Abstract
Background:
Language and the ability to communicate effectively are key factors in mental health and well-being. Despite this critical importance, research on language is limited by the lack of a scalable phenotyping toolkit.
Methods:
Here, we describe and showcase Lingo – a flexible online battery of language and nonverbal reasoning skills based on seven widely used tasks (COWAT, picture narration, vocal rhythm entrainment, rapid automatized naming, following directions, sentence repetition, and nonverbal reasoning). The current version of Lingo takes approximately 30 minutes to complete, is entirely open source, and allows for a wide variety of performance metrics to be extracted. We asked > 1,300 individuals from multiple samples to complete Lingo, then investigated the validity and utility of the resulting data.
Results:
We conducted an exploratory factor analysis across 14 features derived from the seven assessments, identifying five factors. Four of the five factors showed acceptable test-retest reliability (Pearson’s R > 0.7). Factor 2 showed the highest reliability (Pearson’s R = 0.95) and loaded primarily on sentence repetition task performance. We validated Lingo with objective measures of language ability by comparing performance to gold-standard assessments: CELF-5 and the VABS-3. Factor 2 was significantly associated with the CELF-5 ”core language ability” scale (Pearson’s R = 0.77, p-value < 0.05) and the VABS-3 ”communication” scale (Pearson’s R = 0.74, p-value < 0.05). Factor 2 was positively associated with phenotypic and genetic measures of socieconomic status. Interestingly, we found the parents of children with language impairments had lower Factor 2 scores (p-value < 0.01). Finally, we found Lingo factor scores were significantly predictive of numerous psychiatric and neurodevelopmental conditions.
Conclusions:
Together, these analyses support Lingo as a powerful platform for scalable deep phenotyping of language and other cognitive abilities. Additionally, exploratory analyses provide supporting evidence for the heritability of language ability and the complex relationship between mental health and language.
1. Introduction
Language is a complex phenomenon that relies on multiple cognitive domains and brain networks [1,2]. Across individuals there is incredible variability in language ability and speech patterns [3]. Studies of language ability, particularly specific language impairments, have shown language ability is a highly genetic trait (twin based heritability estimates range from 45–60%) [4,5]. Despite evidence for substantial heritability, the relatively low power of candidate gene association studies has led to a small handful of tenuously implicated genes and genetic loci [6]. Successful identification of these language-associated genes depends on our ability to uniformly phenotype individuals across the domains of language, and at the large scale needed for genetic research. Traditionally, gold standard assessment of language ability has depended on paper-and-pencil, in-person proctored tests. These assessments are expensive to administer, time consuming, and require highly trained proctors. Beyond the challenges related to collecting a large enough sample for a genome wide association study (GWAS) with this method, some of these tests are not amenable to genetic analysis in the general population because they are designed to detect clinically relevant language impairment. This may result in a low ceiling, thus limiting their usefulness in assessing inter-individual variation in language ability in population samples. While recent efforts from the GenLang consortium have shown significant GWAS associations with traditional language assessments in a large meta-analysis of more than 30,000 individuals, they also discussed the difficulty of collecting additional data with these assessments and the need for a scalable deep language phenotyping tool to facilitate future research [7]. Recent work has shown short online assessments can accurately measure general intelligence that has a genetic basis [8]. Unfortunately, current online assessments lack the flexibility to capture a variety of speech and language related phenotypes. Development of a language and cognition toolkit that allows for a variety of data to be collected, like audio and selection data, would be incredibly valuable for deep phenotyping in future genetic research.
Language is an important endophenotype in psychiatric research. Children with language impairments are more likely to develop psychiatric disorders later in life [9]. Many individuals with psychiatric and neurodevelopmental disorders show aberrant speech patterns. Schizophrenia is associated with deficits in receptive and expressive language as well as decreased coherence between sentences when describing pictures [10,11]. Major depression has been linked to less frequent use of words with positive meaning and more monotone speech [12]. Autism, a neurodevelopmental condition, is highly comorbid with language impairments. It is estimated that 25–30% of autistic children at the age of 5 do not speak at all or use very few words when speaking [13]. Collecting rich speech data from populations with psychiatric and neurodevelopmental conditions may lead to more efficient diagnosis and monitoring [14].
To begin to remove the methodological barriers to study language, we have developed a battery of language and related cognitive tasks that are administered remotely and unsupervised through a web browser. This web-based language battery records a variety of user-generated data, including item selections (with time-to-selection) and recorded audio, from which several latent phenotypes can be derived. Here, we describe this battery, the rationale for selecting the tasks involved, and several validations of the resulting data across multiple samples. We then conduct exploratory analysis of the derived phenotypes and show significant associations with mental health, genetic risk, and socioeconomic status. Together, these results suggest that this web-based battery is a promising tool for quantitatively phenotyping multiple domains of language and cognitive abilities.
2. Results
2.1. Lingo overview
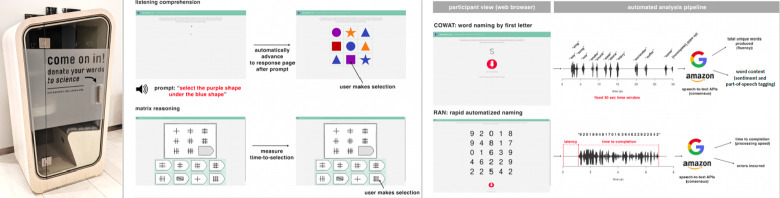
An overview of the Lingo battery can be seen in Figure 1. Prompt images can be seen in the Supplementary Figures and descriptions of each task can be found in the ”Methods and materials section”. Code is available at https://research-git.uiowa.edu/michaelson-lab/language-screener/ScreenerVersion2/-/tree/master.
Figure 1. Schematic overview of Lingo, our online language battery.
Participants were routed to the Lingo website where they completed the Lingo battery using personal computers and laptops. Raw audio, selections, and timing are recorded during Lingo. Some screenshots of the assessment are shown here as well as examples of how the data can be analyzed
2.2. Lingo performance features and their factor structure
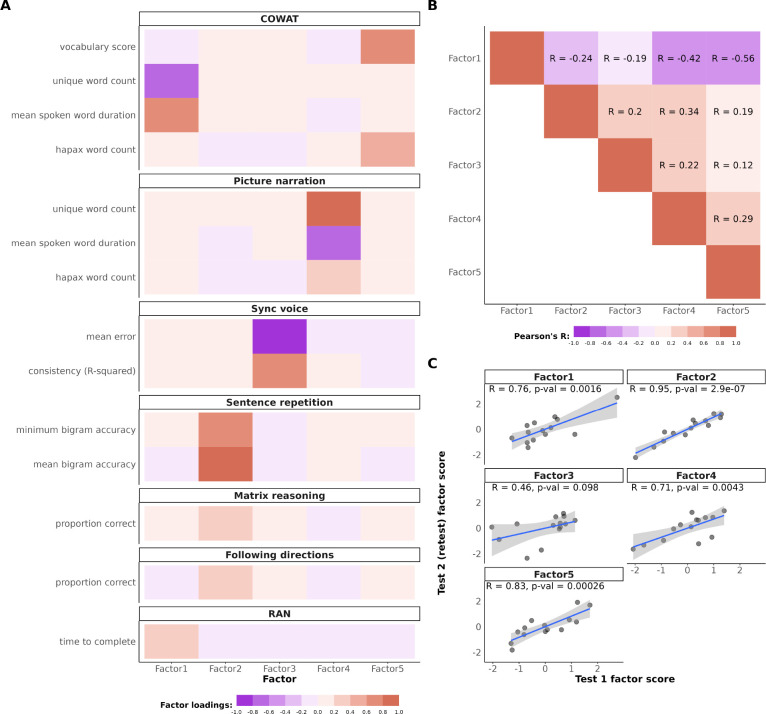
The open-ended nature of many tasks in the Lingo screener offers flexibility for measuring task performance, allowing for both hypothesis-driven and exploratory data analysis. Throughout this manuscript we showcase the utility of 14 features from the Lingo tasks in measuring different aspects of language ability. All derived features used in our analyses are listed in Figure 2A and described in table 2. In brief, we extracted performance metrics related to concepts like verbal memory, verbal fluency, vocabulary, reading speed, receptive language ability, rhythm, and nonverbal ability.
Figure 2. Factor structure of Lingo and test-retest reliability.
2.A. Factor loadings of the 14 Lingo performance features we analyzed. We found a 5 factor model fit our data best. 2.B. Correlation heatmap between the Lingo factor scores for all participants. 2.C. Test-retest reliability analysis of the 5 Lingo factors for the subset of participants who completed the assesment twice (at least 2 weeks apart).
Due to the high intercorrelation of these features, we conducted an exploratory factor analysis across a sample of N = 1,344 unique individuals from multiple samples (descriptions can be found in 1. First, we found the Lingo data is factorable – as measured by Bartlett’s Test of Sphericity (p-value < 2e-16) and the Kaiser-Meyer-Olkin criterion test on the correlation matrix of the Lingo variables (KMO value > 0.6). Next, we identified five distinct factors from our 14 input Lingo features using a data-driven parallel analysis approach (see Figure 2A). Factor 1 (”poor verbal fluency”) loaded negatively onto number of unique words said during the COWAT and positively onto reading time. Factor 2 (”core language”) loaded strongly sentence repetition task performance as well as weaker loading from the following directions and matrix reasoning tasks. Factor 3 (”rhythm/timkeeping”) loaded mostly onto performance metrics from the sync voice task. Factor 4 (”talkativeness/descriptiveness”) loaded onto number of words said during the picture narration task. Finally, Factor 5 (”vocabulary”) loaded primarily onto our vocabulary measure and hapax count, suggesting individuals scoring high on this factor frequently used word others did not. Factor score inter-correlations are shown in Figure 2B. Of note, Factors 1 and 5 had the strongest correlation (Pearson’s R = −0.56, p-value < 0.01).
