mismatch) and lead-exposed samples (74% ML-predicted level, 40% U-to-C mismatch).
Plasticity of pseudouridylation of mRNAs in response to changes in the cellular state
We then sought to compare our 3 cellular states for the same cell type to see which psi positions are static and which are plastic (i.e., cell state dependent psi regulation). We compared the relative occupancy for psi-modified positions across perturbations. We selected orthogonally-validated positions that met the following criteria to evaluate the changes to individual positions between the three conditions: 1. Detected by Mod-p ID as a psi position with p-value < 0.001 in at least one condition; 2. U-to-C mismatch was >40% in at least one of the conditions; 3. Number of reads in all conditions was >10 in DRS and IVT libraries. We rank-ordered these 36 positions based on the standard deviation (SD) over the three conditions (Fig. 3a). Interestingly, 27 out of these 36 positions fall within a TRUB1 motif, and 5 fall within a PUS7 motif. The most similar position between the three conditions is INTS1 (chr7:1476655), with a SD of 0 and >90% U-to-C error for each condition. INTS1 is a component of the integrator complex which has been linked to developmental delays and is involved in in RNA processing and transcriptional regulation55-57. Positions with an SD of >5 are considered to have relatively high. Among these sites, we identify 10 positions with a SD > five, which we consider the most static psi positions. The least similar position is on THY1 (chr11:119418679), with a SD of 19.6 and U-to-C errors of 33.5%, 61.3%, 45.2%, on untreated, differentiated, and Pb2+-treated libraries respectively. THY1 encodes a cell surface glycoprotein involved in cell adhesion processes and modulates neurite outgrowth58.
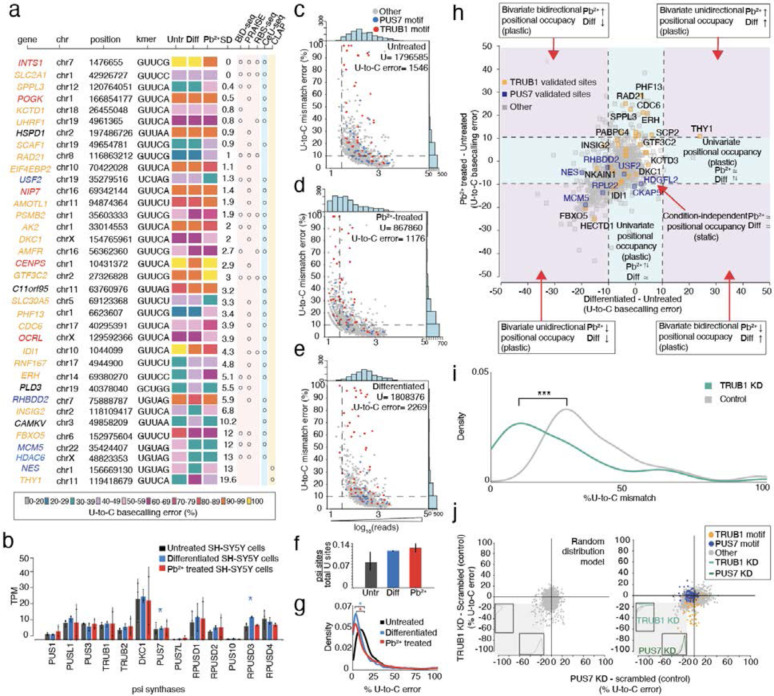
Figure 3. Psi analysis across three cellular states enables the classification of plastic and static sites of modification, transcriptome wide.
a. Heatmap of sites with at least 40% U-to-C basecalling error in one of three conditions. Colors indicate the percentage of U-to-C basecalling errors. For each position the standard deviation (SD) is calculated on all three conditions and used to rank the sites in order, with the most similar at the top and the least similar at the bottom. KD validated substrates for TRUB1 are shown in orange, and TRUB1 motifs that have not yet been validated are shown in red. KD validated substrates for PUS7 are shown in dark blue, and PUS7 motifs that have not been validated are shown in light blue. Orthogonal validation with non-nanopore methods is performed for all the sites.
b. DRS determined the TPM of various PUS enzymes in untreated, differentiated, and Pb2+-treated SH-SY5Y cells. Individual colored bars represent each experimental condition, with error bars describing the standard error of the mean (SEM) across downsampled replicates. Individual replicates are shown as black dots. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
c. Untreated SH-SY5Y number of total DRS reads are plotted against the U-to-C mismatch error of putative Ψ sites detected by Mod-p ID (p-value < 0.001 in at least two biological replicates). Blue dots represent uridine positions within a PUS7 motif, while red dots represent uridine positions within a TRUB1 motif. All the other motifs are shown in grey. Dashed lines represent the criteria used for defining a position as a psi site (number of reads>10; U-to-C error>10).
