Abstract
At least seven viral genes encode proteins (UL6, UL15, UL17, UL25, UL28, UL32, and UL33) that are required for DNA cleavage and packaging of herpes simplex virus type 1 (HSV-1) DNA. Sequence analysis reveals that UL15 shares homology with gp17, the large catalytic subunit of the bacteriophage T4 terminase. Thus, UL15 may play a direct role in the cleavage of viral DNA replication intermediates into monomers. In this study, we asked whether UL15 and other cleavage and packaging proteins could be detected in capsids isolated from infected cells. Consistent with previous studies showing that UL6 and UL25 are minor protein constituents of the capsids, we detected these proteins in both B and C capsids. In contrast, the previously identified full-length version (81 kDa) of UL15 was found predominantly in B capsids and in much smaller amounts in C capsids. In addition, the UL28 protein was found predominantly in B but not C capsids in a distribution similar to that of the 81-kDa version of UL15. These results suggest that UL28 and the 81-kDa form of UL15 are transiently associated with capsid intermediates during the packaging process. Surprisingly, however, a previously unidentified 87-kDa form of UL15 was found in the B and C capsids and in virions. Analysis of cells infected with mutants individually lacking UL6, UL15, UL25, UL28, or UL32 demonstrates that the lack of one cleavage and packaging protein does not affect the expression of the others. Furthermore, this analysis, together with guanidine HCl extraction analysis of purified capsids, indicates that UL6, UL25, and UL28 are able to associate with B capsids in the absence of other DNA cleavage and packaging proteins. On the other hand, the two UL15-related proteins (81 and 87 kDa) do not associate efficiently with B capsids in cells infected with UL6 and UL28 mutants. These results suggest that the ability of the UL15-related proteins to bind to B capsids may be mediated through interactions with UL6 and UL28.
In cells infected with herpes simplex virus type 1 (HSV-1), three types of intracellular capsids have been identified by sucrose gradient sedimentation and electron microscopic analysis: A capsids (empty), B capsids (containing scaffold protein), and C capsids (containing DNA) (14, 18, 31, 39). The shells of all three capsid types have a similar structural composition: they contain VP5 (major capsid protein), VP19C, VP23 (triplex proteins), and VP26 (9, 16, 17, 27, 31, 35, 37, 38, 54). DNA-containing C capsids represent the products of successful DNA packaging events. B capsids were initially thought to be analogous to phage proheads in that B capsids contain a protein scaffold composed mainly of VP22a, which is lost from capsids when DNA is packaged. However, in a cell-free capsid assembly system, a fourth form of capsids has been recently discovered, which is a spherical, unstable structure containing the precursor form of the scaffold protein (30, 48). It has been suggested that these less-angular and more-open structures rather than B capsids are authentic procapsid intermediates. Although B capsids may be a dead-end product of the capsid maturation process, they represent the most closely related structures to procapsids that can be isolated as stable structures. The empty A capsids which lack both DNA and scaffold are thought to result from abortive attempts at DNA encapsidation (34).
At least seven genes encode proteins (UL6, UL15, UL17, UL25, UL28, UL32, and UL33), that are required for the DNA cleavage and packaging process, in which replicated concatemeric DNA is cleaved into unit-size monomers and encapsidated into preformed capsids (41; for a review, see reference 49). The functions of each of the cleavage and packaging proteins have not been elucidated. Mutant viruses defective in UL6, UL15, UL17, UL28, UL32, or UL33 are defective in DNA cleavage and packaging, and cells infected with these mutants produce only B capsids (2, 4, 24, 25, 33, 41, 47, 52). The absence of A and C capsids is taken as evidence that cleavage and packaging was not even attempted in cells infected with these mutants. These results suggest that these proteins function at relatively early stages of the cleavage and packaging process. A recently described mutant virus defective in UL25, on the other hand, is capable of DNA cleavage; the accumulation of A and B capsids in cells infected with this mutant virus indicates that UL25 is required for stable retention of DNA in capsids (26). Thus, it is likely that UL25 functions later in the process than the other known cleavage and packaging proteins (26). The phenotype of the UL25 mutant is somewhat reminiscent of that of UL12 (alkaline nuclease)-null mutants, which are capable of cleavage of viral genomes and which also display an elevated ratio of A to B capsids (42). We have proposed that in cells infected with the UL12-null mutant, abnormally processed DNA is packaged, leading to the accumulation of unstable DNA-containing capsids which fail to bud from the nucleus and disgorge viral DNA to generate elevated levels of A capsids.
One key component of the cleavage and packaging machinery of the better-studied double-stranded DNA bacteriophages is a two-subunit terminase which binds and cleaves concatemeric viral DNA into monomers and translocates the DNA into capsids by using energy from ATP hydrolysis (reviewed in reference 7). In addition to the terminase complex, the bacteriophage cleavage and packaging machinery also includes a prohead with an internal scaffold and a portal vertex through which DNA is taken up (reviewed in reference 7). The portal protein is found as a dodecameric ring at the unique vertex of most bacteriophages and is considered an integral part of the capsid itself (reviewed in reference 5). The terminase complex, however, is only transiently associated with proheads and is not found in mature capsids (28). Other phage packaging proteins, such as lambda gpD, have been reported to form a stable association with mature capsids, but these proteins are not found in procapsids (19, 20). They are believed to function later in the cleavage and packaging process to enhance the stability of DNA-containing capsids (45).
By analogy with bacteriophages, the presence or absence of HSV-1 cleavage and packaging proteins in procapsids and mature virions may provide clues as to their possible functions. UL6 and UL25, two of the HSV-1 DNA cleavage and packaging proteins, are found in capsids and in mature virions (1, 24, 26, 32). We recently showed that UL32, another cleavage and packaging protein, cannot be detected in capsids or in virions (25). We have proposed that this protein may be involved in the efficient localization of procapsids to the location within the cell at which cleavage and packaging occur. We and others have previously demonstrated that two proteins from the UL15 locus are expressed in infected cells: a full-length 81-kDa form and a 30-kDa protein translated separately from the UL15.5 open reading frame (4, 52). In this report, we examined whether any UL15 species could be detected in capsids. We report that the 30-kDa UL15.5 protein cannot be detected specifically in any capsid form, whereas the 81-kDa form of UL15 is detected predominantly in B capsids and in much lower levels in C capsids and in virions. In addition, the UL28 protein was found predominantly in B but not C capsids in a distribution pattern similar to that of the 81-kDa version of UL15. Taken together with the previous report that UL15 shares homology with the ATP binding motif of the catalytic subunit (gp17) of bacteriophage T4 terminase (15), these results suggest that UL15 and UL28 may constitute the two subunits of the putative HSV-1 terminase. Moreover, we found that while the capsid association of UL6, UL25, and UL28 does not require the presence of other DNA cleavage and packaging proteins, cells infected with UL6 or UL28 mutants display altered associations of UL15 with capsids. This suggests that both UL6 and UL28 play a role in mediating the interaction of UL15 with capsids. Models for the possible roles of UL6, UL15, and UL28 in the cleavage and packaging process will be discussed.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
African green monkey kidney cells (Vero; American Type Culture Collection, Rockville, Md.) were propagated and maintained as described previously (50). Cell lines permissive for the individual DNA cleavage and packaging mutant were propagated as described above, but with the addition of 100 μg of the antibiotic G418 (Geneticin, a neomycin analog; GIBCO Laboratories, Grand Island, N.Y.) per ml of medium. The KOS strain of HSV-1 was used as the wild-type virus. Mutant HSV-1 viruses in which the ICP6::lacZ mutagenic cassette was used to disrupt UL6 (hr74), UL15 (hr81-1 and hr81-2), or UL32 (hr64) have been described previously (24, 25, 52). Insertion mutants were propagated on complementing cell line UL6-31 (UL6), C-2 (UL15), or 158 (UL32) (24, 25, 52). Mutant KUL25NS virus containing an in-frame stop codon in the UL25 gene and gCB virus containing a 1,881-bp deletion in the UL28 gene, as well as permissive cell lines (8-1 and A1, respectively), were kindly provided by Fred Homa (Pharmacia and Upjohn, Kalamazoo, Mich.) (26, 47). An anti-ICP8 polyclonal antibody was a kind gift from William Ruyechan (SUNY, Buffalo, N.Y.) (43), and an anti-VP5 polyclonal antibody (NC-1) was a kind gift from Gary H. Cohen and Roselyn J. Eisenberg (University of Pennsylvania, Philadelphia) (13). The polyclonal anti-UL6 and anti-UL15 antibodies were described previously (24, 25, 52). The polyclonal anti-VP16, anti-UL25, and anti-UL28 antibodies were generously provided by Daniel J. Tenney (Bristol-Myers Squibb, Wallingford, Conn.). The anti-UL32 polyclonal antibody was kindly donated by Bernard Roizman (University of Chicago, Chicago, Ill.) (12).
