Abstract
The structures of three different human rhinovirus 14 (HRV14)-Fab complexes have been explored with X-ray crystallography and cryoelectron microscopy procedures. All three antibodies bind to the NIm-IA site of HRV14, which is the β-B–β-C loop of the viral capsid protein VP1. Two antibodies, Fab17-IA (Fab17) and Fab12-IA (Fab12), bind bivalently to the virion surface and strongly neutralize viral infectivity whereas Fab1-IA (Fab1) strongly aggregates and weakly neutralizes virions. The structures of the two classes of virion-Fab complexes clearly differ and correlate with observed binding neutralization differences. Fab17 and Fab12 bind in essentially identical, tangential orientations to the viral surface, which favors bidentate binding over icosahedral twofold axes. Fab1 binds in a more radial orientation that makes bidentate binding unlikely. Although the binding orientations of these two antibody groups differ, nearly identical charge interactions occur at all paratope-epitope interfaces. Nucleotide sequence comparisons suggest that Fab17 and Fab12 are from the same progenitor cell and that some of the differing residues contact the south wall of the receptor binding canyon that encircles each of the icosahedral fivefold vertices. All of the antibodies contact a significant proportion of the canyon region and directly overlap much of the receptor (intercellular adhesion molecule 1 [ICAM-1]) binding site. Fab1, however, does not contact the same residues on the upper south wall (the side facing away from fivefold axes) at the receptor binding region as do Fab12 and Fab17. All three antibodies cause some stabilization of HRV14 against pH-induced inactivation; thus, stabilization may be mediated by invariant contacts with the canyon.
Picornaviruses are among the largest of animal virus families and include the well-known poliovirus, rhinovirus, foot-and-mouth disease virus (FMDV), coxsackievirus, and hepatitis A virus. The rhinoviruses, of which there are more than 100 serotypes subdivided into two groups, are major causative agents of the common cold in humans (42). The viruses are nonenveloped and have an ∼300-Å-diameter protein shell that encapsidates a single-stranded, plus-sense RNA genome of about 7,200 bases. The human rhinovirus 14 (HRV14) capsid exhibits a pseudo-T=3 (P=3) icosahedral symmetry and consists of 60 copies each of four viral proteins, VP1, VP2, VP3, and VP4, with VP4 at the RNA-capsid interface (40). An ∼20-Å deep canyon lies roughly at the junction of VP1 (forming the north rim) with VP2 and VP3 (forming the south rim) and surrounds each of the 12 icosahedral fivefold vertices. The canyon regions of HRV14 and HRV16, both major receptor group rhinoviruses, were shown to contain the binding site of the cellular receptor, intercellular adhesion molecule 1 (ICAM-1) (8, 24a, 37). Four major neutralizing immunogenic (NIm) sites, NIm-IA, NIm-IB, NIm-II, and NIm-III, were identified by studies of neutralization escape mutants with monoclonal antibodies (MAbs) (46, 47) and then mapped to four protruding regions on the viral surface (40).
Several mechanisms of antibody-mediated neutralization have been proposed. Perhaps the simplest is based on aggregation of virions (5, 53, 54), which generally occurs over a narrow range of antibody/virus ratios. This limited range has raised questions about the role of aggregation in vivo. Alternative suggestions are that antibodies may neutralize virions by inducing extensive conformational changes in the capsid (15, 29), abrogate virus attachment to the host cell (8, 14), or prevent uncoating (57). There is no universal acceptance of a single neutralization mechanism, and the various MAbs may neutralize with different combinations of these mechanisms.
Neutralizing MAbs against HRV14 have been divided into three groups: strong, intermediate, and weak neutralizers (26, 34). All strongly neutralizing antibodies bind to the NIm-IA site, which was defined by natural escape mutations at residues D1091 and E1095 of VP1 on the loop between the β-B and β-C strands of the VP1 β-barrel (the letter designates the amino acid, the first digit identifies the viral protein, and the remaining three digits specify the sequence number). Because strongly neutralizing antibodies form stable, monomeric virus-antibody complexes with a maximum stoichiometry of 30 antibodies per virion, it was concluded that they bind bivalently to the virions (26, 34). Weakly neutralizing antibodies form unstable, monomeric complexes with HRV14 and bind with a stoichiometry of ∼60 antibodies per virion (26, 52). The remaining antibodies, all of which precipitate the virions, are classified as intermediate neutralizers (26, 34).