d. Pb2+ treated SH-SY5Y number of total DRS reads are plotted against the U-to-C mismatch error of putative Ψ sites detected by Mod-p ID (p-value < 0.001 in at least two biological replicates).
e. Differentiated SH-SY5Y number of total DRS reads are plotted against the U-to-C mismatch error of putative Ψ sites detected by Mod-p ID (p-value < 0.001 in at least two biological replicates).
f. Colored bars shows the number of significant pseudouridine sites, labeled as U-to-C error in panels d,e,f, normalized by the total number of reads in each of the three conditions. Error bars show the standard error of the mean across biological replicates.
g. Distribution of the occupancy levels calculated on untreated, differentiated, and lead-treated libraries. Statistical significance is calculated using 95% CI, * p < 0.05.
h. Putative psi-positions determined by Mod-p ID are plotted according to the difference in U-to-C basecalling error in the untreated and Pb2+ treated samples against the difference in U-to-C basecalling error between the differentiated and untreated for each position. A dotted line at the +10% and −10% marks indicate the cutoff for a position to be changed in response to perturbation. Condition-independent positional occupancy (static) sites are shown in the center square. Single condition-dependent positional occupancy sites are shown in the blue stripes. Double condition-dependent positional occupancy sites are shown in the purple areas.
i. Distribution of the occupancy levels calculated on PUS7 validated sites before (grey) and after the TRUB1 KD (green). The occupancy levels in the KD library are decreased compared to the control condition without KD. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
j.Co-operation effect of TRUB1 and PUS7 KD on SH-SY5Y cells. (Left) Random distribution model of points constrained by the data parameters. Distributions are calculated for points in the lower left quadrant as a function of each simulated knockdown. (Right) distribution of TRUB1 KD plotted against PUS7 knockdown. Distributions are calculated for points in the lower left quadrant as a function of each knockdown.
The abundance of psi synthase may account for some of the differences in psi percentage within that condition. Hence, we compared mRNA levels of psi synthases across all three conditions and found significant differences in PUS7 and RPUSD3 expression (Fig. 3b). We also hypothesized that for the sites with higher plasticity, mRNA expression of the target may explain the discrepancy (Supplementary Fig. 4c). However, we did not observe an association between mRNA expression levels and the percentage of U-to-C error for targets with >10 SD.
To assess which cell condition had the highest number of sites bearing psi (irrespective of occupancy), we calculated the number of significant psi sites (p-value < 0.001 in at least two biological replicates) with sufficient coverage (>10 reads in DRS and IVT libraries) detected by Mod-p ID across cell conditions and normalized it by the total number of reads in each of the three samples (see Methods). We found that the Pb2+-exposed cells have the higher normalized percentage of psi sites (0.14%), followed by the differentiated (0.13%) and untreated conditions (0.09%; Fig. 3c-f). Interestingly, for the sites shared between the three libraries, we find a statistically significant decrease in occupancy levels (p<0.05; Fig. 3g) in both treated libraries (differentiated and Pb2+) compared to the untreated one.
Finally, we divided the positions into different zones based on the relationship between the conditions: 1. Univariate positional occupancy whereby the relative occupancy of a given site changes for either differentiation or Pb2+ exposure, demonstrating the plasticity of a given site; 2. Bivariate positional occupancy whereby the relative occupancy changes for both differentiation and Pb2+ exposure, demonstrating plasticity; 3. Condition-independent positional occupancy, whereby the occupancy does not change in response to the different conditions (these modifications are present and stable between different perturbations, i.e., static; Figure 3g).
We found that 73% of sites are static (condition 3) and 27% are plastic (condition 1 and/or 2; Supplementary Table 7). Most of the Trub1 targets fell within condition 3 (static). Notably, we found two TRUB1 KD validated sites in the univariate group affected by differentiation and ten TRUB1 KD validated psi sites in the univariate Pb2+-dependent group. These include sites within PHF13, CDC6 and RAD21, which encode proteins that are linked to DNA repair mechanisms and cellular response to toxicants59. These are examples of potentially interesting sites for future analysis as mediators of RNA binding proteins or translational control.
Interestingly, we observed a cluster of targets (541 sites) that were decreased for both Pb2+ exposure and differentiation which we classified as group 2 plasticity. This cluster is interesting because these targets are higher occupancy in untreated cells and decreased in both perturbations. We compared this distribution to a random distribution and found that the two distributions were significantly different (Mann–Whitney–Wilcoxon test, p < 0.001), meaning that the concomitant decrease in both perturbations is inconsistent with a random effect. Among these plastic sites with a bivariate, unidirectional decrease in occupancy, we find sites within MCM5 and FBXO5, involved in cellular stress response; HECTD1, which is known to modulate retinoic acid signaling60 and RPL22, a ribosomal protein linked to common cancer-associated mutations and nucleolar stress response61,62.