Cell fractionation.
Cell fractionation of infected Vero cells was carried out with a modification of a previously described procedure (51). KOS virus-infected Vero cells were lysed in hypotonic buffer (20 mM HEPES [pH 7.9], 10 mM KCl, 20 mM NaF, 0.1 mM Na3VO4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 3 μg of aprotinin per ml, 1 μg of pepstatin per ml, 1 mM dithiothreitol, 10% glycerol) containing 0.2% Nonidet P-40 (NP-40). After 10 min of incubation on ice, cell lysates were subjected to centrifugation in a microcentrifuge (12,400 rpm) at 4°C for 1 min. The supernatant was collected and designated as the first cytoplasmic extract. The pellet was resuspended in hypotonic buffer containing 0.2% NP-40, and the extraction was repeated. The supernatant was saved and designated as the second cytoplasmic extract. The pellet (containing crude nuclei) was resuspended in high-salt buffer (hypotonic buffer with 20% glycerol and 420 mM NaCl) and incubated at 4°C for 30 min. The resuspended nuclei were then subjected to centrifugation (12,400 rpm in microcentrifuge) at 4°C for 10 min. The supernatant was collected as the first nuclear extract. The pellet was subjected to high-salt-buffer extraction a second time, and the supernatant was saved and designated as the second nuclear extract. The pellet was designated as the insoluble fraction.
Analysis of capsids and virions.
HSV-1 capsids were isolated as described by Sherman and Bachenheimer (44) with some modifications. Five 152-cm2 flasks of confluent Vero cells were infected with either KOS virus or mutants defective in individual DNA cleavage and packaging genes at a multiplicity of infection (MOI) of 6 PFU per cell. At 20 h postinfection, cells were collected by centrifugation at 4,000 rpm in a GSA rotor for 10 min, rinsed with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]), resuspended in lysis buffer (a phosphate buffer containing 40 mM phosphate [pH 7.4] and 150 mM NaCl) containing 0.5% NP-40 and proteinase inhibitors (1 mM PMSF, 5 μg of pepstatin A per ml, 5 μg of leupeptin per ml), and incubated at 4°C for 30 min. Cell lysates were then snap frozen and thawed and cleared of cell debris by centrifugation at 6,000 rpm in an SS-34 rotor for 10 min and subsequently at 2,500 rpm in a microcentrifuge for 3 min. The supernatant was then subjected to probe sonication, layered onto a 15 to 45% (wt/wt [in phosphate buffer]) sucrose gradient, and centrifuged at 35,000 rpm for 30 min in an SW40 rotor. All centrifugation steps were carried out at 4°C. Capsid bands were identified by light scattering upon illumination with a halogen fiber optic lamp. The gradients were collected with a piston gradient fractionator (BioComp Instruments, Inc., New Brunswick, Canada) at 1 ml per fraction. Trichloroacetic acid was added to a final concentration of 10%, and the samples were stored at −20°C overnight. The precipitated proteins were collected by centrifugation in a microcentrifuge at 12,400 rpm for 30 min and resuspended in 150 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Mature virions were prepared as described previously (46). Sucrose gradient fractions and purified virions were subjected to SDS-PAGE and Western blot analysis as described below.
Preparation of extracellular extracts.
One 152-cm2 flask of 80% confluent Vero cells was infected with wild-type or mutant strains of HSV-1 at an MOI of 10 PFU per cell for 20 h. The media were carefully decanted into a 50-ml centrifuge bottle and subjected to low-speed centrifugation (4°C) at 5,000 rpm in an SS-34 rotor for 10 min to remove cell debris. The supernatant was collected and subjected to low-speed centrifugation a second time. The resulting supernatant was then subjected to centrifugation (4°C) at 15,000 rpm in an SW28 rotor for 2 h to pellet extracellular particles. The pellet was resuspended in 100 μl of 1× SDS sample buffer and used for analysis.
GuHCl extraction.
Guanidine hydrochloride (GuHCl) extraction was performed as described by Newcomb and Brown (29). In brief, B capsids were purified following sucrose gradient centrifugation and were diluted in lysis buffer (phosphate buffer containing 0.5% NP-40 and the proteinase inhibitor cocktail). A 6.0 M concentration of GuHCl was slowly added to the capsid samples with rigorous stirring to final concentrations of 0, 0.5, 1.0, or 2.0 M. The samples were incubated on ice for 1 h with occasional agitation, and the capsids were recovered by centrifugation through 150 μl of a 25% sucrose cushion (in phosphate buffer) in an SW50.1 rotor at 23,000 rpm for 1 h. The capsids were resuspended in 150 μl of 1× SDS sample buffer for analysis.
Silver staining, Western blotting, and quantitative analysis.
Cell lysates or sucrose gradient fraction samples resuspended in 1× SDS-PAGE sample buffer were heated at 95°C for 5 min, vortexed vigorously for 30 s, and subjected to electrophoresis on an SDS–10% polyacrylamide gel. The gels were then analyzed by silver staining or Western blotting. Silver staining was carried out as described previously (3). The ECL (enhanced chemiluminescence) method of Western blotting analysis was performed for all proteins except UL6 according to the manufacturer’s instructions (Amersham, Buckinghamshire, England) as previously described (53). Because the anti-UL6 antibody did not perform well by the ECL method, UL6 immunoblots were performed with an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Promega, Madison, Wis.) and alkaline phosphatase color development as previously described (24). The anti-UL29, anti-VP16, anti-VP5, anti-UL6, anti-UL15, anti-UL25, anti-UL28, and anti-UL32 polyclonal antibodies were used as the primary antibodies at dilutions of 1:2,000, 1:1,000, 1:10,000, 1:1,000, 1:2,000, 1:2,000, 1:1,000, and 1:2,000 (in 5% nonfat milk in PBST [PBS containing 0.2% Tween 20]), respectively. Protein quantification was performed as suggested by the manufacturer’s instructions with bands which were in the linear response range. The protein bands were quantified with ImageQuaNT, version 1.2 (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Expression of UL15 in mammalian cells.
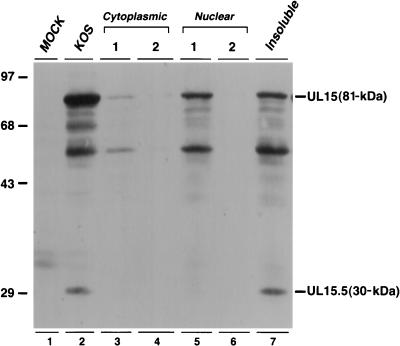
A detailed biochemical analysis of the function of UL15 in the cleavage and packaging process will require the expression of the soluble UL15 protein. Full-length UL15 expressed in insect cells infected with recombinant baculovirus, however, was found to be present in the insoluble fraction (data not shown). We next examined UL15 expression in HSV-1-infected mammalian cells. Vero cells were infected with strain KOS of HSV-1 and fractionated into two cytoplasmic fractions, two nuclear fractions, and an insoluble fraction (see Materials and Methods). Each fraction was examined by immunoblotting with a polyclonal antibody raised against the second exon of UL15 (52). The full-length 81-kDa UL15 protein was detected in both the first nuclear fraction and the insoluble fraction (Fig. 1, lanes 5 and 7). The presence of UL15 in the insoluble fraction was not due to incomplete nuclear extraction, since no soluble UL15 was detected in the second nuclear fraction (Fig. 1, lane 6). Consistent with previous results obtained by immunofluorescence (53), very little UL15 was detected in either of the two cytoplasmic fractions. We estimate that about 50% of the UL15 protein expressed during viral infection is in an insoluble form. Interestingly, UL15.5, an N-terminally truncated gene product of the UL15 open reading frame (52), was found only in the insoluble fraction (Fig. 1, lane 7). Figure 1 also shows a total-cell extract from KOS-infected cells and indicates that in addition to the previously described 81- and 30-kDa UL15 bands, several other bands also react with the UL15 antiserum (lane 2); these may represent proteolytic products of the 81-kDa UL15 protein or other proteins which cross-react with the antiserum. In summary, full-length UL15 is partially soluble in infected-cell extracts. The insoluble fraction of UL15 may represent protein which is associated with capsids or with nuclear matrix.
FIG. 1.
Distribution of UL15 and UL15.5 in infected-Vero-cell extracts. Vero cells were infected with KOS at an MOI of 10 PFU per cell for 16 h. Cell lysates were fractionated into cytoplasmic, nuclear, and insoluble fractions as described in Materials and Methods. Proteins were revealed by ECL Western blot analysis with an anti-UL15 polyclonal antibody. The exposure time for the blot shown here was 5 min. The positions and sizes of UL15 and UL15.5, along with the migration of size markers (in kilodaltons), are indicated.