2.3. Test-retest reliability
To investigate test-retest reliability of Lingo performance, we asked 14 independent adults from the SPARK research match study to take the Lingo battery twice (separated by at least two weeks between tests). Test-retest reliability was measured by calculating Pearson’s correlations between the factor score for the first and second testing sessions, on a factor-wise basis (Figure 2C). Factors 1, 2, 4, and 5 showed acceptable test-retest reliability (Pearson’s R > 0.7). Factor 2 showed particularly high test-retest reliability (Pearson’s R = 0.95, p-value < 0.01). Factor 3 (rhythm/vocal timekeeping) did not show strong reliability (Pearson’s R = 0.46, p-value = 0.1) These results suggest that many of the factors derived from Lingo data are reliable, in particular the verbal memory related variables.
2.4. Agreement with a gold-standard language assessments
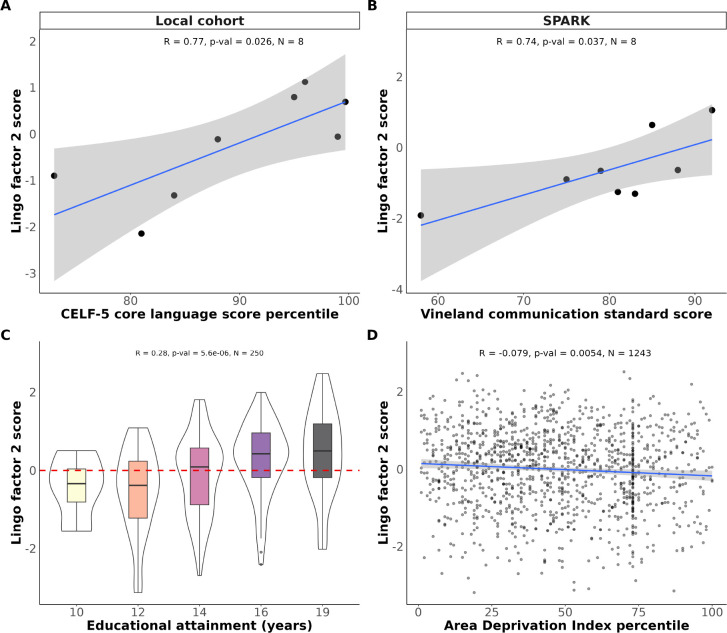
To establish concurrent validity between the Lingo battery and well-established measures of language ability, we compared our factor scores to gold-standard clinical assessments: the CELF-5 and the Vineland Adaptive Behavior Scales 3 (VABS-3) [15, 16]. For the CELF-5 test validity analysis we recruited 8 individuals from a local neurodevelopmental registry (devGenes, Table 1) who completed Lingo and were assessed with the CELF-5 on the same day. Factor scores from Lingo were then correlated with their age-normed CELF-5 clinical subscale scores (Figure 3). Factor 2 had a significant correlation with the ”core language score percentile” from the CELF-5 (Pearson’s R = 0.77, p-value = 0.03, Figure 3A). Next, we conducted a similar analysis in our SPARK sample. Eight individuals in our SPARK sample also had complete VABS-3 data. We found that factor 2 was significantly associated with the VABS-3 ”communication” standard score (Pearson’s R = 0.74, p-value = 0.04, Figure 3B).
Table 1.
Demographic information of samples collected.
| sample | N | Mean age (std. dev.) | Females | Males | ASD |
|---|---|---|---|---|---|
| SPARK | 1336 | 41.43 (10.3) | 1085 (81.2%) | 251 (18.7%) | 423 (31.7%), 132 male |
| SPARK retest | 14 | 43.3 (6.5) | 11 (78.6%) | 3 (21.4%) | 3 (21.4%), 1 male |
| Local cohort (devGenes) | 8 | 13.97 (1.8) | 2 (25%) | 6 (75%) | 2 (25%), 2 male |
Figure 3. Validation of Lingo factors with gold-standard assessments and SES.
3.A. Concurrent validity analysis of the Lingo factor 2 scores with the CELF-5 scores in a small local cohort who completed both assessments. 3.B. Concurrent validity analysis of the Lingo factor 2 scores with the Vineland Adaptive Behavior Scales-3 language score in a small subset of SPARK participants who had data for both assessments available. 3.C. Correlation between educational attainment in years (i.e., 10 = did not complete high school, 19 = graduate or professional degree) and Lingo factor 2 scores. As many young adults are still in school, we subset the sample to individuals > 30 years old to limit the effects of age in our analysis. 3.D. Correlation between the Area Deprivation Index and Lingo factor 2 scores.
2.5. Socioeconomic correlations with Lingo factors
Given the findings that Lingo scores are reliable and valid, we sought to showcase the utility of our Lingo factors in a series of phenotypic and genetic analyses. While previous research has found robust links between socioeconomic status (SES) and language test scores in children [17–19], less is known about how SES influences specific domains of language later in life. Common measurements of SES include educational attainment (highest level of education completed) and the Area Deprivation Index (ADI – a normed measure of socioeconomic disadvantage based on quality of life variables mapped to the zip code of someone’s residence). To determine the relationship of SES with different language and cognitive scores in adults, we investigated possible associations between our Lingo factor scores with educational attainment and ADI. Lingo factor 2 showed a significant positive correlation with educational attainment in adults at least 30 years old (Pearson’s R = 0.28, p-value < 0.01, N = 250, Figure 3C). Similarly, we found ADI was negatively correlated with factor 2 scores (Pearson’s R = −0.08, p-value < 0.01, N = 1243, Figure 3D). The associations between SES and Lingo scores were persistent. All five Lingo factors were significantly correlated with education level and ADI.
2.6. Polygenic score associations with Lingo factors
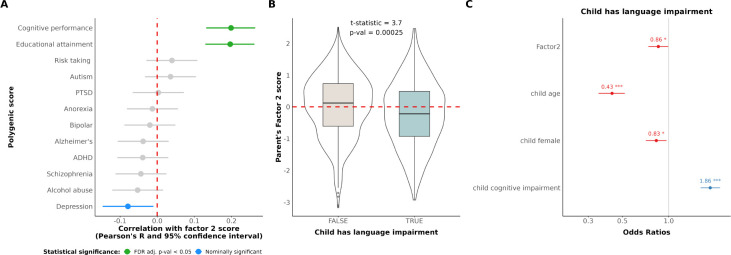
In the previous section we describe evidence linking phenotypic measures of SES to Lingo factors. To investigate whether these findings had support at the biological level, we tested for associations with genetic estimates of SES. We computed 12 different polygenic scores (PGS) for 811 individuals of European descent in our SPARK sample with genotype data to link with our Lingo factors. The 12 PGS we calculated were relevant to cognitive ability, SES, and neuropsychiatric risk. All PGS were corrected for population stratification prior to analysis. We found factor 2 was significantly correlated with educational attainment PGS (Pearson R = 0.20, FDR adjusted p-value < 0.01) and cognitive performance PGS (Pearson R = 0.20, FDR adjusted p-value < 0.01, Figure 4A). Factor 2 also showed a nominally significant association with depression PGS (Pearson R = −0.08, uncorrected p-value = 0.02) Interestingly, factor 4 (talkativeness/descriptiveness) was associated with decreased polygenic risk for schizophrenia (Pearson R = −0.11, FDR adjusted p-value = 0.03) and depression (Pearson R = −0.10, FDR adjusted p-value = 0.04).
Figure 4. Genetic analysis of Lingo factors.
4.A. Brain related polygenic score (PGS) correlations with Lingo factor 2 scores for European SPARK participants with available genetic data (N = 811). Due to the multiple testing burden we used FDR p-value correction. Plotted are the Pearson’s R and 95% confidence interval estimates for correlations between PGS and Lingo factor 2 scores. 4.B. Parental score on Lingo factor 2 was associated with their child’s language impairment. 4.C. Parental score on Lingo factor 2 are an independent predictor of their child’s language impairment after accounting for their child’s cognitive impairment, age, and sex. Odds’ ratios and 95% confidence interval computed from a multivariate logisitc regression model predicting the child’s language impairment are shown. ”***” indicates p-value < 0.001, ”**” indicates p-value < 0.01, and ”*” indicates p-value < 0.05.
2.7. Familiality of language ability
Using our SPARK sample, we investigated whether our factor scores derived from Lingo data showed evidence of familiality, which would support the well-established heritability of language ability [4,5]. Upon enrolling in SPARK, parents are asked to characterize their autistic child’s language ability with Likert-scale responses ranging from: ”Uses longer sentences of his/her own and is able to tell you something that happened” to ”No words/does not speak”. This Likert-scale allowed us to binarize their child’s language ability into an unimpaired language group (N = 693, ”Uses longer sentences of his/her own and is able to tell you something that happened”), and an impaired language group (N = 310, ”Combines 3 words together into short sentences” or ”Uses single words meaningfully” or ”No words/does not speak”). Indeed, we found that parents’ Factor 2 score was significantly associated with their children’s language ability (t-statistic = 3.7, p-value < 0.01, Figure 4B) – suggesting parents of autistic children with language impairments have poorer verbal memory themselves. Notably, parents’ Factor 2 score was independently predictive of their child’s language impairment after controlling for the child’s cognitive impairment, age, and sex (beta = −0.16, Z = −2.05, p-value = 0.04, Figure 4C). These results support the heritability of language ability that has been previously reported, and further underscore the validity of the data yielded through Lingo.