Modeling co-regulation of TRUB1 and PUS7 on mRNA substrates
We observed that a group of psi-sites that were validated as PUS7 substrates showed decreased occupancy levels in the TRUB1KD library (i.e., the TRUB1 KD determines a lower psi occupancy not only on TRUB1 motifs but also within PUS7 motifs; Supplementary Fig. 8d). To test if TRUB1 KD causes an occupancy reduction not just in TRUB1, but also in PUS7 sites, we plotted the distribution of the %U-to-C mismatch of validated PUS7 sites pre- and post-TRUB1 KD. We found a statistically significant shift of PUS7 sites (UNUAR motif) towards lower occupancy levels in the TRUB1 KD library (p=0.0003, Student's t-test; Fig. 3i). This shift towards lower occupancy levels is unexpected, as it occurs in TRUB1 KD samples. In fact, we would expect this to happen upon the KD of PUS7 as this is the writer acting on UNUAR motifs. This suggests that the TRUB1 and PUS7 enzymes have a co-regulatory mechanism.
To explore this observation further, we first assessed the changes in PUS mRNA levels upon TRUB1 and PUS7 KD and observed a significant reduction in TRUB1 mRNA upon PUS7 KD (Student’s t-test, p-value < 0.05) and a reduction in PUS7 upon TRUB1 KD compared to the scrambled control (Supplementary Fig. 8c).To test the hypothesis that TRUB1 KD has a global effect on psi levels within PUS7 motifs, we compared the differences in positional occupancy for the TRUB1 KD with the PUS7 KD to a random distribution model of positions that were constrained on the parameters of the observed sequencing data (Fig. 3j). We found that the data are inconsistent with a random model (Mann–Whitney–Wilcoxon test, p < 0.001) in the third quadrant, a region where the position shows a decreased positional occupancy for both TRUB1 KD and PUS7 KD. We repeated the same analysis on two additional random models, both statistically different from the real dataset (Mann–Whitney–Wilcoxon test, p < 0.01; Supplementary Fig. 9a). To support a potential coregulation of TRUB1 and PUS7 that we observed on SH-SY5Y cells, we applied the same analysis and filtering steps to a HeLa cell dataset on which we performed a siRNA-based knockdown of the same enzymes TRUB1 and PUS7 (Supplementary Fig. 9b). As described before, we randomized the distribution of the real data constraining it to the parameters of the real sequencing data. As for SH-SY5Y cells, we found that HeLa KD data are not consistent with a random model (Mann-Whitney-Wilcoxon test, p < 0.001) in the decreased-occupancy region. These data support a possible coregulation of Trub1 and Pus7.
Discussion
This study assessed the plasticity of psi machinery and modifications in SH-SY5Y cells in response to perturbation of cellular state. We found that, in response to differentiation, the mRNA levels of psi writers PUS7L and RPUSD3 were increased. PUS7L has been shown to have cell type-specific expression in the brain while PUS7 has relatively stable expression across cell types. This observed change is the first experimental evidence that PUS7L is regulated in response to physiological conditions because the same neuronal cell type is compared in 2 different conditions.
Our comparative analysis of the positional occupancy in multiple cellular conditions (untreated, differentiated, and Pb2+ treated) revealed various types of site responses: we found numerous positions with high variable occupancy across conditions, indicating plasticity at those sites. Interestingly, the most static psi positions were frequently targets of TRUB1, which suggests a high degree of conservation both in the positions detected and the occupancy. We also found that 3 out of 4 PUS7 sites were in the plastic group, which may indicate that PUS7 deposited sites are less conserved across conditions although this may be due to a low number of reads. Among these, NES (chr1:156669130) encodes the neuronal marker and cytoskeletal protein Nestin, which plays a role in differentiation and self-renewal. This NES psi site has decreased psi levels following differentiation, while it does not change upon Pb2+ treatment. Noteworthy, mRNA levels for NES are unchanged across all conditions (Supplementary Fig. 4c). In contrast, THY1 (chr11:119418679) exhibits high upregulation of psi occupancy for both Pb2+ treatment and differentiation and encodes for a glycoprotein involved in neuronal processes. Possible functions for sites highly sensitive to environmental factors include translational control in response to cellular stress and maintenance of cellular fitness. Future studies may test cellular fitness in the presence of these stressors in combination with TRUB1/PUS7 knockdown to determine mechanism.
Among the most static positions, some psi sites are found on genes linked to neuronal functions: SLC2A1 is associated with GLUT1 deficiency syndrome, a neurological disorder characterized by seizures, developmental delay, and movement disorders63,64. Dysregulation of UHRF1 has been implicated in various cancers and neurodevelopmental disorders61,62. EIF4EBP2 regulates translation initiation by binding to eukaryotic translation initiation factor 4E (eIF4E) and inhibiting its interaction with the mRNA cap structure. It plays a role in synaptic plasticity and memory formation and in neurodevelopmental disorders such as autism spectrum disorders65-67. Other transcripts, although not relevant to neuronal functions, contain psi static sites. The biological function of these genes is general and related to basic functions and cellular mechanisms. Future studies may test the hypothesis that static sites serve a “housekeeping” function, maintaining homeostasis.
