Abstract
Objectives
Carbapenem-resistance is a challenging healthcare concern and require specific stewardship programs. Monitoring workflows include the identification from surveillance samples, such as rectal swabs. Although culture assays represent the gold standard, data report a significant effectiveness in detecting carbapenemases genes directly from rectal swabs. The aim of this study was to evaluate the REALQUALITY Carba-Screen kit (AB ANALITICA, Padova, Italy) in detecting carbapenemases genes directly from rectal swabs, also comparing its effectiveness to culture assays results. A next-generation sequencing (NGS) was performed to investigate the positive samples about resistance markers and sequence type (ST).
Methods
A number of 136 rectal swabs were collected from the University Hospital Policlinico of Catania critical wards. The samples simultaneously underwent culture and molecular assays (REALQUALITY Carba-Screen kit). The molecular method included two-steps. The first step (1 h and 6 min) rapidly excluded negative samples, while the second one (1 h and 6 min) included only positive samples for a resistance confirmation. All the positive culture samples underwent NGS analysis.
Results
Statistical evaluations demonstrated high sensitivity (100%) and detection rates (92.6%) for the REALQUALITY Carba-Screen kit, which mostly correlated to the standard workflow. All the culture positive results matched the positive molecular results, which were mainly confirmed by the NGS resistome analysis. The identified ST appeared to be diversified and different from the clinically significative strains of the same setting, furnishing interesting epidemiological evidence.
Conclusion
The molecular detection allowed a coordinate approach in a high-prevalence multi-drug-resistance area. The rapid identification with a multi-step procedure accelerated the infection control procedures, while the preliminary negative results reduced the overtreatment episodes. The molecular method efficacy was confirmed through the NGS. In conclusion, the molecular screening could initially lead to a more conservative approach, which may be reevaluated after a culture result about the microorganisms’ identification and susceptibility profile.
Keywords: carbapenem-resistance, infection control measures, molecular assays, resistome analysis, screening protocol
1. Introduction
Carbapenem-resistance prevention is one of the most challenging concerns in healthcare settings, which need to organize stewardship programs and isolation precautions to control these alert microorganisms’ spread (Caliskan-Aydogan and Alocilja, 2023). The European surveillance networks highlight the importance of carefully monitoring carbapenem-resistant Enterobacterales (CRE) and Acinetobacter baumannii, which hardly respond to common antimicrobial therapies and express a massive endemic diffusion capability among hospital settings (CDC, 2019). Literary data often describe the clonal diffusion of identical clones among the same extended health-care setting (CDC, 2019; Ochońska et al., 2021). Italy recently recorded a carbapenem-resistant K. pneumoniae rate of 26.7% and a carbapenem-resistant E. coli percentage of 23.8%. Finally, Acinetobacter spp. reached 86.9% in the same country (EARS-Net, 2023). These data suggest the importance of performing a precise surveillance protocol in the Italian hospital setting. Specifically, Sicily accounted for 51.5% of carbapenem-resistant K. pneumoniae, 1.9% of carbapenem-resistant E. coli, and 88.4% of carbapenem-resistant A. baumannii (Regione siciliana, 2019). In addition, specific surveillance protocols about colistin-resistant strains have recently been planned within European countries to monitor both carbapenem-resistant Enterobacterales and Acinetobacter spp. (Lötsch et al., 2020). Carbapenem-resistant microorganisms may be long-term persistent colonizing agents, thus carbapenem-resistance screening programs need to be settled since patients’ admission. Rectal swabs represent an ideal surveillance specimen due to the possibility of the gastrointestinal tract being a reservoir for carbapenem-resistant microorganisms (Tacconelli et al., 2014; Kim et al., 2018). Culture assay still represents the most reliable method in determining a possible carbapenem resistance on grown colonies from rectal swabs (Adler et al., 2011; Foschi et al., 2020). This conventional workflow includes antimicrobial susceptibility testing (AST) to provide MIC values for carbapenems. Despite the high specificity and reproducibility rates, culture methods require a long turn-around time (TAT), which is not optimal for managing critical patient courting and isolation. Data report a significant effectiveness in detecting carbapenemase genes directly from rectal swabs. Direct detection leads to a TAT reduction and helps critical settings manage colonized patients (Shorten et al., 2016; Del Bianco et al., 2019; Cury et al., 2020; Alqahtani et al., 2021; García-Fernández et al., 2022; EUCAST, 2023).
The aim of this prospective experimental study was to compare the REALQUALITY Carba-Screen kit (AB ANALITICA, Padova, Italy) to standard culture-based methods in detecting carbapenemases genes directly from rectal swabs. The evaluation aimed to demonstrate the reliability of a fast diagnostic workflow for screening protocols, allowing rapid patient courting and alert for fast control purposes. Furthermore, the protocol applied a next-generation sequencing for the grown colonies related to all the REALQUALITY Carba-Screen positive specimens. This ultimate step allowed to evaluate circulating MDR clones in stool samples and confirm molecular results already obtained directly from clinical samples.
2. Materials and methods
The experimental protocol was performed at the University Hospital Policlinico of Catania during a one-month evaluation (April 2023–May 2023), where routinary surveillance rectal swabs integrate a periodical collection as a prevention. A total of 136 rectal swabs derived from Hematology, Intensive Care, and emergency units. All these samples represented a part of conventional antimicrobial resistance surveillance programs. These programs specifically include a rectal screening for CRE microorganisms on patients’ admission and weekly during their recovery period. All the samples were collected using ESwab, a screw-cap tube filled with 1 mL of Liquid Amies Medium, and a regular FLOQSwabs® (COPAN Italia SpA., Brescia, Italy). Rectal swabs were transported to the Laboratory Analysis Unit and processed within 24 h. Specifically, they simultaneously went through the traditional culture-based method and the experimental molecular workflow with the REALQUALITY Carba-Screen® kit (AB ANALITICA, Padova, Italy). On that premise, the study did not involve supplementary samples or direct intervention on patients and only included laboratory processes. Data anonymization protected all the patients whose informed consent was not mandatory for the local Research Ethics Committee. Only residual leftover specimens from the eSwabs sample experienced the molecular technique. Finally, the protocol included next-generation sequencing (NGS) to confirm molecular results and enrich epidemiological evaluations of all the culture-positive specimens. Graph 1 describes all the stages of the experimental workflow.
GRAPH 1.
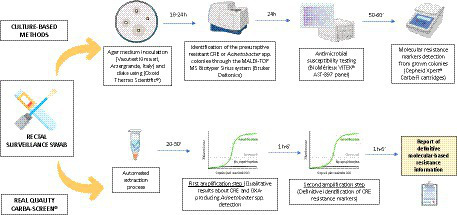
Graphical representation of the applied experimental workflow. Each collected surveillance swab was processed simultaneously through culture-based and molecular methods. The graph was created through the Biorender.com database.