Expression of DNA cleavage and packaging proteins in infected mammalian cells.
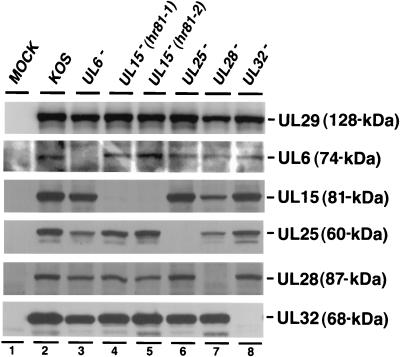
The involvement of at least six genes in the HSV-1 DNA cleavage and packaging process suggests potentially complex protein-protein interactions among various components. It is possible that one or more of these proteins may regulate the expression of the others at the transcriptional, translational, or posttranslational levels. In order to test this possibility, Vero cells were infected with KOS or mutant viruses defective in UL6, UL15, UL25, UL28, or UL32, and the cell lysates were examined by immunoblot analysis (Fig. 2). Since all viruses used in this experiment are expected to exhibit a normal pattern of early protein expression, the levels of UL29 (ICP8) detected by an anti-ICP8 antibody were used to indicate the relative amount of lysate added to each lane of the SDS-PAGE gel. Figure 2 shows that mutant viruses defective in UL15 express other cleavage and packaging proteins (i.e., UL6, UL25, UL28, and UL32) at levels similar to those expressed in cells infected with wild-type virus; as expected, these mutant viruses are not capable of expressing UL15 (lanes 4 and 5). Likewise, the levels of individual cleavage and packaging proteins expressed in cells infected with either KOS, UL25, or UL32 mutant viruses do not vary significantly. In cells infected with the UL6 and UL28 mutants, the slightly lower levels of UL15 and UL25 are likely due to a smaller amount of cell lysate loaded, as indicated by the levels of UL29 (Fig. 2, lanes 3 and 7). In several independent experiments, expression levels of UL6, UL15, UL25, and UL32 were not significantly lower in cells infected with mutant viruses lacking UL6 or UL28 (data not shown). Thus, we have demonstrated that the five DNA cleavage and packaging proteins tested are not coregulated.
FIG. 2.
Analysis of total-cell lysates from cells infected with wild-type and mutant viruses defective in individual DNA cleavage and packaging genes. Vero cells were either mock infected or infected with wild-type KOS virus or mutant UL6 (hr74), UL15 (hr81-1 and hr81-2), UL25 (KUL25NS), UL28 (gCB), or UL32 (hr64) virus at an MOI of 10 PFU per cell for 16 h. Total-cell lysates were examined for the expression of individual proteins (except UL6) as indicated by ECL immunoblotting. All immunoblottings for UL6 in this paper were carried out by alkaline phosphatase color development as described in Materials and Methods.
The 81-kDa version of UL15 is associated predominantly with B but not C capsids.
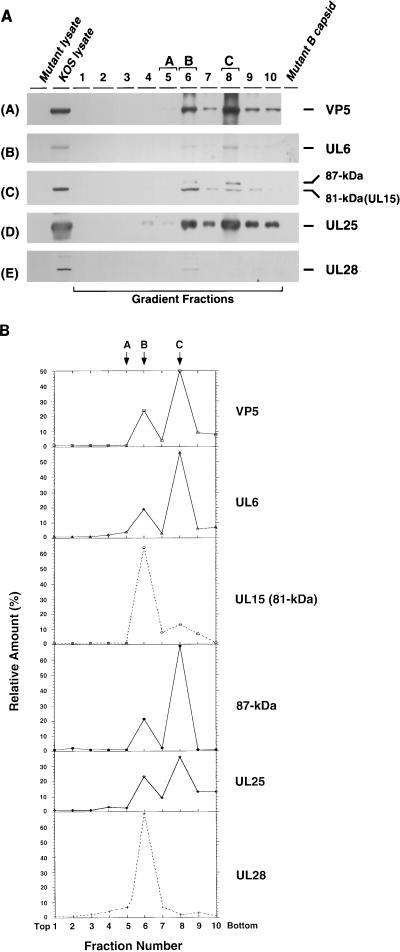
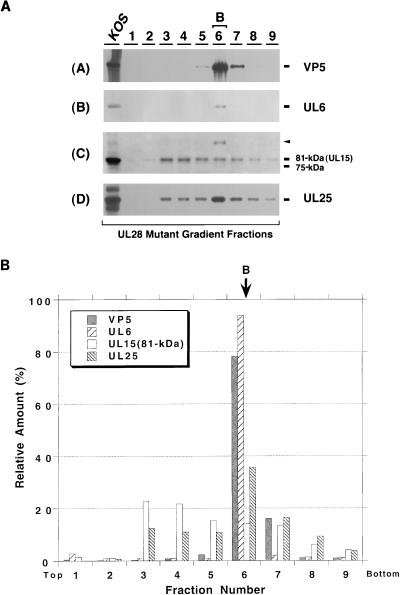
By analogy with the double-stranded DNA bacteriophages, it is expected that components of the HSV-1 cleavage and packaging machinery will exhibit various types of interactions with procapsids. UL6 and UL25 have been detected in all three types of capsids (A, B, and C) and in mature virions (1, 25, 26, 32), whereas UL32 cannot be detected in capsids or virions (25). In this study, we asked whether UL15 and UL28 are associated with capsids. Vero cells were infected with KOS, and cell lysates were loaded onto a 15 to 45% sucrose gradient and subjected to velocity ultracentrifugation. The gradient was then fractionated into 10 fractions (Fig. 3A). Fractions containing A (fraction 5), B (fraction 6), or C (fraction 8) capsids were identified by light scattering as described in Materials and Methods. The assignment of A, B, and C capsids to fractions 5, 6, and 8, respectively, was confirmed by silver staining: bands corresponding to VP5 were detected in fractions 6 and 8, whereas VP22a (scaffold) was detected only in fraction 6 (data not shown). The presence of capsids in these fractions was also confirmed by immunoblot analysis with an antibody against the major capsid protein (VP5) (Fig. 3A, panel A). Only fractions 6 and 8 contain elevated levels of VP5. The failure to detect VP5 in fraction 5 is likely due to the lower abundance of A capsids compared to B and C capsids. Samples were also analyzed for the presence of DNA cleavage and packaging proteins by immunoblot analysis with the appropriate antisera. As previously reported (1, 25, 26, 32), UL6 and UL25 were detected predominantly in fraction 6 (containing B capsids) and fraction 8 (containing C capsids) (Fig. 3A, panels B and D). The failure to detect UL6 and UL25 in A capsids in this experiment is probably due to the small amount of A capsids present. Under the same conditions, VP16 was found in all fractions of the gradient, consistent with previous reports (19) (data not shown). The relative amounts of individual proteins in various fractions were determined by quantitative analysis as described in Materials and Methods (Fig. 3B). The levels of UL6 and UL25 clearly peaked in the B and C capsid-containing fractions. The elevated levels of UL6 and UL25 in C compared to B capsids likely reflect the higher levels of C capsids in this experiment (Fig. 3B).
FIG. 3.
Sucrose gradient analysis of lysates from KOS-infected cells. KOS-infected cell lysates were fractionated by ultracentrifugation on a 15 to 45% (wt/wt) sucrose gradient (in phosphate buffer) as described in Materials and Methods. Ten fractions from the gradient were collected and examined for individual proteins as indicated. (A) Gradient fractions were examined by immunoblotting with the appropriate antibodies for VP5, UL6, UL15, UL25, and UL28 as indicated. The UL15-related bands were revealed by exposure for 20 min. KOS lysate represents a total-cell lysate from KOS-infected cells; Mutant lysate represents a mock-infected-cell lysate (A), or a total-cell lysate from cells infected with the mutant virus defective for UL6 (B), UL15 (C), UL25 (D), or UL28 (E); Mutant B capsid represents the B capsids isolated from cells infected with a mutant virus defective for UL6 (B), UL15 (C), UL25 (D), or UL28 (E). (B) Quantification of the immunoblots from Fig. 3A was carried out as described in Materials and Methods. The y axis shows the relative amount of each protein in each fraction calculated as a percentage of the total amount of protein. The total amount of protein was calculated by summing the amount of protein in each fraction. Fractions 5, 6, and 8 contain the A, B, and C capsids, respectively. The top and bottom of the gradient are indicated.