Finally, a global pseudouridylation level assessment of each cell condition (untreated, differentiated, and Pb2+ treated) showed that the Pb2+ libraries had a higher normalized percentage of psi sites compared to the untreated (Fig. 3f). Interestingly, despite the higher frequency levels, we find a statistically significant decrease of the occupancy levels in differentiated and lead-treated libraries compared to the untreated libraries (p<0.05; Fig. 3g) despite the higher frequency levels (Fig. 3f). We hypothesize that the broader installation of the psi-sites across the transcriptome is used by the cell as a mechanism of protection/response to the treatment and that the enzyme responsible for installing the modification may not work as efficiently as in the untreated cells because of enzymatic alterations (i.e., saturation); however, further studies will be required to assess the hypothesis.
We validated psi sites by siRNA knockdown of the predominant psi synthases acting on human mRNAs, TRUB1 and PUS7. Interestingly, we found that TRUB1 and PUS7 KDs affect other psi writers. This effect is stronger in PUS7 KD cells, as the other six PUS enzymes (RPUSD1, TRUB1, DKC1, PUSL1, RPUSD4, RPUSD3) have a significant reduction in their mRNA levels, while TRUB1 KD only affects the RPUSD1 enzyme. This finding is suggestive that the Pus7 protein may act as a transcription factor for the other psi synthase enzymes. Interestingly, in TRUB1 KD libraries, certain positions displaying TRUB1 motifs with high U-to-C basecalling errors did not exhibit changes following TRUB1 KD, and most of these positions have been validated through chemical-based methods. It is possible that, with only a partial knockdown, there were sufficient levels of enzyme available to modify the site. Alternatively, other PUS enzymes could compensate for the decrease in Trub1 enzyme.
We also found evidence of co-regulation for PUS7 and TRUB1. This was a surprising finding, and we analyzed the KD datasets published by Dai et al.21 and found 21 psi sites to have reduced psi modification levels under the depletion of both TRUB1 and PUS7 enzymes, supporting our finding.
We used DRS-independent methods to confirm the knockdown-validated sites found in the differentiated and Pb2+-exposed libraries. We found that DMAC1, EFEMP2, NES, NKAIN1, THY1, and HDGFL2 transcripts carried a psi site that had not been discovered by other orthogonal methods in the differentiated sample. We found four of these transcripts (TPM3, PIR, HDGFL2, and NES) in the Pb2+-exposed library. The presence of these unconfirmed sites could be because the SH-SY5Y cell line has not been previously analyzed by any other groups. This is further suggested by our CLAP results, which validated five new sites that we uncovered to be psi.
This study was the first to determine the plasticity of psi modifications across cellular states. Future analysis will determine whether static sites play critical roles in the cell's biological function and whether the plastic sites are responses to external cues for fine-tuning gene expression.
STAR Methods
Experimental Model and Subject Details
Cell culture
Human neuroblastoma SH-SY5Y cells were cultured in EMEM/F12 (EMEM from Quality Biological Inc and Cytiva HyClone Ham's Nutrient Mixture F12 Media) supplemented with 10% Fetal Bovine Serum (FisherScientific, FB12999102). For untreated SH-SY5Y cells, the culture remained in this medium for seven days at 37C and 5% CO2, refreshed every three days. For differentiated SH-SY5Y cells, after 24h, the media changed to differentiation media, which is Neurobasal media (Gibco Neurobasal-A Medium, minus phenol red) supplemented with 10uM all-trans-retinoic acid (Fisher, AC207341000), 1X B27(Fisher, A3582801), and 1X Glutamax (Fisher, 35-050-061). The differentiation media was renewed every other day. For lead exposure SH-SY5Y cells, after 24 hours, the culture media was removed, and a 50 μM Pb2+-supplemented (Lead (II) acetate trihydrate, Sigma) untreated media was added to the cells. The media was replaced every three days.
Immunofluorescence (IF)
For fixing SH-SY5Y cells, half of the culture media was removed, and an equal volume of 4% formaldehyde (Fisher, F79500) in PBS was added to each well for the final of 2% formaldehyde. After 2 min incubation at room temperature, the solution was aspirated, replaced by 4% formaldehyde, and incubated for 10 mins. The cells were then washed with PBS and permeabilized by incubating in PBS-Triton (0.1%) for 10 min. The cultures were blocked by incubation in 2% bovine serum albumin (BSA) in BS-Triton (0.1%) for one hour, followed by three times washed with PBS-Tween 20 (0.1%). The cells were then incubated with 1ug/ul primary antibody (For TRUB1 staining, TRUB1 Rabbit anti-human polyclonal, 50-172-8037, Protein tech; for PUS7 staining, PUS7 Rabbit anti-human, HPA024116, Sigma) in 1% BSA/PBS-Triton (0.1%) overnight at 4°C. The following day, the cells were washed with PBS-Tween 20 (0.1%) and incubated for one hour in 1:1000 secondary antibody (Mouse anti-rabbit IgG-PE-Cy7, NC1569698, Fisher) in 1% BSA/PBS-Triton (0.1%) at room temperature and stained using DAPI.