2.1. Culture-based methods and sample storage
A 10 μL aliquot of liquid rectal swabs transport medium was inoculated into a MacConkey agar plate (Vacutest Kima srl, Arzergrande, Italy) using a 10 μg meropenem disk (Oxoid Thermo Scientific®) as an indicator for possible carbapenemases production. The plates faced a 37°C overnight incubation period, and the grown bacteria endured identification protocols.
The eventual meropenem inhibition diameter [European Committee on Antimicrobial Susceptibility Testing (EUCAST)] (3dmedcare, 2015; EUCAST, 2017) allowed the presumptive identification of Enterobacterales or Acinetobacter spp. The MALDI-TOF MS Biotyper Sirius system (Bruker Daltonics) identified the carbapenem-resistant strains and the BioMérieux VITEK® AST-397 panel provide antimicrobial susceptibility testing. The MIC values were analyzed according to an EUCAST expert evaluation (3dmedcare, 2015; EUCAST, 2017). The meropenem, meropenem/vaborbactam, ceftazidime/avibactam, ceftolozano/tazobactam and colistin MIC values were confirmed through a broth microdilution (BMD). The Kirby-Bauer method determined the cefiderocol MIC values. The phenotypically resistant strains grown colonies faced a supplementary molecular process to detect KPC, OXA48, NDM, VIM, or IMP genes, requiring 50–60 min by Cepheid Xpert® Carba-R cartridges.
The entire process required a minimum of 48 h to provide a definitive result. All the grown carbapenem-resistant Enterobacterales and Acinetobacter spp. colonies were stored in 20% glycerol and brain heart infusion (BHI) vials at −80°C.
2.2. Real quality Carba-Screen experimental protocol
Each sample followed the scheme summarized in Graph 1.
A 200 μL quantity after a vortex step and a mixing with 20 μL of K proteinase, was extracted with a semi-automated nucleic acid extraction system (ANDiS 360 Automated Nucleic Acid Extraction System, 3D Medicines Biomedical Technologies), which allowed the simultaneous processing of 16 samples. The extraction process followed the manufacturer’s instructions (ANALITICA, 2022). Briefly, from an aliquot of 60 μL DNA elution, 5 μL was mixed with 20 μL of Master Mix. Then, the AriaDx system (Agilent Technologies) performed a qRT-PCR. The REALQUALITY Carba-Screen kit (AB ANALITICA, Padova, Italy) was validated both on rectal swabs and bacterial colonies. In our specific investigations, the kit was only used directly on rectal swab material. The workflow involved a two-step amplification procedure. The first part was considered as a screening using the REALQUALITY Carba-Screen Real Time mix, which is able to distinguish negative samples from class B, A, and D carbapenemases and/or Acinetobacter spp. OXA positive samples. Specifically, the technology can detect OXA-23 like, OXA-24 like, OXA-51 like with promoter ISAba1 and OXA-58 like.
All the positive samples endured a second differential step, which involves Mix Carba-B (for blaNDM, blaVIM, and blaIMP) and Mix Carba a + D (for blaKPC, blaOXA48, and mcr1,2,4). Each master Mix contains an internal control (IC) based On The amplification of The bacterial 16S RNA gene, which ensures The sample adequacy To amplification processes. The product also includes a dUTP/UNG system To prevent contaminations. The amplification processes were used according To The manufacturer’s instructions, through The Aria dx software (Agilent Technologies) (Bongiorno et al., 2023; QPCR, 2023).
2.3. Next-generation sequencing
A whole genome sequencing (WGS) analysis involved all the grown colonies from positive Carba-Screen samples. The WGS analysis aimed to define the Sequence Type (ST), confirm the carbapenem-resistance through a resistome analysis, and identify the virulome of all the isolates. The QIAGEN QIAamp® DNA Mini Kit (Ref. 51304, QIAGEN, 40724 Hilden, Germany) allowed the colony extraction. The Eppendorf BioPhotometer® D30 and the fluorimeter Qubit dsDNA BR Assay Kit, respectively, evaluated purity and quantity of the initial sample, favoring the DNA quantification (Ref. 32850, Invitrogen, 92008 Carlsbad, CA, United States) (FASTQ, 2021).
The Molecular Biology Laboratory from the University of Catania performed the NGS sequencing, placing 100 ng of each sample on an Illumina MiSeq platform according to the manufacturer’s instructions provided in Watchmaker DNA Library Prep kit with Fragmentation–Watchmaker Genomics® (Ref. 7 K0013-024, 5744 Central Avenue, Suite 100 Boulder, CO 80301, United States). Indexes were provided with Twist Universal Adapter System (16 Indexes, 16 Samples) (Ref. 101307, Twist Bioscience, HQ 681 Gateway Blvd, South San Francisco, CA 94080FAQ).
The fluorometric Qubit dsDNA HS Assay Kit (Ref. Q32851, Invitrogen, Carlsbad, CA 92008, United States) and the Agilent® High Sensitivity DNA Kit (Ref. 5067-4626) allowed the libraries quantification and quality evaluation. Denature and dilute libraries were performed following the “Denature and Dilute Libraries Guide” protocol provided by Illumina®, choosing 8.5 pM as the loading concentration. Finally, sequencing was performed using the MiSeq Reagent Kits v3 (Ref. 15043895, Illumina, Inc., 92122, San Diego, CA, United States). The Sample Sheet was created using the Local Run Manager v3 software, and following the instructions in the Local Run Manager v3 Software Guide provided by Illumina (Martin, 2011; FASTQ, 2021).
The protocol was completed using two different types of analysis with the QIAGEN CLC Genomics Workbench software, following the User Manual for CLC Microbial Genomics Module v22.0, released on January 4, 2022 (QIAGEN, Aarhus, 8000 Denmark). The software assigned resistance, virulence, and MLST genes (FASTQ, 2021). The bioinformatic analysis was also manually performed using further tools. Specifically, TrimGalore (v0.5.0) (Wick et al., 2017; Krueger, 2022) removed the adapter sequence, while de novo bacterial sequence assembly was executed through Unycler (v0.4.8) (Lam et al., 2021) with the Illumina-only assembly modality. The identification of known virulence factors, resistance genes, and capsule loci was performed using Kleborate (v2.2.0), and the Kaptive (Seemann, 2014; Wyres et al., 2016) command for Klebsiella pneumoniae instead of A. baumannii BacPipe. Prokka (v1.13) (Danecek et al., 2021) was used for bacterial annotation. They aligned the output assemblies of Unycicler with several through bwa (0.7.17) to identify punctual mutations in selected genes, while BFTftools (1.3.1) (Li, 2011; Quijada et al., 2019) provided the variants extraction. Specifically, lamB (ARO:3007420), ompk35 (ARO:3003966), ompk36 (ARO:3003968) and ompk37 (ARO:3004122) were used as references.1 Metagenome assemblies were also screened for antimicrobial resistance genes using TORMES (version 1.3.0) (Dhanya et al., 2022). All the commercial kits and interpretation software were applied following the manufacturer’s instructions.