2.8. Lingo factors are predictive of psychiatric diagnoses
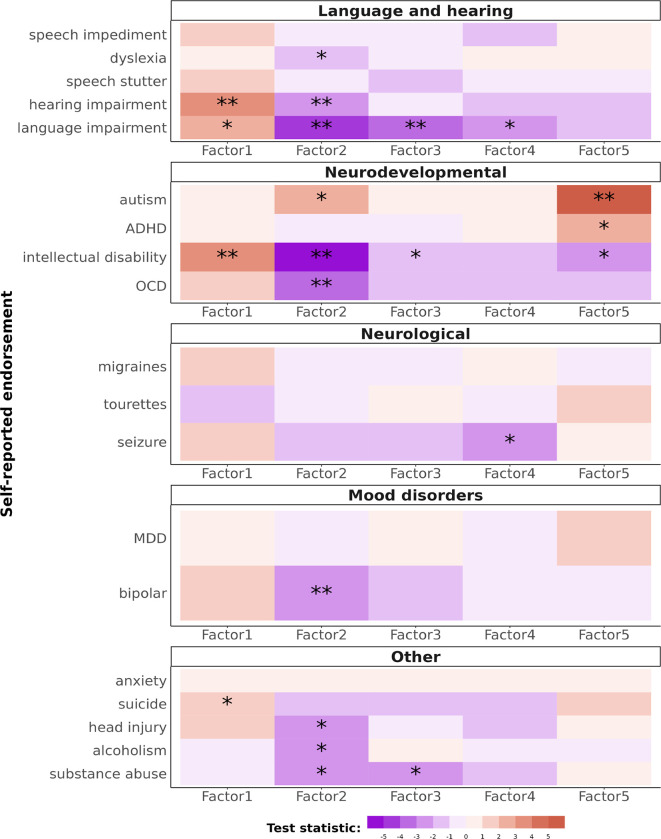
Finally, we tested whether Lingo captures language and cognitive phenotypic variation relevant to psychiatric diagnosis. A subset of SPARK participants (N = 845) completed an additional self-report questionnaire about psychiatric, neurodevelopmental, or neurological diagnoses they have received. We then fit logistic regression models to predict diagnosis from Lingo factor scores while controlling for age and sex of the participant. Lingo factor 2 was negatively associated with endorsement of language impairment (beta = −0.6, Z = −4.1, FDR adjusted p-value < 0.01), intellectual disability (beta = −0.5, Z = −5, FDR adjusted p-value < 0.01), and bipolar disorder (beta = −0.29, Z = −2.84, FDR adjusted p-value = 0.04), Figure 5. Factor 5 was associated with higher endorsement of autism (beta = 0.51, Z = 5.1, FDR adjusted p-value < 0.01, Figure 5).
Figure 5. Lingo factors associations with psychiatric and neurodevelopmental conditions.
Plotted are the test statistics from logistic regression models predicting self-reported diagnosis from the Lingo factor scores after accounting for age and sex (N = 845). Due to the multiple testing burden we used FDR p-value correction. ”**” indicates FDR adjusted p-values are less than 0.05. ”*” indicates nominal statistical significance, unadjusted p-vale < 0.05 but the FDR adjusted p-value > 0.05. Negative values indicate scoring high on the Lingo factor is associated with a lower likelihood of having that diagnosis. Positive values indicate scoring high on the Lingo factor is associated with a higher likelihood of having that diagnosis. Values near zero indicate no significant association.
3. Discussion
Lingo is a powerful platform for scalable deep phenotyping of language ability. With multiple samples collected from a variety of settings we were able to validate the utility of Lingo in quantifying distinct language domains. We identified a factor related to verbal memory that had strong correlations with core language measures of gold-standard clinical language assessments, providing support that our battery can objectively measure language ability. Additionally, the Lingo-derived factors showed high test-retest reliability. Analysis of Lingo factor scores offered insights into the interrelationship that language ability has with genetics, SES, and mental health. While many of these analyses cannot infer causality (i.e., low language abilities early in life lead to poor SES in adulthood or vice versa), these findings speak to the importance of considering language when studying SES and mental health. Taken together, these validations suggest that our battery is a robust platform for measuring individual differences in language/cognitive skills that are relevant to behavioral and social sciences.
The scalability of Lingo makes it a prime candidate for prospective and genetic study designs. Our analyses showing significant associations between PGS and our factor scores suggest there is a biological basis to the variation captured by Lingo, making this assessment amenable to a large-scale GWAS in the future. Finally, the ability to deploy this battery in large population samples, allowing for rapid deep phenotyping, is ideal for genetic analysis like GWAS. Follow up studies, like GWAS or rare variant association tests on Lingo scores could clarify the role genes may play in specific speech and language domains. Based on previous large-scale GWASs on reading phenotypes and the heritability of multiple language domains [4,7], we expect that the different Lingo scores would be genetically distinct.
The recorded audio that Lingo provides is a significant benefit that distinguishes it from traditional language and cognitive assessments. Lingo’s recorded audio and millisecond-scale timing allows for unparalleled feature extraction, allowing researchers to address a variety of research questions with data collected from one session. The richness of the raw recordings allows for a degree of ”future proofing” as new analytic techniques are developed to derive features with greater signal-to-noise and interpretability properties. Beyond the 14 features we examined in the factor analysis, Lingo also offers the ability to assess acoustic speech qualities. Changes in speech acoustics have been linked to a variety of mental health and mood outcomes, like depression and mania in bipolar disorder [14]. It can be extremely difficult to diagnose or treat manic and depressed individuals, but tools like Lingo offer a novel avenue to collect this data without the need for them to come into a clinical setting [14]. Additionally, the nature of the COWAT and picture narration tasks would allow for semantic or sentiment analysis by looking at the specific words participants use. Recent work has shown significant differences in the semantic space of words used by individuals with schizophrenia compared to controls [20]. Ultimately, digital phenotyping tools like Lingo could reduce mental healthcare treatment barriers and allow for improved diagnosis, monitoring, and treatment.
The relatively short time to complete and inexpensive implementation of the Lingo screener makes it a promising tool for longitudinal studies. Participants can complete the Lingo battery multiple times (as long as they have access to a web browser and device with a microphone), which would allow for longitudinal tracking of speech and cognitive phenotypes. This could be particularly useful in speech biomarker studies of neurodegenerative diseases like Alzheimer’s, or on the opposite end of the age spectrum to capture developmental trajectories of language acquisition and use in children.
3.1. Limitations
While we believe the Lingo battery is a significant stride forward in scalable deep phenotyping of language and cognitive abilities, there are a number of limitations with the current implementation. First and foremost, we do not cover all domains of language. For example, written language (including grammar and spelling) are not probed in Lingo - limiting its utility. Still, it may be possible to compute proxies for written grammar from the spoken grammar in the picture narration task. Additionally, while we found machine transcription to be highly accurate, human involvement is still needed for the highest levels of accuracy. Machine transcription is also biased by confounds that may be of interest to language researchers, like accent or dialect, which likely decrease transcription accuracy. Disrupted speech and vocal acoustics seen in psychiatric, neurological, and neurodevelopmental conditions could also decrease automated transcription accuracy and can lead to inaccurate results with clinical implications if they are not corrected or flagged [21]. Consequently, depending on the application, machine mistranscription may be either a feature or a bug.
4. Methods and materials
4.1. Implementation of Lingo
This language battery is a custom built, self-contained, web server. The front end (the website the participant sees and interacts with) was written with Angular (https://angular.io/), the back end was written in NodeJS (https://nodejs.org/en/), and the server is deployed on Amazon Web Services (AWS, https://aws.amazon.com/). This design allows for straightforward customization of the tasks included in the battery, where one can add or remove tasks easily prior to deployment on AWS. Once a participant navigates to the corresponding URL, their device compatibility is tested by ensuring they can listen to and record audio. Following device testing the participant begins the battery, which takes approximately 25–35 minutes for the full battery (depending on time to complete the matrix reasoning and listening comprehension tasks). We record raw audio, timing, and item selections across the battery. The raw audio for each trial undergoes text-to-speech transcription, which was checked manually through random sampling for transcription accuracy. Audio recordings of responses were initially transcribed and timestamped by OpenAI’s Whisper Large-V2 text-to-speech model []. To ensure the machine transcription process was reliable, we completed manual review of >100 randomly selected audio files and found the Whisper transcriptions to be highly accurate (95%).
4.2. Verbal fluency task: word naming by letter
To assess verbal fluency, we prompted participants to name as many words as they could think of that start with a provided prompt letter in 30 seconds. During this 30 second interval, audio was recorded from the participant’s device microphone. The prompt letters were A, C, F, L, and S. No further instruction (e.g., exclusion of proper nouns) was given. Similar word naming tasks have long been used in neurology and psychiatry to assess verbal fluency (see the FAS or COWAT [22,23]).
4.3. Phonological memory task: sentence repetition
Phonological memory was assessed with a sentence repetition task. Prompt sentences were provided via playback of a recording of a female narrator. Prompt sentences were structurally identical to some of those on the sentence repetition task of the CELF-5 [15]. We then augmented this set with several additional sentences generated by us to raise the difficulty ceiling. In total, 12 sentences are presented in the task, in order of increasing complexity. Three seconds after the auditory prompt, the participant’s response was recorded through their device microphone.
Typical scoring of in-person administration of sentence repetition tasks provides little numerical range (i.e., scored 0–3), making it challenging to assess accuracy on easy or difficult prompts where the majority of individuals will be at one extreme score. In our implementation, we provide a continuous score that is based on the bigram accuracy, i.e., the proportion of correct bigrams (consecutive word pairs) included in the participant’s response. Performance in the sentence repetition task is dependent on phonological memory, yet includes other aspects of language as well (i.e., it is a combined receptive/expressive language task). Prompt sentences can be seen in Supplementary Figure S1 (with punctuation and capitalization removed).
