Abstract
The human papillomavirus (HPV) E1 and E2 proteins bind cooperatively to the viral origin of replication (ori), forming an E1-E2-ori complex that is essential for initiation of DNA replication. All other replication proteins, including DNA polymerase α-primase (polα-primase), are derived from the host cell. We have carried out a detailed analysis of the interactions of HPV type 16 (HPV-16) E1 with E2, ori, and the four polα-primase subunits. Deletion analysis showed that a C-terminal region of E1 (amino acids [aa] 432 to 583 or 617) is required for E2 binding. HPV-16 E1 was unable to bind the ori in the absence of E2, but the same C-terminal domain of E1 was sufficient to tether E1 to the ori via E2. Of the polα-primase subunits, only p68 bound E1, and binding was competitive with E2. The E1 region required (aa 397 to 583) was the same as that required for E2 binding but additionally contained 34 N-terminal residues. In confirmation of these differences, we found that a monoclonal antibody, mapping adjacent to the N-terminal junction of the p68-binding region, blocked E1-p68 but not E1-E2 binding. Sequence alignments and secondary-structure prediction for HPV-16 E1 and other superfamily 3 (SF3) viral helicases closely parallel the mapping data in suggesting that aa 439 to 623 constitute a discrete helicase domain. Assuming a common nucleoside triphosphate-binding fold, we have generated a structural model of this domain based on the X-ray structures of the hepatitis C virus and Bacillus stearothermophilus (SF2) helicases. The modelling closely matches the deletion analysis in suggesting that this region of E1 is indeed a structural domain, and our results suggest that it is multifunctional and critical to several stages of HPV DNA replication.
Human papillomaviruses (HPVs) are a family of small DNA viruses which infect epithelial cells, inducing the formation of benign tumors. Over 70 HPV genotypes have been identified, and these cause a range of diseases, including skin warts, anogenital warts, and life-threatening laryngeal papillomas. In addition, certain high-risk anogenital HPV types (particularly HPV types 16, 18, 31, and 33) are implicated in intraepithelial neoplasias which can progress to malignancies (for example, cervical cancer). Papillomaviruses (PVs) encode few proteins and are therefore highly dependent on the host cell for replication and expression of their genomes. Early work with bovine papillomavirus type 1 (BPV-1), whose DNA can be propagated in cell lines as an episome, showed that the viral E1 and E2 genes are required for DNA replication (reviewed by Chow and Broker [9]). Subsequently, it was established that the BPV-1 or HPV E1 and E2 genes are necessary and sufficient for the transient replication of plasmids containing a PV origin of replication (ori) in transfected cells (8, 56). Replication of ori-containing plasmids in cell extracts containing E1 and E2 proteins has also been demonstrated for BPV-1 and HPV (25, 62). These assays have shown that efficient DNA replication requires E1, E2, and a minimal ori which contains binding sites (E1BS and E2BS, respectively) for these proteins.
Sequence similarities within the C-terminal 200 amino acids (aa) originally indicated that E1, like the simian virus 40 (SV40) and polyomavirus T antigens, may be a DNA helicase-ATPase involved in initiating DNA replication (10, 49). Sequence similarities to the parvovirus and human herpesvirus 6 NS-1 proteins have since been found, and these have all been grouped into helicase superfamily 3 (SF3) (18, 61). BPV-1 E1 has been shown to be a nuclear 68- to 72-kDa ATP-binding phosphoprotein (53) that binds specifically to an E1BS within the ori (20, 55) and has 3′-5′ DNA helicase activity (63) capable of unwinding ori-containing DNA in the presence of topoisomerase (25, 46). BPV-1 E1 also forms a specific complex with the 48-kDa viral transactivator E2 (4, 31), whose major role in replication appears to be to strengthen the E1-ori interaction by forming an E1-E2-ori ternary complex (38, 64).
The factors involved in eukaryotic DNA replication have been identified by using a fully reconstituted SV40 in vitro replication system (57, 58). Formation of an initiation complex involves assembly of SV40 T antigen on the origin and unwinding of duplex DNA in the presence of ATP, replication protein A (RPA), and topoisomerase I. Replication additionally requires DNA polymerase α-primase (polα-primase) to initiate DNA synthesis and DNA polymerase δ, replication factor C, and proliferating cell nuclear antigen for elongation. In the elongation phase, T antigen acts as the DNA helicase at the replication fork. In vitro replication studies with BPV-1 and HPV-11 demonstrate a requirement for the same cellular factors, although the viral initiator-helicase function is provided by E1 in association with E2 (25, 36, 64). Since T antigen has been shown to bind RPA (37), polα-primase (14), and topoisomerase I (47) to the SV40 replication complex, it would be predicted that E1 and E2 should between them recruit these factors. Indeed, binding of the polα-primase p180 subunit to BPV-1 E1 (41) and of RPA to E2 (28) has been reported.
Biochemical analysis of HPV E1 proteins has been much more limited than that of BPV-1 E1. HPV-11 and -16 E1 and E2 proteins have been shown to associate (6, 51). For HPV-11 and -18, and possibly other HPVs as well, this association is more critical than it is for BPV-1, since E2 appears to be the primary ori recognition protein (30, 54). Nevertheless, weak E1-ori binding has been shown for HPV-31b (15) and HPV-11 (29). HPV-11 and -16 E1 proteins have been shown to have ATPase activity (6, 51), and HPV-6b E1 has been demonstrated to have DNA helicase-ATPase activity (21, 22).
A number of studies have mapped functional domains of BPV-1 E1 involved in nuclear localization, helicase-ATPase activity, DNA binding, and E2 binding (reviewed by Chow and Broker in reference 9). A Pro-to-Ser mutation within the predicted ATP-binding site of E1 has been shown to impair E1 ATPase activity (53) and, in the case of HPV-6b, ATPase and helicase activities (21); these results confirm the functional homology of the C-terminal region of E1 to the SV40 T-antigen helicase domain. However, there have been conflicting results in the mapping of the E2-binding domain, which has been reported to encompass N-terminal (1, 15) or C-terminal (19, 31, 39, 44) regions of BPV-1 and HPVs. We have therefore undertaken a detailed functional analysis of HPV-16 E1 in order to (i) attempt to resolve the conflicting data on E2-binding domains; (ii) identify any interactions occurring between E1 and HPV-16 ori, polα-primase, and topoisomerase I; and (iii) provide a refined map of the different functional domains. In addition, we have utilized refined sequence alignments with other SF3 helicases, and the recently available X-ray crystallographic structures of two more distantly related (SF2) helicases, to derive a structural model for the helicase domain of E1.
MATERIALS AND METHODS
Escherichia coli E1 and E2 expression vectors.
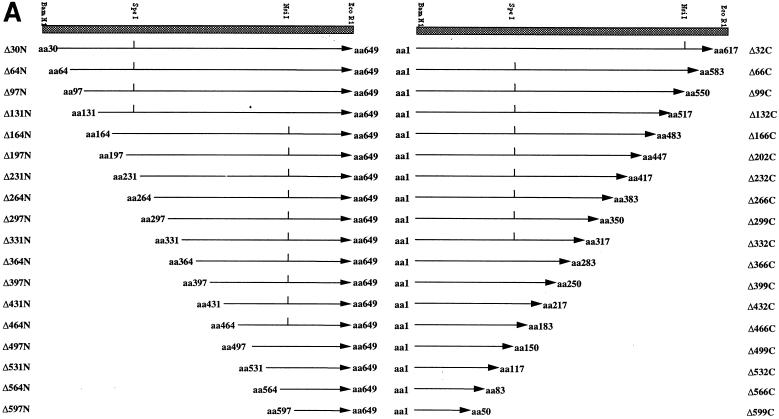
Vectors expressing full-length HPV-16 E1 and E2 fused to glutathione S-transferase (GST) (pGEX-E1 and -E2, respectively) were constructed, using PCR to add 5′ BamHI and 3′ EcoRI sites to these genes, and then cloned into pGEX2T. PCR was used to generate a set of 36 5′ and 3′ deletions of E1, differing by 32 or 33 codons, from pGEX-E1 (see Fig. 2). N-terminal deletion mutants were generated with a set of 5′ deletion primers, each containing a BamHI site, together with a 3′ primer. To keep PCR products short and to minimize errors, we used three different 3′ PCR primers (one corresponding to the NsiI site, one complementary to the SpeI site, and one complementary to the EcoRI site in pGEX2T). C-terminal deletions were generated in a similar way. PCR products for each of the deletion mutants were digested with the relevant enzymes and subcloned back into pGEX2T. The HPV-16 E2 gene was cloned into pMALc, using PCR to add SacI and HindIII sites, for expression of pMAL-E2, full-length E2 fused to maltose-binding protein (MBP) (41a).