2.4. Statistical evaluation and comparisons
All the gathered data experienced a statistical evaluation of agreement rates between conventional and molecular methods. The statistical analysis regarded only valid samples obtained through the application of the REALQUALITY Carba-Screen technique. Specificity and sensitivity rates together with positive and negative predictive values were calculated. A comparison of the two different turn-around time has been also provided. The REALQUALITY Carba-Screen results were also compared to the WGS confirmation and the phenotypical susceptibility profiles of the analyzed strains.
3. Results
A total of 29 (21.3%) samples tested positive after the application of the culture-based methods, which otherwise revealed 107 (78.7%) negative results. Table 1 summarizes these percentages.
Table 1.
Culture-based methods and Carba Screen® assay gathered results.
| Samples | Culture exams | Carba Screen assay |
|---|---|---|
| Tested samples | 136 | 136 |
| Valid samples | 136 (100%) | 126 (92.6%) |
| Invalid samples | 0 | 10 (7.4%) |
| Positive valid samples | 29 (21.3%) | 52 (41.3%) |
| Negative valid samples | 107 (78.7%) | 74 (58.7%) |
Among the 136 tested samples, 126 (92.6%) got a valid REALQUALITY Carba-Screen result, meaning that internal controls and amplification processes were correctly completed. Otherwise, 10 (7.4%) samples resulted in invalid processes and did not enter into the statistical evaluation. A number of 52 (41.3%) valid samples resulted as positive for carbapenem-resistant Enterobacterales or Acinetobacter spp. On the other hand, 74 (58.7%) valid samples tested negative for the same targets. A global detection rate of 92.6% was obtained. Table 1 summarizes the REALQUALITY Carba-Screen gathered results, while Table 2 includes the statistical evaluations of the performance.
Table 2.
Statistical evaluation about the conventional methods and the Carba-Screen results comparison.
| Statistical evaluations | |
|---|---|
| Sensitivity (culture positive samples which also tested positive to Carba-Screen techniques) | 100% |
| Specificity (culture negative samples which also tested negative to Carba-Screen Techniques) | 76.3% |
| Positive predictive value (positive REALQUALITY Carba-Screen kit samples which also tested positive to culture methods) | 55.8% |
| Negative predictive value (negative REALQUALITY Carba-Screen kit samples which also tested negative to culture methods) | 100% |
| Agreement (global percentage indicating the number of REALQUALITY Carba-Screen kit result which matched the culture methods results) | 81.7% |
| Accuracy (capability to match all the true positive among the involved samples) | 100% |
The invalid samples evaded the statistical evaluations. The agreement to the culture-based methods was calculated based on the number of Carba-Screen tests which perfectly matched the culture assays. Consequently, all the positive Carba-Screen results which tested negative after the culture assay were considered as discrepancies. The accuracy percentage accounted for all the true positive samples, also including culture negative results which tested positive after the application of a conventional molecular method (Cepheid GeneXpert Carba-R) and the REALQUALITY Carba-Screen Kit.
The analysis revealed an agreement of 81.7% between the molecular assay and the culture-based methods. Specifically, all (100%) of the culture-positive samples (29) also tested positive through the molecular assay, while 74 (76.3%) of negative samples confirmed the same result with the REALQUALITY Carba-Screen kit. Among the culture-negative samples, 23 (23.7%) results tested positive after the application of the molecular assay due to the higher consolidated sensitivity rate of the Real-Time PCR protocol.
The conventional Cepheid GeneXpert Carba-R cartridge confirmed the REALQUALITY Carba-Screen kit result on this particular subset. Moreover, the molecular assay revealed a sensitivity rate and a negative predictive value of 100%. However, the specificity rate (76.3%) and the positive predictive value (55.8%) were lower than the above-mentioned percentages.
The molecular positive samples revealed significant percentages of KPC and NDM-producing Enterobacterales together with OXA-producing Acinetobacter. Remarkably, OXA48- or mcr-producing Enterobacterales did not result during the time-period of our experimental study. Table 3 indicates all the details about Carba-Screen’s identified targets among all the tested samples.
Table 3.
Details about Carba-Screen identified targets among all the tested samples.
| Detected genes (positive valid samples) | Samples number |
|---|---|
| Enterobacterales KPC | 10 (19.2%) |
| Enterobacterales OXA-48 | 0 |
| Enterobacterales mcr-1,2,3,4 | 0 |
| Enterobacterales NDM | 6 (11.5%) |
| Enterobacterales VIM | 2 (3.8%) |
| Enterobacterales KPC + NDM | 6 (11.5%) |
| Enterobacterales KPC + VIM | 2 (3.8%) |
| Acinetobacter spp. OXA | 6 (11.5%) |
| Acinetobacter OXA + Enterobacterales KPC | 4 (7.7%) |
| Acinetobacter spp. OXA + Enterobacterales KPC + Enterobacterales NDM | 1 (1.92%) |
| Acinetobacter spp. OXA + Enterobacterales NDM | 12 (23.1%) |
| Acinetobacter spp. OXA + Enterobacterales NDM + Enterobacterales VIM | 3 (5.8%) |
A turn-around time (TAT) analysis has been provided. The conventional culture-based method requires a minimum of 18–24 h to obtain a species identification and a presumptive meropenem resistance, prolonging this interval to 48 h to gather a definitive susceptibility report and information about a precise resistance marker. Otherwise, the REALQUALITY Carba-Screen method allowed us to articulate a two-step process after a 20–30-min extraction phase.
Specifically, the first level revealed a qualitative and preliminary result about positive or negative samples requiring 1 h and 6 min, including also information about the eventual presence of OXA-producing Acinetobacter baumannii allowing the activation of infection control procedures. All the positive samples underwent the second level, which allowed us to identify the specific resistance marker, prolonging the process for a further 1 h and 6 min. Globally, the molecular workflow demanded about 160 min. Graph 2 shows a TAT comparison between the culture-based method and the Carba-Screen molecular assay.
GRAPH 2.
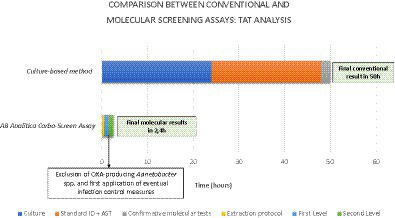
TAT differences between the gold standard method and innovative molecular techniques in detecting resistance markers.
All the samples which tested positive for the culture method related to positive rectal samples on the Carba-Screen molecular assay went through a resistome analysis by using Next generation sequencing (NGS). Specifically, 25 Klebsiella pneumoniae and 4 Acinetobacter baumannii emerged from the assays. K. pneumoniae strains belonged to several sequence type (ST). Notably, the analysis registered ST395 (n. 10), ST101 (n. 4), ST17 (n. 5), ST307 (n. 3), ST46 (n. 2), and ST35 (n. 1). All the K. pneumoniae ST were closely related to specific capsular types. A number of 18 cases (72%) revealed a complete match between WGS analysis and Carba-Screen results about carbapenemase genes. Otherwise, 7 K. pneumoniae strains (28%) showed discrepancies between the two analyses. Specifically, 4 cases described NDM and KPC genes detection after the Carba-Screen application, while the resistome analysis only reported NDM amplification. Furthermore, two strains showed a KPC and NDM Carba-Screen amplification, while the NGS exclusively informed on the KPC presence.
