Abstract
Trained immunity is one of the mechanisms by which BCG vaccination confers persistent nonspecific protection against diverse diseases. Genomic differences between the different BCG vaccine strains that are in global use could result in variable protection against tuberculosis and therapeutic effects on bladder cancer. In this study, we found that four representative BCG strains (BCG-Russia, BCG-Sweden, BCG-China, and BCG-Pasteur) covering all four genetic clusters differed in their ability to induce trained immunity and nonspecific protection. The trained immunity induced by BCG was associated with the Akt-mTOR-HIF1α axis, glycolysis, and NOD-like receptor signaling pathway. Multi-omics analysis (epigenomics, transcriptomics, and metabolomics) showed that linoleic acid metabolism was correlated with the trained immunity–inducing capacity of different BCG strains. Linoleic acid participated in the induction of trained immunity and could act as adjuvants to enhance BCG-induced trained immunity, revealing a trained immunity–inducing signaling pathway that could be used in the adjuvant development.
Linoleic acid metabolism was correlated with the trained immunity–inducing capacity of different BCG strains.
INTRODUCTION
Innate immune responses were considered transient until evidence grew that innate immune cells displayed a memory-like phenotype after exposure to certain stimuli, a property that has become defined as trained immunity (1). Innate immune cells acquire trained immunity through epigenetic modifications and metabolic reprogramming so that they produce a more robust inflammatory immune response upon secondary exposure to extraneous pathogens (1, 2). More precisely, trained immunity confers to innate immune cells nonspecific protection against rechallenge that is characterized by increased glycolysis and cytokine production by the responding cells (3, 4).
The Bacille Calmette–Guérin (BCG) vaccine is the only licensed tuberculosis vaccine, and over 4 billion individuals worldwide have received the BCG vaccine over the past century (5). It has become evident that, in addition to protection against Mycobacterium tuberculosis, it can provide nonspecific protection against other diseases, such as viral diseases (6) and tumors (7, 8). For example, a recent clinical trial found that BCG vaccination provided nonspecific protection against viral respiratory tract infections in the elderly, and laboratory evidence suggested that the nonspecific protection was mediated by BCG-induced trained immunity (9).
From 1921 to 1960, more than 14 BCG strains evolved through unique mutations that arose during culture by different laboratories and manufacturers (10). On the basis of the tandem duplication-2 (DU2) forms present in all BCG strains examined so far, the strains were classified into four classes: DU2 classes I to IV. These numerous extensive sequence polymorphisms resulted in between-strain differences in some important antigens of BCG, such as lipids and proteins (11), and these differences may be leading causes for observed variations in the efficacy of BCG in protection against tuberculosis (12–14). The differences in protective efficacy against tuberculosis may be largely attributed to differences in effects on specific immunity, but there may also be contributions by different effects on trained immunity. BCG strains differed in affording nonspecific therapeutic effects on non–muscle-invasive bladder cancer (15–18). However, whether the diverse nonspecific effects induced by different BCG strains are associated with trained immunity has yet to be elucidated.
In this study, we investigated variations in the capacity for and mechanisms of trained immunity induction among four widely used BCG strains chosen for their range of use (19) and virulence (12), covering all four DU2 genetic classes: class I (BCG-Russia), class II (BCG-Sweden), class III (BCG-China), and class IV (BCG-Pasteur). Our data showed that the different BCG strains induced various levels of trained immunity and nonspecific protection both in vitro and in mice. As expected, the enhanced trained immunity induced by BCG was associated with elevated levels of Akt–mammalian target of rapamycin (mTOR)–hypoxia-inducible factor (HIF) axis, glycolysis, and nonobese diabetic (NOD)–like receptor signaling pathways. The multi-omics (epigenomics, transcriptomics, and metabolomics) showed that the linoleic acid metabolism was correlated with the trained immunity–inducing capacity of the different BCG strains. Further, we found that linoleic acid could act as adjuvants to induce trained immunity and enhanced BCG-induced trained immunity. Together, our data showed that the different BCG strains induced different levels of nonspecific protection mediated by trained immunity and revealed a previously unidentified pathway and potential adjuvants for the induction of trained immunity.
RESULTS
The four BCG strains induced different levels of trained immunity in vivo
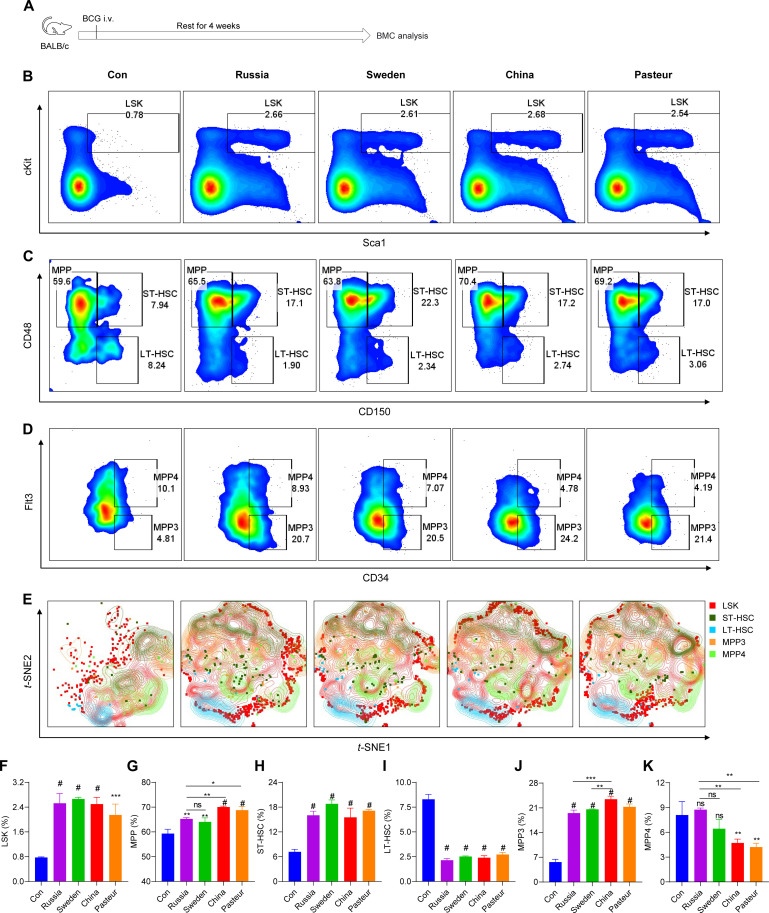
Initially, a mouse model of trained immunity was established to evaluate the induction of trained immunity in vivo by different BCG strains. Modulation of hematopoietic stem cells (HSCs) in the bone marrow has been demonstrated to be an integral component of trained immunity (20, 21). Briefly, mice were trained by intravenous immunization with different BCG strains and subsequently euthanized to collect bone marrow cells (BMCs) for flow cytometry analysis (Fig. 1A and fig. S1, A and B). After BCG intravenous vaccination, the HSC progenitor lineage-Sca-1+c-Kit+ (LSK+) population in BMCs was significantly expanded (Fig. 1, B, E, and F). Further, the proportions of short-term HSCs (ST-HSCs; LKS+CD150+CD48+) and multipotent progenitors (MPPs; LKS+CD150−CD48+) were increased in all BCGs-trained mice, but not long-term HSCs (LT-HSCs; LKS+CD150+CD48−) (Fig. 1, C, E, and G to I). BCG-China and BCG-Pasteur induced more MPPs than BCG-Russia and BCG-Sweden (Fig. 1G). Further analysis revealed that BCG immunization skewed hematopoiesis toward MPP3 (MPP+CD34+Flt3−), especially in BCG-China-trained group (Fig. 1, D, J, and K), indicating that myeloid development was enhanced after BCG vaccination, which is consistent with other reports (22). In addition, increased CD11b+ cells, CD19+ cells, and CD49b+ cells but decreased CD3+ cells were detected in bone marrow (fig. S1, B to F). These data indicate that four BCG strains differed in inducing HSC expansion.
Fig. 1. BCG vaccination induced HSC expansion.
(A) Schema of the in vivo trained immunity experiments for bone marrow cell (BMC) analysis. i.v., intravenous. (B) Representative fluorescence-activated cell sorting (FACS) plots of LKS+ cells in bone marrow of phosphate-buffered saline (PBS)–vaccinated (Con) or BCG-vaccinated BALB/c mice after 4 weeks. (C) Representative FACS plots of multipotent progenitor (MPP) cells, short-term HSCs (ST-HSCs), and long-term HSCs (LT-HSCs) in bone marrow of PBS-vaccinated (Con) or BCG-vaccinated BALB/c mice after 4 weeks. (D) Representative FACS plots of MPP3 and MPP4 cells in bone marrow of PBS-vaccinated (Con) or BCG-vaccinated BALB/c mice after 4 weeks. (E) The t-distributed stochastic neighbor embedding (t-SNE) analysis of LSK cells in normalized Lin− cells. Files containing Lin− cells were down sampled without replacement using the FlowJo DownSample plugin and concatenated to ∼170,000 events suitable for t-SNE analysis. (F) The proportion of LSK cells in Lin− cells. (G) The proportion of MPP cells in LSK cells. (H) The proportion of ST-HSCs in LSK cells. (I) The proportion of LT-HSCs in LSK cells. (J) The proportion of MPP3 cells in MPP cells. (K) The proportion of MPP4 cells in MPP cells. Data represent means ± SD of three independent biological duplicates. One-way analysis of variance (ANOVA) test was used for comparisons between groups: ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.0001.
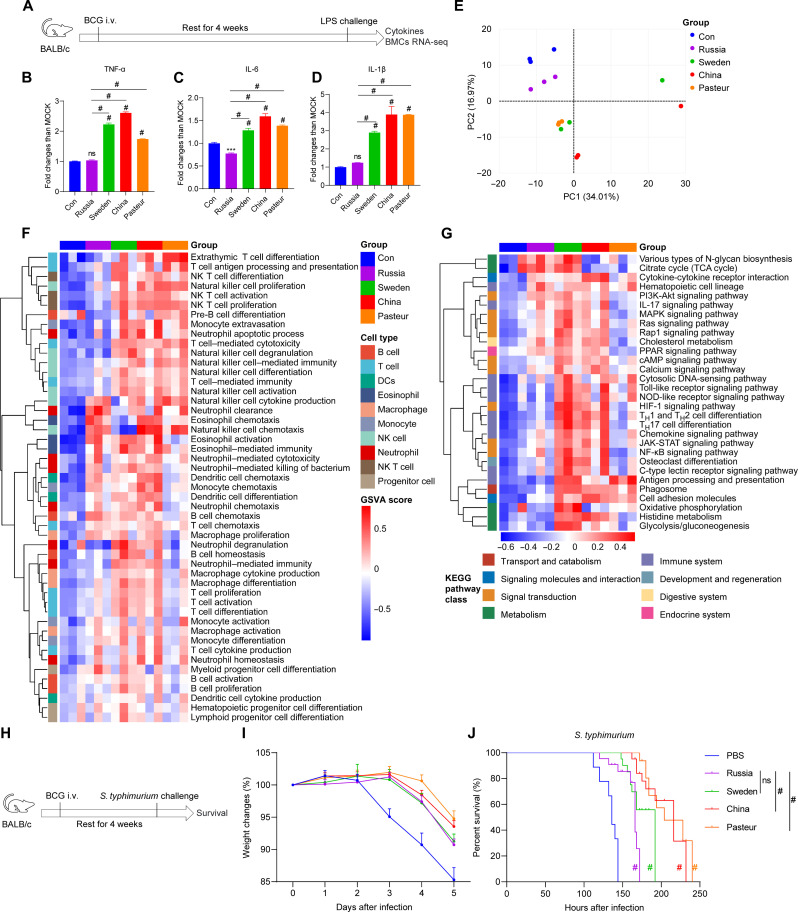
For the nonspecific protection experiment, BCG-immunized mice were challenged 1 month later with either lipopolysaccharide (LPS) (Fig. 2A) or Salmonella typhimurium (Fig. 2H) via intraperitoneal injection. Serum cytokine analyses revealed that BCG-Sweden–, BCG-China–, and BCG-Pasteur–trained mice produced more inflammatory cytokines in response to LPS stimulation than BCG-Russia–trained and non-trained control groups (Fig. 2, B to D). The transcriptome profile of BMCs from BCG-trained mice differed from that of the control group (Fig. 2E). Consistent with flow cytometry analysis, BCG training activated the developmental proliferation and immune function of lymphocytes and myeloid cells in bone marrow, according to the gene set variation analysis (GSVA) for gene ontology biological process (GOBP) analysis. A higher level of activation was observed in the BCG-China and BCG-Pasteur groups compared to the BCG-Russia group (Fig. 2F). Furthermore, the GSVA for Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of BMCs revealed that the BCG-Sweden and BCG-China groups had stronger metabolic and signaling transduction activation than the control and BCG-Russia groups (Fig. 2G). Of utmost significance, BCG-trained mice exhibited reduced weight loss and prolonged survival after infection with S. typhimurium in contrast to the control group. The effects in the BCG-China and BCG-Pasteur groups were significantly superior to those in the BCG-Russia group (Fig. 2, I and J). Thus, the four strains of BCG elicited diverse degrees of bone marrow modification and nonspecific protection against S. typhimurium infection in vivo.