siRNA Knockdown (KD) of TRUB1 and PUS7
SH-SY5Y cells were cultured in untreated media for 24h for KD and control samples. The media was replaced with siRNA delivery media (Horizon, B-005000-500) with 1uM of Accell Non-targeting Control Pool (Horizon, D-001910-10-05), PUS7 siRNA (Horizon, E-015341-00-0050) or TRUB1 siRNA (Horizon, E-016391-00-0050) in delivery media for KD samples and cultured for 72h. Total RNA was extracted after three days for qPCR KD confirmation and cells were fixed for IF imaging. The KD and scrambled control cells were stained with anti-Pus7 and anti-Trub1 antibodies to evaluate protein expression following the knockdown.
Total RNA extraction
Total RNA was extracted from cells by combining a TRIzol (Invitrogen,15596026) RNA extraction and the PureLink RNA Mini Kit (Invitrogen, 12183025). Cells were first washed with ice-cold PBS, followed by incubation for 5 min in TRIzol at RT (2ml for 10mm dishes and 300ul for 8-well plates). Then, the solution was transferred to Eppendorf tubes, and 200ul chloroform (Thermo Scientific Chemicals, AC423555000) was added to each 1ml of TRIzol. The solution was mixed by shaking it for 15 sec and incubated at RT for 3 min, followed by centrifugation for 15 min at 12000 x g at 4°C. The aqueous supernatant was then transferred to a new tube, and the manufacturer's recommended protocol was followed for PureLink RNA Mini Kit RNA extraction. RNA was quantified using the RNA Qubit assay.
Poly-A RNA isolation
According to the manufacturer's protocol, poly-A mRNA was selected using the NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490L). RNA was quantified using the RNA Qubit assay.
RT-qPCR
The extracted total RNA was treated with TURBO DNase (Fisher, AM2238) following the manufacturer’s protocol. The RNA is then reverse transcribed using SuperScript III RT kit (Fisher,18080044) using the target primers and housekeeping gene HPRT. qPCR was performed using Luna qPCR master mix (NEB, M3004).
CMC-RT and ligation assisted PCR analysis of psi modification (CLAP)
The method is adapted from the protocol of Zhang et al. Primer3Plus was used to design primer sequences, which were tested for unspecific binding with BLAST.
Direct RNA library preparation and sequencing
A direct RNA sequencing library (SQK-RNA002) was prepared following the manufacturer’s instructions. Briefly, 500ng poly-A tailed RNA was ligated to ONT RT adaptor (RTA) using T4 DNA ligase (NEB, M0202M) and reverse transcribed by SuperScript III Reverse transcriptase (Invitrogen, 18080044). The product was then purified using Agencourt RNAClean XP beads (Beckman, A63987) ligated to the RNA adaptor (RMX) and purified by Agencourt RNAClean XP beads, followed by washing with wash buffer (WSB) and eluted in elution buffer (ELB). The final product was mixed with an RNA running buffer and loaded into R9.4.1 FLO-MIN106D flow cells from ONT. For the KD samples and scrambled control, the samples were loaded onto PromethION flow cells (ONT, FLO-PRO004RA).
Base-calling and alignment
Fast5 files were basecalled using Guppy version 6.4.2 and aligned to the human reference genome (hg38) using Minimap2 version 2.17 with the “-ax splice -uf -k 14” option. The aligned sam files were converted to .bam and indexed using samtools version 2.8.13.
Paired IVT enrichment and psi detection with Mod-p ID
We generated an unmodified IVT transcriptome paired to the untreated library which we used as a control to identify putative psi positions for the. All the positions with a high number of reads in the DRS sample and insufficient coverage in the paired IVT library were enriched using a pan human IVT which merges IVT libraries from 6 human cell lines68. We apply a conditional approach to determine the baseline: we use the paired IVT when the number of reads is sufficient (>10), and we recover sites with insufficient reads in the paired IVT by using the pan-human IVT as a baseline. After defining the appropriate IVT control for each site, we filtered the putative psi positions detected by the Mod-p ID tool according to multiple criteria. First, we defined a psi site to be significant if at least two biological replicates had a p-value < 0.001. We then filtered all the significant sites for the number of reads in the DRS and IVT samples, to retain positions with at least 10 reads in both libraries. To account for the presence of SNVs, we filtered the significant sites with sufficient coverage to those with a U-to-C basecalling error < 10 in the IVT library.