Finally, 1 case reported a KPC and VIM genes Carba-Screen detection, but the resistome analysis did not reveal carbapenemase genes. The same case was confirmed to carry the OXA-9 gene together with ompK37 alterations. Regarding further resistance information, the resistome analysis highlighted the eventual presence of mutations in lamb genes (encoding for a maltose-inducible membrane protein) and ompK genes (encoding for outer membrane proteins) for all the K. pneumoniae genes. A. baumannii strains belonged to the sequence type 2 (ST2). In these strains, Carba-Screen totally matched (100%) the resistome analysis, which revealed the presence of OXA-23 (100%) and OXA-66 (50%) beyond the presence of the constitutive OXA-51. In conclusion, no carbapenem-resistant isolates were missed by the tested system.
Table 4 shows the colistin and β-lactams susceptibility profiles of K. pneumoniae and A. baumannii strains to support the investigated evidence. Additionally, Tables 5, 6 summarize the comparison between the Carba-Screen and the NGS results about the positive K. pneumoniae and A. baumannii samples.
Table 4.
Phenotypical colistin and β-lactam susceptibility profile of K. pneumoniae (Kp) and A. baumannii (Ab) strains.
| Strain | Meropenem MIC | Meropenem/vaborbactam MIC (mg/L) | Ceftazidime/avibactam MIC (mg/L) | Ceftolozano/tazobactam MIC (mg/L) | Cefiderocol Kirby-Bauer result (mm) | Colistin MIC (mg/L) | |
|---|---|---|---|---|---|---|---|
| ST395 | Kp 229 | 16 | >256 | >256 | 16 | 22 mm | 0.5 |
| ST395 | Kp 322 | 32 | >256 | >256 | 16 | 24 mm | 0.5 |
| ST395 | Kp 326 | 32 | >256 | >256 | 16 | 22 mm | 0.5 |
| ST395 | Kp 233 | 32 | >256 | >256 | 16 | 22 mm | 0.5 |
| ST395 | Kp 236 | 16 | >256 | >256 | 16 | 20 mm | 0.5 |
| ST395 | Kp 273 | 32 | >256 | 16 | 8 | 22 mm | 0.5 |
| ST395 | Kp 304 | 64 | >256 | 16 | 8 | 22 mm | 0.5 |
| ST395 | Kp 281 | 32 | >256 | 16 | 16 | 20 mm | 0.5 |
| ST395 | Kp 267 | 16 | >256 | 64 | 8 | 20 mm | 0.5 |
| ST395 | Kp 300 | 64 | >256 | 64 | 8 | 20 mm | 0.5 |
| ST17 | Kp 234 | 16 | 1 | 0.5 | 8 | 20 mm | 0.5 |
| ST17 | Kp 241 | 16 | 2 | 0.5 | 16 | 20 mm | 0.5 |
| ST17 | Kp 325 | 16 | 0.064 | 0.5 | 8 | 22 mm | 0.5 |
| ST17 | Kp 324 | 16 | 1 | 0.5 | 8 | 20 mm | 0.5 |
| ST17 | Kp 323 | 32 | 0.064 | 0.5 | 8 | 22 mm | 0.5 |
| ST101 | Kp 209 | 16 | 2 | 1 | 16 | 18 mm | 0.5 |
| ST101 | Kp 244 | 16 | 2 | 1 | 16 | 20 mm | 0.5 |
| ST101 | Kp 230 | 64 | 1 | 1 | 16 | 18 mm | 0.5 |
| ST101 | Kp 312 | 64 | 0.064 | 0.5 | 8 | 20 mm | 0.5 |
| ST307 | Kp 316 | 16 | 2 | 1 | 1 | 22 mm | 0.5 |
| ST307 | Kp 341 | 16 | 1 | 1 | 16 | 19 mm | 0.5 |
| ST307 | Kp 285 | 16 | 2 | 2 | 16 | 20 mm | 0.5 |
| ST46 | Kp 327 | 16 | 0.064 | 0.5 | 8 | 20 mm | 0.5 |
| ST46 | Kp 248 | 16 | 1 | 0.5 | 8 | 19 mm | 8 |
| ST35 | Kp 232 | 16 | >256 | >256 | 16 | 22 mm | 0.5 |
| ST2 | Ab 292 | 16 | NA | NA | NA | 17 mm | 0.5 |
| ST2 | Ab 230/2 | 128 | NA | NA | NA | 20 mm | 0.5 |
| ST2 | Ab 250 | 16 | NA | NA | NA | 22 mm | 0.5 |
| ST2 | Ab 311 | 64 | NA | NA | NA | 18 mm | 0.5 |
NA, Not applicable according to EUCAST guidelines.
Table 5.
Comparison between Carba-Screen results and NGS resistome analysis related to K. pneumoniae (Kp) isolates.