The structures of two complexes, the strongly neutralizing antibody MAb17-IA and its Fab fragment, Fab17, bound to HRV14, were determined by means of cryo-transmission electron microscopy (cryo-TEM) and three-dimensional image reconstruction (51, 52) and interpreted on the basis of model-building studies that used the atomic structures of HRV14 (40) and Fab17 (28). These studies showed that no observable conformational changes were induced in the viral capsid upon Fab or MAb binding. Modeling and site-directed mutagenesis studies demonstrated that electrostatic interactions play a key role in the binding of Fab17 to HRV14 (52). In the complex, the loop of the NIm-IA site on HRV14 sits clamped in the cleft between the heavy- and light-chain hypervariable regions and forms complementary electrostatic interactions with Lys58H (on the heavy chain) and Arg91L (on the light chain) of Fab17. In addition, a cluster of lysines on HRV14 (K1236, K1097, and K1085) interact with two acidic residues, Asp45H and Asp54H, in the CDR2 (CDR stands for complementarity-determining region) of the Fab heavy chain (49). Earlier modeling studies also suggested that bidentate binding of MAb17-IA to HRV14 is facilitated by rotation of the Fab constant domains about the elbow axes towards the viral twofold axes (51). This suggested that the flexibility of the elbow region (the junction between the variable and constant domains) plays a role in the bivalent binding process, which in turn increases antibody avidity. Finally, the 4-Å-resolution crystal structure of the Fab17-HRV14 complex clearly showed that the virion does not undergo conformational changes upon Fab binding (49). This crystal structure determination also revealed that the earlier docking of the HRV14 and Fab17 atomic structures into the 22-Å cryo-TEM density map (50) yielded a pseudo-atomic model that was very close to the real structure of the complex.
We have expanded our complementary X-ray crystallography and cryo-TEM microscopy studies to examine the structures of two more Fab-virus complexes, using Fab fragments from two other NIm-IA antibodies, MAb1-IA (MAb1) and MAb12-IA (MAb12), bound to HRV14. MAb1 and MAb12 are weak and strong neutralizing antibodies, respectively. Image reconstructions of these two complexes are interpreted on the basis of pseudo-atomic models, which substantiate the previous hypothesis that neutralizing efficacy and binding valency are interrelated (34). Electrostatic interactions at the epitope-paratope interface are highly conserved and apparently important for the antibody binding to the virion surface. Like Fab17, Fab1 and Fab12 penetrate the canyon. There are, however, differences between the orientations of the strongly and weakly neutralizing antibodies and in the contacts made with the receptor binding region of the canyon. Finally, data suggesting that antibody binding to HRV14 is alone sufficient for neutralization and that other possible mechanisms are not required are presented.
MATERIALS AND METHODS
Preparation of MAb1-IA and MAb12-IA.
MAb1-IA and MAb12-IA were produced as previously described (52) and were found to be immunoglobulin G1 (IgG1) and IgG2a, respectively, by using Screentype (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). High-glucose Dulbecco’s modified Eagle medium (catalog no. 430-2100; GIBCO/Bethesda Research Laboratories, Grand Island, N.Y.) containing 10% fetal bovine serum was used for the hybridoma cell cultures grown in the Cellmax Quad 4 cell culture system (Cellco Corp., Germantown, Md.). Cells were removed from the culture aliquots with 10-min centrifugations at 10,000 × g. Antibodies were precipitated from the cellular supernatant by using a 50% (final concentration) saturated solution of ammonium sulfate and collected with 10-min centrifugations at 10,000 × g. The precipitate was resuspended and dialyzed with 0.1 M sodium phosphate (pH 7.0) buffer. Aliquots were loaded onto a protein G affinity column (Pharmacia/LKB Corp., Piscataway, N.J.) and washed with 0.1 M sodium phosphate (pH 7.0), and antibodies were eluted with 50 mM sodium acetate buffer (pH 2.0). The pH of the eluant was immediately brought back up to neutrality by collecting fractions in tubes containing 1 M sodium phosphate (pH 9.0). The antibody samples were pooled and dialyzed against 0.1 M sodium phosphate (pH 7.0).