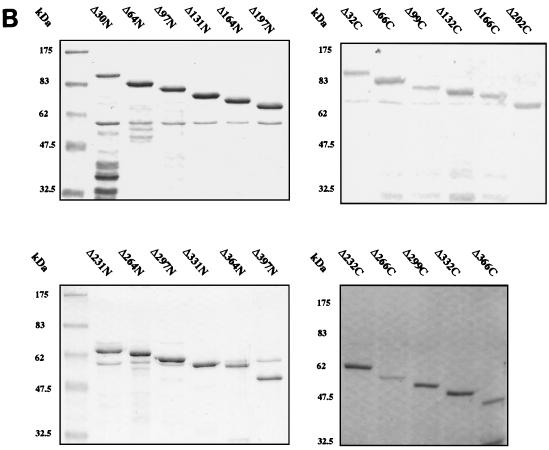
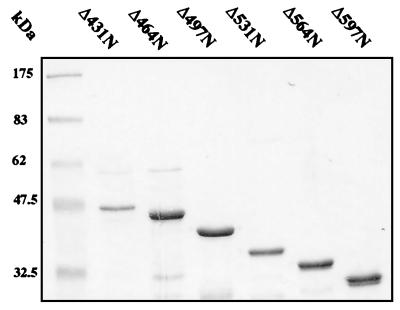
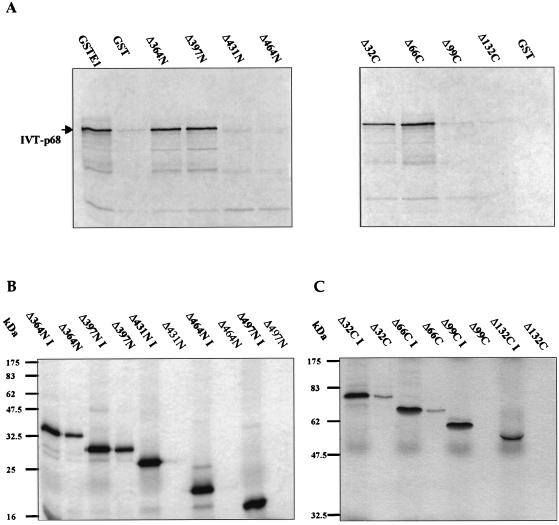
FIG. 2.
(A) Schematic representation of the amino-terminal (N) and carboxyl-terminal (C) GST-E1 deletion series. Mutants are annotated according to the number of amino acids removed from the N or C terminus. Internal restriction sites utilized in construction are indicated where appropriate. (B) SDS-PAGE and Coomassie blue staining of glutathione-agarose-purified GST-E1 deletion proteins. The positions of molecular mass markers are indicated to the left of each gel.
The GST-E1 deletion mutants were assayed in binding assays with in vitro-translated (IVT) E2. However, several E1 deletion mutants were assayed in a reverse binding assay (binding of mature IVT E1 proteins to GST-E2). The E1 deletions were transferred from pGEX2T, using PCR to introduce EcoRV and HindIII sites for subcloning into the SP6 promoter vector pING14.1.
Expression vectors for polα-primase subunits.
The cDNA encoding the human p68 subunit cloned into pGEX2T (11) was kindly provided by T. Kelly (Johns Hopkins University, Baltimore, Md.). For in vitro transcription-translation, we subcloned the cDNA into the BamHI site of pSP64. cDNAs encoding the human p48 and p58 subunits, cloned into pBluescript KS, were kindly provided by H.-P. Nasheuer (Institute of Molecular Biotechnology, Jena, Germany). A baculovirus stock containing the human p180 cDNA (12) was kindly provided by T. Wang (Stanford University School of Medicine, Stanford, Calif.). This was used to infect Sf9 insect cells and extract the viral DNA from which the p180 cDNA (4,389 bp) was isolated by PCR. This was carried out in two sections: from the 5′ end to a unique SalI site (1,988 bp), inserting an XbaI site, and from the SalI site to the 3′ end, inserting an EcoRI site (2,401 bp). The cDNA was cloned in a three-way ligation between the XbaI and EcoRI sites of pSP64 and then authenticated by sequencing.
Expression and purification of GST and MBP fusion proteins.
Expression of E1, E2, p68, and TATA-binding protein (TBP) fusions in E. coli DH5α cells and their purification were performed essentially as described by Hibma et al. (19). Overnight cultures (10 ml each) of each GST (Luria-Bertani [LB] medium with 0.5 M sorbitol, 2.5 mM betaine, and 100 μg of ampicillin per ml) and MBP (LB medium with 100 μg of ampicillin per ml) construct were set up and incubated at 37°C with shaking. Overnight cultures were used to inoculate 800 ml of LB medium plus 1.0 M sorbitol, 2.5 mM betaine, and 100 μg of ampicillin per ml for GST fusions and 800 ml of LB medium plus 100 μg of ampicillin per ml for MBP fusion proteins. Bacteria were grown at 37°C to an A600 of 0.8. Expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM and incubation overnight at 25°C with shaking. Cells were harvested by centrifugation at 2,500 × g for 10 min. Bacteria were resuspended in 40 ml of buffer C (50 mM Tris-HCl [pH 8.5], 100 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol). The suspension was sonicated for four 30-s intervals on ice. Insoluble material was removed by centrifugation for 20 min at 10,000 × g and 4°C and filtered through a 0.2-μm-pore-size filter (Sartorius). GST and MBP fusion proteins were purified by using reduced glutathione-agarose (GSH-agarose) resin (Pharmacia) and amylose resin (New England Biolabs), respectively, and either were used directly in assays as bound proteins or were first eluted from the resin. For some preparations of GST-E1, an extensive 1 M NaCl wash step was included prior to elution in order to remove possible bound contaminating DNA. Purified proteins were assayed for the presence of contaminating DNA by the highly sensitive PicoGreen DNA quantitation assay (Molecular Probes) (48). DNA concentrations were found to be very low in protein preparations tested with or without the 1 M NaCl wash step, with the salt wash resulting in a threefold-lower value (4.12 ng/μg of E1 protein for the phosphate-buffered saline [PBS]–0.1% Tween 20 wash and 1.3 ng/μg of E1 protein for the 1.0 M NaCl wash). These amounts are comparable to the value of 5 ng/μg of GST-E1 protein estimated with a 5′-end labelling technique by Santucci et al. (43).
Transcription-translation of proteins and in vitro association assay.
35S-labelled proteins were produced in vitro by using a coupled transcription-translation system (Promega) as directed by the manufacturer. Binding of 2 μl of IVT protein to 2 μg of GST fusion protein immobilized on GSH-agarose beads was carried out essentially as described by Storey et al. (51). The amount of GST fusion protein bound to 50 μl of GSH-agarose beads was determined (following elution), and then the volume of beads was adjusted to give 2 μg of protein per binding assay. After binding, nonspecifically bound proteins were removed by washing the beads five times in PBS containing 0.1% Nonidet P-40. Proteins specifically bound to the beads were removed by boiling in an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer, electrophoresed on SDS–10% polyacrylamide gels, and analyzed by autoradiography.
McKay precipitation assay for E1 and E2 ori binding.
Precipitation assays were performed essentially as described by Berg and Stenlund (2; see also reference 35). Briefly, 25 ng of full-length GST-E1 or an E1 deletion mutant and 5 ng of MBP-E2 were added to 10 μl of buffer A (20 mM potassium phosphate [pH 7.5], 0.1 M NaCl, 10% glycerol, 5 mM EDTA, 2.5 mM dithiothreitol, 0.7 mg of bovine serum albumin per ml, 0.1% Nonidet P-40) containing 5,000 cpm each of two labelled double-stranded oligonucleotides and 100 ng of competitor DNA [poly(dI-dC)] in buffer A. The two oligonucleotides contained sequences from the HPV ori; the 119-bp oligonucleotide contained the complete HPV-16 ori (nucleotides 7856 to 69), while the 35-bp oligonucleotide contained only the E1BS (nucleotides 7886 to 14). After 20 min of incubation at room temperature, 2.5 μl of glutathione resin and 50 μl of buffer A were added, and the mixture was incubated on a rotating wheel for a further 20 min at room temperature. The beads were washed three times with 200 μl of buffer A before 100 μl of stop buffer B (1% SDS, 50 mM EDTA, 0.1 M NaCl, 50 μg of tRNA per ml) was added. After phenol extraction and ethanol precipitation, the recovered DNA was analyzed on nondenaturing 15% polyacrylamide gels.
Antibody competition assay.
GST-E1 (0.4 μg) bound to glutathione-Sepharose was incubated with anti-HPV-16 E1 monoclonal antibodies (MAbs) (E1-N1 and E1-C1), Camvir 6 MAb (anti-HPV 16L1) (68a), or bovine serum albumin at 15, 1.5, and 0.15 μg/μl for 2 h at 4°C with gentle agitation. IVT p68 or IVT E2 (0.5 μl) was added, and incubation was continued for a further 45 min. Glutathione-Sepharose beads were collected by centrifugation and washed five times with PBS–0.2% Tween 20 (1 ml each time). Following the last wash, the supernatant was completely removed, the pellet was resuspended in 20 μl of 2× SDS sample buffer, and the suspension was boiled for 10 min. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and bound IVT p68 or IVT E2 was detected by autoradiography.