Fig. 2. Four BCG strains induced various levels of cytokines and nonspecific protection in vivo.
(A) Schema of the in vivo trained immunity experiments. (B to D) Cytokine levels in the serum of trained mice upon lipopolysaccharide (LPS) restimulation. MOCK, mean of the control group. Values are expressed as means ± SD. For (B) to (D), data are representative of three independent experiments with three biological duplicates. (E) Principal components analysis (PCA) of the global genes in BMCs. (F) gene set variation analysis (GSVA) of gene ontology biological process (GOBP) related to immune cell development and function. NK, natural killer. (G) GSVA of KEGG pathway related to immune cell metabolism and function. For (E) to (G), data represent means ± SD of three independent biological duplicates. TCA, tricarboxylic acid; MAPK, mitogen-activated protein kinase; PPAR, peroxisome proliferator–activated receptor; cAMP, cyclic adenosine 3′,5′-monophosphate; JAK-STAT, Janus kinase–signal transducer and activator of transcription. (H) Schema of S. typhimurium challenge infection. (I) Changes in the body weight of S. typhimurium–infected mice within 5 days. (J) Survival curves of infected mice, n = 9. PBS, control group. Survival curve was analyzed using log-rank (Mantel-Cox) test: ns, P > 0.05; #P < 0.0001.
The four BCG strains induced various degrees of nonspecific protection in BMDMs
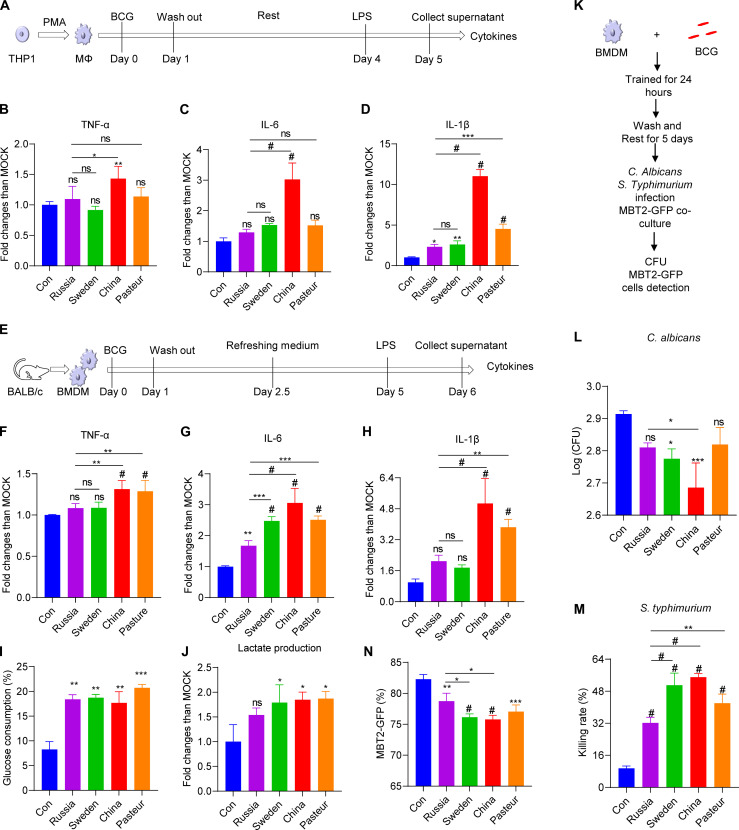
A macrophage-based in vitro model was next established to compare the trained immunity induced by different strains of BCG. In brief, macrophages [bone marrow–derived macrophages (BMDMs) and Tohoku Hospital Pediatrics-1 (THP1) cell–differentiated macrophages] were trained with BCG [multiplicity of infection (MOI) = 1] for 24 hours; then, after 5 days (BMDMs) or 3 days (THP1 cell–differentiated macrophages), they were restimulated with LPS (Fig. 3, A and E). Four BCG strains could activate cytokine secretion of macrophages 24 hours after infection (fig. S2, A to C). Macrophages trained by BCG-China and BCG-Pasteur exhibited higher levels of inflammatory cytokines [tumor necrosis factor–α (TNF-α), interleukin-6 (IL-6), and IL-1β] upon LPS restimulation compared to those of control and BCG-Russia groups (Fig. 3, B to D and F to H). Expectedly, the BCG-trained BMDMs had increased glucose consumption and lactic acid production (Fig. 3, I and J). Furthermore, the trained BMDMs had a reduced fungal load when challenged with Candida albicans in comparison to the control group, with the BCG-China group exhibiting a lower load than the BCG-Russia group (Fig. 3, K and L). BMDMs trained by BCG-China, BCG-Pasteur, and BCG-Sweden showed stronger bactericidal activity against S. typhimurium compared to BCG-Russia and control groups (Fig. 3M). When cocultured with mouse bladder cancer cell line Mouse bladder tumor line-2 (MBT2)–green fluorescent protein (GFP), BMDMs trained by BCG-China and BCG-Sweden had stronger inhibition on the proliferation of tumor cells than the control and BCG-Russia–trained groups (Fig. 3N and fig. S2, D to F). Thus, the four BCG strains differed in trained immunity for different degrees of enhancement of inflammatory cytokine secretion and protection against nonspecific infection/tumor cells proliferation (lowest, BCG-Russia; highest, BCG-China).
Fig. 3. Four BCG strains induced various degrees of cytokines and nonspecific protection in vitro.
(A) Schematic diagram of trained immunity in THP1–differentiated macrophages. (B to D) Cytokine profiles in control and trained THP1 macrophages upon LPS restimulation. (E) Schematic diagram of trained immunity in BMDMs. (F to H) Cytokine profiles in control and trained BMDMs upon LPS restimulation. (I) Glucose consumption of BCG-trained BMDMs upon LPS stimulation. (J) Lactate production of BCG-trained BMDMs upon LPS stimulation. (K) Schematic diagram of nonspecific protection against infection in BMDMs. (L) Total colony-forming units (CFU) of C. albicans quantified after 4 hours of infection. (M) Intracellular killing of S. typhimurium 2 hours after phagocytosis by trained BMDMs. (N) The proportion of MBT2-GFP cells cocultured with BCG-trained BMDMs. One-way ANOVA test was used to compare groups: *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.0001. Values are expressed as means ± SD. Data are representative of three independent experiments with three replicates.
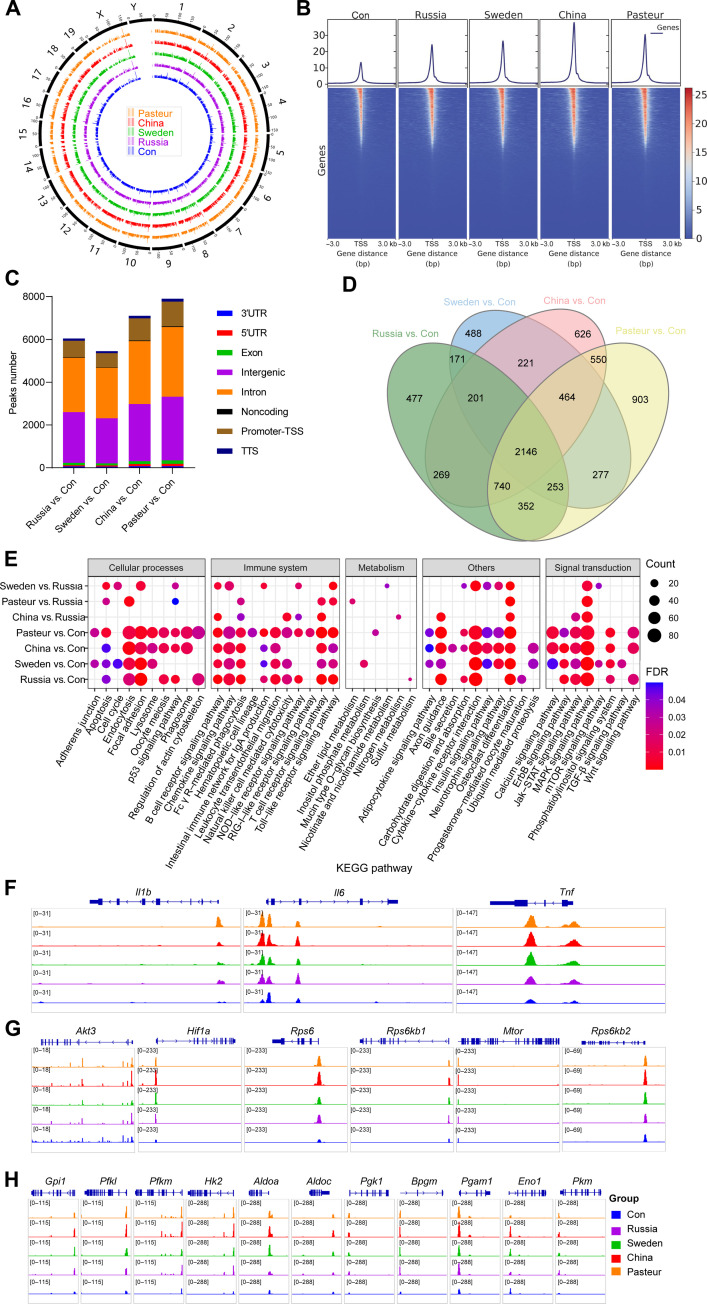
Macrophages trained by the different BCG strains had different transcription profiles
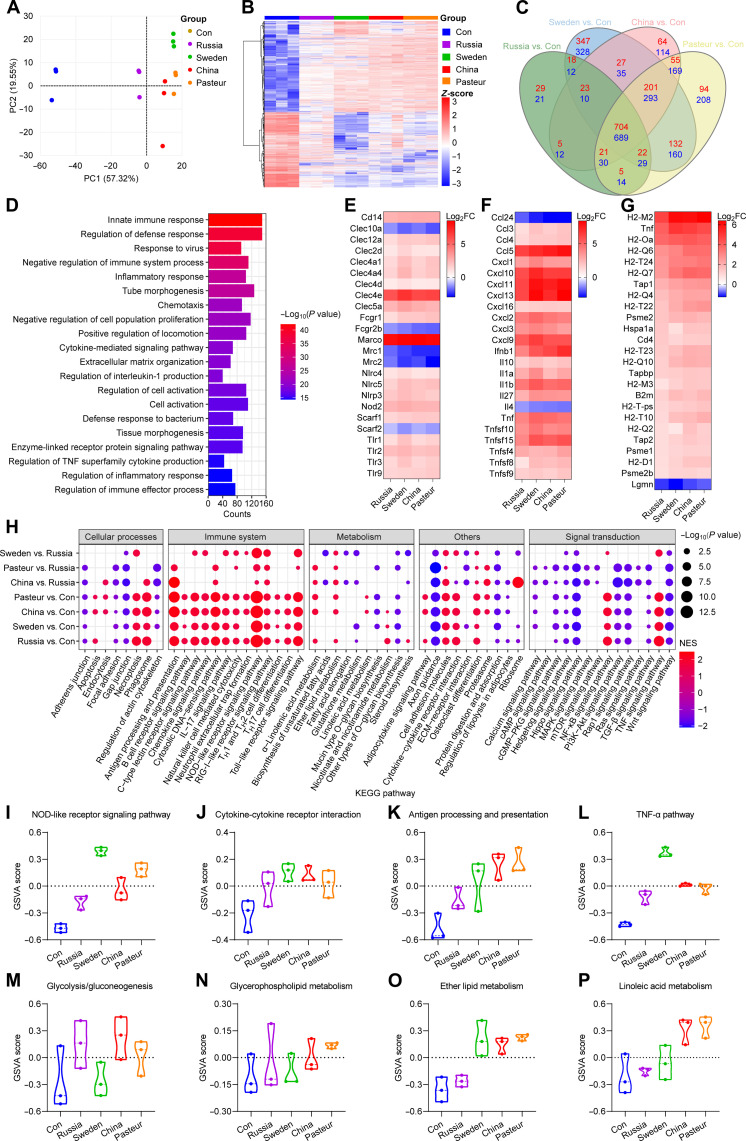
RNA sequencing (RNA-seq) was used to compare the transcriptional characteristics of BMDMs after training. BCG-trained BMDMs displayed significantly different transcriptional patterns than the control (Fig. 4, A and B). In comparison to the control, with BCG-Russia, there were 1644 differentially expressed genes [DEGs; adjusted P value < 0.05, |log2 fold change (log2FC)| > 1] (827 up-regulated and 817 down-regulated); with BCG-Sweden, there were 3030 (1474 up-regulated and 1556 down-regulated); with BCG-China, there were 2452 (1100 up-regulated and 1352 down-regulated); and with BCG-Pasteur, there were 2826 (1234 up-regulated and 1592 down-regulated) (Fig. 4C). Ly6i, Acod1, Lcn2, Marco, Saa3, Fpr1, and Fpr2 genes were significantly up-regulated in BCG-trained BMDMs (fig. S3A). GOBP enrichment analysis showed that the 1338 intersection genes shared by the four groups were primarily enriched for those in innate immune response, defense response, and inflammatory response (Fig. 4D and table S1). After BCG training, the receptors for recognition (Tlr1-3, Nod2, Cd14, etc.) and phagocytosis (Marco, Scarf1, Nlrp3, etc.) were markedly increased, but macrophage mannose receptors were significantly down-regulated (Fig. 4E). Furthermore, with the exception of Ccl24 and Il4, inflammatory cytokines (Tnf, Il1b, Il10, etc.) and chemokines (Ccl3-5, Cxcl1-3, etc.) were elevated (Fig. 4F). Genes related to antigen processing and presentation were also significantly up-regulated after training (Fig. 4G). Gene set enrichment analysis (GSEA) and GSVA were used for genome-wide expression analysis on the RNA-seq data and showed that BCG training activated the immune system-related KEGG pathways [e.g., Toll-like receptor (TLR) signaling pathway, NOD-like receptor signaling pathway, chemokine signaling pathway, antigen processing, and presentation]. Higher levels were seen in the BCG-China and BCG-Pasteur groups than that in the BCG-Russia group (Fig. 4, H to L, and fig. S3, E to L). The pathways that were reported to inhibit inflammation (23–25) (Hedgehog signal pathway, Wnt signal pathway, and transforming growth factor–β signal pathway) were suppressed after training (Fig. 4H and fig. S3, O to Q). The Akt/mTOR/HIF1α-dependent induction of aerobic glycolysis, which was reported to mediate the trained immunity induced by β-glucan (3), was up-regulated in BCG-trained groups (Fig. 4M and fig. S3, B and C). Lipid metabolism (glycerophospholipid metabolism, glycerolipid metabolism, ether lipid metabolism, linoleic acid metabolism, etc.) was enhanced in the trained BMDMs and was higher in the BCG-China and BCG-Pasteur groups than that in the BCG-Russia group (Fig. 4, M to P, and fig. S3, D, M, and N). In summary, metabolic profiling confirmed the activation of innate immune responses and lipid metabolism in BCG-trained BMDMs, and the effects were most evident in the BCG-China–and BCG-Pasteur–trained cells.