SNR Calculation
We modeled the IVT and DRS data separately for each target position with a beta-binomial distribution using Jeffrey’s noninformative prior65-67. We used U and C counts to parameterize the beta of the beta-binomial distribution and calculate the log marginal likelihood of the posterior distribution. Additionally, we modeled a combined distribution of IVT and DRS U and C counts with the same beta-binomial and Jeffrey’s prior. The ratio of these log marginal likelihoods approximates the degree to which the U to C mismatch at a position is better modeled with two independent distributions instead of a single joined distribution. This calculation is important because in DRS the total amount of reads is limited by the flow-cell design, therefore some abundant transcripts have very high coverage while others do not. For a transcript that is very highly abundant (i.e., >100 reads), a lower mismatch error could be considered significantly modified (i.e., <20% mismatch error). For a transcript that is lower abundance (~10 reads), these sites could also be considered significant if most of the reads have a high mismatch error. According to Kass and Raftery's model 68, we selected a value of SNR ≥ 1 to guarantee a strong DRS signal level as compared to the IVT.
SNR analysis and Mod-p ID cutoff selection in KD and perturbated libraries account for depth limitations
A MinION flow cell typically generates 1–2 million aligned reads69. This allows to achieve very high coverage depths of more than 50x in smaller genomes and transcriptomes (i.e. yeast and bacteria) but limits the coverage for the human transcriptome. Opposite to Illumina RNA-seq, the depth of a single DRS replicate cannot be improved with longer runs because of how the flow cell is constructed. When using DRS, we cannot increase the coverage/sequence depth by combining the sequencing output from different flow cells as each technological replicate is loaded on a unique flow cell, therefore merging the data coming from different flow cells cannot solve depth limitations. To leverage these technical depth limitations and guarantee a highly conservative and rigorous selection of psi sites, we propose an SNR-based filtration of the data to be coupled with the previously developed Mod-p ID tool. Therefore, we relax the U-to-C cutoff originally used for Mod-p ID as this is now coupled with the assessment of each site noise ratio, calculated using the unmodified IVT transcript. We apply more stringent cutoffs on the KD libraries as the selection of psi sites will affect the downstream analysis. The differential U-to-C mismatch calculated on KDs vs Scrambled library is particularly conservative as it requires a decrease of at least We then relax the U-to-C mismatch cutoff considering the CI of the perturbated libraries. We further apply a minimum read cutoff (>10) for each putative site in both the DRS and IVT sample. This criterion accounts for differences in depth between the smaller MinION libraries used in the original version of Mod-p ID, and the current PromethION runs. The read cutoff adds up to the original p-value filtration (p-value<0.001), to account for the increased number of reads per site in PromethION libraries. Although the selection of the cutoffs used in this work is determined empirically, we apply multiple filtering steps to account for coverage, SNVs presence, occupancy levels. We further validate the filtered list of sites with genetic and orthogonal methods. All these steps allow us to confidently define a rigorous, although small, list of psi-sites.
Assignment of psi-sites to TRUB1 and PUS7 enzymes.
We define sites as knocked down if they met the SNR ≥ 1 criteria, had sufficient coverage in KD and SCR libraries (>10) and a specified % decrease between U-to-C error in the control and KD sample. For positions with more reads (>30 reads), we set the cutoff at a 15% dicrease in occupancy in the KD-sample. For positions with <30 reads, we set the cutoff at a 30% decrease in occupancy. These cutoffs allow us to call a site as knocked down only when a substantial decrease in occupancy is observed the KD library vs the control but are selected to be more stringent with sites that have fewer reads.
ML training on synthetic standards
We trained a ML model according to Makhamreh et al.29 for 10 synthetic RNA modified standards, of which four (PSMB2,PRPSAP1,MRPS14,MCM5) come from the original paper.
Synthetic sequence design
We constructed six synthetic RNA oligos according to Tavakoli et al.25 and Gamper et al.27,29,70, each for a specific psi-site. Two versions of each RNA were prepared, one with 100% uridine and the other with 100% psi at the center of the synthetic standard.
Resource availability
Lead contact.
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Sara H. Rouhanifard (s.rouhanifard@northeastern.edu).
Accessing Publicly Available Data Sets
All IVT Libraries used in this work were sourced from NIH NCBI SRA under BioProject accession PRJNA947135.
Data and code availability
All fastq data for Direct Libraries generated in this work has been made publicly available in NIH NCBI SRA under the BioProject accession PRJNA1092333.
All bam files used for Mod-p ID can be found at the following Dropbox link: https://www.dropbox.com/scl/fo/4slozy1wmyxftbfzgyhaf/AE1nNQYNOoW-RUQibr0LTX8?rlkey=zqg24v8kpppst041d3oky0uk4&st=zu35f6i1&dl=0
All psi-sites detected by Mod-p ID in the KD libraries are available in Supplementary Table 8. The SNR valued calculated on KD libraries can be found in Supplementary Table 9. Knocked down psi-sites assigned to PUS enzymes are available in Supplementary Table 10.