MAbs E1-N1 and E1-C1 were derived by immunizing BALB/c mice with GST-E1 and generating hybridomas by a standard methodology (16). Using the GST-E1 N- and C-terminal deletions in enzyme-linked immunosorbent assays, the E1-N1 epitope was mapped to aa 50 to 64 and the E1-C1 epitope was mapped to aa 364 to 383 (unpublished data).
Competition assay for binding of E2 and p68 to E1.
E1-N1 MAb, at a concentration of 10 mg/ml, was coupled to protein G-Sepharose (Pharmacia) overnight at 4°C and subsequently washed five times with PBS. GST-E1 (1 μg) and increasing amounts of GST-E2 or GST control proteins (0.2 to 3.2 μg) were mixed and incubated with gentle agitation for 2 h at 4°C in a total volume of 50 μl made up with buffer B (20 mM Tris-HCl, 200 mM NaCl, 2 mM EDTA, 10% glycerol, 1 mM dithiothreitol, pH 7.5). IVT p68 (1 μl) was added, and the mixture was incubated for 45 min, at which time protein G-Sepharose-E1 antibody complex (10 μl of a 50% suspension) was added and the mixture was further incubated for 1 h. The Sepharose was collected by centrifugation and washed 10 times with PBS–0.2% Tween 20 (1 ml each time). Following the last wash, the supernatant was completely removed, the pellet was resuspended in 20 μl of 2× SDS sample buffer, and the suspension was boiled for 10 min. Samples were analyzed by SDS-PAGE, and bound IVT p68 was detected by autoradiography. Densitometric analysis was performed on a Macintosh computer, using the public-domain NIH Image program (developed at the U.S. National Institutes of Health and available on the internet at http://rsb.info.nih.gov/nih-image/).
RESULTS
Expression of fusion proteins in E. coli.
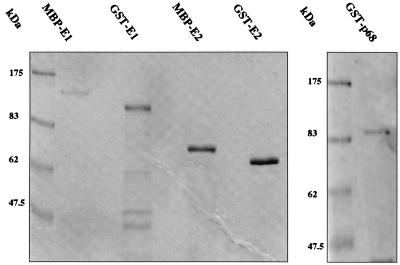
HPV-16 E1 and E2 were expressed in E. coli as GST or MBP fusions for use in protein association assays, as described in Materials and Methods. The polα-primase p68 subunit and TBP were used as GST fusion proteins. We utilized E. coli induction conditions, including the presence of sorbitol and betaine, which had been found to minimize degradation of the highly labile HPV-16 GST-E1 protein (19). Proteins were purified by glutathione-agarose or amylose resin affinity chromatography. SDS-PAGE analysis indicated that GST-E1, GST-E2, MBP-E1, MBP-E2, and GST-p68 generated in this way had only minor levels of contamination (Fig. 1), although GST-E1 contained breakdown products.
FIG. 1.
Fusion proteins purified from E. coli. GST-E1, GST-E2, MBP-E1, MBP-E2, and GST-p68 were expressed and purified as described in Materials and Methods and then separated by SDS-PAGE and stained with Coomassie blue. The positions of molecular mass markers are indicated on the left of each gel.
To map regions of E1 required for E2 binding, we generated a comprehensive set of 36 N-terminal and C-terminal deletions of E1 (in steps of ∼30 codons) as GST fusions (Fig. 2A). These fusion proteins were expressed in E. coli and purified as described for full-length GST-E1. SDS-PAGE showed that the fusion proteins migrated with the predicted molecular masses (Fig. 2B). Although otherwise pure, some of the purified GST-E1 deletion proteins contained breakdown products and some contained low levels of a contaminating 60-kDa polypeptide, which we identified as the E. coli chaperonin GroEL. Since this could potentially perturb association with E2 and other proteins, we confirmed all of our deletion mapping results by using a reciprocal protein association assay in which unfused IVT E1 deletion proteins are bound to GST-E2. This controlled both for possible effects of the GST fusion on E1 and for possible effects of GroEL.
Localization of a domain of the HPV-16 E1 protein that is necessary for E2 binding.
The binding of 35S-labelled IVT E2 to GST-E1 proteins bound to agarose beads was measured by in vitro association assays (Fig. 3). In all of these experiments we used fixed amounts of IVT E2 and controlled for the amount of each GST-E1 protein in the assay (see Materials and Methods).
FIG. 3.
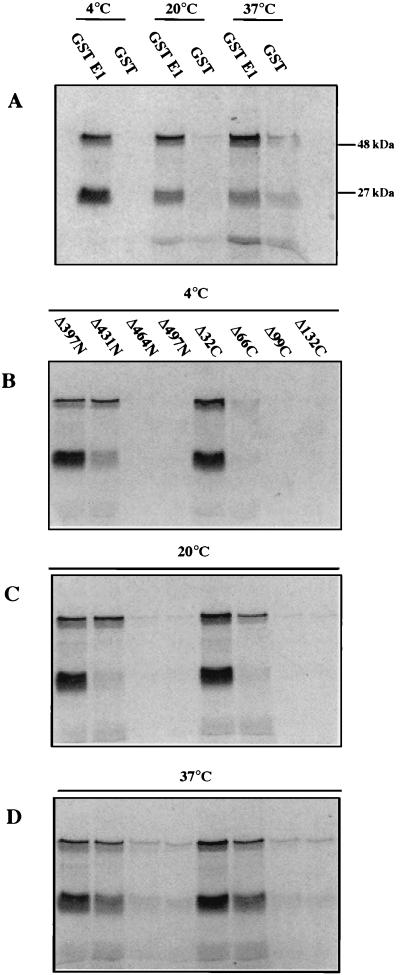
Deletion analysis of GST-E1 binding to IVT E2. In vitro association of bead-bound GST-E1 or GST to radiolabelled IVT E2 was assayed as described in Materials and Methods. Autoradiographs show E2 protein that bound and was eluted from the beads. In addition to full-length E2 (48 kDa), a 27-kDa truncated form was also seen. (A) Binding of E2 to full-length GST-E1 and GST (as a control) at 4, 20, and 37°C. (B to D) Binding to selected GST-E1 N- and C-terminal deletion mutants that span the cutoff points for E2 binding at 4°C (B), 20°C (C), and 37°C (D). These results show that deletion of 432 N-terminal residues significantly affects binding at each assayed temperature while deletion of 66 C-terminal residues significantly affects binding at 4°C but not at the higher temperatures.
Since recent reports suggest that the E1-E2 interaction is affected by temperature (1, 39), we initially investigated the temperature dependence of the binding of E2 to full-length GST-E1. The results showed that IVT E2 binds strongly to GST-E1 at 4, 20, or 37°C (Fig. 3A). However, the level of nonspecific binding to GST was significantly lower at 4°C, so most of the remaining binding assays were carried out at that temperature. In all of these assays, as well as full-length E2 (48 kDa), we consistently detected a 27-kDa polypeptide that also bound GST-E1 strongly (Fig. 3A). Since this reacted with a MAb that maps to aa 18 to 41 of E2 (19), we concluded that it was an N-terminal fragment of E2 (data not shown).
Using the N-terminal deletion series, a clear cutoff was observed: mutants E1Δ30N to E1Δ431N bound strongly to IVT E2 at 4°C, while neither E1Δ464N nor any of the further-N-terminally deleted proteins bound E2. This indicates that residues C terminal of aa 431 in E1 are necessary for interaction with E2. In the C-terminal deletion series, E1Δ32C bound strongly and E1Δ66C bound very weakly to IVT E2 at 4°C, but further deletions resulted in a complete loss of binding. The critical results from these experiments, i.e., those spanning the cutoff points, are shown in Fig. 3B. The N-terminal and C-terminal deletion data together localize the region of HPV-16 E1 essential for E2 binding at 4°C to aa 432 to 617. (This is the maximal extent of the region, since our data actually identify the boundaries as being aa 432 to 464 and 583 to 617. For simplicity, we shall henceforth refer only to the outer coordinates.)
Effect of temperature on the E1 region necessary for association with E2.
To determine whether the E2-binding domain is affected by temperature, we compared binding at 4, 20, and 37°C for GST-E1 deletion mutants which span the cutoff points observed at 4°C (Fig. 3B to D). For the N-terminal E1 deletion series, exactly the same cutoff point in E2 binding was observed at all temperatures. A small change in the cutoff point was seen in the C-terminal series; while only a slight trace of E1Δ66C binding was seen at 4°C, this increased at 20 and 37°C. Therefore, the region of E1 essential for efficient E2 binding is slightly altered at 20 or 37°C (aa 432 to 583) compared to 4°C (aa 432 to 617).
Confirmation of the E2-binding domain of E1 by a reciprocal association assay.