The same gradient fractions were examined for the presence of UL15 by immunoblot analysis. An 81-kDa protein corresponding to the previously identified full-length UL15 protein (52) was found predominantly in fraction 6 (containing B capsids) (Fig. 3A, panel C). While fraction 8 (containing C capsids) contains higher levels of VP5, UL6, and UL25 than does fraction 6, much lower levels of the 81-kDa band was detected in this fraction. The specificity of the antibody was also examined in this blot: the 81-kDa protein band is clearly detected in a total-cell lysate from KOS-infected cells, but not in a total-cell lysate from cells infected with hr81-1 (a UL15 mutant) or in purified B capsids from hr81-1-infected cells (Fig. 3A, panel C). Further indication of the predominant association of UL15 with B but not C capsids comes from the quantification shown in Fig. 3B. The level of UL15 in B capsids is higher than that seen in C capsids, despite the higher level of C capsids compared to B capsids in this experiment. In addition, the 30-kDa product of the UL15.5 open reading frame was detected throughout the gradient, indicating that this protein is not likely to be specifically associated with capsids (data not shown). In summary, we conclude that in contrast to UL6 and UL25, the 81-kDa UL15 protein is predominantly associated with B capsids.
Detection in capsids of a previously unidentified 87-kDa protein which reacts with UL15 antiserum.
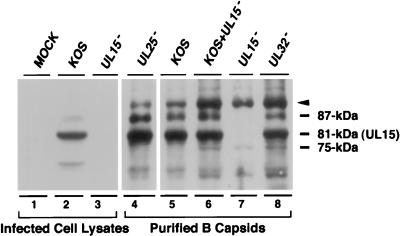
Interestingly, the UL15 immunoblot in Fig. 3A reveals the presence of an 87-kDa band in addition to the full-length 81-kDa UL15 protein band (Fig. 3A, panel C). This 87-kDa band was detected weakly in fraction 6 (B capsids) and more strongly in fraction 8 (C capsids), correlating with the levels of UL6, UL25, and VP5 (Fig. 3B). Thus, while the 81-kDa protein is found predominantly in B capsids, the 87-kDa band is found in both B and C capsids. This 87-kDa protein was also undetectable in either UL15-null mutant-infected-cell lysate or B capsids lacking UL15, and, surprisingly, it was not detected in the KOS-infected cell lysate. In order to address whether the 87-kDa form seen in B and C capsids is related to UL15 rather than a cross-reacting protein, we examined purified B capsids from cells infected with KOS or with mutants lacking UL15, UL25, or UL32 (Fig. 4). In this experiment, larger amounts of infected-cell lysates and purified capsids were loaded in order to detect minor forms of UL15. B capsids from cells infected with KOS (Fig. 4, lane 5) or with mutants defective in UL25 (Fig. 4, lane 4) or UL32 (Fig. 4, lane 8) exhibit multiple bands which react with the UL15 antiserum, including 75-, 81-, and 87-kDa bands and a more slowly migrating band marked with an arrow which corresponds to a protein with a size of 91 kDa. In the blot shown in Fig. 4, lanes 4 and 5, the 75-kDa protein is not visible; however, in other experiments, it can be detected (data not shown). The 91- and 75-kDa bands were also detected in capsids from cells infected with a mutant defective for UL15 (Fig. 4, lane 7), suggesting that these two bands may represent cross-reacting proteins rather than specific UL15-related proteins. The 91-kDa band appears to also react with the VP5 antibody, suggesting that it may represent a partially degraded form of VP5 (data not shown). The 81- and 87-kDa bands were not detected in B capsids from cells infected with a mutant defective for UL15. A strong 81-kDa band and a weak 87-kDa band were also detected in the total KOS-infected-cell lysate (lane 2) but not in mock-infected or UL15 mutant-infected-cell lysates (lanes 1 and 3, respectively). These results indicate that the 81- and 87-kDa bands likely represent distinct forms of UL15 rather than cross-reacting species; however, we cannot rule out the possibility that the 87-kDa protein is not related to UL15 but is fortuitously recognized by the anti-UL15 antibody. The ultimate confirmation of its identity will require amino acid sequence analysis.
FIG. 4.
Detection of UL15-related proteins in cell lysates and purified capsids. Lysates from cells mock infected or infected with KOS or a mutant virus defective in UL15 (hr81-1) for 16 h were prepared as described in Materials and Methods. B capsids from cells infected with either KOS, hr81-1, KUL25NS (lacking UL25), or hr64 (lacking UL32) were purified by sucrose gradient sedimentation as described in Materials and Methods. The UL15-related bands were revealed by exposure for 20 min. Infected-cell lysates as indicated are shown in lanes 1 to 3. Lanes 4 to 8 show B capsids from cells infected as indicated. In lane 6, B capsids from cells infected with KOS were added to a preparation of B capsids from cells infected with hr81-1 to determine whether minor anti-UL15 antibody-reacting bands could be distinguished. Four major bands (91, 87, 81, and 75 kDa) which react with the UL15 antiserum are indicated. The 91-kDa band marked with an arrowhead likely represents a partial degradation product of VP5 (see text).
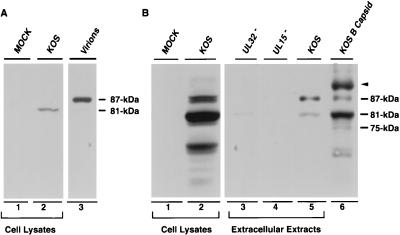
We next asked whether the 81- and 87-kDa proteins could also be detected in mature virions isolated from KOS-infected cells. A prominent 87-kDa band was observed in purified virions from KOS-infected cells, while the 81-kDa form was not detected (Fig. 5A, lane 3); in longer exposures, a faint 81-kDa band was observed (data not shown). Mock- and KOS-infected-cell lysates were subjected to electrophoresis in parallel for comparison; the 81-kDa band is the only band seen in KOS-infected-cell lysates at this exposure (Fig. 5A, lane 2, see figure legend). To confirm that the 87-kDa band is present in the virion, an experiment was performed to analyze extracellular particles from cells infected with KOS or mutants lacking UL15 or UL32 (as described in Materials and Methods). Since mutants lacking UL15 and UL32 would not be expected to produce mature virions, these samples provide a control for cell breakage and release of intracellular proteins. Extracellular particles from KOS-infected cells contain a major 87-kDa band and a minor 81-kDa band which react with the UL15 antiserum (Fig. 5B, lane 5). A small amount of the 81-kDa band was also detected in the media from cells infected with a mutant lacking UL32 (Fig. 5B, lane 3). This result suggests that the 81-kDa band present in extracellular particles from KOS-infected cells may represent intracellular UL15 released into the media by cell breakage. No bands are detected in the extracellular extract from cells infected with mutants lacking UL15 (Fig. 5B, lane 4). Unlike the 81-kDa full-length UL15 protein, which is predominantly associated with B capsids, the 87-kDa protein is present in both B and C capsids (Fig. 3A, panel C). Taken together, these results indicate that the 87-kDa version of UL15 is likely to be virion associated. Figure 5B also shows that the 87-kDa form of UL15 in B capsids from KOS-infected cells runs in an identical position to the 87-kDa band seen in virions (compare lanes 5 and 6). In summary, the 81-kDa version of UL15, which is the predominant species in infected-cell extracts, was detected predominantly in B capsids but was found at much lower levels in C capsids and in virions. A previously unidentified 87-kDa protein which reacts with the UL15 antiserum, however, was detected in B capsids and C capsids and was found to be present in large amounts in virions.
FIG. 5.
Detection of UL15-related proteins in infected-cell extracts, extracellular extracts, and virions. Infected-cell extracts were prepared as described in the legend to Fig. 4. Extracellular extracts and purified virions were prepared as described in Materials and Methods. (A) Detection of UL15-related proteins in purified virions and infected-cell extracts as indicated. Lanes 1 and 2 represent lysates from cells either mock infected or infected with KOS, respectively; lane 3 represents purified virions from KOS-infected cells. The positions of 81- and 87-kDa UL15-related proteins are indicated. The immunoblot was exposed for 1 min. (B) Infected-cell lysates were prepared as described above (lanes 1 and 2). Extracellular extracts from cells infected with KOS (lane 5), hr81-1 (lane 4), or hr64 (lane 3) were prepared as described in Materials and Methods. This immunoblot was exposed for 20 min to detect minor forms of UL15-related proteins. B capsids from cells infected with KOS are shown in lane 6 for comparison. The positions of the 75-, 81-, 87-, and 91-kDa bands are indicated.
UL28 is associated predominantly with B but not C capsids.