All code can be found at github.com/RouhanifardLab/NeuronalEpitranscriptomePlasticity.
Supplementary Material
Acknowledgments
S.H.R and M.W. acknowledge support from the National Institutes of Health (R01HG011087, R01HG012856). Y.M.H acknowledges support from the National Institutes of Health (R35-GM134931).
Footnotes
Declaration of Interest
The authors declare no competing interests.
REFERENCES
- 1.Roundtree I. A., Evans M. E., Pan T. & He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borchardt E. K., Martinez N. M. & Gilbert W. V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 54, 309–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franco M. K. & Koutmou K. S. Chemical modifications to mRNA nucleobases impact translation elongation and termination. Biophys. Chem. 285, 106780 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyler D. E. et al. Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. U. S. A. 116, 23068–23074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlile T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlile T. M. et al. mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol. 15, 966–974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez N. M. et al. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell 82, 645–659.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parr C. J. C. et al. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 48, e35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Li X., Xiong X. & Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Dominissini D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Liu J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortese R., Kammen H. O., Spengler S. J. & Ames B. N. Biosynthesis of pseudouridine in transfer ribonucleic acid. J. Biol. Chem. 249, 1103–1108 (1974). [PubMed] [Google Scholar]
- 15.Kierzek E. et al. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 42, 3492–3501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arroyo J. D. et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 24, 875–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakin A. & Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Zhang M. et al. Quantitative profiling of pseudouridylation landscape in the human transcriptome. Nat. Chem. Biol. 19, 1185–1195 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Lovejoy A. F., Riordan D. P. & Brown P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoddami V. et al. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl. Acad. Sci. U. S. A. 116, 6784–6789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai Q. et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat. Biotechnol. (2022) doi: 10.1038/s41587-022-01505-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begik O. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. (2021) doi: 10.1038/s41587-021-00915-6. [DOI] [PubMed] [Google Scholar]
- 23.Huang S. et al. Interferon inducible pseudouridine modification in human mRNA by quantitative nanopore profiling. Genome Biol. 22, 330 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S., Wylder A. C. & Pan T. Simultaneous nanopore profiling of mRNA m6A and pseudouridine reveals translation coordination. Nat. Biotechnol. (2024) doi: 10.1038/s41587-024-02135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavakoli S. et al. Semi-quantitative detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct long-read sequencing. Nat. Commun. 14, 334 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick C. A. et al. Multicellular, IVT-derived, unmodified human transcriptome for nanopore-direct RNA analysis. GigaByte 2024, gigabyte129 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamper H. et al. Synthesis of Long RNA with a Site-Specific Modification by Enzymatic Splint Ligation. Preprint at 10.1101/2022.09.17.508400. [DOI] [Google Scholar]
- 28.Fleming A. M., Mathewson N. J., Howpay Manage S. A. & Burrows C. J. Nanopore Dwell Time Analysis Permits Sequencing and Conformational Assignment of Pseudouridine in SARS-CoV-2. ACS Cent. Sci. (2021) doi: 10.1021/acscentsci.1c00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makhamreh A. et al. Nanopore signal deviations from pseudouridine modifications in RNA are sequence-specific: quantification requires dedicated synthetic controls. Sci. Rep. 14, 22457 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W., Eckwahl M. J., Zhou K. I. & Pan T. Sensitive and quantitative probing of pseudouridine modification in mRNA and long noncoding RNA. RNA 25, 1218–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaheen R. et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 135, 707–713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung Y. & Goldman D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 17, e12444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shipley M. M., Mangold C. A. & Szpara M. L. Differentiation of the SH-SY5Y Human Neuroblastoma Cell Line. J. Vis. Exp. 53193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janesick A., Wu S. C. & Blumberg B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 72, 1559–1576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Påhlman S., Ruusala A. I., Abrahamsson L., Mattsson M. E. & Esscher T. Retinoic acid-induced differentiation of cultured human neuroblastoma cells: a comparison with phorbolester-induced differentiation. Cell Differ. 14, 135–144 (1984). [DOI] [PubMed] [Google Scholar]
- 36.Sanders T., Liu Y., Buchner V. & Tchounwou P. B. Neurotoxic effects and biomarkers of lead exposure: a review. Rev. Environ. Health 24, 15–45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santa Maria M. P., Hill B. D. & Kline J. Lead (Pb) neurotoxicology and cognition. Appl. Neuropsychol. Child 8, 272–293 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Shih R. A., Hu H., Weisskopf M. G. & Schwartz B. S. Cumulative lead dose and cognitive function in adults: a review of studies that measured both blood lead and bone lead. Environ. Health Perspect. 115, 483–492 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’souza H. S., Dsouza S. A., Menezes G. & Venkatesh T. Diagnosis, evaluation, and treatment of lead poisoning in general population. Indian J. Clin. Biochem. 26, 197–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Safra M., Nir R., Farouq D., Vainberg Slutskin I. & Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 27, 393–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korecka J. A. et al. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One 8, e63862 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovalevich J. & Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 1078, 9–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harasym E., McAndrew N. & Gomez G. Sub-micromolar concentrations of retinoic acid induce morphological and functional neuronal phenotypes in SK-N-SH neuroblastoma cells. In Vitro Cell. Dev. Biol. Anim. 53, 798–809 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Khazeem M. M. et al. TOP2B Is Required to Maintain the Adrenergic Neural Phenotype and for ATRA-Induced Differentiation of SH-SY5Y Neuroblastoma Cells. Mol. Neurobiol. 59, 5987–6008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonicka H. et al. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 18, 28–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Human Protein Atlas. https://www.proteinatlas.org/.