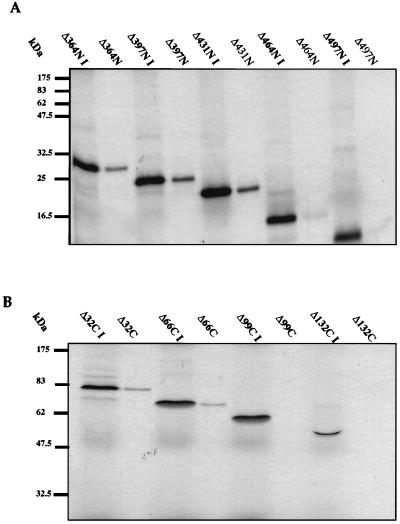
To confirm that the E1 mapping data were not affected by the use of GST fusions, we carried out reciprocal assays in which IVT E1 proteins were tested for binding to GST-E2. In these assays we used a fixed amount of GST-E2 and fixed volumes of the IVT E1 protein solutions. Six E1 deletion mutants, spanning the cutoff points for E2 binding (E1Δ364N, E1Δ397N, E1Δ431N, E1Δ464N, E1Δ497N, E1Δ32C, E1Δ66C, E1Δ99C, and E1Δ132C), were tested at 20°C by comparing the amount of truncated IVT E1 protein bound to GST-E2 beads to the total input protein. This experiment confirmed the previous result, in that no binding was observed when N-terminal deletions of >431 aa or C-terminal deletions of >66 aa were made while partial binding was seen on removal of 66 C-terminal residues (Fig. 4).
FIG. 4.
Deletion analysis of IVT E1 binding to GST-E2. In vitro association of bead-bound GST-E2 to radiolabelled IVT E1 N- and C-terminal deletion mutants was assayed as described in Materials and Methods. Autoradiographs show E1 protein that bound to and then was eluted from the beads. Input IVT protein is denoted I. (A) Binding of selected IVT E1 N-terminal deletion mutants to GST-E2 at 20°C. (B) Binding of selected IVT E1 C-terminal deletion mutants to GST-E2 at 20°C. This reciprocal binding experiment confirms the deletion analysis data obtained with IVT E2 and GST-E1 at 20°C. The positions of molecular mass markers are shown to the left of each gel.
A minimal fragment of E1 is sufficient for E2 binding.
We next expressed the aa 432 to 583 region of E1 both as a GST fusion and as an IVT protein to test its ability to bind E2. The DNA encoding this region of E1 was cloned into pGEX2T for expression as a GST fusion protein and into pING14.1 for in vitro transcription and translation. Although this fragment was stable as either a GST fusion or an IVT polypeptide, no binding was observed in either assay at 4 or 20°C (data not shown). Thus, the smallest fragment of E1 necessary and sufficient for binding of E2 is the region consisting of aa 432 to 649, i.e., the C-terminal 217 residues of E1 (Fig. 4A), suggesting that it may correspond to a stable structural domain of E1. Any C-terminal deletions may adversely affect the stability of this region of E1, and the results presented above suggest that this is modulated by temperature.
E1 binding to the HPV-16 ori in the presence or absence of E2.
We next examined the ability of HPV-16 E1 to bind to the viral ori in the presence or absence of E2. This was measured by a version of the McKay precipitation assay recently described by Berg and Stenlund (2). GST-E1 protein was incubated with two labelled oligonucleotides corresponding to native HPV-16 ori sequence, one containing only the E1BS and the other containing the E1BS and three E2BSs (in their native configuration), in the presence or absence of purified MBP-E2 fusion protein (Fig. 5A). GST-E1 (and complexes containing GST-E1) was recovered by using glutathione-agarose beads, and the bound oligonucleotides were analyzed by gel electrophoresis. Thus, oligonucleotides bound directly or indirectly to GST-E1 should be detected. As expected, no oligonucleotide was recovered with MBP-E2 alone. In the presence of GST-E1 alone, we saw no recovery of either oligonucleotide (confirmed over a range of GST-E1 concentrations [data not shown]), indicating that HPV-16 E1 does not detectably bind the E1BS in this assay. Addition of MBP-E2 resulted in a good recovery of the larger oligonucleotide but not of the shorter oligonucleotide (which lacks the E2BS), indicating that under these assay conditions HPV-16 E1 ori binding occurs entirely through E2 and the E2BS.
FIG. 5.
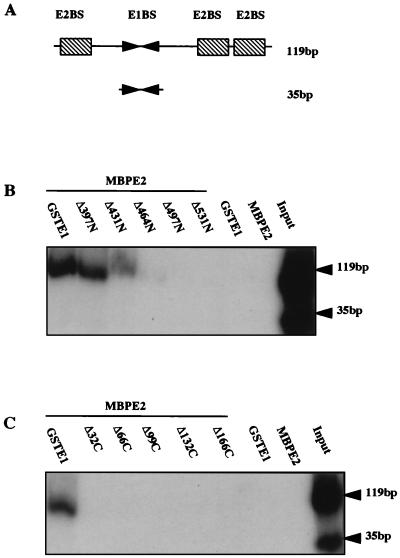
Binding of E1 and E2 to the HPV-16 ori. McKay precipitation assays were performed as described in Materials and Methods. MBP-E2, in the presence of GST-E1 or GST-E1 N- or C-terminal deletion mutants, was incubated with two ori probes. Following incubation with glutathione-agarose beads and washes, bead-bound probes were recovered and analyzed by SDS-PAGE. (A) Schematic representation of HPV ori probes, the larger (119 bp) containing the entire ORI-binding E1BS and E2BS and the smaller (35 bp) containing only the E1BS. (B) Binding of selected GST-E1 N-terminal deletion mutants to the ori probes in the presence of MBP-E2. (C) Binding of selected GST-E1 C-terminal deletion mutants to the ori probes in the presence of MBP-E2. Input lanes show the ratios of the two probes.
It has been reported that recombinant BPV-1 E1 may form DNA complexes which, unless purified by a lengthy process, have reduced ori-binding activity (45, 46). In contrast, others have used BPV-1 GST-E1 purified in the standard manner (33), or even E1 in crude bacterial extracts, and readily detected E1-ori binding (27). We purified GST-E1 by glutathione-agarose chromatography but included a 1 M NaCl wash step to remove contaminating DNA. This type of preparation, for BPV-1 GST-E1, has been shown to be essentially free of contaminating DNA and active both in DNA binding assays and DNA replication assays (5, 33, 43). Using the highly sensitive PicoGreen assay, we detected very low levels of DNA contamination in our GST-E1 purified by standard glutathione-agarose chromatography, whether or not a 1 M NaCl wash step was included (4.12 ng/μg of E1 protein and 1.3 ng/μg of E1 protein, respectively). Indeed, both GST-E1 preparations failed to bind HPV-16 ori oligonucleotide in the McKay precipitation assay, suggesting that contaminating DNA was not the reason for the negative result.
To determine the regions of E1 required for formation of this E1-E2-ori complex, we assayed the N- and C-terminal GST-E1 deletions. The results show that N-terminal deletions beyond E1Δ431N and all C-terminal deletions prevent binding (Fig. 5B and C). This maps the region involved to aa 432 to 649, compared to aa 432 to 583 or 617 in the E1-E2 association assay. However, the region is identical to that found to be sufficient to bind E2 (see above), supporting the suggestion that any C-terminal deletions of E1 can reduce stability, and this may depend on the precise assay conditions.
These experiments suggest that direct binding of HPV-16 E1 to the E1BS is at best very weak and that E2 therefore has a major role in the initial targeting of E1 to the ori. Deletion analysis suggests that this initial targeting could occur purely via the C-terminal domain of E1, which is sufficient for the formation of a stable complex with E2 bound to ori DNA. However, it is unlikely that a productive initiation complex can be formed without subsequent direct E1-DNA interactions, which probably involve additional regions of E1 (see Discussion). This analysis does not address the precise nature of the E1-E2-ori complex formed or whether it differs if full-length E1 is used instead of just the C-terminal domain.
E1 binds the DNA polymerase α p68 subunit.
To determine which, if any, of the four subunits of human polα-primase interact with HPV-16 E1, we assayed the binding of each IVT subunit (p180, p68, p58, and p48) to GST-E1 attached to agarose beads, using beads to which GST alone was attached as a control for nonspecific binding.
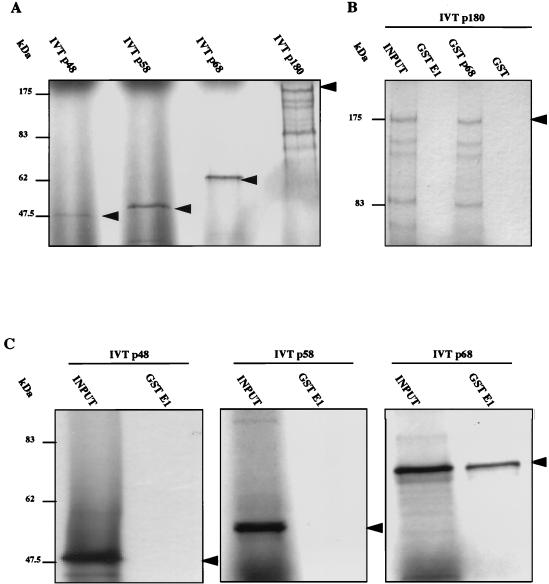
Specific binding of GST-E1 to p68 was detected, while no binding to p180, p58, or p48 was seen under the same assay conditions (Fig. 6). Since a previous study reported an interaction between BPV-1 E1 and p180 (41), we investigated the expected association between p180 and p68; this was easily detected, suggesting that IVT p180 is active and that the lack of binding to GST-E1 is real (Fig. 6B).