| Realquality carbascreen screening (1° level) | Realquality carbascreen identification (2° level) | NGS (resistome analysis related to β-lactams resistance) | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class B | Class A + D | Acinetobacter OXA | Carba B | Carba A + D | |||||||||||||||||||||||
| Strain | Carbapenem-resistant phenotype | Identification | MLST | k-locus | o-locus | NDM | VIM | IMP | KPC | OXA-48 | mcr | blaNDM | blaKPC | blaSHV | blaCTX | blaTEM | blaOXA | LamB mutants | ompK35/36/37 mutants | ||||||||
| Kp 229 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Positive | Negative | blaNDM | Negative | Negative | blaKPC | Negative | Negative | NDM-1 | SHV-187 | CTX-M-15 | OXA-1 | Yes | Yes (ompK36, ompK37) | |||||||
| Kp 322 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Positive | Negative | blaNDM | Negative | Negative | blaKPC | Negative | Negative | NDM-1 | SHV-187 | CTX-M-15 | OXA-1 | Yes | Yes (ompK36, ompK37) | |||||||
| Kp 326 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Positive | Negative | blaNDM | Negative | Negative | blaKPC | Negative | Negative | NDM-1 | SHV-61 | SHV-152 | CTX-M-15 | OXA-1 | Yes | Yes (ompK36, ompK37) | ||||||
| Kp 233 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Negative | blaNDM | Negative | Negative | Negative | Negative | Negative | NDM-1 | SHV-187 | CTX-M-15 | OXA-1 | Yes | Yes (ompK36, ompK37) | |||||||
| Kp 236 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Negative | blaNDM | Negative | Negative | Negative | Negative | Negative | NDM-1 | SHV-187 | CTX-M-15 | OXA-1 | Yes | Yes (ompK36, ompK37) | |||||||
| Kp 273 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Positive | blaNDM | Negative | Negative | Negative | Negative | Negative | NDM-1 | CTX-M-15 | TEM-181 | OXA-1 | No | No | |||||||
| Kp 304 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Positive | blaNDM | Negative | Negative | Negative | Negative | Negative | NDM-1 | CTX-M-15 | TEM-122 | OXA-1 | No | Yes (ompK36) | |||||||
| Kp 281 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Positive | blaNDM | Negative | Negative | Negative | Negative | Negative | NDM-1 | CTX-M-15 | TEM-181 | OXA-1 | Yes | Yes (ompK36) | |||||||
| Kp 267 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Positive | blaNDM | Negative | Negative | Negative | Negative | Negative | NDM-1 | SHV-152 | CTX-M-15 | TEM-181 | OXA-1 | Yes | Yes (ompK35) | ||||||
| Kp 300 | Positive | K. pneumoniae | ST395 | 2 | O1/O2v1 | Positive | Negative | Positive | blaNDM | blaVIM | Negative | Negative | Negative | Negative | NDM-1 | CTX-M-15 | TEM-181 | OXA-1 | Yes | Yes (ompK36) | |||||||
| Kp 234 | Positive | K. pneumoniae | ST17 | 25 | O5 | Positive | Positive | Negative | Negative | blaVIM | Negative | blaKPC | Negative | Negative | OXA-9 | No | Yes (ompK37) | ||||||||||
| Kp 241 | Positive | K. pneumoniae | ST17 | 25 | O5 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-61 | SHV-152 | SHV-187 | OXA-9 | No | No | ||||||
| Kp 325 | Positive | K. pneumoniae | ST17 | 25 | O5 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-152 | TEM-181 | Yes | Yes (ompK36) | ||||||||
| Kp 324 | Positive | K. pneumoniae | ST17 | 25 | O5 | Positive | Positive | Negative | Negative | blaVIM | Negative | blaKPC | Negative | Negative | KPC-3 | TEM-181 | TEM-122 | OXA-9 | No | Yes (ompK37) | |||||||
| Kp 323 | Positive | K. pneumoniae | ST17 | 25 | O5 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-187 | TEM-181 | OXA-9 | No | Yes (ompK36) | |||||||
| Kp 209 | Positive | K. pneumoniae | ST101 | 17 | O1/O2v1 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-187 | SHV-212 | No | Yes (ompK36, ompK37) | ||||||||
| Kp 244 | Positive | K. pneumoniae | ST101 | 17 | O1/O2v1 | Positive | Positive | Negative | blaNDM | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-187 | SHV-212 | Yes | Yes (ompK36, ompK37) | ||||||||
| Kp 230 | Positive | K. pneumoniae | ST101 | 17 | O1/O2v1 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-187 | SHV-212 | No | Yes (ompK36) | ||||||||
| Kp 312 | Positive | K. pneumoniae | ST101 | 17 | O1/O2v1 | Negative | Positive | Positive | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | TEM-181 | OXA-9 | No | No | ||||||||
| Kp 316 | Positive | K. pneumoniae | ST307 | 102 | O1/O2v2 | Negative | Positive | Positive | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | TEM-181 | OXA-1 | No | No | ||||||||
| Kp 341 | Positive | K. pneumoniae | ST307 | 102 | O1/O2v2 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | CTX-M-15 | TEM-122 | OXA-1 | OXA-9 | No | No | ||||||
| Kp 285 | Positive | K. pneumoniae | ST307 | 102 | O1/O2v2 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | CTX-M-15 | TEM-181 | TEM-122 | OXA-1 | OXA-9 | No | No | |||||
| Kp 327 | Positive | K. pneumoniae | ST46 | 64 | O1/O2v1 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | No | Yes (ompK37) | ||||||||||
| Kp 248 | Positive | K. pneumoniae | ST46 | 64 | O1/O2v1 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-61 | SHV-152 | TEM-181 | OXA-9 | No | Yes (ompK36, ompK37) | ||||||
| Kp 232 | Positive | K. pneumoniae | ST35 | 22 | O1/O2v1 | Negative | Positive | Negative | Negative | Negative | Negative | blaKPC | Negative | Negative | KPC-3 | SHV-187 | TEM-181 | No | Yes (ompK36, ompK37) | ||||||||
Table 6.
Comparison between Carba-Screen results and NGS resistome analysis related to A. baumannii (Ab) isolates.
| NGS (resistome analysis related to β-lactam resistance) | |||||||
|---|---|---|---|---|---|---|---|
| Realquality carbascreen screening (1° level) | blaOXA | ||||||
| Strain | Carbapenem-resistant phenotype | Identification | MLST | Acinetobacter OXA | OXA-51 | OXA-23 | OXA-66 |
| Ab 292 | Positive | A. baumannii | ST2 | Positive | Positive | Positive | Negative |
| Ab 230/2 | Positive | A. baumannii | ST2 | Positive | Positive | Positive | Negative |
| Ab 250 | Positive | A. baumannii | ST2 | Positive | Positive | Positive | Positive |
| Ab 311 | Positive | A. baumannii | ST2 | Positive | Positive | Positive | Positive |
4. Discussion
Surveillance protocols represent an essential resource in facing and containing the antimicrobial resistance increase. According to surveillance networks e recent literary data, rectal swabs may be an optimal candidate to detect multi-drug microorganisms as initial gut commensals (Adler et al., 2011; Tacconelli et al., 2014; Lee et al., 2015; Foschi et al., 2020; Fasciana, 2023). The molecular technologies may ensure a resistance markers detection directly from rectal swabs, demonstrating convincing agreement rates with conventional culture-based methods (Lerner et al., 2013; Viale et al., 2015). Our experimental study aimed to evaluate the REALQUALITY Carba-Screen kit (AB ANALITICA, Padova, Italy) in detecting carbapenems and colistin resistance markers of Enterobacterales and Acinetobacter spp. The study compared its molecular performance to the traditional culture-based method. Furthermore, the additional next-generation sequencing technology further elaborated and confirmed the presence of resistant isolated and their markers, outlining an important epidemiological context.