Next, we examined gradient fractions from cells infected with KOS for the presence of UL28 (Fig. 3A, panel E). Interestingly, UL28 can only be detected in fraction 6 containing B capsids, although fraction 8 has higher levels of VP5, UL6, and UL25. The specificity of the antibody (47) used was also examined in this blot: while the 87-kDa band can be detected in KOS-infected total Vero cell lysate and in gradient fraction 6, it is absent in UL28 mutant-infected-cell lysate and in B capsids purified from UL28 mutant-infected cells. Quantification of the signals confirms that levels of UL28 peaked in fraction 6, which contains B capsids (Fig. 3B). Therefore, we conclude that like the 81-kDa form of UL15, UL28 is associated predominantly with B capsids; much smaller amounts of the 81-kDa forms of UL15 and UL28 were detected in C capsids.
UL6 and UL28 are required for UL15 to associate with B capsids.
We next asked whether other cleavage and packaging proteins are essential for the observed association of UL15 with B capsids. Sucrose gradient fractions from lysates of Vero cells infected with mutants lacking individual cleavage and packaging genes were analyzed by immunoblotting as described above. Capsids were identified by light scattering and by the presence of VP5. The blotted membranes were intentionally overexposed in order to reveal any possible association of the cleavage and packaging protein with capsids. Although long exposures may detect specific as well as nonspecific associations of cleavage and packaging proteins with capsids, we reasoned that a comparison of relative signal strengths in capsid-containing fractions with neighboring fractions would indicate whether a protein is specifically associated with capsids. Figure 6A shows the fractionation pattern from cells infected with a mutant defective in UL6, previously shown to produce only B capsids (24, 33). In these cells, the 81-kDa form of UL15 was found in most of the fractions across the gradient; quantification of the bands seen in Fig. 8A indicates, however, that fraction 6 containing B capsids does not contain significantly higher levels of this form of the UL15 protein than do neighboring fractions (Fig. 6B). Thus, although UL15 is expressed at wild-type levels in cells infected with a mutant lacking UL6 (Fig. 2), it does not appear to associate specifically with B capsids in these cells. In addition, under these conditions, we failed to detect the 87-kDa UL15-related protein in the B capsid fraction, whereas the 91-kDa VP5 cross-reacting species (marked with an arrowhead in Fig. 6A) can be seen. In contrast, UL25 and UL28 were detected predominantly in fraction 6 (Fig. 6A, panels C and D, and 6B). These results suggest that UL6 may be required for the efficient association of the 81- and 87-kDa UL15-related proteins with B capsids, while the capsid associations of UL25 and UL28 are independent of the presence of UL6. Further evidence that UL15 does not specifically associate with B capsids in cells infected with mutants lacking UL6 is presented below.
FIG. 6.
Sucrose gradient analysis of lysates from cells infected with hr74 lacking UL6. Gradients similar to those described in the legend to Fig. 3 were collected into nine fractions. Fraction 6 contains B capsids. (A) Immunoblot analysis was performed to detect VP5 (panel A), UL15 (panel B), UL25 (panel C), and UL28 (panel D). A long (20 min) exposure of the UL15 immunoblot is shown. The first lane represents a KOS-infected-cell lysate. (B) Quantitative analysis was performed as described in the legend to Fig. 3.
FIG. 8.
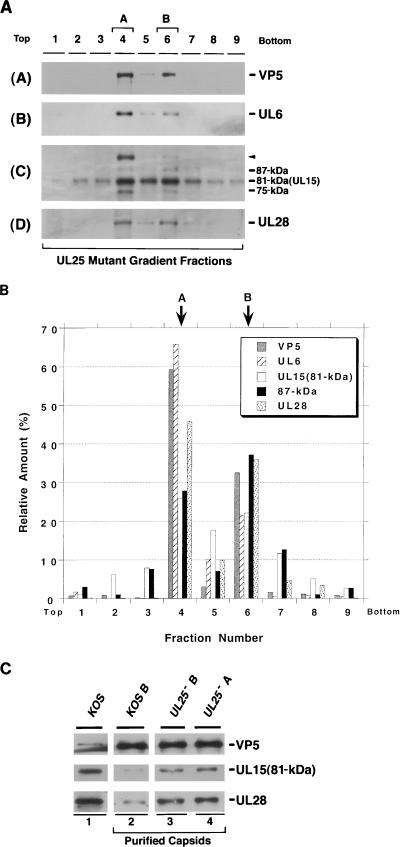
Sucrose gradient analysis of lysates from cells infected with KUL25NS lacking UL25 (UL25−). Gradients similar to those described in the legend to Fig. 3 were collected into nine fractions. Fractions 4 and 6 contain A and B capsids, respectively. (A) Immunoblot analysis was performed to detect VP5 (panel A), UL6 (panel B), UL15 (panel C), and UL28 (panel D). A long (20 min) exposure of the UL15 immunoblot is shown. (B) Quantitative analysis was performed as described in the legend to Fig. 3. (C) Analysis of the 81-kDa UL15 and UL28 levels in capsids from cells infected with the wild type or KUL25NS. A or B capsids were purified by sucrose gradient sedimentation. The amount of capsids in each sample was determined by normalization to the amount of VP5 present. Roughly equal amounts of capsids were subjected to immunoblot analysis with antisera against VP5, UL15, and UL28. Lysates of cells infected with KOS were also included to indicate the position of each of the proteins (lane 1).
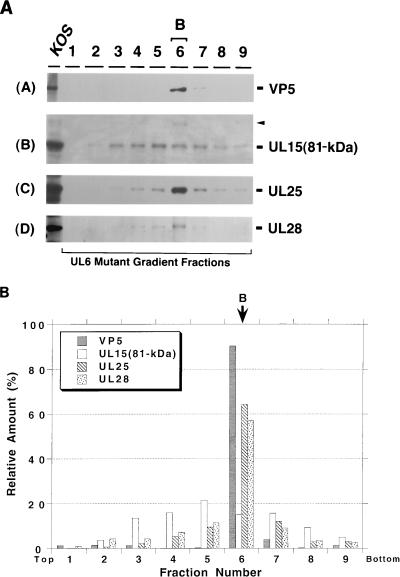
The lysates from cells infected with mutant viruses lacking UL15, UL25, UL28, or UL32 were also analyzed by sucrose gradient sedimentation. In gradient fractions from cells infected with a UL15 mutant virus, hr81-1, UL6, UL25, and UL28 appear to be specifically associated with the B capsid fraction 6 (Fig. 7). This result indicates that the lack of UL15 does not affect the ability of these proteins to bind to B capsids. In gradient fractions from cells infected with a UL25 mutant virus, both A and B capsids were observed, consistent with previous reports (26). In this experiment, UL6, the 81- and 87-kDa forms of UL15, and UL28 were detected in both A and B capsids (Fig. 8A and B), indicating that UL6, UL15, and UL28 are able to bind to capsids in the absence of UL25. Interestingly, capsids in UL25 mutant-infected cells appear to contain elevated levels of the 81-kDa forms of UL15 and UL28 compared to the levels seen in previous experiments. To confirm this observation, B capsid fractions from cells infected with wild-type virus were directly compared with A and B capsids from cells infected with the mutant virus lacking UL25 (Fig. 8C). While the levels of VP5 in these capsid samples are roughly similar, the levels of the 81-kDa versions of UL15 and UL28 are clearly higher in the capsids from cells infected with the UL25 mutant virus. The potential significance of this observation will be discussed below.
FIG. 7.
Sucrose gradient analysis of lysates from cells infected with hr81-1 lacking UL15. Gradients similar to those described in the legend to Fig. 3 were collected into nine fractions. Fraction 6 contains B capsids. (A) Immunoblot analysis was performed to detect VP5 (panel A), UL6 (panel B), UL25 (panel C), and UL28 (panel D). (B) Quantitative analysis was performed as described in the legend to Fig. 3.