Antibody neutralization plaque assays.
Two different protocols were used to ascertain the efficacy of the antibodies on virus neutralization prior to uncoating (method A) and the ability of the antibodies to prevent plaque formation (method B).
(i) Method A.
Samples of purified HRV14 (at ∼5 × 106 PFU/ml) were incubated with various concentrations of purified antibodies in phosphate-buffered saline with bovine serum albumin (PBSA) for 1 h at room temperature and then overnight at 4°C. These samples were then serially diluted in PBSA, and 200-μl aliquots were added to monolayers of HeLa cells. After incubation for 1 h at room temperature to allow for viral attachment to the cells, each plate was rinsed with 2.5 ml of PBSA. The monolayers were then covered with 2.5 ml of 0.8% agar in medium P6. The hardened agar was then covered with 2.5 ml of medium P6 supplemented with 4 mM glutamine, 4 mM oxaloacetic acid, 2 mM pyruvate, and 0.2% glucose. Plates were incubated in 5% CO2 for 48 h at 35°C, and plaques were visualized by removing the overlays, staining the cell monolayers with 0.5% crystal violet in 20% ethanol, and rinsing with water.
(ii) Method B.
The ability of antibodies to inhibit plaque formation was tested by maintaining a high concentration of antibodies in the plaque assay. Serially diluted samples of HRV14 were allowed to attach to monolayers of HeLa cells for 1 h at room temperature. The monolayers were then covered with the agar and medium overlays as described above with the exception that antibodies were added to the overlays (∼28-μg/ml final concentration) rather than being washed away. Subsequent incubation and visualization were performed as in method A.
Generation and purification of Fab1 and Fab12.
Fab fragments were generated by papain cleavage at a 1:50 (wt/wt) enzyme-to-antibody ratio for 12 h at 37°C in the presence of 30 mM β-mercaptoethanol. The reaction was quenched by adding iodoacetamide to give a final concentration of 75 mM. After extensive dialysis (>3 changes of buffer every 8 h) against 20 mM Tris (pH 7.5), the digested sample was purified by using a Mono-Q anion-exchange column on a fast protein liquid chromatography system (Pharmacia/LKB). Pure Fab fragments eluted in the void volume, whereas Fc fragments and intact antibodies eluted at 0.1 to 0.2 M NaCl. The Fabs were pooled and concentrated with Centricon 10 microconcentrators (Amicon Corporation, Beverly, Mass.).
Preparation of HRV14-Fab1 and HRV14-Fab12 complexes.
Fabs and HRV14 (prepared as previously described [16]) were mixed at a ratio of four Fab molecules per NIm-IA site (240 Fabs per virion). The mixture was incubated at 4°C overnight and then passed through a Superose 6 column (Pharmacia/LKB) equilibrated with 50 mM sodium phosphate buffer (pH 7.0) to separate the complex and the unbound Fab molecules. Pure complexes were concentrated to ∼10 mg/ml with Centricon 10 microconcentrators (Amicon Corp.).
Cryo-TEM and image reconstructions of HRV14-Fab1 and HRV14-Fab12 complexes.
Cryo-TEM of the HRV14-Fab complexes was performed essentially as previously described (3, 7, 36, 49). Micrographs were recorded at a magnification of ×49,000, at ∼1.2 μm for Fab1 and ∼1.3 μm underfocus for Fab12, and under minimal dose conditions (∼20 e−/Å2), on an EM420 electron microscope (Philips Electronic Instruments, Mahwah, N.J.) equipped with a 626 cryotransfer holder (Gatan, Warrendale, Pa.). Twenty-six images of the Fab1 complexes and 41 images of the Fab12 complexes were used to calculate 3D reconstructions according to established protocols (2, 17). Effective resolutions of 30 and 27 Å were achieved for the Fab1 and Fab12 reconstructions, respectively. The calculated eigenvalues of each data set exceeded 10.0, which indicated that random and unique data were used for each reconstruction (11). The reconstructions were then corrected for effects of the phase-contrast transfer function as described previously (52).