4.4. Reading task: rapid automatized naming
Reading fluency was tested using the well-validated Rapid Automatized Naming (RAN) task. Briefly, participants are shown a grid of numbers and asked to read them aloud as quickly as they can. Our implementation includes 4 total trials, with number grid lengths of either 25 (5×5) or 36 (6×6) integer numbers. During this task, audio from the participant reading off the grid of numbers was recorded, allowing us to calculate the time spent reading and correctness through each number grid. Actual number grids used in this task can be seen in Supplementary Figure S2
4.5. Timing task: sync voice
This task aimed to evaluate participants’ abilities in vocal timekeeping and rhythm. Participants were instructed to mimic a series of ”La’s” pronounced at fixed intervals (e.g., every 1 second) and continue the pattern for 10 seconds. The task comprised separate trials with intervals set at 0.5, 0.75, 1, and 1.25 seconds. To analyze participant performance, we examined the raw audio recordings to pinpoint instances of vocalization during the task. From these recordings, we extracted timestamps, allowing us to calculate the time intervals between each uttered ”La”. Subsequently, we computed several features related to consistency, error, and overall accuracy based on these timestamps. Consistency-related features were determined by fitting linear models to each participant’s response data, predicting actual prompt period times based on the intervals between their ”La” utterances. Participant-level R-squared values were then extracted from these models, indicating the degree to which participants’ performances across trials accurately predicted the prompt period times.
4.6. Expressive language task: picture narration
During this task, participants were prompted with pictures and asked to describe what they saw for 30 seconds. No further instruction was given. During this 30 second interval, audio was recorded from the participant’s device microphone. Participants completed 4 trials, and were shown the pictures in Supplementary Figure S3
4.7. Spatial receptive language task: following directions
In order to quantify the participants’ ability to interpret and follow verbal directions we implemented a task similar to the “Following Directions” task found in the CELF assessment [15]. Participants were given 8 prompts of increasing difficulty, where they heard a narrator give directions to select a specific shape in a grid of shapes. We record both the time to select an item and whether that item was correct for each trial. For example, prior to seeing the prompt in Supplementary Figure S4, participants might hear “select the blue triangle underneath the red square”.
4.8. Nonverbal reasoning task: matrix reasoning
Matrix reasoning is a visual-spatial problem solving task often used as a measure of nonverbal intelligence. The prompt assets for the matrix reasoning task used here were used with permission from the well-validated UK Biobank version [24], which was presented in a comparable fashion. In brief, the matrix reasoning problems involve seeing an incomplete pattern, then selecting the option that would correctly complete the pattern from six choices. This task had 14 trials of increasing difficulty. We record both the time to select an item and whether that item was correct for each trial. An example prompt is shown in Supplementary Figure S5
4.9. Sample description: SPARK
Through a research match for the SPARK for Autism study [25,26] we recruited 1,336 adults (mean age = 41.4 years old, SD = 10.3) with rich phenotypic data. Sample demographics can be seen in Table 1 and Supplementary Figure S6. In order to establish test-retest reliability, 14 participants retook the battery two weeks after the initial test. Individuals who reported being blind or deaf were excluded due possible issues with tasks involving visual or audio stimuli (like matrix reasoning or the spatial receptive language tasks). Approximately 1/3 of this sample had been diagnosed with autism (31.2%), the other 68.8% were parents of children with autism. Many of these individuals and/or their children had extensive genetic, cognitive, and behavioral phenotype data which were used in downstream analysis to better understand the relationship relevant phenotypes and our Lingo performance results. We utilized SPARK V10 phenotype data for downstream analysis.
4.10. Sample description: in-person
We recruited 8 children from a local neurodevelopmental condition registry (devGenes, 1) who completed a gold-standard language assessment, the CELF-5, as well as our battery. We then compared CELF-5 and Lingo scores to evaluate the suitability of the included tasks. The CELF-5 was administered by trained research assistants. Due to limitations in the valid age range for the CELF-5 (valid in ages 5–21), this sample was substantially younger than the SPARK sample. All participants were younger than 21 years old.
4.11. Imputation
There was sporadically missing data in the matrix reasoning and following directions tasks due to server issues in part of the data collection phase for the SPARK sample (we believe excessive server load led to failure to save user data; no such data loss was experienced in a second wave of recruitment). Since the other five tasks recorded in full, and many participants had the majority of their data recorded, we decided to impute the missing data to improve statistical power in downstream analyses. The imputation procedure consisted of subsetting our data to individuals missing less than 33.3% of features. We then imputed the missing values using the ”missRanger” R package [27], a random forest and predictive mean matching based method. We used the following parameters: ”num.trees = 500”, ”pmm.k = 10” and a seed value for reproducibility. Imputation converged after 4 iterations, and provided a final dataset of 1,344 samples with 14 features each.
4.12. Factor analysis
Prior to determining the factor structure with exploratory factor analysis of the Lingo screener, we residualized for the effects of age and sex for each input variable. Then, we fit the initial factor model using 604 samples who had complete Lingo data (i.e., did not suffer any data loss and were not repeats). To determine the optimal factor number, we used the ”fa.parallel” function from the ”psych” package in R [27]. The ”fa.parallel” function uses parallel analysis to identify the number of factors in a dataset by comparing eigenvalues of the observed data to eigenvalues of a random dataset with the same dimensionality. Using this data-driven method, we identified five factors in our data. We then fit our five factor model with the ”fa” function from the ”psych” package. After computing initial factor scores, we removed 36 outliers (i.e., samples whose absolute value of a factor score was greater than 3 * median absolute deviation + median value of that factor), and refit the five factor model. Once factors were derived, we predicted factor scores for all of the held out data (i.e., the samples with imputed data or retest data). In total, we computed Lingo factor scores for 1,336 unique SPARK participants (14 of these samples had data from 2 Lingo sessions for test-retest reliability validation) and 8 participants from our local cohort.
4.13. Genetic data and polygenic score analysis
Much of this methodology has been previously described in our prior work [28]. We describe a few small updates to our methodology and ensured we followed current best practices with the PGS calculation tool used here.
4.13.1. Genotype Quality Control and Imputation
For the SPARK reseach match cohort, many participants had genetic data available. We used the genotype arrays from SPARK integrated whole-exome sequencing (iWES1) 2022 release and SPARK whole-genome sequencing (WGS) releases 2, 3, and 4. iWES1 (n = 69,592) was quality controlled on release, including removing samples due to heterozygosity or high missingness, so no quality control was performed by us before imputation. iWES1 provided genetic ancestry assignments based on 1000 Genomes populations (26). WGS release 2 (n = 2365), release 3 (n = 2871), and release 4 (n = 3684) were not quality controlled on release, so we performed quality control using PLINK (27) before imputation. First, we removed participants from WGS if they were in iWES1. Second, we removed variants with missingness > 0.1 and participants with missingness > 0.2. Third, we merged the 3 releases and removed any participant whose heterozygosity (F statistic) was not within 3 standard deviations of the mean. We used the TopMed reference panel to identify strand flips [29]. The final sample size for WGS 2 to 4 was n = 8152. iWES1 and WGS 2 to 4 were then imputed to TopMed [29] using the Michigan Imputation Server [30] with phasing and quality control steps included and to output variants with imputation quality r2 ¿ 0.3. After imputation, variants were filtered to single nucleotide polymorphisms with imputation quality r2 ¿ 0.8 and a minor allele frequency > 0.1% in our sample using bcftools [31]. They were lifted over from hg38 to hg19 using the VCF-liftover tool (https://github.com/hmgu-itg/VCF-liftover) and normalized to the hg19 reference genome. Finally, files were merged, subset to HapMap3 Plus variants [32], and variants with 0% missingness were retained (final number of variants = 1,147,121).
4.13.2. Genetic Ancestry
Genetic principal components (PCs) were calculated using bigsnpr [33], specifically following the authors’ recommendations [34] and tutorial: https://privefl.github.io/bigsnpr/articles/bedpca.html. In summary, we 1) used the ”snpplinkKINGQC” function to identify and remove related participants at KING threshold of 2–3.5, 2) performed PC analysis using the bedautoSVD with unrelated participants, 3) detected and removed PC outliers, 4) recalculated PCs, and 5) projected PCs onto the entire cohort using the bedprojectSelfPCA function. We used 40 PCs and performed k-means clustering with K = 5 (for the 5 populations of 1000 Genomes [35] and used the genetic ancestry labels from iWES1 to assign labels to the genetic population clusters. Due to the lack of diversity in many GWASs, PGS based on these results tend to perform much better in individuals with predominantly European ancestry. To ensure our PGS were valid and accurate, for the PGS analysis we subset to individuals who clustered with the Europeans samples (final N = 811 with QC passing genetic data, were European, and had Lingo data).
4.13.3. Polygenic Scores
PGSs were calculated using the ”LDpred2” [36] and ”bigsnpr” tools [33] in R. Because SPARK is family based, an external linkage disequilibrium reference based on 362,320 participants in the UK Biobank (provided by the authors of LDpred2) was used to calculate infinitesimal beta weights. PGSs were calculated from the following GWASs conducted by the Psychiatric Genetics Consortium: alcohol abuse [37], Alzheimer’s disease [38], attention-deficit/hyperactivity disorder (ADHD) [39], anorexia nervosa [40], autism [41], bipolar disorder [42], major depression [43], post traumatic stress disorder (PTSD) [44], and schizophrenia [45]. As well as the following GWASs conducted by the the Social Science Genetic Association Consortium: cognitive performance [46], educational attainment [47], and risk taking [48]. To correct for population stratification we residualized out the main effects of the first 20 genetic PCs from each PGS and z-scaled the scores. Finally, we tested for PGS associations with Lingo factor scores using Pearson’s correlations, as both the PGS and Lingo scores were normally distributed. P-values were adjusted for multiple testing burden by using the False Discovery Rate (FDR) method with the ”p.adjust” function in R.
4.14. Statistical analysis
All statistical analysis was conducted in R version 4.3.0 [49].