FIG. 6.
Investigation of polα-primase subunit binding to E1. In vitro association of bead-bound GST-E1 to radiolabelled p180, p68, p58, or p48 subunits was assayed as described previously. (A) In vitro translation products of each polα-primase subunit. (B) Binding of p180 to full-length GST-E1, GST-p68, and GST, demonstrating clear interaction with p68 but no interaction with GST-E1 above background levels. (C) Binding of p48, p58, and p68 to full-length GST-E1, revealing strong interaction with only the p68 subunit. The positions of molecular mass markers are indicated. The arrowheads indicate full-length IVT protein products.
In analogous binding assays, IVT topoisomerase I showed no detectable association with GST-E1 (data not shown).
Mapping of the minimal E1 domain necessary for interaction with p68.
The panel of 36 N- and C-terminal E1 deletion mutants was used to map regions of HPV-16 E1 necessary for association with p68. Deletions of >397 aa from the N terminus or >66 aa from the C terminus of E1 resulted in the abolition of p68 binding (Fig. 7A). Thus, aa 397 to 583 are essential for interaction with the p68 subunit. This 186-aa region almost completely matches that involved in the interaction with E2 (aa 432 to 583). However, an additional 34 aa (residues 397 to 431) at the N terminus are necessary for a strong association with p68.
FIG. 7.
(A) Binding of p68 to GST-E1 and selected N- and C-terminal deletion mutants, demonstrating cutoff points for binding in each deletion series. (B and C) Reciprocal assays of binding of GST-p68 to IVT E1 N-terminal (B) and C-terminal (C) deletion mutants. The positions of molecular mass markers are indicated.
We confirmed the mapping of the p68-binding domain by using a reciprocal binding assay (binding of IVT E1 deletion proteins to GST-p68 [Fig. 7B and C]).
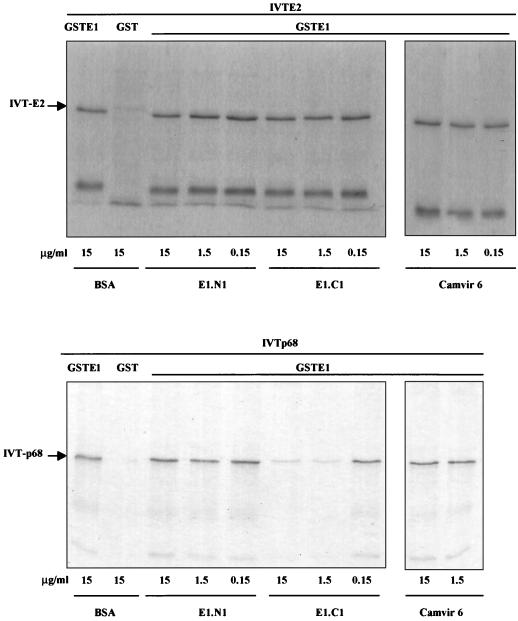
Effects of E1 MAbs on E1-E2 and E1-p68 binding.
We tested the effects of two anti-E1 MAbs on the E1-E2 and E1-p68 associations: E1-N1, which maps to aa 50 to 64, and E1-C1, which maps to aa 364 to 383 (see Materials and Methods). We found that at antibody concentrations of up to 15 μg/ml, E1-N1 had no effect on the association of GST-E1 with E2 or p68 while E1-C1 specifically inhibited binding of p68, but not E2, in a dose-dependent fashion (Fig. 8). The E1-C1 epitope lies adjacent to the N-terminal junction of the p68-binding region mapped by deletion analysis (aa 397), and therefore the antibody must sterically hinder p68 binding. This result confirms the differences in the p68- and E2-binding regions determined by deletion analysis in the context of full-length E1 and provides further support for the specificity of the E1-p68 interaction.
FIG. 8.
Antibody analysis of E1-E2 and E1-p68 interaction. The cross-blocking effect of two anti-E1 antibodies (E1-N1 and E1-C1) on the association of bead-bound GST-E1 with IVT E2 (A) and IVT p68 (B) was assayed as described in Materials and Methods. Antibodies were tested over a threefold range of concentrations. E1-N1 and Camvir 6 (anti-HPV 16L1) did not affect interaction of E1 with either E2 or p68, while E1-C1 significantly reduced only E1-p68 association.
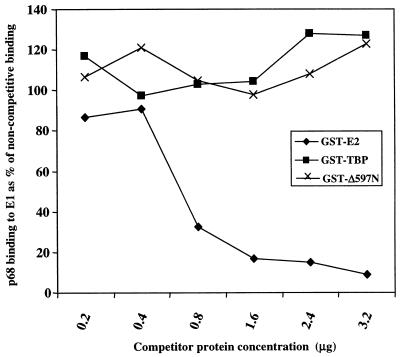
E2 and p68 compete for binding to E1.
Having demonstrated the binding of E2 and p68 to an almost identical domain of E1, assays were performed to establish whether the two proteins competed for binding to E1. In this assay, E1-N1 antibody bound to protein G-Sepharose was used to immunoprecipitate GST-E1–p68 complexes. Titration of GST-E1 and IVT p68 in the assay established the minimum quantity of each protein necessary to visualize binding above background levels, and these amounts were used in subsequent assays (data not shown). Competition was determined by coincubation with increasing amounts of purified GST-E2. The results (Fig. 9) show that GST-E2 effectively inhibits binding of p68 to GST-E1 in a dose-dependent fashion. The specificity of the competition was confirmed by the inability of two unrelated GST fusion proteins (Fig. 9) to block the GST-E1–p68 binding. Further controls confirmed that (i) GST-E2 does not bind IVT p68 and (ii) neither of the control GST fusion proteins was able to bind IVT p68, GST-E1, or E1-N1–protein G-Sepharose (data not shown).
FIG. 9.
Competitive binding of E2 and p68 to E1. Competition between IVT p68 and GST-E2 for binding to GST-E1 was assayed as described in Materials and Methods. The results demonstrate that GST-E2 and p68 compete for E1 binding with p68 in a dose-dependent manner, compared to GST-TBP and GST-E1Δ597N control proteins. Autoradiograms were scanned densitometrically to quantify the data.
A structural model for E1: the helicase domain.
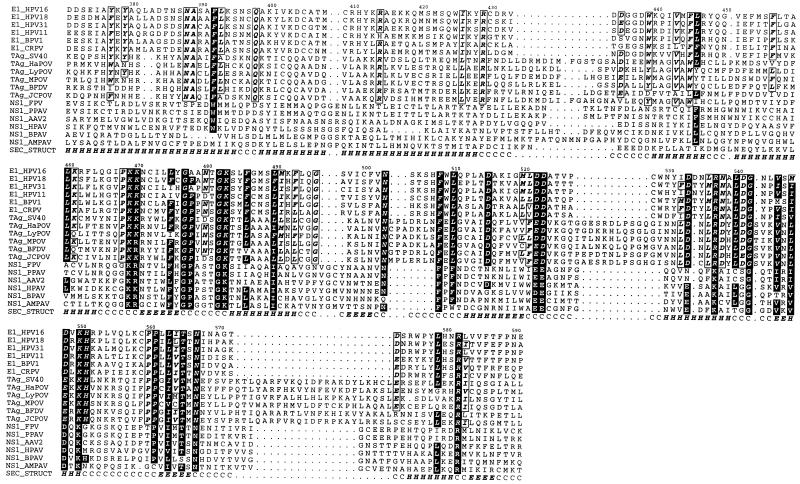
Our mapping data appeared to identify the presence of a discrete domain in a region of E1 implicated in helicase activity. We therefore examined predicted structural features and sequence homology in this region more closely. Secondary-structure predictions (Fig. 10) for the C-terminal helicase homology region (aa 371 to 623) of HPV-16 E1 and those of related helicases suggest that there are two large α-helices in the first 58 aa (aa 371 to 428), previously described as region A (10), followed by a short unstructured region and an α/β structure in the remainder of the region (aa 439 to 623). A number of observations from sequence analysis suggest that aa 439 to 623 constitute a discrete physical domain. First, the region A α-helical stretch of E1 shows similarities to the T antigens but not to parvovirus or human herpesvirus 6 NS-1 helicases, while the α/β region has similarities to both groups of helicases (Fig. 10). Second, protein domains are generally found to be predominantly either α, β, or α/β (40), suggesting that the α-helical region A may be a domain separate from the α/β region. Third, the unstructured region between region A and the α/β region is extremely variable in sequence and size in different SF3 helicases, suggesting that it may be a linker region between structural domains.
FIG. 10.