Sequence determinations of Fab1 and Fab12.
The amino acid sequences of the variable domains of Fab1 and Fab12 were derived from cDNAs by using the Mouse Ig-Prime kit (Novagen, Madison, Wis.). Total RNAs from MAb1 and MAb12 hybridoma cells were isolated by the rapid GuSCN method, which is similar to conventional phenol-chloroform extraction. cDNAs for both light chains were synthesized and amplified by PCR with the primer pair MuIgkVL5′-G–MuIgkVL3′-1, whereas the primer pairs MuIgVH5′-B–MuIgGVH3′-2 and MuIgVH5′-C–MuIgGVH3′-2 were used in the synthesis and PCR amplification of the heavy chains of Fab1 and Fab12, respectively. The PCR products were cloned into a T-A vector (pT7BlueT) and then transformed into competent Escherichia coli (NovaBlue). E. coli containing plasmids with cDNA inserts produced white colonies after the transformed competent cells were plated onto ampicillin–X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates. These plasmids were extracted and purified from the minipreps grown out of the white colonies. Double-stranded DNA sequencing was then performed with a Sequenase version 2.0 DNA sequencing kit (United States Biochemical, Cleveland, Ohio). Each sequence was determined from at least three independent PCRs. The amino acid sequences were derived from the nucleotide sequences and aligned to the Fab17 sequence according to previously published nomenclature (23).
Fab1 crystal structure determination.
Details of the X-ray crystallographic structure determination of Fab1 will be published elsewhere. Briefly, Fab1 crystals were obtained by the sitting-drop method with 18 to 22% polyethylene glycol 8000, 0.1 M sodium phosphate buffer (pH 6.0), and 1% 2-methyl-2,4-pentanediol and an Fab1 concentration of 18 mg/ml. Crystals appeared within a day and continued growing for a week to maximum dimensions of 0.5 to 0.7 mm. Oscillation X-ray diffraction data were collected from two crystals, using an R-axis imaging detector and a Rigaku X-ray generator. The oscillation angle was 1°, and the exposure time was 10 min for each image. The intensities were integrated and merged by using the programs DENZO and SCALEPACK (38). The final data set extended to a 2.7-Å resolution. The crystal belonged to space group P21212, with unit cell dimensions of a = 92.17, b = 135.95, and c = 81.08 Å and two Fab molecules in the asymmetric unit. The structure was determined by the molecular replacement method (41) with the Fab17 structure (28) as the initial phasing model. Rotation and translation functions were calculated with a combination of X-PLOR (6) and AMORE (34a) in the CCP4 program suite (1). The model was built by using “O” (22), taking into account the differences in Fab17 and Fab1 sequences, and was refined with X-PLOR with all reflections in the range of 6 to 2.7 Å. The final R factor was 16.9% (Rfree = 26.6%), with root mean square deviations in bond lengths and angles of 0.010 Å and 1.45°, respectively. From Ramachandran analysis, 85.2% of the residues were in the most favored regions and none of the residues were in disallowed regions.
Modeling of the HRV14-Fab1 complex.
Initial modeling studies of the HRV14-Fab1 complex were performed with the program “O” (22) by fitting the atomic structures of HRV14 and Fab1 into the electron density map of the complex generated by cryo-TEM and image reconstruction. The SFALL program in the CCP4 program suite (1) was used to generate structure factors from the cryo-TEM maps. The rigid-body refinement algorithm within X-PLOR (6) was then used to refine the initial models against the computed structure factors. The HRV14 structure was constrained, and only the position and orientation of Fab1 were refined.
Effect of antibodies on acid inactivation.