The Carba-Screen molecular assay demonstrated optimal sensitivity rates, confirming the possibility of detecting resistance markers despite the occasional presence of low clinical material quantity. Additionally, a 100% negative predictive value revealed the high probability of rapidly excluding the presence of carbapenem-resistant microorganisms. A discrete number of culture-negative samples with a molecular positive result emerged from the study. This molecular positive result was confirmed after the application of the Cepheid GeneXpert Carba-R methodology on the same rectal swabs, whose cartridges are normally used to confirm resistance markers on grown colonies within our diagnostic routine. On one side, the presence of these culture-negative and molecular positive results slightly affected specificity, positive predictive value rates, and statistical agreement. On the other side, this aspect may be considered an advantage, highlighting the molecular assay’s capability to detect resistance markers within low-inoculum positive samples or previously broad-spectrum treated patients. Moreover, previously published studies about molecular technologies documented similar results, enhancing occasional low positive predictive values but high sensitivity and overall agreement rates (EUCAST, 2023). On that premise, the high sensitivity rate is the most important feature clinicians require to better manage colonized patients. A restricted percentage of the analyzed samples revealed an invalid result. We hypothesized a low bacterial count and further investigations by fluorometric assays (data not shown) demonstrated the hypothesis of a poor or absent microbial DNA quantification. Moreover, the culture exams related to these specific samples confirmed a totally negative result. No relationships between invalid results and specific clinical setting have been reported.
The Carba-Screen assay perfectly matched the phenotypical results about β-lactams MIC values of the corresponding isolated strains. The assay efficiency reflected the actual epidemiological condition (Regione siciliana, 2019; Ochońska et al., 2021; EARS-Net, 2023), which describes increasing antimicrobial resistance levels among specific European areas. Notably, the two-step procedure allows organizing a specific and prompt patient’ coorting, due to the rapid qualitative response about eventual positive results. Our experience really appreciated the possibility of detecting both Acinetobacter OXA (first level) and mcr genes (second level), which are uncommon targets for current molecular diagnostic technologies. The restricted time interval to produce a resistance marker result suggests the hypothesis to extend the surveillance protocols to all the possible inter-wards patients’ transfers. This option could lead to better infection control measures, enhancing antimicrobial resistance awareness within all the hospital setting units. In addition, fast surveillance procedures may contain overtreatment predicting actual resistance markers. Insufficient evidence supports the effectiveness of decontamination treatments in colonized patients, especially within health-care settings with a high antimicrobial resistance prevalence (Shaidullina et al., 2023). The possibility to noticeably exclude resistant microorganisms and encourage a prompt infection control procedure within critical wards reduce the indiscriminate antibiotics spread against colonizing strains. In our opinion, the rapidity and sensitivity of the molecular assay may be an interesting solution in containing overtreatment about non-symptomatic colonized patients, which only need isolation protocols and infection control measures application.
The NGS analysis highlighted interesting epidemiological and resistance data. First of all, K. pneumoniae strains belonged to really diversified ST, which appeared different from the clinically significative K. pneumoniae strains (FASTQ, 2021). Ten out of 26 isolates showed the ST395, including emerging K. pneumoniae carbapenem-resistant high-risk clones (Maida et al., 2018). The ST395 has been extensively described among European countries, but its presence within the Southern Italy regions is relatively recent (FASTQ, 2021). Moreover, literary data document an OXA-48 and ST395 frequent combination, while our results reported a strict association between the ST395 and NDM carbapenemases genes. Otherwise, our ST395 strains showed the recurring detection of SHV-187, supporting previous data which described the association between this sequence type and SHV alleles (De Koster et al., 2022).
Finally, all the reported ST395 K. pneumoniae strains showed the capsular type k2, which has already been recognized as an alert type due to the simultaneous presence of high-risk virulence and resistance markers (Hetland et al., 2023).
A number of 4 K. pneumoniae strains belonged to ST101, while three isolates showed the ST307. Both these STs incorporate carbapenem-resistance high-risk clones frequently described as systemic infections etiological agents. Five strains reported the ST17, which is already known for gastro-intestinal commensalism and intensive care unit persistence (Frenk et al., 2020).
Two strains belonged to ST46, already described as an extended β-lactamases (ESBL) and OXA producer (Marcade et al., 2013). One of our isolates only revealed KPC and porine gene alterations, while the other reported SHV, TEM, KPC and OXA genes together with a non-mcr-1 related colistin resistance (MIC = 8 mg/L). These considerations will probably deserve further investigations in the future, with the aim to establish the actual colistin resistance mechanism.
One single strain belonged to the ST35, globally known for to its ubiquitous distribution. Literary data often associate the ST35 with ESBL genes such as SHV and CTX-M (Nawfal Dagher et al., 2019). According to this assumption, our strain reveals the SHV-187 and the less common TEM-181, which has not already described within this ST.
As regards A. baumannii, all the strains belonged to the ST2, described as the main European pandemic A. baumannii lineage. Most literary data report the ST2 as the crucial carbapenem-resistance OXA gene carrier. According to this evidence, our results perfectly match the global epidemiological condition (Osman et al., 2020; Rezaei et al., 2020; Fonseca et al., 2023). Two analyzed strains revealed the detection of OXA-66, described as an OXA-51 family member (Gupta et al., 2022).
The WGS analysis performed on culture-positive isolates (29 samples) not always matched the Carba-Screen detection (7 cases). Specifically, 6 cases reported a Carba-Screen double carbapenem-resistance markers (NDM and KPC) detection, while the NGS analysis confirmed only one marker (NDM or KPC). On the other hand, one strain revealed a Carba-Screen VIM and KPC detection, but the NGS analysis retracted both carbapenemases genes presence. However, the carbapenem-resistance phenotype was explained by the presence of the OXA-9 gene and porine gene alterations.
Certainly, our protocol suffers from some weaknesses. First, a short experimental period (1 month) has been documented. This disadvantage may lead to a restricted sample number and a limited circulating strains spectrum. Additionally, the intention to compare the molecular assay to the culture method affected the statistical evaluation, resulting in low specificity and predictive positive volumes rates. It would be interesting to extend the experimental workflow to longer time intervals, also comparing the REALQUALITY performance to other molecular technologies routinary used into the diagnostic workflow as previous authors described (EUCAST, 2023).
The fundamental purpose of our work is to strongly recommend the introduction of a fast molecular screening in extensive healthcare settings within the principal endemic areas for multi-drug resistant microorganisms. The efficacy of the molecular screening in terms of sensitivity, specificity and detection, has been extensively demonstrated through the application of a universal investigation method such as the next-generation sequencing. The possibility of a multi-step procedure is useful to rapidly provide preliminary results, resulting in coordinated infection control procedures which are subsequently confirmed by the molecular identification of precise resistance markers. The two-levels screening strategy is an added value comparing the analyzed kit to other molecular technologies. Indeed, the first level allows to exclude all the negative results from unuseful and expensive further laboratory procedures, also early alerting clinicians about positive samples. Some limitations of the two-levels procedures may be related to the need of a dedicated manual labor. Globally, the protocol could require a restricted time interval, ensuring a rapid patients’ coorting and an interesting sensitivity rate. Clinical settings such as inter-wards patients’ transfers or pre-surgical screening are ideal healthcare context to apply a molecular screening directly from biological samples.