Fig. 4. Transcriptome characteristics of BMDMs after BCG training.
(A) PCA of the global gene changes. (B) Heatmap of pooled differentially expressed genes (DEGs) [adjusted P < 0.05, log2 fold change (log2FC) > 1] between BCG-trained groups and Con group. (C) Venn diagram of DEGs between Russia versus Con, Sweden versus Con, China versus Con, and Pasteur versus Con groups. (D) Top 20 GOBP pathway of 1393 shared DEGs in (C). (E) Heatmap of genes involved in phagocytosis and cell activation. (F) Heatmap of cytokines and chemokines genes. (G) Heatmap of genes involved in antigen processing and presentation. (H) Heatmap comparison of gene set enrichment analysis (GSEA) results (P < 0.05) among BCG-trained groups. NES, Normalized enrichment score; ECM, extracellular matrix; cGMP, cyclic guanosine 3′,5′-monophosphate. (I to L) Violin plots of the GSVA of factors involved in cell activation and inflammation. (M to P) Violin plots of the GSVA of pathways involved in cell metabolism. Data represent means ± SD of three independent biological duplicates.
Macrophages trained by the different BCG strains had diverse epigenetic landscapes
In parallel, we assessed the alteration of BMDMs chromosome structure after BCG training by assay for transposase accessible chromatin (ATAC) sequencing (ATAC-seq). A total of 26,207, 38,139, 41,897, 49,520, and 46,541 high confidence open chromatin regions (or peaks) were identified in three biological replicates samples from the control, BCG-Russia, BCG-Sweden, BCG-China, and BCG-Pasteur groups, respectively. Chromosome accessibility increased generally following BCG training compared to the control (Fig. 5A). We observed higher ATAC signals on the transcriptional start site (TSS) of BCG-trained BMDMs genes, with the highest in the BCG-China group (Fig. 5B). Compared with the control group, the BCG-Russia, BCG-Sweden, BCG-China, and BCG-Pasteur groups, respectively, had 6044, 5451, 7105, and 7902 significantly different peaks, and these peaks were mainly distributed in the intron, intergenic, and TSS regions of the genes (Fig. 5C and table S2). Differential peaks in the BCG-Russia, BCG-Sweden, BCG-China, and BCG-Pasteur groups were, respectively, annotated to 4609, 4221, 5217, and 5682 genes, and these had an intersection of 2146 genes (Fig. 5D), which mainly enriched in cell activation and response to external stimulus (table S3). There were higher ATAC signals in the genes of inflammatory signal receptors (Tlr2, Tlr4, Tlr9, Nod2, etc.), chemokines (Ccl2-4, Cxcl1, Cxcl10, and Cxcl12), and cytokines (Tnf, Il1b, Il6, Il10, Il16, etc.) after BCG training (Fig. 5F and fig. S4, A to C). In similarity to the transcriptome results, genes with differential ATAC peaks (GAPs) were mainly enriched in the immune system pathway (Fig. 5E). The genes involved in pathways of the Akt-mTOR-HIF axis, glycolysis, and NOD-like receptor signaling showed higher chromatin accessibility after BCG training. This was more pronounced in the BCG-China and BCG-Pasteur groups (Fig. 5, G and H, and fig. S4D). Integrated analysis with ATAC-seq and RNA-seq data found 1448 shared genes between pooled DEGs in RNA-seq and pooled GAPs in ATAC-seq (fig. S4E), which were primarily enriched for genes involved in cell activation and immune system responses (fig. S4F and table S4). KEGG enrichment analysis showed that inflammatory-related pathways such as the TNF signaling pathway, NOD-like receiver signaling pathway, PI3K-Akt signaling pathway, and chemokine signaling pathway were significantly enriched (fig. S4G and table S4). Evidently, BCG training could extensively alter the chromosomal accessibility of genes pertinent to trained immunity in BMDMs, with the most pronounced changes occurring in the BCG-China–and BCG-Pasteur–trained cells.
Fig. 5. The epigenetic landscape of BMDM cells after BCG training.
(A) Genome-wide chromatin accessibility of trained BMDMs. (B) Heatmaps of differential assay for transposase accessible chromatin (ATAC) peak (P < 0.05) densities at transcriptional start site (TSS) regions. bp, base pairs. (C) Differential ATAC peak distributions on gene loci. 3′UTR, untranslated region; 5′UTR, untranslated region; TTS, transcription termination site. (D) Venn diagram of genes with differential ATAC peaks. (E) Heatmap comparison between BCG-trained groups, KEGG enrichment results of genes with differential ATAC peaks [false discovery rate (FDR) < 0.05]. (F) ATAC signals of Il1b, Il6, and Tnf gene. (G) ATAC signals of genes involved in the HIF-1 signaling pathway. (H) ATAC signals of genes involved glycolysis. Data represent three independent biological duplicates.
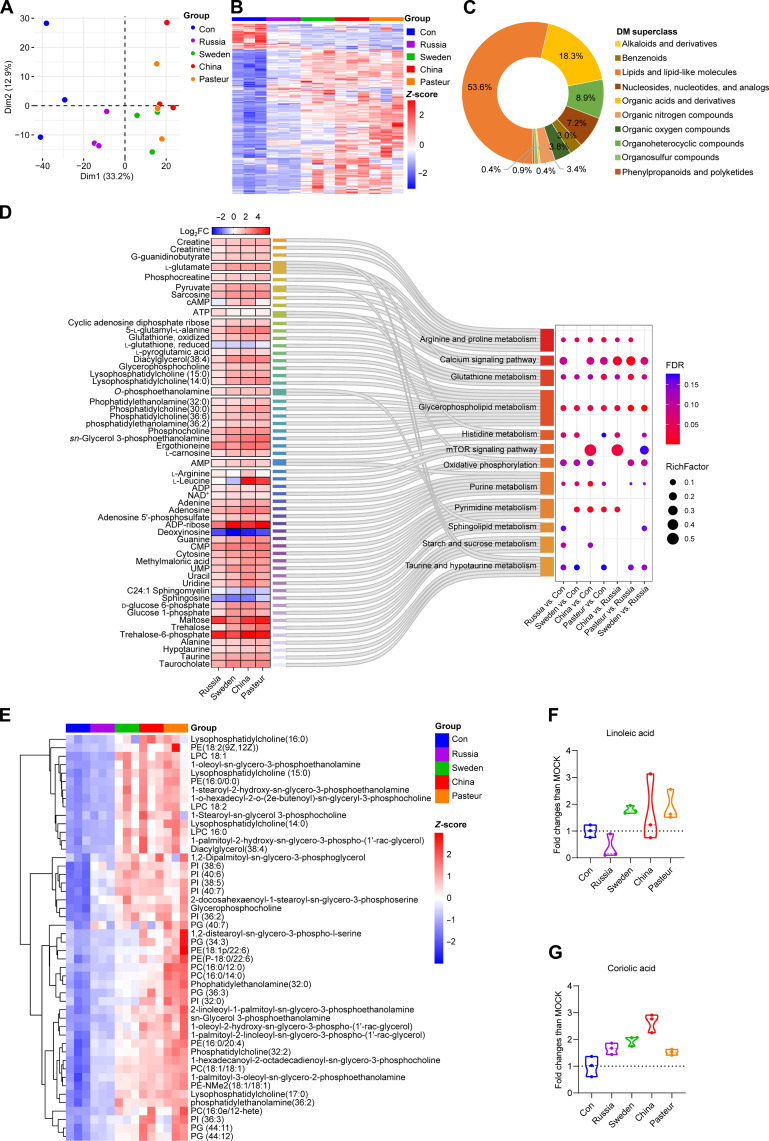
The macrophages trained by different BCG strains had different metabolic profiles
Nontarget liquid chromatography–tandem mass spectrometry (LC-MS/MS)–based metabolomics detection was carried out to characterize the metabolic profile of BCG-trained BMDMs. Principal components analysis (PCA) of qualitative metabolite and heatmap data of 235 pooled differential metabolites (DMs) [variable’s significance in the projection (VIP) > 1, P < 0.05] showed that the BCG-trained BMDMs had different metabolic profiles than the control (Fig. 6, A and B). The DMs mainly included lipid and lipid-like molecules (53.6%), organic acids and derivatives (18.3%), organoheterocyclic compounds (8.9%), and nucleosides/nucleotides/analogues (7.2%) (Fig. 6C). Metabolites trehalose, trehalose-6-phosphate, itaconic acid, maltose, galactinol, lysophosphatidylcholine (18:1) (LPC18:1), and LPC18:2 were elevated markedly after BCG training (fig. S5A). KEGG enrichment analysis revealed that most DMs in the calcium signal pathway, glycolysis, starch and sucrose metabolism, glycerophospholipid metabolism, purine metabolism, and pyrimidine metabolism were up-regulated (Fig. 6D, fig. S5D, and table S5). Glycerophospholipid and linoleic acid metabolisms were prominently enriched in pooled DMs (fig. S5, B and C). The expression of glycerophospholipids in the BCG-Sweden, BCG-China, and BCG-Pasteur groups was significantly higher than that in the control and Russia groups (Fig. 6E and table S6). When compared with the BCG-Russia group, the BCG-Sweden, BCG-China, and BCG-Pasteur groups had 135, 172, and 169 DMs, respectively, and 92 DMs were shared, while 42 DMs were exclusive to the BCG-China group (fig. S5E). The 42 unique DMs were enriched in pyrimidine and linoleic acid metabolites (fig. S5F). Furthermore, LC-MS/MS results showed that linoleic acid and its derivative, coriolic acid [13(S)-HODE], were elevated in BCG-Sweden, BCG-China, and BCG-Pasteur groups (Fig. 6, F and G, and table S5). To conclude, the metabolic profiles of BMDMs had changed after BCG training; most notably, the glycerophospholipid and linoleic acid metabolism of the cells trained with BCG-Sweden, BCG-China, and BCG-Pasteur was different from those trained with BCG-Russia.