Supplementary Material
Acknowledgments
We are grateful to all of the participants and families in SPARK, the SPARK research match team and Tempus, the SPARK clinical sites, and SPARK staff. We appreciate obtaining access to genetic and phenotypic data for SPARK data on SFARI Base. We are very appreciative of time and effort of the research assistants who helped quality control the data. Some of the study data were collected and managed using REDCap electronic data capture tools hosted at the Univerity of Iowa.
Funding
This work was supported by the National Institutes of Health (DC014489 to JJM) as well as grants from the Simons Foundation (SFARI 516716 to JJM) and the Clinical and Translational Science Award (KL2TR001877 to JJM). This work was also supported by the Roy J. Carver Charitable Trust through a professorship to JJM. This work was supported by the University of Iowa Hawkeye Intellectual and Developmental Disabilities Research Center (Hawk-IDDRC) through the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P50HD103556).
Footnotes
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability
The SPARK genetic and phenotypic data can be obtained at SFARI Base: https://base.sfari.org
The SPARK Research Match data will be available to qualified, approved researchers through SFARI Base upon publication of this article.
References
- 1.Fedorenko E., “The role of domain-general cognitive control in language comprehension,” Frontiers in Psychology, vol. 5, 4 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deldar Z., Gevers-Montoro C., Khatibi A., and Ghazi-Saidi L., “The interaction between language and working memory: a systematic review of fmri studies in the past two decades.,” AIMS neuroscience, vol. 8, pp. 1–32, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu A. C. and Zellou G., “Individual differences in language processing: Phonology,” Annual Review of Linguistics, vol. 5, pp. 131–150, 1 2019. [Google Scholar]
- 4.Bishop D. V. M., Laws G., Adams C., and Norbury C. F., “High heritability of speech and language impairments in 6-year-old twins demonstrated using parent and teacher report,” Behavior Genetics, vol. 36, pp. 173–184, 3 2006. [DOI] [PubMed] [Google Scholar]
- 5.Tomblin J. B. and Buckwalter P. R., “Heritability of poor language achievement among twins,” Journal of Speech, Language, and Hearing Research, vol. 41, pp. 188–199, 2 1998. [DOI] [PubMed] [Google Scholar]
- 6.Graham S. A. and Fisher S. E., “Understanding language from a genomic perspective,” Annual Review of Genetics, vol. 49, pp. 131–160, 11 2015. [DOI] [PubMed] [Google Scholar]
- 7.Eising E., Mirza-Schreiber N., de Zeeuw E. L., Wang C. A., Truong D. T., Allegrini A. G., Shapland C. Y., Zhu G., Wigg K. G., Gerritse M. L., Molz B., Alagöz G., Gialluisi A., Abbondanza F., Rimfeld K., van Donkelaar M., Liao Z., Jansen P. R., Andlauer T. F. M., Bates T. C., Bernard M., Blokland K., Bonte M., Børglum A. D., Bourgeron T., Brandeis D., Ceroni F., Csépe V., Dale P. S., de Jong P. F., DeFries J. C., Démonet J.-F., Demontis D., Feng Y., Gordon S. D., Guger S. L., Hayiou-Thomas M. E., Hernández-Cabrera J. A., Hottenga J.-J., Hulme C., Kere J., Kerr E. N., Koomar T., Landerl K., Leonard G. T., Lovett M. W., Lyytinen H., Martin N. G., Martinelli A., Maurer U., Michaelson J. J., Moll K., Monaco A. P., Morgan A. T., Nöthen M. M., Pausova Z., Pennell C. E., Pennington B. F., Price K. M., Rajagopal V. M., Ramus F., Richer L., Simpson N. H., Smith S. D., Snowling M. J., Stein J., Strug L. J., Talcott J. B., Tiemeier H., van der Schroeff M. P., Verhoef E., Watkins K. E., Wilkinson M., Wright M. J., Barr C. L., Boomsma D. I., Carreiras M., Franken M.-C. J., Gruen J. R., Luciano M., Müller-Myhsok B., Newbury D. F., Olson R. K., Paracchini S., Paus T., Plomin R., Reilly S., Schulte-Körne G., Tomblin J. B., van Bergen E., Whitehouse A. J. O., Willcutt E. G., Pourcain B. S., Francks C., and Fisher S. E., “Genome-wide analyses of individual differences in quantitatively assessed reading- and language-related skills in up to 34,000 people,” Proceedings of the National Academy of Sciences, vol. 119, 8 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malanchini M., Rimfeld K., Gidziela A., Cheesman R., Allegrini A. G., Shakeshaft N., Schofield K., Packer A., Ogden R., McMillan A., Ritchie S. J., Dale P. S., Eley T. C., von Stumm S., and Plomin R., “Pathfinder: a gamified measure to integrate general cognitive ability into the biological, medical, and behavioural sciences,” Molecular Psychiatry, vol. 26, pp. 7823–7837, 12 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson C. J., Beitchman J. H., Young A., Escobar M., Atkinson L., Wilson B., Brownlie E. B., Douglas L., Taback N., Lam I., and Wang M., “Fourteen-year follow-up of children with and without speech/language impairments,” Journal of Speech, Language, and Hearing Research, vol. 42, pp. 744–760, 6 1999. [DOI] [PubMed] [Google Scholar]
- 10.Tan E. J., Yelland G. W., and Rossell S. L., “Characterising receptive language processing in schizophrenia using word and sentence tasks,” Cognitive Neuropsychiatry, vol. 21, pp. 14–31, 1 2016. [DOI] [PubMed] [Google Scholar]
- 11.Morgan S. E., Diederen K., Vértes P. E., Ip S. H. Y., Wang B., Thompson B., Demjaha A., Micheli A. D., Oliver D., Liakata M., Fusar-Poli P., Spencer T. J., and McGuire P., “Natural language processing markers in first episode psychosis and people at clinical high-risk,” Translational Psychiatry, vol. 11, p. 630, 12 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumus M., DeSouza D. D., Xu M., Fidalgo C., Simpson W., and Robin J., “Evaluating the utility of daily speech assessments for monitoring depression symptoms.,” Digital health, vol. 9, p. 20552076231180523, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson D. K., Lord C., Risi S., DiLavore P. S., Shulman C., Thurm A., Welch K., and Pickles A., “Patterns of growth in verbal abilities among children with autism spectrum disorder.,” Journal of consulting and clinical psychology, vol. 75, pp. 594–604, 8 2007. [DOI] [PubMed] [Google Scholar]
- 14.Low D. M., Bentley K. H., and Ghosh S. S., “Automated assessment of psychiatric disorders using speech: A systematic review,” Laryngoscope Investigative Otolaryngology, vol. 5, pp. 96–116, 2 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiig E. H., Secord W. A., and Semel E., Clinical evaluation of language fundamentals: CELF-5. Pearson, 2013. [Google Scholar]
- 16.Sparrow S. S., Ciccheti D. V., and Saulnier C. A., Vineland Adaptive Behavior Scales, third edition. Pearson, 2013. [Google Scholar]
- 17.Tomasi D. and Volkow N. D., “Associations of family income with cognition and brain structure in usa children: prevention implications,” Molecular Psychiatry, vol. 26, pp. 6619–6629, 11 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noble K. G., Norman M. F., and Farah M. J., “Neurocognitive correlates of socioeconomic status in kindergarten children,” Developmental Science, vol. 8, pp. 74–87, 1 2005. [DOI] [PubMed] [Google Scholar]
- 19.Burneo-Garcés C., Cruz-Quintana F., Pérez-García M., Fernández-Alcántara M., Fasfous A., and Pérez-Marfil M. N., “Interaction between socioeconomic status and cognitive development in children aged 7, 9, and 11 years: A cross-sectional study,” Developmental Neuropsychology, vol. 44, pp. 1–16, 1 2019. [DOI] [PubMed] [Google Scholar]
- 20.Nour M. M., McNamee D. C., Liu Y., and Dolan R. J., “Trajectories through semantic spaces in schizophrenia and the relationship to ripple bursts,” Proceedings of the National Academy of Sciences, vol. 120, 10 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soroski T., da Cunha Vasco T., Newton-Mason S., Granby S., Lewis C., Harisinghani A., Rizzo M., Conati C., Murray G., Carenini G., Field T. S., and Jang H., “Evaluating web-based automatic transcription for alzheimer speech data: Transcript comparison and machine learning analysis,” JMIR Aging, vol. 5, p. e33460, 9 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman J. R., Fatima H., Lacritz L. H., and Cullum C. M., “Utility of a short-form phonemic fluency task.,” Archives of clinical neuropsychology : the official journal of the National Academy of Neuropsychologists, 3 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruff R. M., Light R. H., Parker S. B., and Levin H. S., “Benton controlled oral word association test: reliability and updated norms.,” Archives of clinical neuropsychology : the official journal of the National Academy of Neuropsychologists, vol. 11, pp. 329–38, 1996. [PubMed] [Google Scholar]