The high sensitivity may occasionally identify resistance markers which are not confirmed by colonies growth on culture exams. In conclusion, the molecular screening could initially lead to a more conservative approach, which may be reevaluated after a definitive conventional result about the microorganisms’ identification and susceptibility profile.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/bioproject/; PRJNA1063661.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki and the best clinical practice (D.M. 15/07/1997). The present study does not directly involve patient management or drug administration. The institutional document n.101/CECT2 approved the investigations on patients’ biological samples. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because The study did not directly involve human beings or supplementary biological samples apart from the diagnostic routine.
Author contributions
MC: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. GMi: Data curation, Investigation, Methodology, Writing – original draft. GMa: Data curation, Investigation, Methodology, Writing – original draft. DB: Conceptualization, Investigation, Methodology, Supervision, Writing – review & editing. CB: Data curation, Investigation, Methodology, Writing – original draft. EN: Data curation, Investigation, Methodology, Writing – original draft. GS: Supervision, Writing – review & editing. SS: Conceptualization, Supervision, Writing – review & editing.
Acknowledgments
The authors wish to thank AB ANALITICA (Padova, Italy) for providing the REALQUALITY Carba-Screen kit, all the automated applied technologies, and their logistic support during the experimental protocol. Moreover, the authors would like to thank Dr. Grete Francesca Privitera (Department of Clinical and Experimental Medicine, Bioinformatics Unit, University of Catania, Catania, Italy) for her essential contribution.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was partially supported by EU funding with the MUR PNRR Extended Partnership Initiative of Emerging Infection Disease Project n. PE 00000007 INF-ACT.
Footnotes
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 3dmedcare . (2015). Available at: https://www.3dmedcare.com/productdetail/601.html (Accessed April 2023).
- Adler A., Navon-Venezia S., Moran-Gilad J., Marcos E., Schwartz D., Carmeli Y. (2011). Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J. Clin. Microbiol. 49, 2239–2242. doi: 10.1128/JCM.02566-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani M., Tickler I. A., Al Deesi Z., AlFouzan W., Al Jabri A., Al Jindan R., et al. (2021). Molecular detection of carbapenem resistance genes in rectal swabs from patients in gulf cooperation council hospitals. J. Hosp. Infect. 112, 96–103. doi: 10.1016/j.jhin.2021.03.027, PMID: [DOI] [PubMed] [Google Scholar]
- ANALITICA . (2022). Available at: https://www.abanalitica.com/it/catalogo/prodotto/realquality-carba-screen/ (Accessed April 2023).
- Bongiorno D., Bivona D. A., Cicino C., Trecarichi E. M., Russo A., Marascio N., et al. (2023). Omic insights into various ceftazidime-avibactam-resistant Klebsiella pneumoniae isolates from two southern Italian regions. Front. Cell. Infect. Microbiol. 12:1010979. doi: 10.3389/fcimb.2022.1010979, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan-Aydogan O., Alocilja E. C. (2023). A review of Carbapenem resistance in Enterobacterales and its detection techniques. Microorganisms 11:1491. doi: 10.3390/microorganisms11061491, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . (2019). Available at: https://www.cdc.gov/hai/organisms/acinetobacter.html (Accessed September 8, 2023).
- Cury A. P., Almeida Junior J. N., Costa S. F., Salomão M. C., Boszczowski Í., Duarte A. J. S., et al. (2020). Diagnostic performance of the Xpert Carba-R™ assay directly from rectal swabs for active surveillance of carbapenemase-producing organisms in the largest Brazilian university hospital. J. Microbiol. Methods 171:105884. doi: 10.1016/j.mimet.2020.105884, PMID: [DOI] [PubMed] [Google Scholar]
- Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koster S., Rodriguez Ruiz J. P., Rajakani S. G., Lammens C., Glupczynski Y., Goossens H., et al. (2022). Diversity in the characteristics of Klebsiella pneumoniae ST101 of human, environmental, and animal origin. Front. Microbiol. 13:838207. doi: 10.3389/fmicb.2022.838207, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bianco F., Morotti M., Zannoli S., Dirani G., Fantini M., Pedna M. F., et al. (2019). Comparison of four commercial screening assays for the detection of blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA48 in rectal secretion collected by swabs. Microorganisms 7:704. doi: 10.3390/microorganisms7120704, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanya R., Agarwal R. K., Ramprakash S., Trivedi D., Shah V., Bhat N., et al. (2022). Do weekly surveillance cultures contribute to antibiotic stewardship and correlate with outcome of HSCT in children? A multicenter real-world experience of 5 years from the Indian subcontinent. Transplant Cell Ther. 28, 170.e1–170.e7. doi: 10.1016/j.jtct.2021.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- EARS-Net . (2023). Antimicrobial resistance surveillance report in Europe 2023–2021 data, European antimicrobial resistance surveillance network (EARS-net). Available at: https://www.ecdc.europa.eu/en/about-us/networks/disease-networks-and-laboratory-networks/ears-net-data (Accessed September 7, 2023).
- EUCAST Guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. (2017). Available at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (Accessed September 6, 2023).
- EUCAST . The European committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 13.0 (2023). Available at: http://www.eucast.org, https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf (Accessed April and May 2023).
- Fasciana T. (2023). Multicenter study on the prevalence of colonization due to carbapenem-resistant Enterobacterales strains before and during the first year of COVID-19, Italy 11:1270924. doi: 10.3389/fpubh.2023.1270924, [DOI] [PMC free article] [PubMed] [Google Scholar]
- FASTQ . (2021). Local run manager generate FASTQ analysis module workflow guide (1000000003344), 13.
- Fonseca É. L., Morgado S. M., Freitas F., Oliveira P. P. C., Monteiro P. M., Lima L. S., et al. (2023). Persistence of a carbapenem-resistant Acinetobacter baumannii (CRAB) international clone II (ST2/IC2) sub-lineage involved with outbreaks in two Brazilian clinical settings. J. Infect. Public Health 16, 1690–1695. doi: 10.1016/j.jiph.2023.08.014, PMID: [DOI] [PubMed] [Google Scholar]
- Foschi C., Gaibani P., Lombardo D., Re M. C., Ambretti S. (2020). Rectal screening for carbapenemase-producing Enterobacteriaceae: a proposed workflow. J. Glob. Antimicrob. Resist. 21, 86–90. doi: 10.1016/j.jgar.2019.10.012, PMID: [DOI] [PubMed] [Google Scholar]
- Frenk S., Rakovitsky N., Temkin E., Schechner V., Cohen R., Kloyzner B. S., et al. (2020). Investigation of outbreaks of extended-Spectrum Beta-lactamase-producing Klebsiella Pneumoniae in three neonatal intensive care units using whole genome sequencing. Antibiotics (Basel) 9:705. doi: 10.3390/antibiotics9100705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Fernández S., Simner P. J., Thomson G., Faron M., Bosboom R., van Griethuijsen A., et al. (2022). Rapid identification from rectal swabs of the clinically most relevant carbapenemase genes from gram-negative bacteria using the BD MAX check-points CPO assay. Diagn. Microbiol. Infect. Dis. 102:115554. doi: 10.1016/j.diagmicrobio.2021.115554, PMID: [DOI] [PubMed] [Google Scholar]
- Gupta N., Angadi K., Jadhav S. (2022). Molecular characterization of Carbapenem-resistant Acinetobacter baumannii with special reference to Carbapenemases: a systematic review. Infect. Drug Resist. 15, 7631–7650. doi: 10.2147/IDR.S386641, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetland M. A. K., Hawkey J., Bernhoff E., Bakksjø R. J., Kaspersen H., Rettedal S. I., et al. (2023). Within-patient and global evolutionary dynamics of Klebsiella pneumoniae ST17. Microb. Genom. 9:mgen001005. doi: 10.1099/mgen.0.001005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., Song S. A., Lee J. N., Oh M., Jo K. M., Kim H. J., et al. (2018). Clinical factors predicting persistent carriage of Klebsiella pneumoniae carbapenemase-producing carbapenem-resistant Enterobacteriaceae among patients with known carriage. J. Hosp. Infect. 99, 405–412. doi: 10.1016/j.jhin.2017.10.017, PMID: [DOI] [PubMed] [Google Scholar]
- Krueger F. Trim Galore [Internet]. (2022). Available at: https://github.com/FelixKrueger/TrimGalore.