Fig. 6. Metabolic profiles of BMDMs after BCGs training.
(A) PCA plot of the qualitative metabolite changes. (B) Heatmap of pooled DMs (VIP > 1, P < 0.05) between BCG-trained groups and control group. (C) DM classification donut chart. (D) KEGG enrichment results of DMs (VIP > 1, P < 0.05). ATP, adenosine 5′-triphosphate; NAD+, nicotinamide adenine dinucleotide (oxidized form); UMP, uridine 5′-monophosphate. (E) Heatmap of differentially expressed glycerophospholipids (VIP > 1, P < 0.05, |log2FC| ≥ 1). (F) Relative abundance of linoleic acid identified by liquid chromatography–tandem mass spectrometry (LC-MS/MS). (G) Relative abundance of coriolic acid identified by LC-MS/MS. Data represent means ± SD of three independent biological duplicates.
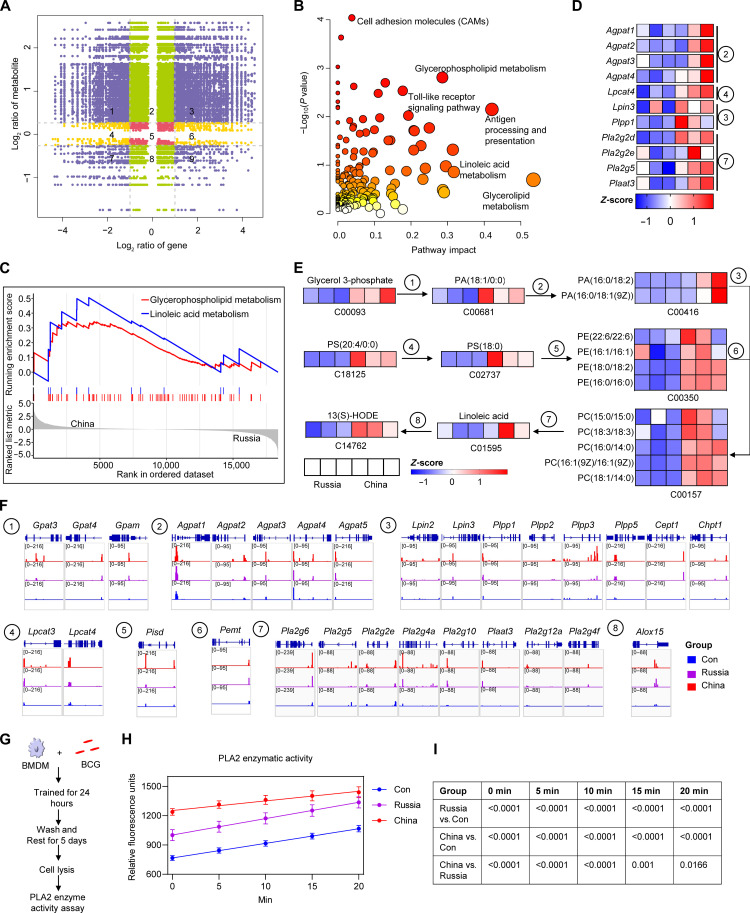
Glycerophospholipid–linoleic acid metabolism was involved in mediating the difference in trained immunity induced by BCG-China and BCG-Russia
The above data indicated that, among the four BCG strains, BCG-China and BCG-Russia induced the strongest and weakest trained immunity, respectively. A combined multi-omics analysis was performed to explore the mechanism contributing to heterogeneity. We found that the up-regulated genes and metabolites were significantly enriched in glycerophospholipid and linoleic acid metabolism (Fig. 7, A and B, and figs. S6, A, B, D, and E). GSEA analysis showed that both glycerophospholipid and linoleic acid metabolism were activated in the BCG-China group compared with that in the BCG-Russia group (Fig. 7C and fig. S6, C and F). The genes (Fig. 7D) and metabolites (Fig. 7E) involved in glycerophospholipid metabolism and linoleic acid metabolism in the BCG-China group were at higher levels of expression than those in the BCG-Russia group; the ATAC peaks that signaled genes involved in glycerophospholipid metabolism and linoleic acid metabolism were more elevated in BCG-China–trained BMDMs (Fig. 7F), and the elevation in linoleic acid of BCG-China–and BCG-Pasteur–trained macrophages was significantly greater than that of the BCG-Russia group (fig. S5A).
Fig. 7. Comparative multi-omics analysis of the high (China) and low (Russia) trained immunity groups.
(A) Nine quadrant charts of mRNA expression ratios and metabolite expression ratios between BCG-China and BCG-Russia groups. Purple dots, significant changes in both metabolite (DMs: VIP > 1, P < 0.05, |log2FC| > 0.26) and mRNA (DEGs: adjusted P value < 0.05, |log2FC| > 1); green dots, significant changes in metabolite only; yellow dots, significant changes in mRNA only. (B) Integrated analysis of metabolites and genes in part 3 of (A). (C) GSEA enrichment plot of glycerophospholipid and linoleic acid metabolic pathways of BCG-China versus BCG-Russia. (D) Heatmap of genes involved in glycerophospholipid and linoleic acid metabolism. (E) Glycerophospholipid and linoleic acid metabolic pathways with a heatmap of normalized data. Blue and red colors represent low and high concentrations, respectively, scaled by color intensity. (F) ATAC signals of genes involved in glycerophospholipid metabolism and linoleic acid metabolism. ① to ⑧: Genes involved in glycerophospholipid and linoleic acid metabolism. (G) Schematic diagram of PLA2 enzyme activity assay in BCG-trained BMDMs. (H) PLA2 enzymatic activity was shown as relative fluorescence units with time. Data represent means ± SD of five independent biological duplicates. (I) The table shows the P values (one-way ANOVA) for each time point in (H).
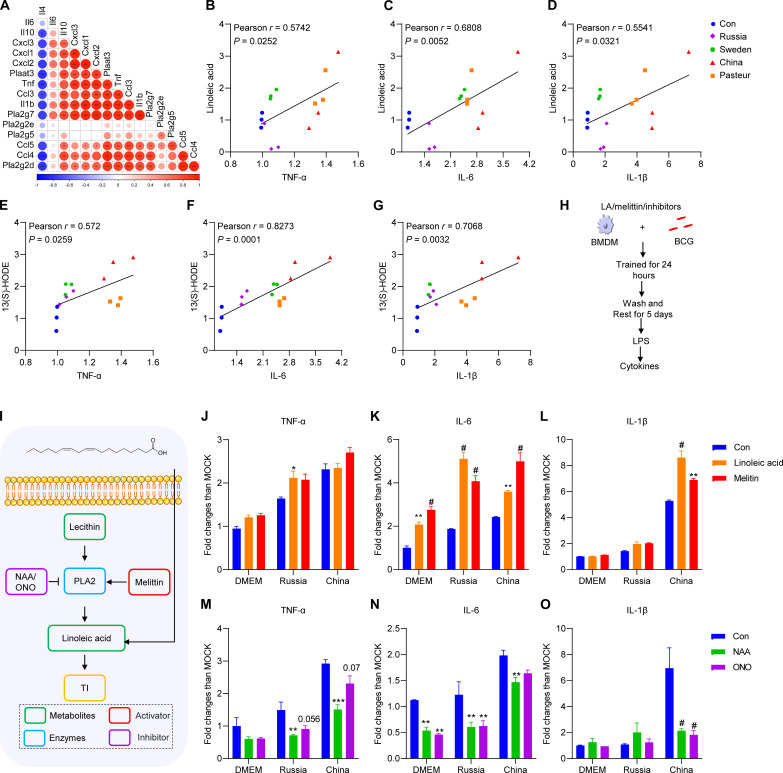
Cholesterol metabolism has been reported to be involved in the trained immunity of macrophages (26). Phospholipase A2 (PLA2) cleaves lecithin, a crucial metabolite in glycerophospholipid metabolism, to generate linoleic acid (27–30). Consistent with the results of the combined multi-omics analysis, the PLA2 enzyme activity of BCG-China–trained macrophages was significantly higher than that of the control and BCG-Russia groups (Fig. 7, G to I). Pearson correlation analysis showed a positive correlation between PLA2 genes (Pla2g2d, Pla2g7, Plaat3, etc.) and inflammatory cytokines (Tnf, Il1b, Ccl3, etc.) expression and a negative correlation with Il4 expression (Fig. 8A). Further, the production of TNF-α, IL-6, and IL-1β from trained macrophages was positively correlated with the concentration of linoleic acid and 13(S)-HODE (Fig. 8, B to G). To assess whether the glycerophospholipid–linoleic acid metabolism is involved in BCG-induced trained immunity, linoleic acid was used as supplement when BMDMs were trained by BCG (Fig. 8, H and I). As expected, linoleic acid could induce trained immunity and enhanced BCG-induced trained immunity (Fig. 8, J to L). As an activator of PLA2 (31), melittin also enhanced BCG-induced trained immunity (Fig. 8, J to L). In contrast, the PLA2 inhibitors, ONO-RS-082 (ONO) and 1-naphthylacetic acid (NAA) (32, 33), significantly attenuated BCG-induced trained immunity (Fig. 8, M to O). Thus, BCG-China–trained BMDM had higher linoleic metabolism than the BCG-Russia group, and linoleic acid supplementation enhanced BCG-induced trained immunity.
Fig. 8. Linoleic acid metabolism enhanced BCG-induced trained immunity.
(A) Correlation between cytokines genes and PLA2 genes. (B) Correlation between TNF-α fold change and linoleic acid fold change. (C) Correlation between IL-6 fold change and linoleic acid fold change. (D) Correlation between IL-1β fold change and linoleic acid fold change. (E) Correlation between TNF-α fold change and 13(S)-HODE fold change. (F) Correlation between IL-6 fold change and 13(S)-HODE fold change; (G) Correlation between IL-1β fold change and 13(S)-HODE fold change. Correlations were performed using Spearman correlation analysis. (H) Schematic diagram of trained immunity supplementation with linoleic acid (LA) and melittin. (I) Schematic diagram of glycerophospholipid–linoleic acid metabolism. (J to L) Cytokines of BCG-trained BMDMs supplementation with linoleic acid (10 μM) and melittin (0.25 μg/ml) upon LPS restimulation. (M to O) Cytokines of BCG-trained BMDMs supplementation with ONO-RS-082 (ONO; 50 μM) and 1-naphthylacetic acid (NAA; 50 μM) upon LPS restimulation. Data represent means ± SD of three independent biological duplicates. Data are representative of two independent experiments. Two-way ANOVA was used to compare groups; *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.0001.
Together, the observations revealed that training by different BCG strains led to various degrees of alterations in macrophage epigenomics, transcriptomics, and metabolomics and thereby induced distinctly different degrees of nonspecific protective immunity. By comparing macrophage responses to the different BCG strains, we found that enhanced linoleic acid metabolism facilitated the induction of trained immunity.
DISCUSSION
Since the approval of BCG for non–muscle-invasive bladder cancer treatment, BCG intravesical instillation has become one of the essential bladder cancer treatments (34). The nonspecific effect of BCG on treating bladder cancer is mediated, at least partially, by trained immunity induction (35, 36). Although the question of whether strain selection influences the efficacy of intravesical BCG remains controversial (37–39), differences in the efficacy of BCG strains in the treatment of bladder cancer have been reported in several studies (15–18). Little is known, however, about the mechanisms underlying the differences in treatment efficacy (40, 41). In this study, we found that four genetically different strains of BCG induced different degrees of trained immunity to nonspecifically protect against challenges with pathogenic microbes. This supported the contention that the genetic differences between the strains might contribute to the diversity in treatment efficacy. From this, it follows that an optimal BCG strain for induction of trained immunity should be selected for bladder cancer therapy, which may be further enhanced by designing a reengineered BCG that targets trained immunity induction. However, head-to-head comparative clinical trials in treating non–muscle-invasive bladder cancer are warranted to better understand the importance of strain selection and trained immunity for BCG efficacy and to establish whether there is an optimal strain.