- 24.Fawns-Ritchie C. and Deary I. J., “Reliability and validity of the uk biobank cognitive tests,” PLOS ONE, vol. 15, p. e0231627, 4 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feliciano P., Daniels A. M., Snyder L. G., Beaumont A., Camba A., Esler A., Gulsrud A. G., Mason A., Gutierrez A., Nicholson A., Paolicelli A. M., McKenzie A. P., Rachubinski A. L., Stephens A. N., Simon A. R., Stedman A., Shocklee A. D., Swanson A., Finucane B., Hilscher B. A., Hauf B., O’Roak B. J., McKenna B., Robertson B. E., Rodriguez B., Vernoia B. M., Metre B. V., Bradley C., Cohen C., Erickson C. A., Harkins C., Hayes C., Lord C., Martin C. L., Ortiz C., Ochoa-Lubinoff C.,Peura C., Rice C. E., Rosenberg C. R., Smith C. J., Thomas C., Taylor C. M., White L. C., Walston C. H., Amaral D. G., Coury D. L., Sarver D. E., Istephanous D., Li D., Nugyen D. C., Fox E. A., Butter E. M., Berry-Kravis E., Courchesne E., Fombonne E. J., Hofammann E., Lamarche E., Wodka E. L., Matthews E. T., O’Connor E., Palen E., Miller F., Dichter G. S., Marzano G., Stein G., Hutter H., Kaplan H. E., Li H., Lechniak H., Schneider H. L., Zaydens H., Arriaga I., Gerdts J. A., Cubells J. F., Cordova J. M., Gunderson J., Lillard J., Manoharan J., McCracken J. T., Michaelson J. J., Neely J., Orobio J., Pandey J., Piven J., Scherr J., Sutcliffe J. S., Tjernagel J., Wallace J., Callahan K., Dent K., Schweers K. A., Hamer K. E., Law J. K., Lowe K., O’Brien K., Smith K., Pawlowski K. G., Pierce K. L., Roeder K., Abbeduto L. J., Berry L. N., Cartner L. A., Coppola L. A., Carpenter L., Cordeiro L., DeMarco L., Grosvenor L. P., Higgins L., Huang-Storms L. Y., Hosmer-Quint L., Herbert L. M., Kasparson L., Prock L. M., Pacheco L. D., Raymond L., Simon L., Soorya L. V., Wasserburg L., Lazar M., Alessandri M., Brown M., Currin M. H., Gwynette M. F., Heyman M., Hale M. N., Jones M., Jordy M., Morrier M. J., Sahin M.,Siegel M. S., Verdi M., Parlade M. V., Yinger M., Bardett N., Hanna N., Harris N., Pottschmidt N., Russo-Ponsaran N., Takahashi N., Ousley O. Y., Juarez A. P., Manning P., Annett R. D., Bernier R. A., Clark R. D., Landa R. J., Goin-Kochel R. P., Remington R., Schultz R. T., Brewster S. J., Booker S., Carpenter S., Eldred S., Francis S., Friedman S. L., Horner S., Hepburn S., Jacob S., Kanne S., Lee S. J., Mastel S. A., Plate S., Qiu S., Sandhu S., Thompson S., White S., Myers V. J.,Singh V., Yang W. S., Warren Z., Amatya A., Ace A. J., Chatha A. S., Lash A. E., Negron B., Rigby C., Ridenour C., Stock C. M., Schmidt D., Fisk I., Acampado J., Nestle J. L., Nestle J. A., Layman K., Butler M. E., Kent M., Mallardi M. D., Carriero N., Lawson N., Volfovsky N., Edgar R., Marini R., Rana R., Ganesan S., Shah S., Ramsey T., Chin W., Jensen W., Krentz A. D., Gruber A. J., Sabo A., Salomatov A., Eng C., Muzny D., Astrovskaya I., Gibbs R. A., Han X., Shen Y., Reichardt L. F., and Chung W. K., “Spark: A us cohort of 50,000 families to accelerate autism research,” Neuron, vol. 97, pp. 488–493, 2 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudnawa K. K. and Chung W. K., “Sparking new insight into autism across the lifespan,” American Journal on Intellectual and Developmental Disabilities, vol. 129, pp. 91–95, 3 2024. [DOI] [PubMed] [Google Scholar]
- 27.Mayer M., missRanger: Fast Imputation of Missing Values, 2024. R package version 2.4.0, https://mayer79.github.io/missRanger/. [Google Scholar]
- 28.Thomas T. R., Tener A. J., Pearlman A. M., Imborek K. L., Yang J. S., Strang J. F., and Michaelson J. J., “Polygenic scores clarify the relationship between mental health and gender diversity,” Biological Psychiatry Global Open Science, vol. 4, p. 100291, 3 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taliun D., Harris D. N., Kessler M. D., Carlson J., Szpiech Z. A., Torres R., Taliun S. A. G., Corvelo A., Gogarten S. M., Kang H. M., et al. , “Sequencing of 53,831 diverse genomes from the nhlbi topmed program,” Nature, vol. 590, pp. 290–299, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S., Forer L., Schönherr S., Sidore C., Locke A. E., Kwong A., Vrieze S. I., Chew E. Y., Levy S., McGue M., Schlessinger D., Stambolian D., Loh P.-R., Iacono W. G., Swaroop A., Scott L. J., Cucca F., Kronenberg F., Boehnke M., Abecasis G. R., and Fuchsberger C., “Next-generation genotype imputation service and methods.,” Nature genetics, vol. 48, pp. 1284–1287, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., Whitwham A., Keane T., McCarthy S. A., Davies R. M., et al. , “Twelve years of SAMtools and BCFtools,” Gigascience, vol. 10, no. 2, p. giab008, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Privé F., Albiñana C., Arbel J., Pasaniuc B., and Vilhjálmsson B. J., “Inferring disease architecture and predictive ability with ldpred2-auto,” The American Journal of Human Genetics, vol. 110, pp. 2042–2055, 12 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Privé F., Aschard H., Ziyatdinov A., and Blum M. G. B., “Efficient analysis of large-scale genome-wide data with two r packages: bigstatsr and bigsnpr,” Bioinformatics, vol. 34, pp. 2781–2787, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Privé F., Luu K., Blum M. G., McGrath J. J., and Vilhjálmsson B. J., “Efficient toolkit implementing best practices for principal component analysis of population genetic data,” Bioinformatics, vol. 36, no. 16, pp. 4449–4457, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium G. P. et al. , “A global reference for human genetic variation,” Nature, vol. 526, no. 7571, p. 68, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Privé F., Arbel J., and Vilhjálmsson B. J., “Ldpred2: better, faster, stronger,” Bioinformatics, vol. 36, pp. 5424–5431, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters R. K., Polimanti R., Johnson E. C., McClintick J. N., Adams M. J., Adkins A. E., Aliev F., Bacanu S.-A., Batzler A., Bertelsen S., Biernacka J. M., Bigdeli T. B., Chen L.-S., Clarke T.-K., Chou Y.-L., Degenhardt F., Docherty A. R., Edwards A. C., Fontanillas P., Foo J. C., Fox L., Frank J., Giegling I., Gordon S., Hack L. M., Hartmann A. M., Hartz S. M., Heilmann-Heimbach S., Herms S., Hodgkinson C., Hoffmann P., Hottenga J. J., Kennedy M. A., Alanne-Kinnunen M., Konte B., Lahti J., Lahti-Pulkkinen M., Lai D., Ligthart L., Loukola A., Maher B. S., Mbarek H., McIntosh A. M., McQueen M. B., Meyers J. L., Milaneschi Y., Palviainen T., Pearson J. F., Peterson R. E., Ripatti S., Ryu E., Saccone N. L., Salvatore J. E., Sanchez-Roige S., Schwandt M., Sherva R., Streit F., Strohmaier J., Thomas N., Wang J.-C., Webb B. T., Wedow R., Wetherill L., Wills A. G., Boardman J. D., Chen D., Choi D.-S., Copeland W. E., Culverhouse R. C., Dahmen N., Degenhardt L., Domingue B. W., Elson S. L., Frye M. A., Gäbel W., Hayward C., Ising M., Keyes M., Kiefer F., Kramer J., Kuperman S., Lucae S., Lynskey M. T., Maier W., Mann K., Männistö S., Müller-Myhsok B., Murray A. D., Nurnberger J. I., Palotie A., Preuss U., Räikkönen K., Reynolds M. D., Ridinger M., Scherbaum N., Schuckit M. A., Soyka M., Treutlein J., Witt S., Wodarz N., Zill P., Adkins D. E., Boden J. M., Boomsma D. I., Bierut L. J., Brown S. A., Bucholz K. K., Cichon S., Costello E. J., de Wit H., Diazgranados N., Dick D. M., Eriksson J. G., Farrer L. A., Foroud T. M., Gillespie N. A., Goate A. M., Goldman D., Grucza R. A., Hancock D. B., Harris K. M., Heath A. C., Hesselbrock V., Hewitt J. K., Hopfer C. J., Horwood J., Iacono W., Johnson E. O., Kaprio J. A., Karpyak V. M., Kendler K. S., Kranzler H. R., Krauter K., Lichtenstein P., Lind P. A., McGue M., MacKillop J., Madden P. A. F., Maes H. H., Magnusson P., Martin N. G., Medland S. E., Montgomery G. W., Nelson E. C., Nöthen M. M., Palmer A. A., Pedersen N. L., Penninx B. W. J. H., Porjesz B., Rice J. P., Rietschel M., Riley B. P., Rose R., Rujescu D., Shen P.