- 49.Playfoot C. J. et al. Transposable elements and their KZFP controllers are drivers of transcriptional innovation in the developing human brain. Genome Res. 31, 1531–1545 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rege T. A. & Hagood J. S. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J. 20, 1045–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Gorokhova S., Bibert S., Geering K. & Heintz N. A novel family of transmembrane proteins interacting with beta subunits of the Na,K-ATPase. Hum. Mol. Genet. 16, 2394–2410 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Abadin H., Ashizawa A., Llados F. & Stevens Y.-W. Toxicological profile for lead. (2007). [PubMed] [Google Scholar]
- 53.Gülden M. & Seibert H. In vitro-in vivo extrapolation of toxic potencies for hazard and risk assessment—problems and new developments. ALTEX 23, e225 (2006). [Google Scholar]
- 54.Krall M. et al. Biallelic sequence variants in INTS1 in patients with developmental delays, cataracts, and craniofacial anomalies. Eur. J. Hum. Genet. 27, 582–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung H.-R. et al. PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borlado L. R. & Méndez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29, 237–243 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Xu H. et al. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS One 5, e12112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugrue K. F., Sarkar A. A., Leatherbury L. & Zohn I. E. The ubiquitin ligase HECTD1 promotes retinoic acid signaling required for development of the aortic arch. Dis. Model. Mech. 12, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang J. et al. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct Target Ther 6, 323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Messana T. et al. Glucose Transporter Type 1 Deficiency Syndrome: Developmental Delay and Early-Onset Ataxia in a Novel Mutation of the SLC2A1 Gene. J. Pediatr. Neurosci. 13, 496–499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reardon E. S. et al. UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 16, 89–103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komada M. & Nishimura Y. Epigenetics and Neuroinflammation Associated With Neurodevelopmental Disorders: A Microglial Perspective. Front Cell Dev Biol 10, 852752 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banko J. L. et al. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J. Neurosci. 25, 9581–9590 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gkogkas C. G. et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tuyl F., Gerlach R. & Mengersen K. A Comparison of Bayes–Laplace, Jeffreys, and Other Priors. Am. Stat. (2008) doi: 10.1198/000313008X267839. [DOI] [Google Scholar]
- 66.Jeffreys H. An invariant form for the prior probability in estimation problems. Proc. R. Soc. Lond. A Math. Phys. Sci. (1946) doi: 10.1098/rspa.1946.0056. [DOI] [PubMed] [Google Scholar]
- 67.GitHub - nanoporetech/modkit: A bioinformatics tool for working with modified bases. GitHub https://github.com/nanoporetech/modkit. [Google Scholar]
- 68.Kass R. E. & Raftery A. E. Bayes Factors. J. Am. Stat. Assoc. 90, 773 (1995). [Google Scholar]
- 69.Jain M., Abu-Shumays R., Olsen H. E. & Akeson M. Advances in nanopore direct RNA sequencing. Nat. Methods 19, 1160–1164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Enzymatic synthesis of RNA standards for mapping and quantifying RNA modifications in sequencing analysis. in Methods in Enzymology vol. 692 127–153 (Academic Press, 2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All fastq data for Direct Libraries generated in this work has been made publicly available in NIH NCBI SRA under the BioProject accession PRJNA1092333.
All bam files used for Mod-p ID can be found at the following Dropbox link: https://www.dropbox.com/scl/fo/4slozy1wmyxftbfzgyhaf/AE1nNQYNOoW-RUQibr0LTX8?rlkey=zqg24v8kpppst041d3oky0uk4&st=zu35f6i1&dl=0
All psi-sites detected by Mod-p ID in the KD libraries are available in Supplementary Table 8. The SNR valued calculated on KD libraries can be found in Supplementary Table 9. Knocked down psi-sites assigned to PUS enzymes are available in Supplementary Table 10.
All code can be found at github.com/RouhanifardLab/NeuronalEpitranscriptomePlasticity.