Centrally trained immunity was defined as the result of training immune progenitor cells in the bone marrow to generate long-term innate immune-mediated protection against heterologous infections (42). Stimulation with BCG could prompt transcriptomic, epigenomic, and functional reprogramming of HSCs and peripheral monocytes to induce trained immunity in humans (21); in contrast, M. tuberculosis reprogramed HSCs via an interferon I response that suppressed myelopoiesis and impaired the development of trained immunity (43), suggesting that bacterial virulence might be a key influence on trained immunity induction. Here, we found that differential modification of bone marrow immune cells by different BCG strains induced various degrees of trained immunity and resistance to S. typhimurium infection in vivo. The less virulent BCG genetic strains (BCG-DU1 and -DU2) afforded lower levels of trained immunity and nonspecific protection compared with more virulent strains (BCG-DU3and -DU4) (12); however, the capacity for trained immunity induction was not well matched with the degree of virulence, suggesting that virulence is not a key influence. Some studies categorized BCG into two clusters based on IS6110 and mpt64: early (obtained before 1927 included BCG-Russia and BCG-Sweden) and later (obtained after 1927 included BCG-China and BCG-Pasteur) strains (10, 44). The early BCG strains (BCG-Russia and BCG-Sweden) induced lower levels of trained immunity compared with the later ones (BCG-China and BCG-Pasteur) in our study, but the performance of other early and later BCG strains in inducing trained immunity remains to be investigated.
The transcriptomic, epigenetic, and metabolomic profiles of macrophages following BCG training were characterized in the current work. Trained immunity could confer a long-term increased response of innate immune cells (macrophages, natural killer cells, dendritic cells, etc.) through epigenetic modifications (45). The results of RNA-seq and ATAC-seq showed that BCG training markedly activated the defense response of macrophages, including antigen processing/presentation and inflammatory response signaling pathways. The mTOR–HIF-1α pathway–mediated increased glycolysis was reported to be essential for the induction of trained immunity (46), and, in the current study, improved accessibility and expression of the genes (Mtor, Hif1a, Hk2, Eno1, Pkg1, etc.) involved in glycolysis were observed. The synthesis of metabolites of glycolysis, such as pyruvate, d-glucose 1-phosphate, 3-phospho-d-glycerate, and thiamin diphosphate, was also increased in BCG-trained macrophages. Moreover, it was reported that BCG could induce NOD2-dependent (not TLRs and dectin-1) nonspecific protection from reinfection via epigenetic reprogramming of monocytes (47), and our RNA-seq and ATAC-seq results showed that all four BCGs activated the NOD-like receptor signaling pathway of macrophages, with more robust activation by BCG-China and BCG-Pasteur than by BCG-Russia. These data are consistent with the current understanding of BCG-induced trained immunity mechanisms and motivated us to further explore the mechanism contributing to heterogeneity in trained immunity induction among different BCG strains.
Ultimately, the LC-MS/MS, ATAC-seq, and RNA-seq combined multi-omics analysis showed that itaconic acid, a metabolite with both anti-inflammatory and bactericidal effects (48), and its synthase gene (Acod1/Irg1) were all markedly elevated in BCG-trained macrophages (figs. S3A and S5A). This suggests a possible association with the nonspecific protective efficacy afforded by BCG strains. It was reported that elevated itaconic acid in macrophages after M. tuberculosis infection was critical for limiting intracellular proliferation and pathological inflammation (49, 50). More recently, dimethyl itaconic acid, a derivative of itaconic acid, has been found to induce trained immunity in human monocytes, and basal plasma itaconic acid has been associated with increased human BCG trained immunity inducing capacity (51). Correlation analysis showed that Acod1 expression and itaconic acid content were positively correlated with inflammatory cytokines (fig. S6, G to L); thus, it is tempting to speculate that itaconic acid may contribute to BCG-induced trained immunity.
Lipid metabolism is crucial in the interaction between M. tuberculosis and macrophages (52), but few studies have been made on the involvement of macrophage lipid metabolism in BCG-induced trained immunity. Oxidized low-density lipoprotein, aldosterone, and low-density lipoprotein cholesterol have been proven to induce trained immunity (53–55). BCG immunization of infants perturbed the lipidome present in plasma, and the changes in LPCs were correlated with cytokine responses recalled in vitro 4 weeks after vaccination (56). Here, we showed that BCG-induced trained immunity could be improved by activating linoleic acid synthesis or directly supplementing with linoleic acid, suggesting that this signaling is a potential target of trained immunity–based immunotherapy and that linoleic acid or linoleic acid metabolism agonists have the potential to be used as vaccine adjuvants. Recently, it was shown that addition of exogenous linoleic acid activated lipid modulator-mediated innate immunity, enhanced macrophage bactericidal capacity, and reduced bacterial load in organs in a mouse bacteremia model (57). Linoleic acid has also been found to improve CD8+ T cell metabolism, prevent exhaustion, and stimulate memory-like phenotypes with superior effector functions, resulting in greater antitumor potency in vitro and in mouse model (58). However, it was shown that elevated dietary linoleic acid increased gastric carcinoma cell invasion and metastasis in mice (59, 60). Considering that gut microbes can convert linoleic acid into other compounds (61, 62), different ways of supplementing linoleic acid (e.g., orally and intraperitoneally) might explain the variation in results across studies. Linoleic acid has been widely studied clinically as a nutritional supplement (63, 64), higher linoleic acid intake, as assessed by dietary surveys or biomarkers, was associated with a lower risk of death from all causes, especially cardiovascular diseases and cancers (63, 65). It has been shown that the western diet, thought to be abundant in linoleic acid, triggered trained immunity in mice via NOD-like receptor thermal protein domain associated protein 3 (NLRP3)–dependent pathway (66). Excessive intake of linoleic acid, however, led to the formation of oxidized linoleic acid metabolites, which impaired mitochondrial function through substandard cardiolipin composition and may contribute to many chronic diseases (67). Thus, the background of linoleic acid intake in the population and the supplemental dose should be carefully considered in the future studies. Nevertheless, recognizing the role of linoleic acid in trained immunity could provide previously unidentified insights for clinical applications.
Although only four BCG strains were compared due to practical constraints, we found substantial differences in the ability of different strains to induce trained immunity, and the possibility that variations in the training dose accounted for the differences was excluded (fig. S7, A and B). Whether varying the BCG dose and revaccination could enhance trained immunity remains controversial. In bladder cancer treatment, there was an association of reduced BCG dose with recurrence but not with tumor progression, cancer-specific survival, and overall survival (68, 69). Two clinical trials of BCG revaccination in children (who received neonatal BCG vaccination) aged 19 months and 7 to 14 years have shown that BCG revaccination provided greater protection against tuberculosis than a single dose (70, 71). However, a clinical trial in adults (small sample size, 51 volunteers) reported that the induction of a trained immunity profile is essentially dose- and frequency-independent (72). It must be stressed, however, that the immune system of humans varies with age and sex (73, 74), and trained immunity can be maintained over time (75). Thus, the time of revaccination is critical. A better clinical trial is warranted to further test for associations between BCG dose, revaccination, and trained immunity.
Muramyl dipeptide, a cell wall component of BCG, has been shown to induce trained immunity via NOD2 (47). Recently, mycolic acid, a lipid component abundant in the cell wall of the mycobacteria, has been shown to cause epigenetic alterations in the promoter regions of the Tnf and Il6 genes in macrophages and to strongly induce pro-inflammatory cytokines (76). BCG has numerous antigens and other bioactive products (proteins, lipids, glycolipids, etc.) (77, 78), which varies between different strains (11, 44). The exploration of their roles in trained immunity may explain the differences in trained immunity induced by various BCG strains and guide the subsequent development and clinical application of recombinant BCG vaccines targeting trained immunity. Transcriptomics showed differences in fatty acid catabolism, transcriptional regulation, and mammalian cell entry genes among BCG strains (79). Thus, after avoiding artificial differences in training dose (fig. S7, A and B), we found that BCG-Russia entered fewer macrophages than the other groups, but there was no difference among the BCG-Sweden, BCG-China, and BCG-Pasteur groups (fig. S7C), which may partially explain the weakest ability of BCG-Russia to induce trained immunity, but not the differences between the other strains (e.g., BCG-China versus BCG-Sweden). Here, we provide evidence for a previously unidentified signaling pathway in linoleic acid metabolism as a potential target of immunotherapies to manipulate trained immunity in macrophage; thus, the genome, metabolome, and lipid composition of different BCGs may provide us with clues to subsequently explore the mechanisms by which different BCGs induce various levels of trained immunity at the bacteriological level.
There are several limitations to the current study. First, although we showed that the PLA2 enzyme in the linoleic acid metabolism pathway in macrophages was activated and the linoleic acid concentration increased after BCG training, whether the linoleic acid synthesized by BCG (80) was involved in the host linoleic acid metabolism pathway remains unknown. Second, although in vitro experiment has shown that linoleic acid supplementation or linoleic acid metabolism activation can enhance BCG-induced trained immunity (enhanced inflammatory cytokine secretion upon nonspecific stimulation), the efficiency of linoleic acid supplementation in inhibiting pathogens infection, and the in vivo dosage or inoculation route still need future investigations.
In conclusion, we showed substantial differences in the transcriptomic, epigenomic, and metabolomic profiles of macrophages after training with different strains of BCG and that four major BCG strains differed in the pathways associated with induction of nonspecific protection both in vitro and in vivo. We also revealed the involvement of glycerophospholipid–linoleic acid metabolism in BCG-induced trained immunity and that linoleic acid could induce trained immunity.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The BCG strains used in this study were presented by J. Liu from University of Toronto, with clear genetic and phenotypic background (12, 81). They were grown at 37°C in liquid Middlebrook 7H9 broth (BD Difco, USA) supplemented with 10% (v/v) oleic acid–albumin–dextrose–catalase enrichment (OADC; BD Difco, USA), 0.5% glycerol, and 0.05% Tween 80, or on solid Middlebrook 7H11 (BD Difco, USA) supplemented with 10% OADC and 0.5% glycerol. C. albicans was grown in Sabouraud broth medium (1% peptone and 4% dextrose) or on Sabouraud agar medium (1% peptone, 4% dextrose, and 2% agar) at 37°C. S. typhimurium was cultured in Luria Bertani (Solarbio) medium.
Preparation of single bacterial suspensions of BCG
One milliliter of BCG culture grown to the logarithmic growth phase [optical density at 600 nm (OD600) = ~1] was collected, centrifuged at 12,000g for 30 s, and washed twice with 1 ml of PBS containing 0.05% Tween-80 (PBST). Last, the pellet was gently resuspended by 1 ml of PBST and then centrifuged at 500g for 2 min, and the supernatant (single bacterial suspension) was used for experiments after standardization of concentration based on OD600.
Cell culture
Six- to 8-week-old specific pathogen–free grade BALB/c female mice were euthanized to provide hind leg BMCs. BMDMs were differentiated from isolated BMCs by culture for 5 days in Dulbecco’s modified Eagle’s medium (DMEM)/Nutrient Mixture F12 (BI) medium containing 10% fetal bovine serum (Gibco) and macrophage colony-stimulating factor (50 ng/ml; PeproTech). THP1 cells (American Type Culture Collection) were grown at 37°C with 5% CO2 in RPMI 1640 (Biological Industries) with 10% fetal bovine serum (Gibco). The mouse bladder cancer MBT2-GFP cell line was purchased from Oricell (Cyagen Biosciences) and cultured in DMEM (BI) containing 10% fetal bovine serum (Gibico, USA) with puromycin dihydrochloride (4 μg/ml; Beyotime).