Aliquots of 4 × 1010 virions were incubated with 2 × 1012 antibodies in 200 μl of PBSA at room temperature for 3 to 4 h, except for MAb3, -4, -6, and -7, whose concentrations are unknown because they could not be purified by protein-A affinity chromatography and precipitated upon purification by ion-exchange chromatography. In these cases, enough antibody was added to neutralize >99.99% of infectivity. Ten microliters of this mixture was diluted into 490 μl of 10 mM citrate buffer containing 0.4% BSA at various pH values. After incubation at room temperature for 10 min, 50 μl of each treated sample was diluted 10-fold into 450 μl of PBSA to neutralize the pH. Each sample was then serially diluted with PBSA and frozen at −70°C to dissociate the antibodies from the virions. After 1 h or more, each diluted sample was thawed, and the surviving infectivity was measured on HeLa cell monolayers.
Calculation of Fab-virus surface contacts.
The buried surfaces within the Fab-virus contact interfaces were determined by using the programs MSPDB, MS, MSSEP, and MSAV (9) and ATMSRF (45) and a solvent probe radius of 1.7 Å. The Fab17 portion of the complex crystal structure (49) and the Fab1 structure from the cryo-TEM fit were used with the surface of HRV14 around the epitope region that had been generated by using the native HRV14 structure (40) and icosahedral symmetry.
Atomic coordinates.
The atomic coordinates for Fab1-IA have been deposited in the Brookhaven Protein Database (accession no. 1a6t).
RESULTS
Antibody neutralization properties.
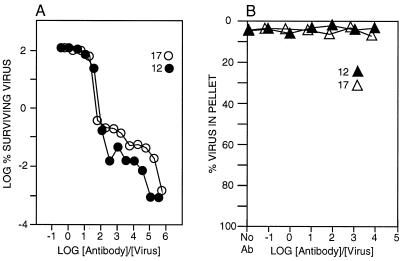
Experiments were performed to ascertain the relative neutralizations (neutralization assay method A) and aggregation efficacies of the NIm-IA antibodies MAb17, MAb12, and MAb1. Both MAb17 and MAb12 were highly efficacious at inhibiting HRV14 infection, with MAb12 being slightly better than MAb17 (Fig. 1). Under optimal neutralization conditions, both antibodies inhibited plaque initiation by about 5 orders of magnitude. However, neither antibody formed appreciable amounts of precipitate over the same antibody/virus ratios. These results, and previous results demonstrating a maximal antibody/virus stoichiometry of ∼30, strongly suggest that both MAb12 and MAb17 (26, 52) bind bivalently to the virion surface and that virion aggregation cannot account for neutralization.
FIG. 1.
Fab12 and Fab17 neutralization and precipitation properties. (A) The abilities of MAb12 and MAb17 to inhibit plaque formation were measured (method A) at various antibody concentrations. (B) The abilities of the antibodies MAb12 and MAb17 to precipitate the virions were measured over a comparable range of antibody concentrations.
In contrast, MAb1 only weakly inhibited plaque formation, with a maximum inhibition of ∼2 orders of magnitude and with a slight enhancement in neutralization at intermediate antibody/virus ratios (Fig. 2). Unlike the other two MAbs, MAb1 strongly aggregated the virions over the same range of ratios at which neutralization enhancement was observed. At very high antibody/virus ratios, there was little precipitation but significant (∼1.5-log-unit) neutralization. Therefore, MAb1 does not neutralize HRV14 solely by precipitating it, but neutralization is enhanced by aggregation at intermediate antibody/virus ratios. Since MAb1 strongly aggregates HRV14, cannot form stable, monomeric virus-antibody species, and binds with a maximal stoichiometry of ∼60 antibodies/virion, this antibody binds monovalently to the virion surface. It is important to note, however, that when antibodies are maintained in the plaque assay overlays (method B), all three antibodies neutralize HRV14 infectivity with comparable efficacies. This latter result may more accurately represent in vivo conditions where antibodies are not removed after the first round of viral attachment.
FIG. 2.
Neutralization (○) and precipitation (▵) properties of the NIm-IA antibodies MAb1 (strong precipitator, weak neutralizer), MAb23 (strong precipitator, strong neutralizer), and MAb14 (strong precipitator, weak neutralizer).