- Lam M. M. C., Wick R. R., Watts S. C., Cerdeira L. T., Wyres K. L., Holt K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12:4188. doi: 10.1038/s41467-021-24448-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. D., Adie K., McNabb A., Purych D., Mannan K., Azana R., et al. (2015). Rapid detection of KPC, NDM, and OXA-48-like Carbapenemases by real-time PCR from rectal swab surveillance samples. J. Clin. Microbiol. 53, 2731–2733. doi: 10.1128/JCM.01237-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A., Romano J., Chmelnitsky I., Navon-Venezia S., Edgar R., Carmeli Y. (2013). Rectal swabs are suitable for quantifying the carriage load of KPC-producing carbapenem-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 57, 1474–1479. doi: 10.1128/AAC.01275-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötsch F., Albiger B., Monnet D. L., Struelens M. J., Seifert H., Kohlenberg A. (2020). European antimicrobial resistance genes surveillance network (EURGen-net) carbapenem-resistant Acinetobacter baumannii capacity survey group; EURGen-net carbapenem-resistant Acinetobacter baumannii capacity survey group. Epidemiological situation, laboratory capacity and preparedness for carbapenem-resistant Acinetobacter baumannii in Europe, 2019. Euro Surveill. 25:2001735. doi: 10.2807/1560-7917.ES.2020.25.45.2001735, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida C. M., Bonura C., Geraci D. M., Graziano G., Carattoli A., Rizzo A., et al. (2018). Outbreak of ST395 KPC-producing Klebsiella pneumoniae in a neonatal intensive care unit in Palermo, Italy. Infect. Control Hosp. Epidemiol. 39, 496–498. doi: 10.1017/ice.2017.267, PMID: [DOI] [PubMed] [Google Scholar]
- Marcade G., Brisse S., Bialek S., Marcon E., Leflon-Guibout V., Passet V., et al. (2013). The emergence of multidrug-resistant Klebsiella pneumoniae of international clones ST13, ST16, ST35, ST48 and ST101 in a teaching hospital in the Paris region. Epidemiol. Infect. 141, 1705–1712. doi: 10.1017/S0950268812002099, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Nawfal Dagher T., Al-Bayssari C., Chabou S., Antar N., Diene S. M., Azar E., et al. (2019). Investigation of multidrug-resistant ST2 Acinetobacter baumannii isolated from Saint George hospital in Lebanon. BMC Microbiol. 19:29. doi: 10.1186/s12866-019-1401-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochońska D., Klamińska-Cebula H., Dobrut A., Bulanda M., Brzychczy-Włoch M. (2021). Clonal dissemination of KPC-2, VIM-1, OXA-48-producing Klebsiella pneumoniae ST147 in Katowice, Poland. Pol. J. Microbiol. 70, 107–116. doi: 10.33073/pjm-2021-010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M., B Halimeh F., Rafei R., Mallat H., Tom J. E., Raad E. B., et al. (2020). Investigation of an XDR-Acinetobacter baumannii ST2 outbreak in an intensive care unit of a Lebanese tertiary care hospital. Future Microbiol. 15, 1535–1542. doi: 10.2217/fmb-2020-0079 [DOI] [PubMed] [Google Scholar]
- QPCR . (2023). Available at: http://www.agilent.com/en/product/real-time-pcr-(qpcr)/real-time-pcr-(qpcr)-instruments/ariadx-real-time-pcr-system-software-pc (Accessed April 2023).
- Quijada N. M., Rodríguez-Lázaro D., Eiros J. M., Hernández M. (2019). TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics 35, 4207–4212. doi: 10.1093/bioinformatics/btz220, PMID: [DOI] [PubMed] [Google Scholar]
- Regione siciliana . (2019). Available at: https://qlik.qualitasiciliassr.it/anonimo/single/?appid=85ada16c-4b41-4bc6-9ca1-405b8243d0c2&sheet=6ad6f3ac-3369-41c5-bd72-792243f9091b&opt=ctxmenu,currsel (Accessed September 8, 2023).
- Rezaei A., Fazeli H., Faghri J. (2020). Investigation of carbapenem resistant Acinetobacter baumannii ST2 in Iran. Acta Microbiol. Immunol. Hung. 68, 20–26. doi: 10.1556/030.2020.01164, PMID: [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153, PMID: [DOI] [PubMed] [Google Scholar]
- Shaidullina E. R., Schwabe M., Rohde T., Shapovalova V. V., Dyachkova M. S., Matsvay A. D., et al. (2023). Genomic analysis of the international high-risk clonal lineage Klebsiella pneumoniae sequence type 395. Genome Med. 15:9. doi: 10.1186/s13073-023-01159-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorten R. J., Ashcroft P., Dodgson A. R. (2016). Rectal swab screening for the detection of carriage of carbapenemase-producing Enterobacteriaceae. J. Hosp. Infect. 94, 131–132. doi: 10.1016/j.jhin.2016.07.009, PMID: [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Cataldo M. A., Dancer S. J., De Angelis G., Falcone M., Frank U., et al. (2014). ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 20, 1–55. doi: 10.1111/1469-0691.12427, PMID: [DOI] [PubMed] [Google Scholar]
- Viale P., Giannella M., Bartoletti M., Tedeschi S., Lewis R. (2015). Considerations about antimicrobial stewardship in settings with epidemic extended-Spectrum β-lactamase-producing or Carbapenem-resistant Enterobacteriaceae. Infect. Dis. Ther. 4, 65–83. doi: 10.1007/s40121-015-0081-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres K. L., Wick R. R., Gorrie C., Jenney A., Follador R., Thomson N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2:e000102. doi: 10.1099/mgen.0.000102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/bioproject/; PRJNA1063661.