In vitro trained immunity experiments
THP1 cells (2 × 105 per well) were seeded into 48-well cell culture plates and differentiated with 100 nM phorbol 12-myristate 13-acetate (PMA; Beyotime) for 48 hours to obtain macrophages. After resting for 12 hours in DMEM without PMA, macrophages were trained with BCG (MOI = 1) for 24 hours, followed by 3 days of rest after washing off the BCG. Last, the supernatants were collected after stimulation with Escherichia coli LPS (25 ng/ml; InvivoGen, China) for 24 hours and frozen at −80°C for cytokine assays. For the BMDM training model, the BMDMs were seeded to flat-bottom 48-well cell culture plates at a concentration of 1 × 105 cells per well and trained with DMEM/F12 or different BCG strains (MOI = 1) by 24-hour incubation at 37°C. The supernatant was discarded, cells were washed twice, and the medium was replaced with a fresh complete medium. The cells were rested for 5 days, with a change of culture medium in the interval. Next, the cells were stimulated with E. coli LPS (25 ng/ml; InvivoGen, China) for an additional 24 hours, and the supernatants were stored at −80°C until assayed for cytokines (IL-1β, IL-6, and TNF-α) with commercial enzyme-linked immunosorbent assay kits (Invitrogen, Austria), glucose, and lactate with the Glucose and Lactate Content Assay Kit (Solarbio). For rechallenge experiments, the trained BMDMs were infected with C. albicans [500 colony-forming units (CFU) per well] and S. typhimurium (MOI = 1) by centrifugation at 600g for 5 min and cultured at 37°C. The supernatant was collected 5 hours after the cells were infected, the cells were lysed with sterile water, then the lysate and supernatant were combined and plated for CFU counting. For the intracellular bactericidal assay, BMDMs were infected by S. typhimurium for 30 min, extracellular bacteria were washed off with 1 ml of phosphate-buffered saline (PBS), and then cells in some wells were lysed and plated for counting as background. The cells in other wells were cultured in complete medium with gentamicin (20 μg/ml) for a further 2 hours, rinsed, and then lysed, and the CFU in the lysates were counted. For the coculture experiment, the trained BMDMs were cocultured with MBT2-GFP cells (BMDM:MBT2-GFP = 20:1) for 3 days and subsequently analyzed the proliferation of MBT2-GFP cells by fluorescence microscopy (Olympus) and flow cytometry (BD Biosciences). For PLA2 activity assay, 1 × 106 cells were seeded into six-well cell culture plates and trained as described above; the PLA2 Activity Assay Kit (Abcam) was used to determine PLA2 activity. Briefly, the trained cells were lysed with 100 μl of PLA2 Assay Buffer on ice for 10 min and then centrifuged at 10,000g for 15 min at 4°C to collect the supernatant. Supernatant (10 μl) was added to 96-Well Black Opaque Plates, followed by 40 μl of PLA2 Assay Buffer, 10 μl of PLA2 probe, and 40 μl of PLA2 substrate sequentially and then mixed. Last, the fluorescence (excitation/emission = 388/513 nm) was detected using the Cell Imaging Multimode Reader (Agilent BioTek Cytation5) in kinetic mode. Activity was expressed as relative fluorescence units for the 10 μl of cell lysate. For the linoleic acid metabolism pathway intervention experiments, BMDMs were trained as described above and supplemented with linoleic acid, activator (melittin), and inhibitors (ONO and NAA) as required, and trained BMDMs were stimulated with LPS for 24 hours. Supernatants were collected for cytokines determination.
RNA-seq library preparation and sequencing
The BMDMs were seeded onto six-well cell culture plates at a concentration of 1 × 106 cells per well and trained as described above. Five days later, the culture medium was removed, cells were washed twice with cold PBS and then lysed by adding 1 ml of TRIzol (TaKaRa), and the lysates were collected and stored frozen at −80°C. Extracted RNA was quantified by a Nanodrop ND-2000 system (Thermo Fisher Scientific), and the RNA integrity number was determined by an Agilent Bioanalyzer 4150 system (Agilent Technologies). RNA-seq libraries were prepared using an ABclonal mRNA-seq Lib Prep Kit (ABclonal). Library quality was assessed on an Agilent Bioanalyzer 4150 system, and sequencing was performed with an Illumina Novaseq 6000 instrument.
Data analysis
The raw data were first cleaned up by removing the adapter sequence and filtering out low-quality data (low quality, the number of lines with a string quality value less than or equal to 25 accounts for more than 60% of the entire reading) and N ratio (N, base information that cannot be determined) greater than 5% reads. Then, clean reads were separately aligned to the reference genome with orientation mode using HISAT2 software (http://daehwankimlab.github.io/hisat2/) to obtain mapped reads. Differential expression analysis was performed using the DESeq2 (http://bioconductor.org/packages/release/bioc/html/DESeq2.html), and DEGs with |log2FC| > 1 and adjusted P < 0.05 were considered to be significantly DEGs.
Metabolomics experiments
BMDM cells were seeded onto 10-cm cell culture dishes (5 × 106 cell per well) and trained as described above. After 5 days, the cells were washed twice with cold PBS, 1 ml of cold methanol/acetonitrile/water (2:2:1, v/v) was added, and then the cells were scraped off and stored frozen at −80°C until metabolite analysis. Analysis was performed using a Ultra-high-performance liquid chromatography (UHPLC) (1290 Infinity LC, Agilent Technologies) coupled to a quadrupole time of flight analyzer (AB SCIEX Triple TOF 6600). The samples were slowly thawed at 4°C, vortexed, sonicated at low temperature for 30 min, left at −20°C for 10 min, and then centrifuged at 14,000g for 20 min at 4°C. The supernatant was dried under vacuum. For LC-MS analysis, the dried extracts were redissolved in 100 μl of acetonitrile/water (1:1, v/v) and centrifuged at 14,000g at 4°C for 15 min, and then the supernatant was injected onto a 2.1 mm–by–100 mm ACQUIY UPLC BEH Amide 1.7-μm column (Waters). In both electrospray ionization (ESI) positive and negative modes, mobile phase A contained 25 mM ammonium acetate and 25 mM ammonium hydroxide in water, and mobile phase B was acetonitrile. The gradient was 95% phase B for 0.5 min, linearly reduced to 65% in 6.5 min, then reduced to 40% in 1 min, kept at 40% for 1 min, then increased to 95% in 0.1 min, and kept at 95% for 3 min. The ESI source temperature was 600°C, and the ion spray floating voltage was ±5500 V (positive and negative modes). The product ion scan was acquired using information-dependent acquisition with high sensitivity mode selected.
Data processing
MSConvert was used to convert the raw MS data to MzXML files (82), which were then imported into the publicly accessible XCMS program. For isotope and adduct annotation, CAMERA (Collection of Algorithms for Metabolite Profile Annotation) was used (83). Metabolite identification was accomplished by comparing accuracy mass/charge ratio values (10 parts per million) and MS/MS spectra to authentic standards. To confirm metabolite identification, the processed data were evaluated for quality of integration and compared to recognized standards.
Statistical analysis
Following sum-normalization, the processed data were submitted to multivariate data analysis using the R package (ropls) (84), including Pareto-scaled PCA and orthogonal partial least-squares discriminant analysis (OPLS-DA). The model’s robustness was assessed using sevenfold cross-validation and response permutation testing. Each VIP value of the OPLS-DA model was calculated to indicate its contribution to categorization. The importance of differences between the two groups of independent samples was determined using the student’s t test. VIP > 1 and P value < 0.05 were used to search for significantly changed metabolites. Pearson’s correlation analysis was used to determine the relationship between two variables.
ATAC-seq library preparation and sequencing
BMDMs were seeded onto 24-well cell culture plates at a concentration of 2 × 105 cells per well and trained as described above. After 5 days, the cells were harvested for ATAC-seq analysis by an ATAC-seq kit (Epibiotek). Cells were pretreated with deoxyribonuclease for 30 min at 37°C to remove free-floating DNA and resuspended in cold lysis buffer. After lysis, crude nuclei were harvested and resuspended in 50 μl of transposition mix (10 μl of 5× Tagment DNA (TD) buffer, 5 μl of Tn5 transposase, and 35 μl of water) with 30 min of incubation at 37°C. The DNA was purified and then amplified by polymerase chain reaction. Libraries were quantified with Bioptic Qsep100 Analyzer (Bioptic lnc.) and paired-end sequenced with read lengths of 150.
Data processing
The ENCODE ATAC-seq pipeline was used for quality control and statistical signal processing of the short-read sequencing data (85), producing alignments and measures of enrichment. One hundred and fifty–base pair paired-end reads were mapped to the reference genome build (mouse, mm10). Differentially expressed sites were detected from the ATAC-seq experiments using the DiffBind R package (86). The differential enrichment peaks from different samples were visualized by IGV (87–89).
Enrichment analysis and data visualization
Analysis of RNA-seq data, KEGG, GO enrichment, and GSEA data was conducted by clusterProfiler R package (90). The GSVA R package was used to perform GSVA (91). Enrichment analysis of DEGs for RNA-seq and ATAC-seq was done with Metascape (92). MetaboAnalyst (www.metaboanalyst.ca/) was used to conduct metabolite enrichment and genes and metabolites joint analysis. The data were visualized using the ggplot2 (https://ggplot2.tidyverse.org/) package and GraphPad Prism v.8.
In vivo trained immunity experiments
The animal study was reviewed and approved by Laboratory Animal Welfare and Ethics Committee of Shanghai Public Health Clinical Center (no. 2021-A047-01). Female BALB/c mice, 6 to 8 weeks old, were vaccinated by the intravenous route with 1 × 106 CFU of a BCG strain. Four weeks later, mice were rechallenged using LPS [10 μg per mouse, intraperitoneal (i.p.)] and euthanized to collect the blood and BMCs after 4 hours. Blood was collected by cardiac puncture after anesthesia, and the serum was frozen at −80°C for cytokine determination. Hind leg BMCs were collected, lysed with TRIzol, and frozen at −80°C for RNA-seq. For the survival experiment, mice under anesthesia were challenged with S. typhimurium (300 CFU per mouse, i.p.).
Flow cytometry
BMCs (2 × 106 cells) after RBC lysis were stained with fixable viability dye Zombie Aqua (BioLegend) at the concentration of 1:500 for 30 min (room temperature). Subsequently, the cells were washed with PBS supplemented with 2% bovine serum albumin (BSA; Gibco) and incubated with anti-CD16/32 (clone 2.4G2, BD Pharmingen) at a concentration of 1:100 in PBS/2% BSA at 4°C for 20 min. A panel of fluorescein isothiocyanate (FITC)–conjugated antibodies including anti–Ter-119–FITC (clone Ter119, BD Pharmingen), anti–CD11b-FITC (clone M1/70, BD Pharmingen), anti–immunoglobulin M–FITC (BioLegend), anti–CD3e-FITC (clone 53-6.7, BD Pharmingen), anti–CD45R-FITC (clone RA3-6B2, BD Pharmingen), and anti–Ly6G/C-FITC (clone RB6-8C5, BD Pharmingen) were mixed as cocktail of lineage (Lin) markers. For HSC staining, the following antibodies were used: Lin cocktail, anti–c-Kit–allophycocyanin (clone 2B8, BD Pharmingen), anti–Stem cell antigen-1 (Sca-1)–phycoerythrin (PE)–Cyanine7(clone D7, BD Pharmingen), anti–CD150-BV421 (clone Q38-480, BD Pharmingen), anti–CD48-BB700 (clone HM48-1, BD Pharmingen), anti–Flt3-PE (clone A2F10.1, BD Pharmingen), and anti–CD34-BV711 (clone RAM34, BD Pharmingen) (all 1:100) were added and incubated at 4°C for 30 min. For staining for innate and adaptive immune cells, antibodies for the panel: anti–CD11b–Pacific Blue (clone M1/70, eBioscience), anti–CD11c–PE-Cy7 (clone HL3, BD Bioscience), anti–CD3-PE (clone 145-2C11, eBioscience), and anti–CD19–PE-Cy7 [clone eBio1D3 (1D3), eBioscience] were added and incubated at 4°C for 30 min. All cells were subsequently washed with PBS/2% BSA and resuspended in 1% paraformaldehyde. Cells were acquired on the LSRFortessa (BD Biosciences) and analyzed using FlowJo software (version 10.8.1). All percentages are of single viable frequency, unless otherwise indicated.
Statistical analysis
All experiments were performed in at least three independent biological replicates. Data are presented as means ± SD, as indicated in the legend of each figure, and the experimental condition was compared with the control condition, unless otherwise stated. The significance of the differences between groups was evaluated using One-way or two-way analysis of variance (ANOVA), as shown in the figure legends. Data were judged to be statistically significant when P < 0.05. Survival was analyzed by the log-rank (Mantel-Cox) test.