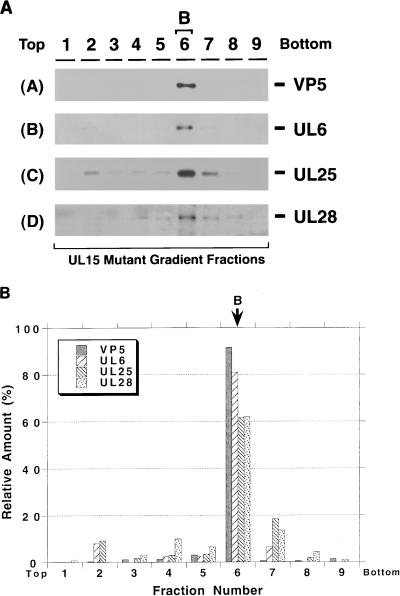
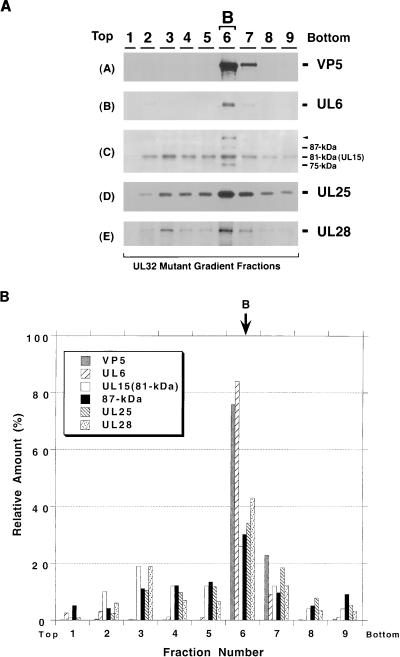
When gradient fractions from cells infected with a mutant lacking UL28 were examined, UL6 and UL25 were found predominantly in B capsids (Fig. 9A and B). The 81-kDa form of UL15, however, can be detected across the gradient and does not appear to peak in the B capsid fraction, indicating that it is not specifically associated with B capsids (Fig. 9B). The 87-kDa UL15-related protein cannot be detected in any of the gradient fractions (Fig. 9A, panel C) in this blot. Thus although UL15 is expressed at wild-type levels in cells infected with the UL28 mutant (Fig. 2), it does not appear to associate specifically with B capsids in these cells. These results suggest that UL28 may be required for the 81- and 87-kDa UL15-related proteins to bind to B capsids. In gradient fractions from cells infected with a UL32 mutant, UL6, UL25, UL28, and the same four UL15-reactive bands seen in previous figures appear to peak in the fraction containing B capsids (Fig. 10). Thus UL32 is not required for the capsid association of the other DNA cleavage and packaging proteins. The results from the wild-type and mutant capsids tested are summarized in Table 1. In conclusion, our results suggest that UL6 and UL28 are required for the 81- and 87-kDa UL15-related proteins to form a specific association with B capsids. UL6, UL25, and UL28, on the other hand, associate with capsids in a manner which is independent of the presence of the previously described essential DNA cleavage and packaging proteins.
FIG. 9.
Sucrose gradient analysis of lysates from cells infected with gCB lacking UL28. Gradients similar to those described in the legend to Fig. 3 were collected into nine fractions. Fraction 6 contains B capsids. (A) Immunoblot analysis was performed to detect VP5 (panel A), UL6 (panel B), UL15 (panel C), and UL25 (panel D). A long (20 min) exposure of the UL15 immunoblot is shown. The first lane represents a KOS-infected-cell lysate. (B) Quantitative analysis was performed as described in the legend to Fig. 3.
FIG. 10.
Sucrose gradient analysis of lysates from cells infected with hr64 lacking UL32. Gradients similar to those described in the legend to Fig. 3 were collected into nine fractions. Fraction 6 contains B capsids. (A) Immunoblot analysis was performed to detect VP5 (panel A), UL6 (panel B), UL15 (panel C), UL25 (panel D), and UL28 (panel E). A long (20 min) exposure of the UL15 immunoblot is shown. (B) Quantitative analysis was performed as described in the legend to Fig. 3.
TABLE 1.
Summary of capsid association of DNA cleavage and packaging proteins
| Virus type | Capsid or virion | Signal strength of DNA cleavage and packaging proteina:
|
||||
|---|---|---|---|---|---|---|
| UL6 | UL15 (81 kDa) | UL15 (87 kDa) | UL25 | UL28 | ||
| Wild type | B | +++ | ++ | + | +++ | ++ |
| Wild type | C | +++ | +b | ++ | +++ | − |
| UL6 mutant | B | − | +b | − | +++ | ++ |
| UL15 mutant | B | +++ | − | − | +++ | ++ |
| UL25 mutant | A | +++ | +++ | + | − | +++ |
| UL25 mutant | B | +++ | +++ | + | − | +++ |
| UL28 mutant | B | +++ | +b | − | +++ | − |
| UL32 mutant | B | +++ | ++ | + | +++ | ++ |
| Wild type | Virion | NDc | +/− | +++ | ND | ND |
Treatment with 2 M GuHCl cannot completely extract UL15 from wild-type B capsids.
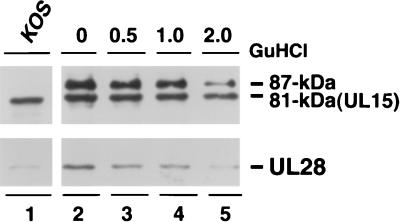
In order to assess the stability of the capsid association of UL15 and UL28, purified B capsids were treated with GuHCl at various concentrations (0, 0.5, 1.0, or 2 M), and the GuHCl-treated capsids were analyzed by SDS-PAGE and immunoblotting. It has been reported previously that GuHCl extraction selectively removes some components of B capsids, such as VP22a (scaffold), VP24 (protease), and VP5 pentons from capsids, while other structural proteins, such as VP19C and VP23 (triplex proteins), for the most part remain in the capsids (29). As previously reported, 2 M GuHCl treatment removed almost all VP24 and a substantial amount of VP22a from the capsids, while most of the VP5, VP19C, and VP23 proteins remain in capsids (data not shown). Immunoblot analysis indicates that a significant amount of the 81- and the 87-kDa proteins in the B capsids was resistant to 2 M GuHCl treatment (Fig. 11B, lane 5). Some UL28 was still associated with capsids after 2 M GuHCl treatment; however, the signal is much weaker than in untreated cells. The significance of this result is not clear; however, it is possible that UL28 associates with capsids through interactions with a capsid structural component, such as the penton, which is removed by GuHCl treatment. In summary, we conclude that 2 M GuHCl treatment cannot completely disrupt the capsid association of the 81- or 87-kDa form of UL15.
FIG. 11.
GuHCl extraction analysis of purified wild-type B capsids. B capsids from KOS-infected Vero cells were purified by sucrose gradient sedimentation and treated with GuHCl at 0, 0.5, 1.0, or 2.0 M as described in Materials and Methods. Equal amounts of capsids treated with various concentrations of GuHCl were analyzed by immunoblotting for the presence of UL15 and UL28. A long (20 min) exposure of the UL15 immunoblot is shown. Lane 1 represents a KOS-infected-cell lysate.
Treatment with 2 M GuHCl disrupts the association of UL15 with capsids lacking UL6 or UL28.
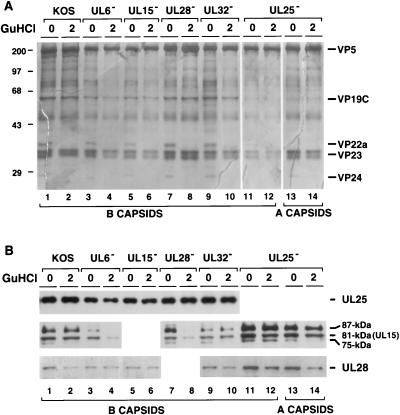
In the sucrose gradients shown in Fig. 6A and 9A, UL15 was detected in many fractions across the gradient. In order to test whether the presence of UL15 in these B capsids is due to nonspecific aggregation of UL15, purified B capsids lacking the individual cleavage and packaging proteins were treated with 2 M GuHCl and examined by immunoblot analysis (Fig. 12). Treated and untreated B capsid extracts were subjected to electrophoresis, and the gels were either silver stained (Fig. 12A) or analyzed by Western blotting (Fig. 12B). Figure 12A shows that 2 M GuHCl removed VP22a and VP24 from wild-type and mutant capsids (Fig. 12A). We found that as previously reported (26), UL25 remains associated with capsids after GuHCl treatment (Fig. 12B). Figure 12B also shows that GuHCl extraction did not remove significant amounts of the 81- and 87-kDa UL15-related proteins from B capsids from cells infected with KOS (lanes 1 and 2) and mutants lacking UL32 (lanes 9 and 10) or UL25 (lanes 11 to 14). On the other hand, GuHCl treatment removed much larger amounts of UL15-related proteins from B capsids from cells infected with mutants lacking UL6 (lanes 3 and 4) or UL28 (lanes 7 and 8). These results support the notion that the UL15-related proteins detected in capsids lacking UL6 or UL28 are not specifically associated with capsids. Although GuHCl treatment removed a substantial amount of UL28 from wild-type and mutant B capsids, we did not observe a significant difference between wild-type and mutant capsids with respect to the amount remaining in capsids after treatment. In summary, the GuHCl results are consistent with the notion that the specific association of UL15 with B capsids requires the presence of UL6 and UL28, while the association of UL28 with B capsids is independent of other cleavage and packaging proteins.
FIG. 12.