-H., Silberg J., Stallings M. C., Tarter R. E., Vanyukov M. M., Vrieze S., Wall T. L., Whitfield J. B., Zhao H., Neale B. M., Gelernter J., Edenberg H. J., and Agrawal A., “Transancestral gwas of alcohol dependence reveals common genetic underpinnings with psychiatric disorders,” Nature Neuroscience, vol. 21, pp. 1656–1669, 12 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wightman D. P., Jansen I. E., Savage J. E., Shadrin A. A., Bahrami S., Holland D., Rongve A., Børte S., Winsvold B. S., Drange O. K., Martinsen A. E., Skogholt A. H., Willer C., Bråthen G., Bosnes I., Nielsen J. B., Fritsche L. G., Thomas L. F., Pedersen L. M., Gabrielsen M. E., Johnsen M. B., Meisingset T. W., Zhou W., Proitsi P., Hodges A., Dobson R., Velayudhan L., Heilbron K., Auton A., Agee M., Aslibekyan S., Babalola E., Bell R. K., Bielenberg J., Bryc K., Bullis E., Cameron B., Coker D., Partida G. C., Dhamija D., Das S., Elson S. L., Filshtein T., Fletez-Brant K., Fontanillas P., Freyman W., Gandhi P. M., Hicks B., Hinds D. A., Huber K. E., Jewett E. M., Jiang Y., Kleinman A., Kukar K., Lane V., Lin K.-H., Lowe M., Luff M. K., McCreight J. C., McIntyre M. H., McManus K. F., Micheletti S. J., Moreno M. E., Mountain J. L., Mozaffari S. V., Nandakumar P., Noblin E. S., O’Connell J., Petrakovitz A. A., Poznik G. D., Schumacher M., Shastri A. J., Shelton J. F., Shi J., Shringarpure S., Tian C., Tran V., Tung J. Y., Wang X., Wang W., Weldon C. H., Wilton P., Sealock J. M., Davis L. K., Pedersen N. L., Reynolds C. A., Karlsson I. K., Magnusson S., Stefansson H., Thordardottir S., Jonsson P. V., Snaedal J., Zettergren A., Skoog I., Kern S., Waern M., Zetterberg H., Blennow K., Stordal E., Hveem K., Zwart J.-A., Athanasiu L., Selnes P., Saltvedt I., Sando S. B., Ulstein I., Djurovic S., Fladby T., Aarsland D., Selbæk G., Ripke S., Stefansson K., Andreassen O. A., and Posthuma D., “A genome-wide association study with 1,126,563 individuals identifies new risk loci for alzheimer’s disease,” Nature Genetics, vol. 53, pp. 1276–1282, 9 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demontis D., Walters R. K., Martin J., Mattheisen M., Als T. D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Bækvad-Hansen M., et al. , “Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder,” Nature Genetics, vol. 51, no. 1, pp. 63–75, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson H. J., Yilmaz Z., Thornton L. M., Hübel C., Coleman J. R., Gaspar H. A., Bryois J., Hinney A., Leppä V. M., Mattheisen M., et al. , “Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa,” Nature Genetics, vol. 51, no. 8, pp. 1207–1214, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grove J., Ripke S., Als T. D., Mattheisen M., Walters R. K., Won H., Pallesen J., Agerbo E., Andreassen O. A., Anney R., et al. , “Identification of common genetic risk variants for autism spectrum disorder,” Nature Genetics, vol. 51, no. 3, pp. 431–444, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullins N., Forstner A. J., O’Connell K. S., Coombes B., Coleman J. R. I., Qiao Z., Als T. D., Bigdeli T. B., Børte S., Bryois J., Charney A. W., Drange O. K., Gandal M. J., Hagenaars S. P., Ikeda M., Kamitaki N., Kim M., Krebs K., Panagiotaropoulou G., Schilder B. M., Sloofman L. G., Steinberg S., Trubetskoy V., Winsvold B. S., Won H.-H., Abramova L., Adorjan K., Agerbo E., Eissa M. A., Albani D., Alliey-Rodriguez N., Anjorin A., Antilla V., Antoniou A., Awasthi S., Baek J. H., Bækvad-Hansen M., Bass N., Bauer M., Beins E. C., Bergen S. E., Birner A., Pedersen C. B., Bøen E., Boks M. P., Bosch R., Brum M., Brumpton B. M., Brunkhorst-Kanaan N., Budde M., Bybjerg-Grauholm J., Byerley W., Cairns M., Casas M., Cervantes P., Clarke T.-K., Cruceanu C., Cuellar-Barboza A., Cunningham J., Curtis D., Czerski P. M., Dale A. M., Dalkner N., David F. S., Degenhardt F., Djurovic S., Dobbyn A. L., Douzenis A., Elvsåshagen T., Escott-Price V., Ferrier I. N., Fiorentino A., Foroud T. M., Forty L., Frank J., Frei O., Freimer N. B., Frisén L., Gade K., Garnham J., Gelernter J., Pedersen M. G., Gizer I. R., Gordon S. D., Gordon-Smith K., Greenwood T. A., Grove J., Guzman-Parra J., Ha K., Haraldsson M., Hautzinger M., Heilbronner U., Hellgren D., Herms S., Hoffmann P., Holmans P. A., Huckins L., Jamain S., Johnson J. S., Kalman J. L., Kamatani Y., Kennedy J. L., Kittel-Schneider S., Knowles J. A., Kogevinas M., Koromina M., Kranz T. M., Kranzler H. R., Kubo M., Kupka R., Kushner S. A., Lavebratt C., Lawrence J., Leber M., Lee H.-J., Lee P. H., Levy S. E., Lewis C., Liao C., Lucae S., Lundberg M., MacIntyre D. J., Magnusson S. H., Maier W., Maihofer A., Malaspina D., Maratou E., Martinsson L., Mattheisen M., McCarroll S. A., McGregor N. W., McGuffin P., McKay J. D., Medeiros H., Medland S. E., Millischer V., Montgomery G. W., Moran J. L., Morris D. W., Mühleisen T. W., O’Brien N., O’Donovan C., Loohuis L. M. O., Oruc L., Papiol S., Pardiñas A. F., Perry A., Pfennig A., Porichi E., Potash J. B., Quested D., Raj T., Rapaport M. H., DePaulo J. R., Regeer E. J., Rice J. P., Rivas F., Rivera M., Roth J., Roussos P., Ruderfer D. M., Sánchez-Mora C., Schulte E. C., Senner F., Sharp S., Shilling P. D., Sigurdsson E., Sirignano L., Slaney C., Smeland O. B., Smith D. J., Sobell J. L., Hansen C. S., Artigas M. S., Spijker A. T., Stein D. J., Strauss J. S., Światkowska B., Terao C., Thorgeirsson T. E., Toma C., Tooney P., Tsermpini E.-E., Vawter M. P., Vedder H., Walters J. T. R., Witt S. H., Xi S., Xu W., Yang J. M. K., Young A. H., Young H., Zandi P. P., Zhou H., Zillich L., Adolfsson R., Agartz I., Alda M., Alfredsson L., Babadjanova G., Backlund L., Baune B. T., Bellivier F., Bengesser S., Berrettini W. H., Blackwood D. H. R., Boehnke M., Børglum A. D., Breen G., Carr V. J., Catts S., Corvin A., Craddock N., Dannlowski U., Dikeos D., Esko T., Etain B., Ferentinos P., Frye M., Fullerton J. M., Gawlik M., Gershon E. S., Goes F. S., Green M. J., Grigoroiu-Serbanescu M., Hauser J., Henskens F., Hillert J., Hong K. S., Hougaard D. M., Hultman C. M., Hveem K., Iwata N., Jablensky A. V., Jones I., Jones L. A., Kahn R. S., Kelsoe J. R., Kirov G., Landén M., Leboyer M., Lewis C. M., Li Q. S., Lissowska J., Lochner C., Loughland C., Martin N. G., Mathews C. A., Mayoral F., McElroy S. L., McIntosh A. M., McMahon F. J., Melle I., Michie P., Milani L., Mitchell P. B., Morken G., Mors O., Mortensen P. B., Mowry B., Müller-Myhsok B., Myers R. M., Neale B. M., Nievergelt C. M., Nordentoft M., Nöthen M. M., O’Donovan M. C., Oedegaard K. J., Olsson T., Owen M. J., Paciga S. A., Pantelis C., Pato C., Pato M. T., Patrinos G. P., Perlis R. H., Posthuma D., Ramos-Quiroga J. A., Reif A., Reininghaus E. Z., Ribasés M., Rietschel M., Ripke S., Rouleau G. A., Saito T., Schall U., Schalling M., Schofield P. R., Schulze T. G., Scott L. J., Scott R. J., Serretti A., Weickert C. S., Smoller J. W., Stefansson H., Stefansson K., Stordal E., Streit F., Sullivan P. F., Turecki G., Vaaler A. E., Vieta E., Vincent J. B., Waldman I. D., Weickert T. W., Werge T., Wray N. R., Zwart J.-A., Biernacka J. M., Nurnberger J. I., Cichon S., Edenberg H. J., Stahl E. A., McQuillin A., Florio A. D., Ophoff R. A., and Andreassen O. A., “Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology,” Nature Genetics, vol. 53, pp. 817–829, 6 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard D. M., Adams M. J., Clarke T.-K., Hafferty J. D., Gibson J., Shirali M., Coleman J. R., Hagenaars S. P., Ward J., Wigmore E. M., et al. , “Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions,” Nature Neuroscience, vol. 22, no. 3, pp. 343–352, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nievergelt C. M., Maihofer A. X., Klengel T., Atkinson E. G., Chen C.-Y., Choi K. W., Coleman J. R. I., Dalvie S., Duncan L. E., Gelernter J., Levey D. F., Logue M. W., Polimanti R., Provost A. C., Ratanatharathorn A., Stein M. B., Torres K., Aiello A. E., Almli L. M., Amstadter A. B., Andersen S. B., Andreassen O. A., Arbisi P. A., Ashley-Koch A. E., Austin S. B., Avdibegovic E., Babić D., Bækvad-Hansen M., Baker D. G., Beckham J. C., Bierut L. J., Bisson J. I., Boks M. P., Bolger E. A., Børglum A. D., Bradley B., Brashear M., Breen G., Bryant R. A., Bustamante A. C., Bybjerg-Grauholm J., Calabrese J. R., de Almeida J. M. C., Dale A. M., Daly M. J., Daskalakis N. P., Deckert J., Delahanty D. L., Dennis M. F., Disner S. G.,Domschke K., Dzubur-Kulenovic A., Erbes C. R., Evans A., Farrer L. A., Feeny N. C., Flory J. D., Forbes D., Franz C. E., Galea S., Garrett M. E., Gelaye B., Geuze E., Gillespie C., Uka A. G., Gordon S. D., Guffanti G., Hammamieh R., Harnal S., Hauser M. A., Heath A. C., Hemmings S. M. J., Hougaard D. M., Jakovljevic M., Jett M., Johnson E. O., Jones I., Jovanovic T., Qin X.