Acknowledgments
We thank J. Liu from University of Toronto and L. Zhang from Fudan University for providing of BCG strains.
Funding: This work was supported by the grants from the National Key Research and Development Program of China (2022YFC2302900 and 2021YFC2301503), National Natural and Science Foundation of China (82171815 and 82171739), Shanghai Municipal Health Bureau (2022XD060), and Shanghai Science and Technology Commission (19XD1403100).
Author contributions: Conceptualization: X.-Y.F., Z.H., and S.-h.L. Methodology: J.-C.X., X.-J.H., Z.-Y.C., J.W., H.H., L.-F.N., H.-L.W., and J.-H.L. Data analysis: J.-C.X., Z.H., and Z.-Y.C. Writing: J.-C.X. Review and editing: Z.H., J.-C.X., X.-Y.F., and D.-B.L. Supervision and finding: X.-Y.F., Z.H., and S.-h.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq (PRJNA992960 and PRJNA993298) and ATAC-seq (PRJNA993603) data of this study have been deposited in the NCBI Sequence Read Archive. Raw metabolomics data are available on Metabolights under accession number MTBLS8166.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Netea M. G., Joosten L. A. B., Latz E., Mills K. H. G., Natoli G., Stunnenberg H. G., O’Neill L. A. J., Xavier R. J., Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fanucchi S., Domínguez-Andrés J., Joosten L. A. B., Netea M. G., Mhlanga M. M., The intersection of epigenetics and metabolism in trained immunity. Immunity 54, 32–43 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Cheng S.-C., mTOR- and HIF-1 alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Z., Lu S. H., Lowrie D. B., Fan X. Y., Trained immunity: A Yin-Yang balance. MedComm 3, e121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whittaker E., Goldblatt D., McIntyre P., Levy O., Neonatal immunization: Rationale, current state, and future prospects. Front. Immunol. 9, 532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moorlag S., Arts R. J. W., van Crevel R., Netea M. G., Non-specific effects of BCG vaccine on viral infections. Clin. Microbiol. Infect. 25, 1473–1478 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Jiang S., Redelman-Sidi G., BCG in bladder cancer immunotherapy. Cancers (Basel) 14, 1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kremenovic M., Schenk M., Lee D. J., Clinical and molecular insights into BCG immunotherapy for melanoma. J. Intern. Med. 288, 625–640 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Giamarellos-Bourboulis E. J., Tsilika M., Moorlag S., Antonakos N., Kotsaki A., Domínguez-Andrés J., Kyriazopoulou E., Gkavogianni T., Adami M. E., Damoraki G., Koufargyris P., Karageorgos A., Bolanou A., Koenen H., van Crevel R., Droggiti D. I., Renieris G., Papadopoulos A., Netea M. G., Activate: Randomized clinical trial of BCG vaccination against infection in the elderly. Cell 183, 315–323.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdallah A. M., Behr M. A., Evolution and strain variation in BCG. Adv. Exp. Med. Biol. 1019, 155–169 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Chen J. M., Islam S. T., Ren H., Liu J., Differential productions of lipid virulence factors among BCG vaccine strains and implications on BCG safety. Vaccine 25, 8114–8122 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Zhang L., Ru H. W., Chen F. Z., Jin C. Y., Sun R. F., Fan X. Y., Guo M., Mai J. T., Xu W. X., Lin Q. X., Liu J., Variable virulence and efficacy of BCG vaccine strains in mice and correlation with genome polymorphisms. Mol. Ther. 24, 398–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangtani P., Abubakar I., Ariti C., Beynon R., Pimpin L., Fine P. E. M., Rodrigues L. C., Smith P. G., Lipman M., Whiting P. F., Sterne J. A., Protection by BCG vaccine against tuberculosis: A systematic review of randomized controlled trials. Clin. Infect. Dis. 58, 470–480 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Schaltz-Buchholzer F., Bjerregaard-Andersen M., Øland C. B., Golding C., Stjernholm E. B., Monteiro I., Aaby P., Benn C. S., Early vaccination with Bacille Calmette-Guérin-Denmark or BCG-Japan versus BCG-Russia to healthy newborns in Guinea-Bissau: A randomized controlled trial. Clin. Infect. Dis. 71, 1883–1893 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Del Giudice F., Efficacy of three BCG strains (Connaught, TICE and RIVM) with or without secondary resection (re-TUR) for intermediate/high-risk non-muscle-invasive bladder cancers: Results from a retrospective single-institution cohort analysis. J. Cancer Res. Clin. Oncol. 147, 3073–3080 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Giudice F., Efficacy of different Bacillus of Calmette-Guérin (BCG) strains on recurrence rates among intermediate/high-risk non-muscle invasive bladder cancers (NMIBCs): Single-arm study systematic review, cumulative and network meta-analysis. Cancers (Basel) 15, 10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentsch C. A., Birkhäuser F. D., Biot C., Gsponer J. R., Bisiaux A., Wetterauer C., Lagranderie M., Marchal G., Orgeur M., Bouchier C., Bachmann A., Ingersoll M. A., Brosch R., Albert M. L., Thalmann G. N., Bacillus Calmette-Guérin strain differences have an impact on clinical outcome in bladder cancer immunotherapy. Eur. Urol. 66, 677–688 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Witjes J. A., The efficacy of BCG TICE and BCG Connaught in a cohort of 2,099 patients with T1G3 non-muscle-invasive bladder cancer. Urol. Oncol. 34, e419–484.e425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancione S., Alvarez J. V., Alsdurf H., Pai M., Zwerling A. A., Tracking changes in national BCG vaccination policies and practices using the BCG World Atlas. BMJ Glob. Health 7, e007462 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitroulis I., Ruppova K., Wang B., Chen L. S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I., Chatzigeorgiou A., Schlitzer A., Beyer M., Joosten L. A. B., Isermann B., Lesche M., Petzold A., Simons K., Henry I., Dahl A., Schultze J. L., Wielockx B., Zamboni N., Mirtschink P., Coskun Ü., Hajishengallis G., Netea M. G., Chavakis T., Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirovic B., de Bree L. C. J., Groh L., Blok B. A., Chan J., van der Velden W. J. F. M., Bremmers M. E. J., van Crevel R., Händler K., Picelli S., Schulte-Schrepping J., Klee K., Oosting M., Koeken V. A. C. M., van Ingen J., Li Y., Benn C. S., Schultze J. L., Joosten L. A. B., Curtis N., Netea M. G., Schlitzer A., BCG vaccination in humans elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe 28, 322–334.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H., Zheng Z., Cheng T., New paradigms on hematopoietic stem cell differentiation. Protein Cell 11, 34–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song D., Zhang X., Chen J., Liu X., Xue J., Zhang L., Lan X., Wnt canonical pathway activator TWS119 drives microglial anti-inflammatory activation and facilitates neurological recovery following experimental stroke. J. Neuroinflammation 16, 256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam A., Choudhury M. E., Kigami Y., Utsunomiya R., Matsumoto S., Watanabe H., Kumon Y., Kunieda T., Yano H., Tanaka J., Sustained anti-inflammatory effects of TGF-β1 on microglia/macrophages. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 721–734 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Zacharias W. J., Hedgehog is an anti-inflammatory epithelial signal for the intestinal lamina propria. Gastroenterology 138, 2377.e2361–2377.e2364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bekkering S., Arts R. J. W., Novakovic B., Kourtzelis I., van der Heijden C. D. C. C., Li Y., Popa C. D., ter Horst R., van Tuijl J., Netea-Maier R. T., van de Veerdonk F. L., Chavakis T., Joosten L. A. B., van der Meer J. W. M., Stunnenberg H., Riksen N. P., Netea M. G., Metabolic induction of trained immunity through the mevalonate pathway. Cell 172, 135–146.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Nagao T., Kubo T., Fujimoto R., Nishio H., Takeuchi T., Hata F., Ca(2+)-independent fusion of secretory granules with phospholipase A2-treated plasma membranes in vitro. Biochem. J. 307, 563–569 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehfeldt W., Resch K., Goppelt-Struebe M., Cytosolic phospholipase A2 from human monocytic cells: Characterization of substrate specificity and Ca(2+)-dependent membrane association. Biochem. J. 293, 255–261 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruzanski W., Lambeau L., Lazdunsky M., Cho W., Kopilov J., Kuksis A., Differential hydrolysis of molecular species of lipoprotein phosphatidylcholine by groups IIA, V and X secretory phospholipases A2. Biochim. Biophys. Acta 1736, 38–50 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Bomalaski J. S., Steiner M. R., Simon P. L., Clark M. A., IL-1 increases phospholipase A2 activity, expression of phospholipase A2-activating protein, and release of linoleic acid from the murine T helper cell line EL-4. J. Immunol. 148, 155–160 (1992). [PubMed] [Google Scholar]
- 31.Yaacoub C., Wehbe R., Roufayel R., Fajloun Z., Coutard B., Bee venom and its two main components—Melittin and phospholipase A2-As promising antiviral drug candidates. Pathogens 12, 1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bechler M. E., Brown W. J., Gβ1γ2 activates phospholipase A(2)-dependent Golgi membrane tubule formation. Front. Cell Dev. Biol. 2, 0004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dileep K. V., Remya C., Tintu I., Mandal P. K., Karthe P., Haridas M., Sadasivan C., Crystal structure of phospholipase A(2) in complex with 1-naphthaleneacetic acid. IUBMB Life 70, 995–1001 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Roumiguié M., Kamat A. M., Bivalacqua T. J., Lerner S. P., Kassouf W., Böhle A., Brausi M., Buckley R., Persad R., Colombel M., Lamm D., Palou-Redorta J., Soloway M., Brothers K., Steinberg G., Lotan Y., Sylvester R., Alfred Witjes J., Black P. C., International Bladder Cancer Group consensus statement on clinical trial design for patients with Bacillus Calmette-Guérin–exposed high-risk non-muscle-invasive bladder cancer. Eur. Urol. 82, 34–46 (2022). [DOI] [PubMed] [Google Scholar]
- 35.van Puffelen J. H., Keating S. T., Oosterwijk E., van der Heijden A. G., Netea M. G., Joosten L. A. B., Vermeulen S. H., Trained immunity as a molecular mechanism for BCG immunotherapy in bladder cancer. Nat. Rev. Urol. 17, 513–525 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Singh A. K., Praharaj M., Lombardo K. A., Yoshida T., Matoso A., Baras A. S., Zhao L., Srikrishna G., Huang J., Prasad P., Powell J. D., Kates M., McConkey D., Pardoll D. M., Bishai W. R., Bivalacqua T. J., Re-engineered BCG overexpressing cyclic di-AMP augments trained immunity and exhibits improved efficacy against bladder cancer. Nat. Commun. 13, 878 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Andrea D., Comparative effectiveness of intravesical BCG-Tice and BCG-Moreau in patients with non-muscle-invasive bladder cancer. Clin. Genitourin. Cancer 18, 20–25.e2 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Chen Y. K., Huang E. Y. H., Chang Y. H., Kuo J. Y., Chung H. J., Wu H. H. H., Lin T. P., Lin C. C., Fan Y. H., Huang I. S., Lin A. T. L., Huang W. J., The comparison of different BCG strains in the intravesical treatment of non-muscle invasive urothelial carcinoma of urinary bladder-A real-world practice. J. Chin. Med. Assoc. 85, 928–934 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Boehm B. E., Cornell J. E., Wang H., Mukherjee N., Oppenheimer J. S., Svatek R. S., Efficacy of bacillus Calmette-Guérin strains for treatment of nonmuscle invasive bladder cancer: A systematic review and network meta-analysis. J. Urol. 198, 503–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettenati C., Ingersoll M. A., Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat. Rev. Urol. 15, 615–625 (2018). [DOI] [PubMed] [Google Scholar]