GuHCl extraction analysis of purified B capsids lacking individual cleavage and packaging proteins. Capsids from Vero cells infected with KOS, hr74 (UL6−), hr81-1 (UL15−), KUL25NS (UL25−), gCB (UL28−), or hr64 (UL32−) were purified from sucrose gradient and either mock treated or treated with 2.0 M GuHCl. In this experiment, twice as much of the GuHCl-treated capsid extract as the untreated extract was loaded in each lane. (A) A silver-stained SDS-PAGE gel shows capsid structural proteins. Assignment of bands was based on previous reports (29, 44). The molecular masses of protein size markers (in kilodaltons) are given on the left. (B) Immunoblot analysis for the presence of UL15, UL25, and UL28. A long (20 min) exposure of the UL15 immunoblot is shown.
DISCUSSION
Several observations were made in this report. (i) Approximately 50% of the UL15 expressed in Vero cells during viral infection is present in the soluble nuclear fraction; the remainder is insoluble. This is in contrast to the situation in insect cells infected with UL15-expressing recombinant baculovirus, in which almost 100% of the UL15 was found in the insoluble fraction. (ii) Previous reports that UL6 and UL25 are constituents of A, B, and C capsids (1, 24–26, 33) were confirmed. (iii) An 81-kDa version of the UL15 protein was found predominantly in B capsids and in much smaller amounts in C capsids and in virions. (iv) Another cleavage and packaging protein, UL28, was also found predominantly in B but not C capsids in a distribution similar to that of the 81-kDa version of UL15. (v) A previously unidentified 87-kDa protein which reacts with the UL15 antiserum was detected in B capsids and as a very prominent band in C capsids and in virions. (vi) Analysis of mutant virus-infected cells demonstrated that UL6, UL15, UL25, UL28, and UL32 are not coregulated. (vii) UL6, UL25, and UL28 are able to associate with B capsids in the absence of other DNA cleavage and packaging proteins; however, specific capsid association of the 81- and 87-kDa forms of UL15 does not occur efficiently in the absence of UL6 or UL28. (viii) Elevated levels of the 81- and 87-kDa forms of UL15 and UL28 were detected in B capsids from cells infected with a mutant lacking UL25.
The roles of HSV-1 cleavage and packaging proteins have not been elucidated. The insolubility of UL15 in insect cells infected with recombinant baculoviruses has hampered attempts to carry out biochemical analyses. In this report, we have used the ability of various cleavage and packaging proteins to associate with capsids to address the possible functions of these proteins in the encapsidation process. The data in this paper indicate that UL28 and the 81-kDa version of UL15 exhibit similar distribution patterns in capsids, in that both appear to associate specifically with B capsids but not C capsids. These results suggest that the interactions of UL15 and UL28 with capsids may be transient during packaging. By analogy with phage, the transient association of these two proteins with capsid intermediates is consistent with the hypothesis that they function as a two-subunit terminase. Several lines of indirect evidence support this hypothesis. (i) UL15 and UL28 are both essential for cleavage and packaging as determined by the analysis of mutants lacking these proteins (4, 47, 52). (ii) UL15 exhibits sequence homology with the T4 terminase (15). (iii) The putative ATP binding site of UL15 is essential for its function (53). (iv) Bogner et al. have recently reported that the human cytomegalovirus (HCMV) homolog of HSV UL28 binds the pac motif and may have specific nuclease activity (8). (v) UL28, when expressed on its own, is localized to the cytoplasm in transfected cells but can enter the nucleus when expressed in the presence of UL15, suggesting a possible interaction between these two proteins (22). (vi) Finally, genetic evidence suggests a possible association between the HCMV homologs of UL15 and UL28 (23). Biochemical confirmation that UL15 and UL28 function as a terminase complex will await the development of an expression system which produces soluble protein.
In phage systems, cleavage and packaging proteins exhibit several distinct patterns of association with procapsids, mature capsids, and virions (5, 7, 19, 20, 28). Portal proteins are found as integral capsid components in both procapsids and mature capsids, while scaffold and terminase proteins are only associated with procapsids and are not present in mature capsids. In phage, the terminase interacts with portal proteins in the procapsid; in herpesviruses, however, a unique portal vertex has not been identified. It is possible that herpesviruses do not have a unique portal vertex and that these structures are only required in bacteriophages which also have a tail. Even if HSV-1 does not have a unique vertex, it is still necessary for the terminase complex to bind DNA and dock at the capsid. The docking process may require an interaction between the terminase and specific protein components in the capsid.
In this report, we demonstrate that UL6, UL25, and UL28 associate with capsids in cells infected with mutant viruses individually lacking UL6, UL15, UL25, UL28, or UL32. We previously showed that UL6 and UL25 associate with capsids in cells infected with a mutant virus lacking UL32 (25). The ability of UL6 and UL25 to associate with B capsids in the absence of other cleavage and packaging proteins is consistent with the observation that both can specifically associate with capsids assembled from insect cells infected with recombinant baculoviruses expressing the six capsid proteins and either UL6 or UL25 (26). The absence of specific capsid association of UL15 in mutants lacking UL6 may indicate that UL6 is required for docking of the putative terminase complex. The presence of UL6 in all forms of capsids examined, including B capsids treated with GuHCl, indicates that it may be an integral component of the capsid itself (32). It will be of considerable interest to determine the precise location of the UL6 protein within the capsid, because this may shed light on the question of whether HSV capsids contain a unique portal vertex. The observation that UL15 does not specifically associate with capsids in cells infected with the UL28 mutant suggests that the interaction with UL28 may also facilitate the binding of UL15 to the procapsid. Results from GuHCl extraction experiments are consistent with the suggestion that UL15 is specifically associated with B capsids and that this association requires the presence of UL6 and UL28. The transient yet stable interaction between UL15 and capsids is reminiscent of the lambda DNA packaging system: stable packaging intermediates containing the terminase and prohead can be isolated by sucrose gradient fractionation; however, the terminase is not found in mature capsids (6, 28).
The ability of UL28 to associate with B capsids in cells infected with mutants lacking individual cleavage and packaging proteins suggests that it is able to associate with one of the structural components of the capsid itself or with an unidentified component of the DNA cleavage and packaging machinery. The lower abundance of UL28 in C capsids compared to B capsids may indicate that its association with capsids may be altered in response to conformational changes during capsid maturation.
The identification of proteins in addition to the previously described 81-kDa full-length version and the 30-kDa truncated version of UL15 which react with the UL15 antiserum and associate specifically with capsids was unexpected. Two of these bands (75 and 91 kDa) are apparent in B capsids from mutants lacking UL15 and thus may represent proteins which cross-react with the UL15 antiserum. The 87-kDa protein, on the other hand, is not present in B capsids from mutants lacking UL15. This UL15-related protein was detected specifically in B capsids and as a prominent band in C capsids and in virions. Interestingly, the 87-kDa protein is present in much smaller amounts in total-infected-cell lysates than in purified capsids. This result suggests that the 87-kDa protein may be specifically enriched in capsids and in virions. Further experiments will be required to determine whether the 87-kDa band represents a posttranslationally modified version of UL15.
While this report was being prepared, Salmon and Baines reported the detection of 79-, 80-, and 83-kDa forms of UL15 gene-encoded proteins in B capsids (40). In that report, all three forms of UL15 gene-encoded proteins were detected in B capsids, C capsids, and virions. Furthermore, the 79- and 80-kDa proteins were absent in B capsids isolated from cells infected with UL6, UL17, or UL28 viral mutants. Although some discrepancies exist between these two reports in the relative amounts of various UL15 species present in different types of capsids and virions, the overall conclusions are similar. Differences in experimental procedures and antibodies used may account for the discrepancies, and the correspondence between the forms of UL15 seen by Salmon and Baines (40) and those reported in this paper is not clear at this time. Further experiments will be required to clarify the remaining inconsistencies between these two reports.
Recent evidence suggests that UL25 plays a role in a late stage of the cleavage and packaging process, perhaps functioning to retain DNA in the capsid following its cleavage from replicating DNA (26). UL25 may play a role analogous to the lambda phage proteins gpD and gpF1 (21, 36, 45). Many lines of evidence indicate that gpD functions to stabilize the DNA-filled head (36). In addition, gpD may also play a role in the dissociation of terminase from filled proheads (10). The lambda gpF1 protein may enhance the turnover rate of the terminase from capsids (11). An intriguing observation made in this report is that UL15 and UL28 are present in somewhat higher levels in capsids from cells infected with the UL25 mutant than in cells infected with wild-type or other cleavage-packaging mutants. If UL25 performs functions similar to those of gpD and gpF1, capsids produced in cells infected with a UL25 mutant may represent trapped intermediates in the cleavage and packaging process. The elevated levels of UL15 and UL28 in these capsids are consistent with this hypothesis.