-J., Junglen A. G., Karstoft K.-I., Kaufman M. L., Kessler R. C., Khan A., Kimbrel N. A., King A. P., Koen N., Kranzler H. R., Kremen W. S., Lawford B. R., Lebois L. A. M., Lewis C. E., Linnstaedt S. D., Lori A., Lugonja B., Luykx J. J., Lyons M. J., Maples-Keller J., Marmar C., Martin A. R., Martin N. G., Maurer D., Mavissakalian M. R., McFarlane A., McGlinchey R. E., McLaughlin K. A., McLean S. A., McLeay S., Mehta D., Milberg W. P., Miller M. W., Morey R. A., Morris C. P., Mors O., Mortensen P. B., Neale B. M., Nelson E. C., Nordentoft M., Norman S. B., O’Donnell M., Orcutt H. K., Panizzon M. S., Peters E. S., Peterson A. L., Peverill M., Pietrzak R. H., Polusny M. A., Rice J. P., Ripke S., Risbrough V. B., Roberts A. L., Rothbaum A. O., Rothbaum B. O., Roy-Byrne P., Ruggiero K., Rung A., Rutten B. P. F., Saccone N. L., Sanchez S. E., Schijven D., Seedat S., Seligowski A. V., Seng J. S., Sheerin C. M., Silove D., Smith A. K., Smoller J. W., Sponheim S. R., Stein D. J., Stevens J. S., Sumner J. A., Teicher M. H., Thompson W. K., Trapido E., Uddin M., Ursano R. J., van den Heuvel L. L., Hooff M. V., Vermetten E., Vinkers C. H., Voisey J., Wang Y., Wang Z., Werge T., Williams M. A., Williamson D. E., Winternitz S., Wolf C., Wolf E. J., Wolff J. D., Yehuda R., Young R. M., Young K. A., Zhao H., Zoellner L. A., Liberzon I., Ressler K. J., Haas M., and Koenen K. C., “International meta-analysis of ptsd genome-wide association studies identifies sex- and ancestry-specific genetic risk loci,” Nature Communications, vol. 10, p. 4558, 10 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ripke S., Walters J. T., O’Donovan M. C., of the Psychiatric Genomics Consortium S. W. G., et al. , “Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia,” MedRxiv, 2020. [Google Scholar]
- 46.Lee J. J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T. A., Bowers P., Sidorenko J., Linnér R. K., et al. , “Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals,” Nature genetics, vol. 50, pp. 1112–1121, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okbay A., Wu Y., Wang N., Jayashankar H., Bennett M., Nehzati S. M., Sidorenko J., Kweon H., Goldman G., Gjorgjieva T.,Jiang Y., Hicks B., Tian C., Hinds D. A., Ahlskog R., Magnusson P. K. E., Oskarsson S., Hayward C., Campbell A., Porteous D. J., Freese J., Herd P., Agee M., Alipanahi B., Auton A., Bell R. K., Bryc K., Elson S. L., Fontanillas P., Furlotte N. A., Hinds D. A., Huber K. E., Kleinman A., Litterman N. K., McCreight J. C., McIntyre M. H., Mountain J. L., Northover C. A. M., Pitts S. J., Sathirapongsasuti J. F., Sazonova O. V., Shelton J. F., Shringarpure S., Tung J. Y., Vacic V., Wilson C. H., Fontana M. A., Pers T. H., Rietveld C. A., Chen G.-B., Emilsson V., Meddens S. F. W., Pickrell J. K., Thom K., Timshel P., de Vlaming R., Abdellaoui A., Ahluwalia T. S., Bacelis J., Baumbach C., Bjornsdottir G., Brandsma J. H., Concas M. P., Derringer J., Galesloot T. E., Girotto G., Gupta R., Hall L. M., Harris S. E., Hofer E., Horikoshi M., Huffman J. E., Kaasik K., Kalafati I. P., Karlsson R., Lahti J., van der Lee S. J., de Leeuw C., Lind P. A., Lindgren K.-O., Liu T., Mangino M., Marten J., Mihailov E., Miller M. B., van der Most P. J., Oldmeadow C., Payton A., Pervjakova N., Peyrot W. J., Qian Y., Raitakari O., Rueedi R., Salvi E., Schmidt B., Schraut K. E., Shi J., Smith A. V., Poot R. A., Pourcain B. S., Teumer A., Thorleifsson G., Verweij N., Vuckovic D., Wellmann J., Westra H.-J., Yang J., Zhao W., Zhu Z., Alizadeh B. Z., Amin N., Bakshi A., Baumeister S. E., Biino G., Bønnelykke K., Boyle P. A., Campbell H., Cappuccio F. P., Davies G., Neve J.-E. D., Deloukas P., Demuth I., Ding J., Eibich P., Eisele L., Eklund N., Evans D. M., Faul J. D., Feitosa M. F., Forstner A. J., Gandin I., Gunnarsson B., Halldórsson B. V., Harris T. B., Heath A. C., Hocking L. J., Holliday E. G., Homuth G., Horan M. A., Hottenga J.-J., de Jager P. L., Joshi P. K., Jugessur A., Kaakinen M. A., Kähönen M., Kanoni S., Keltigangas-Järvinen L., Kiemeney L. A. L. M., Kolcic I., Koskinen S., Kraja A. T., Kroh M., Kutalik Z., Latvala A., Launer L. J., Lebreton M. P., Levinson D. F., Lichtenstein P., Lichtner P., Liewald D. C. M., Loukola A., Madden P. A., Mägi R., Mäki-Opas T., Marioni R. E., Marques-Vidal P., Meddens G. A., McMahon G., Meisinger C., Meitinger T., Milaneschi Y., Milani L., Montgomery G. W., Myhre R., Nelson C. P., Nyholt D. R., Ollier W. E. R., Palotie A., Paternoster L., Pedersen N. L., Petrovic K. E., Räikkönen K., Ring S. M., Robino A., Rostapshova O., Rudan I., Rustichini A., Salomaa V., Sanders A. R., Sarin A.-P., Schmidt H., Scott R. J., Smith B. H., Smith J. A., Staessen J. A., Steinhagen-Thiessen E., Strauch K., Terracciano A., Tobin M. D., Ulivi S., Vaccargiu S., Quaye L., van Rooij F. J. A., Venturini C., Vinkhuyzen A. A. E., Völker U., Völzke H., Vonk J. M., Vozzi D., Waage J., Ware E. B., Willemsen G., Attia J. R., Bennett D. A., Berger K., Bertram L., Bisgaard H., Boomsma D. I., Borecki I. B., Bültmann U., Chabris C. F., Cucca F., Cusi D., Deary I. J., Dedoussis G. V., van Duijn C. M., Eriksson J. G., Franke B., Franke L., Gasparini P., Gejman P. V., Gieger C., Grabe H.-J., Gratten J., Groenen P. J. F., Gudnason V., van der Harst P., Hoffmann W., Hyppönen E., Iacono W. G., Jacobsson B., Järvelin M.-R., Jöckel K.-H., Kaprio J., Kardia S. L. R., Lehtimäki T., Lehrer S. F., Martin N. G., McGue M., Metspalu A., Pendleton N., Penninx B. W. J. H., Perola M., Pirastu N., Pirastu M., Polasek O., Posthuma D., Power C., Province M. A., Samani N. J., Schlessinger D., Schmidt R., Sørensen T. I. A., Spector T. D., Stefansson K., Thorsteinsdottir U., Thurik A. R., Timpson N. J., Tiemeier H., Uitterlinden A. G., Vitart V., Vollenweider P., Weir D. R., Wilson J. F., Wright A. F., Conley D. C., Krueger R. F., Smith G. D., Hofman A., Laibson D. I., Medland S. E., Yang J., Esko T., Watson C., Jala J., Conley D., Koellinger P. D., Johannesson M., Laibson D., Meyer M. N., Lee J. J., Kong A., Yengo L., Cesarini D., Turley P., Visscher P. M., Beauchamp J. P., Benjamin D. J., and Young A. I., “Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals,” Nature Genetics, vol. 54, pp. 437–449, 4 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linnér R. K., Biroli P., Kong E., Meddens S. F. W., Wedow R., Fontana M. A., Lebreton M., Tino S. P., Abdellaoui A., Hammerschlag A. R., Nivard M. G., Okbay A., Rietveld C. A., Timshel P. N., Trzaskowski M., de Vlaming R., Zünd C. L., Bao Y., Buzdugan L., Caplin A. H., Chen C.-Y., Eibich P., Fontanillas P., Gonzalez J. R., Joshi P. K., Karhunen V., Kleinman A., Levin R. Z., Lill C. M., Meddens G. A., Muntané G., Sanchez-Roige S., van Rooij F. J., Taskesen E., Wu Y., Zhang F., Auton A., Boardman J. D., Clark D. W., Conlin A., Dolan C. C., Fischbacher U., Groenen P. J. F., Harris K. M., Hasler G., Hofman A., Ikram M. A., Jain S., Karlsson R., Kessler R. C., Kooyman M., MacKillop J., Männikkö M., Morcillo-Suarez C., McQueen M. B., Schmidt K. M., Smart M. C., Sutter M., Thurik A. R., Uitterlinden A. G., White J., de Wit H., Yang J., Bertram L., Boomsma D. I., Esko T., Fehr E., Hinds D. A., Johannesson M., Kumari M., Laibson D., Magnusson P. K. E., Meyer M. N., Navarro A., Palmer A. A., Pers T. H., Posthuma D., Schunk D., Stein M. B., Svento R., Tiemeier H., Timmers P. R. H. J., Turley P., Ursano R. J., Wagner G. G., Wilson J. F., Gratten J., Lee J. J., Cesarini D., Benjamin D. J., Koellinger P. D., and Beauchamp J. P., “Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences,” Nature Genetics, vol. 51, pp. 245–257, 2 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. C. Team, “R: A language and environment for statistical computing,” 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SPARK genetic and phenotypic data can be obtained at SFARI Base: https://base.sfari.org
The SPARK Research Match data will be available to qualified, approved researchers through SFARI Base upon publication of this article.