- 41.D'Andrea D., Gontero P., Shariat S. F., Soria F., Intravesical bacillus Calmette-Guérin for bladder cancer: Are all the strains equal? Transl. Androl. Urol. 8, 85–93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochando J., Mulder W. J. M., Madsen J. C., Netea M. G., Duivenvoorden R., Trained immunity—Basic concepts and contributions to immunopathology. Nat. Rev. Nephrol. 19, 23–37 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan N., Downey J., Sanz J., Kaufmann E., Blankenhaus B., Pacis A., Pernet E., Ahmed E., Cardoso S., Nijnik A., Mazer B., Sassetti C., Behr M. A., Soares M. P., Barreiro L. B., Divangahi M., M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell 183, 752–770.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behr M. A., Small P. M., A historical and molecular phylogeny of BCG strains. Vaccine 17, 915–922 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Netea M. G., Ziogas A., Benn C. S., Giamarellos-Bourboulis E. J., Joosten L. A. B., Arditi M., Chumakov K., van Crevel R., Gallo R., Aaby P., van der Meer J. W. M., The role of trained immunity in COVID-19: Lessons for the next pandemic. Cell Host Microbe 31, 890–901 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sohrabi Y., Godfrey R., Findeisen H. M., Altered cellular metabolism drives trained immunity. Trends Endocrinol. Metab. 29, 602–605 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Kleinnijenhuis J., Quintin J., Preijers F., Joosten L. A. B., Ifrim D. C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H. G., Xavier R. J., van der Meer J. W. M., van Crevel R., Netea M. G., Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U.S.A. 109, 17537–17542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A., Buttini M., Linster C. L., Medina E., Balling R., Hiller K., Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U.S.A. 110, 7820–7825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gidon A., Louet C., Røst L. M., Bruheim P., Flo T. H., The tumor necrosis factor alpha and interleukin 6 auto-paracrine signaling loop controls Mycobacterium avium infection via induction of IRF1/IRG1 in human primary macrophages. MBio 12, e0212121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nair S., Huynh J. P., Lampropoulou V., Loginicheva E., Esaulova E., Gounder A. P., Boon A. C. M., Schwarzkopf E. A., Bradstreet T. R., Edelson B. T., Artyomov M. N., Stallings C. L., Diamond M. S., Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med. 215, 1035–1045 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreira A. V., Kostidis S., Groh L. A., Koeken V. A. C. M., Bruno M., Baydemir I., Kilic G., Bulut Ö., Andriopoulou T., Spanou V., Synodinou K. D., Gkavogianni T., Moorlag S. J. C. F. M., Charlotte de Bree L., Mourits V. P., Matzaraki V., Koopman W. J. H., van de Veerdonk F. L., Renieris G., Giera M., Giamarellos-Bourboulis E. J., Novakovic B., Domínguez-Andrés J., Dimethyl itaconate induces long-term innate immune responses and confers protection against infection. Cell Rep. 42, 112658 (2023). [DOI] [PubMed] [Google Scholar]
- 52.Laval T., Chaumont L., Demangel C., Not too fat to fight: The emerging role of macrophage fatty acid metabolism in immunity to Mycobacterium tuberculosis. Immunol. Rev. 301, 84–97 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Stiekema L. C. A., Willemsen L., Kaiser Y., Prange K. H. M., Wareham N. J., Boekholdt S. M., Kuijk C., de Winther M. P. J., Voermans C., Nahrendorf M., Stroes E. S. G., Kroon J., Impact of cholesterol on proinflammatory monocyte production by the bone marrow. Eur. Heart J. 42, 4309–4320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bekkering S., Quintin J., Joosten L. A. B., van der Meer J. W. M., Netea M. G., Riksen N. P., Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler. Thromb. Vasc. Biol. 34, 1731–1738 (2014). [DOI] [PubMed] [Google Scholar]
- 55.van der Heijden C., Keating S. T., Groh L., Joosten L. A. B., Netea M. G., Riksen N. P., Aldosterone induces trained immunity: The role of fatty acid synthesis. Cardiovasc. Res. 116, 317–328 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Diray-Arce J., Angelidou A., Jensen K. J., Conti M. G., Kelly R. S., Pettengill M. A., Liu M., van Haren S., McCulloch S., Michelloti G., Idoko O.; EPIC Consortium, Kollmann T. R., Kampmann B., Steen H., Ozonoff A., Lasky-Su J., Benn C. S., Levy O., Bacille Calmette-Guérin vaccine reprograms human neonatal lipid metabolism in vivo and in vitro. Cell Rep. 39, 110772 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan B., Fung K., Ye S., Lai P. M., Wei Y. X., Sze K. H., Yang D., Gao P., Kao R. Y. T., Linoleic acid metabolism activation in macrophages promotes the clearing of intracellular Staphylococcus aureus. Chem. Sci. 13, 12445–12460 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nava Lauson C. B., Tiberti S., Corsetto P. A., Conte F., Tyagi P., Machwirth M., Ebert S., Loffreda A., Scheller L., Sheta D., Mokhtari Z., Peters T., Raman A. T., Greco F., Rizzo A. M., Beilhack A., Signore G., Tumino N., Vacca P., McDonnell L. A., Raimondi A., Greenberg P. D., Huppa J. B., Cardaci S., Caruana I., Rodighiero S., Nezi L., Manzo T., Linoleic acid potentiates CD8(+) T cell metabolic fitness and antitumor immunity. Cell Metab. 35, 633–650.e9 (2023). [DOI] [PubMed] [Google Scholar]
- 59.Matsuoka T., Adair J. E., Lih F. B., Hsi L. C., Rubino M., Eling T. E., Tomer K. B., Yashiro M., Hirakawa K., Olden K., Roberts J. D., Elevated dietary linoleic acid increases gastric carcinoma cell invasion and metastasis in mice. Br. J. Cancer 103, 1182–1191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishioka N., Matsuoka T., Yashiro M., Hirakawa K., Olden K., Roberts J. D., Linoleic acid enhances angiogenesis through suppression of angiostatin induced by plasminogen activator inhibitor 1. Br. J. Cancer 105, 1750–1758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morito K., Shimizu R., Kitamura N., Park S. B., Kishino S., Ogawa J., Fukuta T., Kogure K., Tanaka T., Gut microbial metabolites of linoleic acid are metabolized by accelerated peroxisomal β-oxidation in mammalian cells. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1619–1628 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Devillard E., McIntosh F. M., Duncan S. H., Wallace R. J., Metabolism of linoleic acid by human gut bacteria: Different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol. 189, 2566–2570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J., Guasch-Ferré M., Li Y., Hu F. B., Dietary intake and biomarkers of linoleic acid and mortality: Systematic review and meta-analysis of prospective cohort studies. Am. J. Clin. Nutr. 112, 150–167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez J. A., Skiba M. B., Chow H. H. S., Chew W. M., Saboda K., Lance P., Ellis N. A., Jacobs E. T., A protective role for arachidonic acid metabolites against advanced colorectal adenoma in a phase III trial of selenium. Nutrients 13, 3877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yousefi M., Dietary intake and biomarkers of linoleic acid and risk of prostate cancer in men: A systematic review and dose-response meta-analysis of prospective cohort studies. Crit. Rev. Food Sci. Nutr. 1-17, 10.1080/10408398.2023.2200840 (2023). [DOI] [PubMed] [Google Scholar]
- 66.Christ A., Günther P., Lauterbach M. A. R., Duewell P., Biswas D., Pelka K., Scholz C. J., Oosting M., Haendler K., Baßler K., Klee K., Schulte-Schrepping J., Ulas T., Moorlag S. J. C. F. M., Kumar V., Park M. H., Joosten L. A. B., Groh L. A., Riksen N. P., Espevik T., Schlitzer A., Li Y., Fitzgerald M. L., Netea M. G., Schultze J. L., Latz E., Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–175.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mercola J., D'Adamo C. R., Linoleic acid: A narrative review of the effects of increased intake in the standard american diet and associations with chronic disease. Nutrients 15, 3129 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verri P., Baboudjian M., Diana P., Gallioli A., Territo A., Gaya J. M., Huguet J., Rodriguez-Faba O., Palou J., Breda A., Reduced- vs full-dose BCG in bladder cancer: A systematic review and meta-analysis. Actas Urol Esp (Engl Ed) 47, 4–14 (2023). [DOI] [PubMed] [Google Scholar]
- 69.Choi S. Y., Ha M. S., Kim J. H., Chi B. H., Kim J. W., Chang I. H., Kim T. H., Myung S. C., Low-dose versus standard-dose bacille Calmette-Guérin for non-muscle-invasive bladder cancer: Systematic review and meta-analysis of randomized controlled trials. Invest. Clin. Urol. 63, 140–150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersen A., Roth A., Jensen K. J., Erikstrup C., Lisse I. M., Whittle H., Sartono E., Yazdanbakhsh M., Aaby P., Benn C. S., The immunological effect of revaccination with Bacille Calmette-Guérin vaccine at 19 months of age. Vaccine 31, 2137–2144 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Barreto M. L., Pereira S. M., Pilger D., Cruz A. A., Cunha S. S., Sant’Anna C., Ichihara M. Y., Genser B., Rodrigues L. C., Evidence of an effect of BCG revaccination on incidence of tuberculosis in school-aged children in Brazil: Second report of the BCG-REVAC cluster-randomised trial. Vaccine 29, 4875–4877 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Debisarun P. A., Kilic G., de Bree L. C. J., Pennings L. J., van Ingen J., Benn C. S., Aaby P., Dijkstra H., Lemmers H., Domínguez-Andrés J., van Crevel R., Netea M. G., The impact of BCG dose and revaccination on trained immunity. Clin. Immunol. 246, 109208 (2023). [DOI] [PubMed] [Google Scholar]
- 73.Wilkinson N. M., Chen H. C., Lechner M. G., Su M. A., Sex Differences in Immunity. Annu. Rev. Immunol. 40, 75–94 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Márquez E. J., Chung C. H., Marches R., Rossi R. J., Nehar-Belaid D., Eroglu A., Mellert D. J., Kuchel G. A., Banchereau J., Ucar D., Sexual-dimorphism in human immune system aging. Nat. Commun. 11, 751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kleinnijenhuis J., Quintin J., Preijers F., Benn C. S., Joosten L. A. B., Jacobs C., van Loenhout J., Xavier R. J., Aaby P., van der Meer J. W. M., van Crevel R., Netea M. G., Long-lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J. Innate Immun. 6, 152–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarkar S., Mishra A., Periasamy S., Dyett B., Dogra P., Ball A. S., Yeo L. Y., White J. F., Wang Z., Cristini V., Jagannath C., Khan A., Soni S. K., Drummond C. J., Conn C. E., Prospective subunit nanovaccine against Mycobacterium tuberculosis infection─Cubosome lipid nanocarriers of cord factor, trehalose 6,6′ dimycolate. ACS Appl. Mater. Interfaces 15, 27670–27686 (2023). [DOI] [PubMed] [Google Scholar]
- 77.Ly A., Liu J., Mycobacterial virulence factors: Surface-exposed lipids and secreted proteins. Int. J. Mol. Sci. 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rahlwes K. C., Dias B. R. S., Campos P. C., Alvarez-Arguedas S., Shiloh M. U., Pathogenicity and virulence of Mycobacterium tuberculosis. Virulence 14, 2150449 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdallah A. M., Hill-Cawthorne G. A., Otto T. D., Coll F., Guerra-Assunção J. A., Gao G., Naeem R., Ansari H., Malas T. B., Adroub S. A., Verboom T., Ummels R., Zhang H., Panigrahi A. K., McNerney R., Brosch R., Clark T. G., Behr M. A., Bitter W., Pain A., Genomic expression catalogue of a global collection of BCG vaccine strains show evidence for highly diverged metabolic and cell-wall adaptations. Sci. Rep. 5, 15443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beccaria M., Franchina F. A., Nasir M., Mellors T., Hill J. E., Purcaro G., Investigation of mycobacteria fatty acid profile using different ionization energies in GC-MS. Anal. Bioanal. Chem. 410, 7987–7996 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Leung A. S., Tran V., Wu Z., Yu X., Alexander D. C., Gao G. F., Zhu B., Liu J., Novel genome polymorphisms in BCG vaccine strains and impact on efficacy. BMC Genomics 9, 413 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., Mallick P., A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuhl C., Tautenhahn R., Böttcher C., Larson T. R., Neumann S., CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 84, 283–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thévenot E. A., Roux A., Xu Y., Ezan E., Junot C., Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335 (2015). [DOI] [PubMed] [Google Scholar]
- 85.B. C. Hitz, The ENCODE uniform analysis pipelines. bioRxiv [Preprint] (2023).
- 86.R. Stark, G. Brown, DiffBind: Differential binding analysis of ChIP-Seq peak data (Cancer Research, UK, 2011).
- 87.Robinson J. T., Thorvaldsdóttir H., Wenger A. M., Zehir A., Mesirov J. P., Variant review with the Integrative Genomics Viewer. Cancer Res. 77, e31–e34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G., ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., Benner C., Chanda S. K., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S6
Tables S1 to S6