Previous reports indicate that the protein composition of A capsids is similar to that of C capsids (14, 18, 31); however, minor capsid proteins have not been looked at. We report herein that the 81-kDa forms of UL15 and UL28 are present in A capsids lacking UL25 but not in wild-type C capsids, suggesting a possible difference in protein composition between the A and C capsids. Alternatively, it is possible that the elevated levels of UL15 and UL28 in mutant A capsids may reflect the lack of UL25 in these capsids as discussed above.
A model for the possible roles of the various cleavage and packaging proteins can be proposed based on the results presented in this paper taken together with other reports. We have recently proposed that cleavage and packaging occur within replication compartments at least at early times postinfection (25). Furthermore, we suggested that UL32 plays a role at the early stages of the cleavage and packaging process by promoting the efficient localization of preformed capsids to replication compartments (25). It is not clear whether UL6 and UL25 are present during the capsid assembly process or are added at a later stage; however, we propose that at least UL6 is present in the procapsids in replication compartments prior to cleavage and packaging. According to this scenario, UL15 and UL28 (two-subunit terminase) may recognize and bind viral DNA at the a sequence and mediate docking of this complex to the procapsid. The data shown in this report suggest that UL6 may be required for this docking event. The association of UL15 with the capsid is apparently also stabilized by the presence of UL28. Thus, the ability of the terminase to dock may depend on interactions between the UL15 and UL6 proteins and on interactions between UL28 and another unidentified protein constituent of the capsid. The recent report that UL15 is not associated with B capsids in cells infected with mutants lacking UL17 suggests that UL17 may also play a role in stabilizing the interactions of UL15 with capsids (40). After docking, site-specific cleavage of concatemeric viral DNA and translocation of monomeric units of DNA into the capsid occur, presumably mediated by the UL15-UL28 complex. Since only small amounts of the 81-kDa versions of UL15 and UL28 were detected in C capsids or in virions compared to those detected in B capsids, we propose that after cleavage and packaging have occurred, these two proteins may disassociate from capsids. The presence of the 87-kDa version of UL15 in B capsids and in virions is intriguing; however, it is not clear whether this potentially modified form of UL15 plays an additional role in the cleavage and packaging process. After viral DNA is packaged, UL25 may not only stabilize the DNA-filled capsids but also enhance the turnover rate of the terminase by disassociating it from the capsids. DNA-containing capsids would then be competent to bud from the nuclear membrane. Refinement and confirmation of this working model will require further experimentation.
ACKNOWLEDGMENTS
We are grateful to all of the members of our laboratory for critical discussions during this work and in preparation of the manuscript. We especially thank Carmela Lamberti for helpful discussions and assistance in capsid preparation. We thank Fred Homa (Pharmacia and Upjohn, Kalamazoo, Mich.) for providing mutant KUL25NS virus and gCB virus and complementing cell lines 8-1 and A1; William Ruyechan (SUNY, Buffalo, N.Y.) for the anti-ICP8 polyclonal antibody; Gary H. Cohen and Roselyn J. Eisenberg (University of Pennsylvania, Philadelphia) for the anti-VP5 polyclonal antibody (NC-1); Amy K. Sheaffer, Todd Wilson, and Daniel J. Tenney (Bristol-Myers Squibb, Wallingford, Conn.) for providing the anti-UL15, anti-VP16, anti-UL25, and anti-UL28 antibodies; and Bernard Roizman (University of Chicago, Chicago, Ill.) for the anti-UL32 polyclonal antibody.
This work was supported by National Institutes of Health grant AI 37549.
REFERENCES
- 1.Ali M A, Forghani B, Cantin E M. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology. 1996;216:278–283. doi: 10.1006/viro.1996.0061. [DOI] [PubMed] [Google Scholar]
- 2.Al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 4.Baines J D, Cunningham C, Nalwanga D, Davison A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazinet C, King J. The DNA translocating vertex of DSDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- 6.Becker A, Marko M, Gold M. Early events in the in vitro packaging of bacteriophage lambda DNA. Virology. 1977;78:291–305. doi: 10.1016/0042-6822(77)90100-3. [DOI] [PubMed] [Google Scholar]
- 7.Black L. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 8.Bogner E, Radsak K, Stinski M F. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol. 1998;72:2259–2264. doi: 10.1128/jvi.72.3.2259-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway F F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catalano C E, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 11.Catalano C E, Tomka M A. Role of gpF1 protein in DNA packaging by bacteriophage lambda. Biochemistry. 1995;34:10036–10042. doi: 10.1021/bi00031a027. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y E, Poon A P W, Roizman B. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J Virol. 1996;70:3938–3946. doi: 10.1128/jvi.70.6.3938-3946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen G H, Ponce de Leon M, Diggelmann H, Lawrence W C, Vernon S K, Eisenberg R J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dargan D J. The structure and assembly of herpesviruses. In: Harris J, Horne R, editors. Electron microscopy of proteins. Vol. 5. London, England: Academic Press, Inc.; 1986. pp. 359–437. [Google Scholar]
- 15.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 16.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 17.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrix R W, Casjens S R. Assembly of bacteriophage lambda heads: protein processing and its genetic control in petit lambda assembly. J Mol Biol. 1975;91:187–199. doi: 10.1016/0022-2836(75)90159-x. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix R W, Casjens S R. Protein fusion: a novel reaction in bacteriophage lambda head assembly. Proc Natl Acad Sci USA. 1974;71:1451–1455. doi: 10.1073/pnas.71.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii T, Yanagida M. Molecular organization of the shell of the T even bacteriophage head. J Mol Biol. 1975;97:655–660. doi: 10.1016/s0022-2836(75)80065-9. [DOI] [PubMed] [Google Scholar]
- 22.Koslowski K M, Shaver P R, Wang X-Y, Tenney D J, Pederson N E. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71:9118–9123. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krosky P M, Underwood M R, Turk S R, Feng K W-H, Jain R K, Ptak R G, Westerman A C, Biron K K, Townsend L B, Drach J C. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J Virol. 1998;72:4721–4728. doi: 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamberti C, Weller S K. The herpes simplex type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 25.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNabb D S, Courtney R J. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J Virol. 1992;66:2653–2663. doi: 10.1128/jvi.66.5.2653-2663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murialdo H, Siminovitch L. Morphogenesis of bacteriophage lambda. IV. Identification of gene products and control of the expression of the morphogenetic information. Virology. 1972;48:785–823. doi: 10.1016/0042-6822(72)90162-6. [DOI] [PubMed] [Google Scholar]
- 29.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsids: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:431–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 31.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 32.Patel A H, MacLean J B. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206:465–478. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 33.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 34.Perdue M L, Cohen J C, Randall C C, O’Callaghan D J. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology. 1976;74:194–208. [PubMed] [Google Scholar]
- 35.Pertuiset B, Boccara M, Cebrian J, Berthelot N, Chousterman S, Puvion-Dutilleul F, Sisman J, Sheldrick P. Physical mapping and nucleotide sequence of a herpes simplex virus type 1 gene required for capsid assembly. J Virol. 1989;63:2169–2179. doi: 10.1128/jvi.63.5.2169-2179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perucchetti R, Parris W, Becker A, Gold M. Late stages in bacteriophage lambda head morphogenesis: in vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology. 1988;165:103–114. doi: 10.1016/0042-6822(88)90663-0. [DOI] [PubMed] [Google Scholar]
- 37.Rixon F J. Structure and assembly of herpes viruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 38.Rixon F J, Davison M D, Davison A J. Identification of the genes encoding two capsid proteins of herpes simplex virus type 1 by direct amino acid sequencing. J Gen Virol. 1990;71:1211–1214. doi: 10.1099/0022-1317-71-5-1211. [DOI] [PubMed] [Google Scholar]
- 39.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, editors. Fields’ virology. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1796–1841. [Google Scholar]
- 40.Salmon B, Baines J D. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao L, Rapp L M, Weller S K. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 43.Shelton L S G, Albright A G, Ruyechan W T, Jenkins F J. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J Virol. 1994;68:521–525. doi: 10.1128/jvi.68.1.521-525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman G, Bachenheimer S L. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 45.Sternberg N, Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J Mol Biol. 1977;117:733–759. doi: 10.1016/0022-2836(77)90067-5. [DOI] [PubMed] [Google Scholar]
- 46.Szilagyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 47.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 49.Weller S K. Herpes simplex virus DNA replication and genome maturation. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin’s scientific legacy. Washington, D.C: ASM Press; 1995. pp. 189–213. [Google Scholar]
- 50.Weller S K, Lee K J, Sabourin D J, Schaffer P A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen Z, Zhong Z, Darnell J. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 52.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu D, Weller S K. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology. 1998;243:32–44. doi: 10.1006/viro.1998.9041. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]