Multiple-sequence alignment of the C-terminal regions (HPV-16 aa 371 to 591) of a number of SF3 helicases. E1, E1 protein; TAg, large T antigen; NS1, NS-1 protein; CRPV, cottontail rabbit PV; HaPOV, hamster polyomavirus; LyPOV, lymphotropic polyomavirus; MPOV, mouse polyomavirus; BFDV, budgerigar fledgling disease virus; JCPOV, polyomavirus JC; FPV, feline panleukopenia virus; PPAV, porcine parvovirus; AAV2, adeno-associated virus type 2; HPAV, human parvovirus; BPAV, bovine parvovirus; AMPAV, Aleutian mink disease parvovirus. Residues conserved across the complete alignment are displayed as white boldfaced letters on a black background, while residues conserved across E1 and T antigen sequences are boxed, italicized, and in boldface. A secondary-structure prediction for HPV-16 E1 is labelled SEC-STRUCT in the alignment. The prediction is a consensus of the predictions from three methods, DSC (23), PHDsec (42), and ZPRED (69). H, alpha helix; E, beta strand; C, other (no regular secondary structure).
The recent publication of the X-ray crystallographic structures of the E. coli Rep helicase (24) and two SF2 helicases, hepatitis C virus RNA helicase (65) and the Bacillus stearothermophilus PcrA DNA helicase (52), has revealed similarities in their folds which suggest a unifying structure for all helicases (3). Moreover, the ATP-binding domain of PcrA shows structural similarities to the crystal structure of RecA (52), a homolog of hexameric helicases such as SV40 T antigen and the NS-1 proteins (68), which are related to E1. These observations, along with our identification of a helicase domain by deletion analysis, provide the rationale for the modelling of the E1 helicase domain based on the structures of the hepatitis C virus and PcrA helicases.
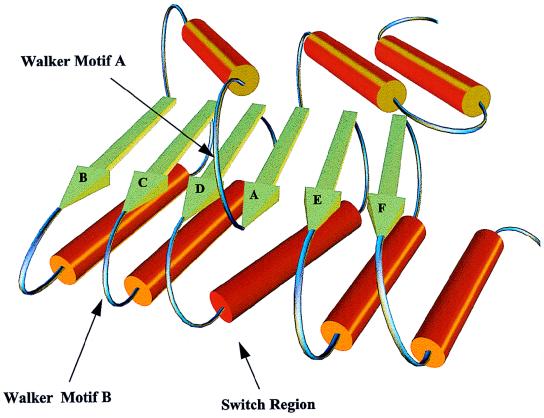
The helicase nucleoside triphosphate (NTP)-binding domains are both three-layer (αβα) sandwiches which appear to share a common β-sheet core with β-strands ordered nBCDA, where n is a variable number of strands following strand B (one in PcrA and two in the hepatitis C virus helicase). The Walker motif A (59), which binds the terminal NTP phosphate, the Walker motif B, containing the Asp-Glu responsible for chelating Mg2+ in the Mg-NTP complex, and the switch region responsible for transmission of conformational changes following NTP hydrolysis (67) are located on the loops following strands A, C, and D, respectively. In the SF3 helicases, the Walker motif A is found after the first predicted β-strand of the C-terminal region and an Asp/Glu-Asp/Glu pair, which is conserved across this superfamily and is thus most likely to be involved in Mg2+ chelation, follows the third strand. Assuming a common fold for the NTP-binding domains of SF2 and SF3 helicases, a topological model for the C-terminal domain of E1 can be postulated (Fig. 11). Note that, as is conventional for models of this type, it is drawn to demonstrate the ordering and general topological relationship between secondary-structural elements (i.e., β-strands and α-helices); thus, the secondary-structure elements are not drawn to scale and are not in their exact proper orientations but instead are drawn to make the figure easy to interpret.
FIG. 11.
A simplified model for the NTP-binding domain of E1 based on the X-ray crystallographic structures of the hepatitis C virus and B. stearothermophilus helicases. Note that this model is not to scale but is intended to display the order and relative positioning of structural elements of the domain.
The strands are ordered BCDAEF, and therefore the variable strand(s) n from the structural core proposed above is absent. An h-Asp–Glu-X-Arg motif (where h is any hydrophobic amino acid), conserved across the SF3 helicase superfamily, is present in the region proposed to be the switch region. As in the PcrA helicase structure, the first β-strand is preceded by two short α-helices, in agreement with our N-terminal boundary prediction.
In conclusion, the E1 helicase domain boundaries predicted from sequence analysis agree closely with our fine deletion mapping analysis of the E2-binding domain, and these boundaries are then consistent with a hypothetical structural model for the domain which takes into account conserved features in helicase domains of known structure. The p68-binding region requires additional residues N terminal to this domain, from the predicted α-helical region A, which is only conserved in the T antigens.
DISCUSSION
Our results of studies using N- and C-terminal deletion mutants revealed that a C-terminal domain of HPV-16 E1, encompassing aa 432 to 583 or 617, is necessary for interaction with E2 (for simplicity we refer only to the outer boundaries; see Results). The smallest fragment of E1 that was able to bind E2 was aa 432 to 649, and therefore this region appears to be independently stable. In contrast to previous studies with BPV-1 and HPV-31b (1, 15), we detected no interaction through the N terminus of E1. Our results are consistent with those for BPV-1 (31), HPV-33 (39), and HPV-16 (19, 66) suggesting that C-terminal regions of E1 are essential for binding E2. However, Yasugi et al. (66) reported that a much larger region (aa 144 to 649) of HPV-16 E1 was required for E2 binding at physiological temperatures.
It is possible that some of the published differences reflect true variations among the different PVs. Two recent studies have confirmed an interaction between E2 and an N-terminal as well as a C-terminal region of BPV-1 E1 (2, 7). Berg and Stenlund (2) reported that the interaction with the N-terminal region of BPV-1 E1 is conditional and is necessary only in the context of an ori in which the E2BS is proximal to the E1BS, a situation that does not occur in HPV oris. In contrast, two previous studies with HPV-16 (19, 66) identified interactions only in C-terminal regions of E1, consistent with our study. Moreover, a comparable analysis of E1-E2 protein interactions in the yeast two-hybrid system identified interactions with N-terminal sequences for BPV-1 but with C-terminal sequences for HPV-16 (1, 66), supporting the view that the viruses differ.
Some differences might be due to effects on E1 or E2 phosphorylation; the proteins we have used are unphosphorylated, whereas the baculovirus-derived E1 and E2 used in several studies are phosphorylated. However, it would be surprising if phosphorylation affected which domains of E1 interact with E2. There is evidence that the mapping data may be affected by temperature and the type of assay used, but the picture is inconsistent. Thorner et al. (55) found that with BPV-1 the interaction was mediated by E1 aa 458 to 605 at 4°C but saw a shift to N-terminal regions at elevated temperatures. Muller and Sapp (39) showed that the C-terminal portion of HPV-33 E1 (aa 312 to 644) was sufficient for E2 binding at 4°C whereas aa 312 to 450 were sufficient at 22°C. Yasugi et al. (66) identified a C-terminal region of HPV-16 E1 (aa 421 to 649) at 4°C that increased to a larger domain (aa 144 to 649) at physiological temperatures. These results cannot be explained easily by the involvement of N-terminal regions of E1 that are known to mediate ori binding and E1-E2-ori complex formation (44), since they were all observed in the absence of ori DNA. However, HPV E1 can show a high level of nonspecific DNA binding (29), and it is conceivable that this can mediate artifactual protein interactions with other DNA-binding proteins via contaminating DNA.
In contrast, we found no changes with temperature except for a small decrease in the size of the region mapped by deletion analysis at 20 and 37°C (aa 432 to 583) compared to 4°C (aa 432 to 617). It is possible that at least some of the differences reported previously can be explained by the effect of specific deletions on domain stability, possibly combined with the effect of temperature. Previous structural predictions identified a clear interdomain linker in the aa 120 to 170 region of E1 and suggested the existence of a very large C-terminal domain (21, 44). Our present results show that a smaller C-terminal region behaves as a structural domain, but it is possible that this is not stable independently under all conditions, and this might explain the results of Yasugi et al. (66), who concluded that at physiological temperatures the E2 interaction required E1 aa 144 to 649.
The collective data for BPV-1 and various HPV subtypes suggest a model for ori recognition in which three interactions—E1-E2, E1-ori, and E2-ori—are involved cooperatively in E1-E2-ori ternary complex formation (reviewed in reference 50). Our results suggest that for HPV-16, E2 is of primary importance for initial ori recognition, since we could not detect E1-ori binding in the absence of E2. In addition, we found that the E2-binding domain of E1 could be sufficient to target E1 to the ori in our assays. Of course these assays do not address the precise molecular interactions that occur within the complexes, and it is likely that direct E1-DNA interaction, involving other regions of E1, would be required for a productive initiation complex. This is not a totally surprising result, since the E1-ori interactions for HPV-11 and -18 also appear to be very weak and, indeed, a duplicated E2BS alone (with no E1BS) can serve as an ori in the transient replication of either viral DNA (30, 54). Nevertheless, since the E1BS is conserved in the ori of each of these viruses, it is likely that its direct interaction with E1 has an important role in the complex, perhaps in enhancing specificity or in progression to a productive initiation complex. In contrast to that of these viruses, replication of HPV-1a can occur efficiently with E1 protein alone, suggesting that in this case E1 has a much greater role in initial ori binding (17). In BPV-1, all of the interactions are important in ori recognition; Sarafi and McBride (44) demonstrated the requirement of both a C-terminal E2-binding domain of E1 (aa 200 to 605) and additional N-terminal residues, involved in ori binding (aa 162 to 205), for efficient E1-E2-ori complex formation.
