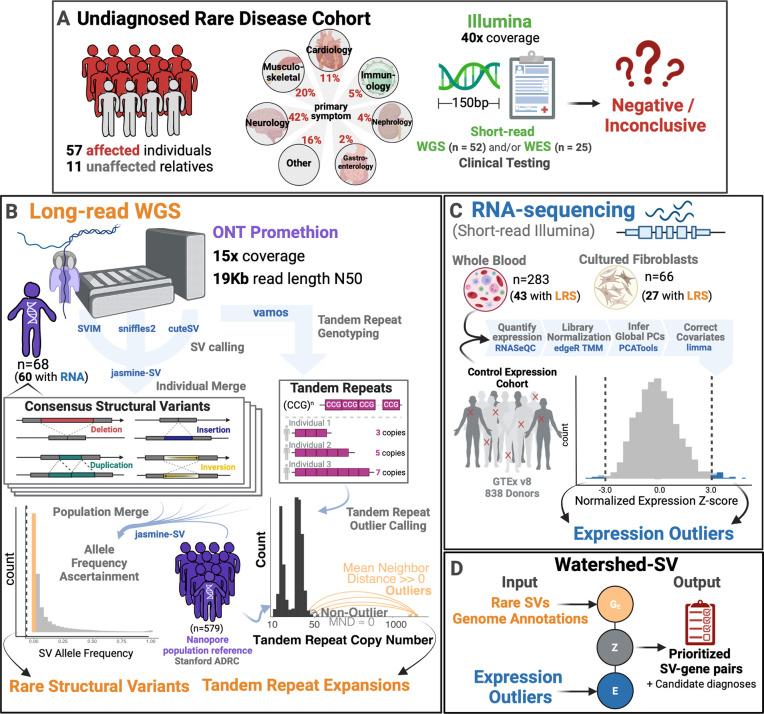
Figure 1. Undiagnosed patient cohort description and pipeline overview. Cohort Description:
A Patients were recruited from the Undiagnosed Disease Network for a long read sequencing (LRS) study. These included 57 affected individuals and 11 unaffected family members from a wide range of primary symptom categories, including Neurology, musculoskeletal, and cardiology. Patients had previous short-read genetic testing with Illumina that was negative or inconclusive. B Long-read Pipeline Overview: individuals were sequenced on R9.4 flowcells on the ONT PromethION. Consensus structural variants were called by merging SVs across individual callers and keeping those that showed multi-algorithm support. A population merge of the UDN genomes together with Stanford ADRC population reference of 579 nanopore genomes, allowed ascertainment of robust allele frequencies for structural variants. Rare structural variants were filtered and intersected with overlapping genome annotations to input into Watershed. Vamos was used on a catalog of polymorphic tandem repeats to genotype tandem repeat copy numbers. A mean neighbor distance based outlier calling method was used to define extreme repeat expansions. C RNA-sequencing expression outlier pipeline: transcriptome data from the UDN was processed by quantifying expression, combining with tissue-matched controls from GTEx, normalizing for library size and composition bias, and correcting for batch effects and hidden factors. Expression outliers of the normalized data were input into Watershed. D Watershed-SV integrates signals from rare SVs and overlapping genome annotations to predict variants with large functional effects. High scoring watershed variants are prioritized and curated per patient for disease relevance.