Recruitment of polα-primase to the ori is critical since this is the only polymerase able to initiate new DNA chains. It is thought to initiate DNA synthesis on the leading strand as well as priming each Okazaki fragment on the lagging strand. The core enzyme consists of two subunits, p180 and p68; p180 contains the catalytic activity, while the function of p68 is unclear (reviewed in reference 60). The primase function resides in the p58 and p48 subunits; p48 has primase activity, while p58 may mediate or stabilize the interaction with p180. We investigated the ability of HPV-16 E1 to associate with each subunit and detected a strong association only with p68, suggesting that this subunit has the function of bringing the polα-primase complex to the HPV ori. We found a C-terminal region of E1 (aa 397 to 583), closely matching the E2-binding domain, to be necessary for the interaction. Previous studies have demonstrated a physical interaction between p180 and SV40 T antigen (13), herpes simplex virus UL9 (26), and BPV-1 E1 (41). SV40 T antigen has also been shown to bind strongly to p68 (11). Bonne-Andrea et al. (5) demonstrated an interaction between BPV-1 E1 and polα-primase but did not identify the interacting subunit. We found no detectable interaction between HPV-16 E1 and p180. We cannot readily explain the difference between our results and those of Park et al. (41), who reported binding of BPV-1 E1 to p180, other than that they suggest a true difference between the viruses or perhaps a difference in the phosphorylation state of the E1 protein (since the BPV-1 E1 protein was produced from insect cells while our protein was E. coli derived). Since p180 and E1 both bind DNA nonspecifically, it is also conceivable that a nonspecific interaction arose from contaminating DNA in the assay.
There are notable differences in the interactions of E2 and p68 with E1. First, E1-E2 binding is highly salt resistant (>2 M NaCl) (data not shown), suggesting a largely hydrophobic interaction, while E1-p68 binding is disrupted by salt washes consisting of 0.5 M NaCl (data not shown). Second, we have shown that a monoclonal antibody, E1-C1, that maps to a site (aa 364 to 383) within region A and adjacent to the N-terminal junction of the p68-binding region mapped by deletion analysis (aa 397) blocks p68 binding but not E2 binding. This result supports our deletion mapping data, which show that E2 binding requires aa 432 to 583 while p68 binding requires additional N-terminal residues (i.e., aa 397 to 583 were required). It also provides further confirmation of the specificity of the E1-p68 interaction. Since the E1-p68 interaction is likely to be essential in viral replication, we would expect the antibody to inhibit in vitro replication of HPV-16 DNA.
Despite these differences between the binding of E2 and p68 to E1, we have found that E2 is able to block E1-p68 binding. This suggests that E1-E2-p68 complexes may not be able to form, although the multimerization of E1 may complicate this interpretation. Nevertheless, it is possible to hypothesize that during replication a switch from a ternary E1-E2-ori complex to a productive initiation complex occurs in which p68 (or the polα-primase complex) replaces E2. This would be consistent with the results of Lusky et al. (32), who identified two classes of BPV-1 E1-ori complexes formed in the presence of E2. Initially, and at high E2/E1 ratios, a complex containing both E1 and E2 (c1) is formed. Subsequently, in the presence of ATP, a much larger, replication-competent complex (c2) which contains E1 (probably in a hexameric form) but lacks E2 is formed. Thus, E2 appears to assist in the loading of E1 on the ori but is itself absent from the preinitiation complex. In the elongation stage, E1 most likely remains with the replication forks, unwinding the template through its helicase activity. polα-primase is required for lagging-strand synthesis at each fork and presumably remains associated with E1. Liu et al. (29) have shown that the elongation stage of HPV-11 in vitro DNA replication requires E1 but not E2. During elongation, E1 was protected against inhibition by a polyclonal antiserum against its C-terminal portion, suggesting that this region is buried, either by multimerization or by complexing to host replication factors.
Both the sequence alignments between the SF3 helicases and the secondary-structure predictions suggest that the aa 439 to 623 region of HPV-16 E1 corresponds to a discrete helicase domain. This region closely matches the E2-binding domain defined by deletion analysis and includes residues previously shown to be required for ATPase and helicase activities (9). The ∼58 preceding residues show similarity to the T antigens, but not to other SF3 helicases, and are predicted to form two large α-helices (Fig. 10). It is noteworthy that this region contains residues (aa 397 to 432) required for p68 binding but not E2 binding, possibly suggesting a similar role in SV40 T antigen. The striking agreement among our mapping data, the sequence alignments, and the secondary-structure predictions supports the view that we have identified a discrete domain and is illustrated graphically in Fig. 12.
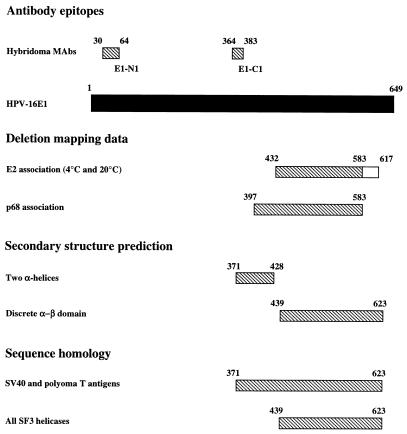
FIG. 12.
Domains of E1 identified by deletion mapping and sequence analysis. Included are the following: epitopes of two novel HPV-16 E1 MAbs mapped by using the deletion series, deletion mapping data for E2 and p68 interaction sites (the larger E2 region is required at 4°C), secondary-structure prediction made by using multiple alignments as input, and sequence homology. Note that the deletion mapping coordinates given refer only to the outer boundaries for simplicity, so the true domains could be several residues shorter at either end. Note the close correspondence of deletion mapping and sequence analysis data, suggesting that the core helicase–E2-binding domain is aa 432 to 583 or 617 whereas an additional region (aa 397 to 432; equivalent to region A, conserved for papovavirus helicases only) is required for p68 binding.
Recent observations suggest that diverse helicases may have a common structural fold (3). We have therefore postulated a novel topological model for the E1 helicase domain (Fig. 11), using the precise domain boundaries that we have identified and the recently available structures of the SF2 helicases from hepatitis C virus and B. stearothermophilus (three-layer αβα structures). The approach of prediction by fold recognition, or threading, has proven increasingly successful in recent years, even when the level of sequence similarity is low (34).
In summary, the C-terminal domain of E1 is multifunctional, having helicase activity and interacting with E2 and polα-primase (via p68). It is therefore crucial in ori recognition, formation of the initiation complex, and DNA replication during both the initiation and the elongation phases. In addition, we have obtained preliminary evidence (unpublished) that the same domain is involved in E1 multimerization. We have already attempted to dissect the functions of the domain by generating mutations that substitute conserved charged residues for Ala but have failed to identify any that had more than two- to threefold-selective effects on E2 versus p68 binding (data not shown). Although speculative, the structural model may help direct further functional analysis of this region of E1 by point-mutational and antibody studies.
ACKNOWLEDGMENTS
We thank Tom Kelly for providing the plasmid pGEX2Tp68, Teresa Wang for AcHDPα baculovirus stock containing the human p180 cDNA, and Heinz-Peter Nasheuer for pBluescriptKS p48 and p58 cDNA clones. We also thank Bill Phelps, Pia Thommes, and Lawrence Banks for helpful discussions.
P.J.M. was funded by the Medical Research Council (CS94/12).
REFERENCES
- 1.Benson J D, Howley P M. Amino-terminal domains of the bovine papillomavirus type 1 E1 and E2 proteins participate in complex formation. J Virol. 1995;69:4364–4372. doi: 10.1128/jvi.69.7.4364-4372.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird L E, Subramanya H S, Wigley D B. Helicases: a unifying structural theme? Curr Opin Struct Biol. 1998;8:14–19. doi: 10.1016/s0959-440x(98)80004-3. [DOI] [PubMed] [Google Scholar]
- 4.Blitz I L, Laimins L A. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J Virol. 1991;65:649–656. doi: 10.1128/jvi.65.2.649-656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase α but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bream G L, Ohmstede C-A, Phelps W C. Characterization of human papillomavirus type 11 E1 and E2 proteins expressed in insect cells. J Virol. 1993;67:2655–2663. doi: 10.1128/jvi.67.5.2655-2663.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Stenlund A. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J Virol. 1998;72:2567–2576. doi: 10.1128/jvi.72.4.2567-2576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow L T, Broker T R. Papillomavirus DNA replication. Intervirology. 1994;37:150–158. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 10.Clertant P, Seif I. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature. 1984;311:276–279. doi: 10.1038/311276a0. [DOI] [PubMed] [Google Scholar]
- 11.Collins K L, Russo A A, Tseng B Y, Kelly T J. The role of the 70 kDa subunit of human DNA polymerase alpha in DNA replication. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copeland W C, Wang T S. Catalytic subunit of human DNA polymerase alpha overproduced from baculovirus-infected insect cells. Structural and enzymological characterization. J Biol Chem. 1991;266:22739–22748. [PubMed] [Google Scholar]
- 13.Dornreiter I, Copeland W C, Wang T S-F. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dornreiter I, Hoss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frattini M G, Laimins L A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 16.Galfre G, Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 1981;73B:3–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- 17.Gopalakrishnan V, Khan S A. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc Natl Acad Sci USA. 1994;91:9597–9601. doi: 10.1073/pnas.91.20.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 19.Hibma M H, Raj K, Ely S J, Stanley M, Crawford L. The interaction between human papillomavirus type 16 E1 and E2 proteins is blocked by an antibody to the N-terminal region of E2. Eur J Biochem. 1995;229:517–525. doi: 10.1111/j.1432-1033.1995.0517k.x. [DOI] [PubMed] [Google Scholar]
- 20.Holt S E, Schuller G, Wilson V G. DNA binding specificity of the bovine papillomavirus E1 protein is determined by sequences contained within an 18-base-pair inverted repeat element at the origin of replication. J Virol. 1994;68:1094–1102. doi: 10.1128/jvi.68.2.1094-1102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes F J, Romanos M A. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 1993;21:5817–5823. doi: 10.1093/nar/21.25.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins O, Earnshaw D, Sarginson G, Del-Vecchio A, Tsai J, Kallender H, Amegadzie B, Browne M. Characterization of the helicase and ATPase activity of human papillomavirus type 6b E1 protein. J Gen Virol. 1996;77:1805–1809. doi: 10.1099/0022-1317-77-8-1805. [DOI] [PubMed] [Google Scholar]
- 23.King, R. D., M. A. S. Saqi, R. A. Sayle, and M. J. E. Sternberg. DSC—public domain secondary structure prediction. CABIOS, in press. [DOI] [PubMed]
- 24.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waskman G. Major domain swivelling revealed by the crystal structure of complexes of E. coli Rep helicase bound to single-strand DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 25.Kuo S R, Liu J S, Broker T R, Chow L T. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J Biol Chem. 1994;269:24058–24065. [PubMed] [Google Scholar]
- 26.Lee S S, Dong Q, Wang T S, Lehman I R. Interaction of herpes simplex virus 1 origin-binding protein with DNA polymerase alpha. Proc Natl Acad Sci USA. 1995;92:7882–7886. doi: 10.1073/pnas.92.17.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leng X, Ludes-Meyers J H, Wilson V G. Isolation of an amino-terminal region of bovine papillomavirus type 1 E1 protein that retains origin binding and E2 interaction capacity. J Virol. 1997;71:848–852. doi: 10.1128/jvi.71.1.848-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Yang L, Fouts E, Botchan M R. Site-specific DNA-binding proteins important for replication and transcription have multiple activities. Cold Spring Harbor Symp Quant Biol. 1993;58:403–413. doi: 10.1101/sqb.1993.058.01.047. [DOI] [PubMed] [Google Scholar]
- 29.Liu J S, Kuo S R, Broker T R, Chow L T. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell free replication. J Biol Chem. 1995;270:27283–27291. doi: 10.1074/jbc.270.45.27283. [DOI] [PubMed] [Google Scholar]
- 30.Lu J Z-J, Sun Y-N, Rose R C, Bonnez W, McCance D J. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J Virol. 1993;67:7131–7139. doi: 10.1128/jvi.67.12.7131-7139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lusky M, Fontane E. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequences within the carboxyl terminus of E1. Proc Natl Acad Sci USA. 1991;88:6363–6367. doi: 10.1073/pnas.88.14.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lusky M, Hurwitz J, Seo Y S. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc Natl Acad Sci USA. 1994;91:8895–8899. doi: 10.1073/pnas.91.19.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansky K C, Batiza A, Lambert P F. Bovine papillomavirus type 1 E1 and simian virus 40 large T antigen share regions of sequence similarity required for multiple functions. J Virol. 1997;71:7600–7608. doi: 10.1128/jvi.71.10.7600-7608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A, Bryant S H. A measure of success in fold recognition. Trends Biochem Sci. 1997;22:236–240. doi: 10.1016/s0968-0004(97)01078-5. [DOI] [PubMed] [Google Scholar]
- 35.McKay R D G. An immuno assay for the interaction between an SV40 T antigen related protein and DNA. J Mol Biol. 1981;145:471–482. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- 36.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 38.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 39.Muller F, Sapp M. Domains of the E1 protein of human papillomavirus type 33 involved in binding to the E2 protein. Virology. 1996;219:247–256. doi: 10.1006/viro.1996.0242. [DOI] [PubMed] [Google Scholar]
- 40.Murzin A G, Brenner S E, Hubbard T, Chothia C. Scop: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 41.Park P, Copeland W, Yang L, Wang T, Botchan M R, Mohr I J. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci USA. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Raj K. Ph.D. thesis. Cambridge, United Kingdom: University of Cambridge; 1994. [Google Scholar]
- 42.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santucci S, Bonne-Andrea C, Clertant P. Bovine papillomavirus type 1 E1 ATPase activity does not depend on binding to DNA nor to viral E2. J Gen Virol. 1995;76:1129–1140. doi: 10.1099/0022-1317-76-5-1129. [DOI] [PubMed] [Google Scholar]
- 44.Sarafi T R, McBride A A. Domains of the BPV-1 E1 replication protein required for origin-specific DNA binding and interaction with the E2 transactivator. Virology. 1995;211:385–396. doi: 10.1006/viro.1995.1421. [DOI] [PubMed] [Google Scholar]
- 45.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo Y S, Muller F, Lusky M, Hurwitz J. Bovine papillomavirus (BPV) encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmons D T, Melendy T, Usher D, Stillman B. Simian virus 40 large T antigen binds to topoisomerase I. Virology. 1996;222:365–374. doi: 10.1006/viro.1996.0433. [DOI] [PubMed] [Google Scholar]
- 48.Singer V L, Jones L J, Yue S T, Haugland R P. Characterisation of PicoGreen reagent and development of a fluorescence-based solution assay for double stranded DNA quantitation. Anal Biochem. 1997;249:228–238. doi: 10.1006/abio.1997.2177. [DOI] [PubMed] [Google Scholar]
- 49.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenlund A. Papillomavirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 679–698. [Google Scholar]
- 51.Storey A, Piccini A, Massimi P, Bouvard V, Banks L. Mutations in the human papillomavirus type 16 E2 protein identify a region of the protein involved in binding to E1 protein. J Gen Virol. 1995;76:819–826. doi: 10.1099/0022-1317-76-4-819. [DOI] [PubMed] [Google Scholar]
- 52.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Crystal structure of a DEXX box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 53.Sun S, Thorner L, Lentz M, MacPherson P, Botchan M. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J Virol. 1990;64:5093–5105. doi: 10.1128/jvi.64.10.5093-5105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sverdrup F, Khan S A. Two E2 binding sites alone are sufficient to function as the minimal origin of replication of human papillomavirus type 18 DNA. J Virol. 1995;69:1319–1323. doi: 10.1128/jvi.69.2.1319-1323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorner L K, Lim D A, Botchan M R. DNA-binding domain of bovine papillomavirus type 1 E1 helicase: structural and functional aspects. J Virol. 1993;67:6000–6014. doi: 10.1128/jvi.67.10.6000-6014.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 58.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 59.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthetase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang T S F. Cellular DNA polymerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 461–494. [Google Scholar]
- 61.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 63.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M R. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang L, Mohr I, Li R, Nottoli T, Sun S, Botchan M. Transcription factor E2 regulates BPV-1 DNA replication in vitro by direct protein-protein interaction. Cold Spring Harbor Symp Quant Biol. 1991;56:335–346. doi: 10.1101/sqb.1991.056.01.040. [DOI] [PubMed] [Google Scholar]
- 65.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 66.Yasugi T, Benson J D, Sakai H, Vidal M, Howley P M. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J Virol. 1997;71:891–899. doi: 10.1128/jvi.71.2.891-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida M, Amano T. A common topology of proteins catalyzing ATP-triggered reactions. FEBS Lett. 1995;359:1–5. doi: 10.1016/0014-5793(94)01438-7. [DOI] [PubMed] [Google Scholar]
- 68.Yu X, Egelman E H. The RecA hexamer is a structural homologue of ring helicases. Nat Struct Biol. 1997;4:101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 68a.Zhang W. Ph.D. thesis. Cambridge, United Kingdom: University of Cambridge; 1996. [Google Scholar]
- 69.Zvelebil M J, Barton G J, Taylor W R, Sternberg M J E. Prediction of protein secondary structure and active sites using the alignment of homologous sequences. J Mol Biol. 1987;195:957–961. doi: 10.1016/0022-2836(87)90501-8. [DOI] [PubMed] [Google Scholar]