For comparison, results for MAb23-IA (MAb23) and MAb14-IA (MAb14) are also shown in Fig. 2. Both of these antibodies precipitate HRV14 over a wider range of antibody concentrations than MAb1. Since MAb23 is also a strong neutralizer, bivalent binding is not a prerequisite for efficient neutralization. This can be explained if antibody avidity mainly dictates neutralization efficacy and the intrinsic affinity of MAb23 for HRV14 is greater than that of either MAb1 or MAb14.
Image reconstructions of strongly and weakly neutralizing antibody-virus complexes.
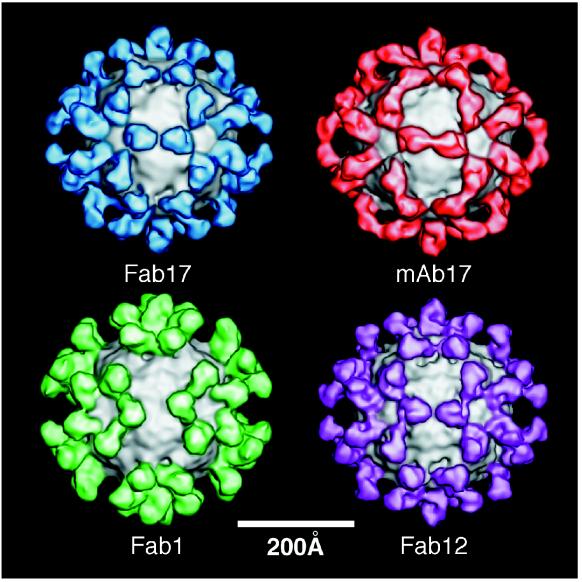
The cryo-TEM structures of three Fab-HRV14 complexes and one MAb-HRV14 complex were determined to explore possible correlations between the orientations and locations of antibody binding and neutralization efficacy (Fig. 3). The orientation of Fab17 on the viral surface suggested that MAb17 would bind bivalently across icosahedral twofold axes (Fig. 3). Bivalent binding of MAb17 was clearly demonstrated in the subsequent image reconstruction of the MAb17-HRV14 complex (Fig. 3) (51). To test whether this binding mode is common to other strongly neutralizing antibodies, the cryo-TEM image reconstruction of the Fab12-HRV14 complex was determined (Fig. 3). Even though both MAb12 and MAb17 are IgG2a antibodies from the same mouse, it was assumed that they were unique since MAb12 is more soluble (data not shown) and a stronger neutralizer (Fig. 1) than MAb17. Nonetheless, the image reconstructions of the respective virus-Fab complexes appeared to be virtually identical. Any differences in these reconstructions were accounted for by the higher resolution of the Fab12-HRV14 density map. As was demonstrated for the Fab17-HRV14 complex, the binding orientation of Fab12 strongly supports the contention that MAb12 binds to virions in a bivalent manner.
FIG. 3.
Shaded, surface representations of the cryo-TEM image reconstructions of HRV14-Fab17, HRV14-MAb17, HRV14-Fab1, and HRV14-Fab12. Fab17, MAb17, Fab1, and Fab12 are blue, brown, green, and mauve, respectively, with the surface of HRV14 shown in gray.
The cryo-TEM image reconstruction of the Fab1-HRV14 complex (Fig. 3) clearly differs from those of the other virus-Fab complexes. The Fab arms of the weakly neutralizing, strongly aggregating MAb1 bind in a more radially directed orientation on the capsid, and they are rotated ∼25° about their long axes compared to Fab12 and Fab17. In this orientation, the constant domains of the symmetry-related, bound Fab1 fragments point away from each other. Unlike the case for MAb17 (51), rotation of the constant domain about the elbow axis cannot generate a bivalently bound model of MAb1. The large separation of neighboring Fab1 molecules and the more radial orientation of the constant region in Fab1 presumably favor monovalent binding. Although the hinge region of antibodies is known to be highly flexible (55, 58), the distance between icosahedral twofold-related NIm-IA sites is sufficiently large to cause this relatively small difference between the Fab17 and Fab1 binding orientations to have a profound effect on binding valency. A similar distance constraint on bivalent binding has been demonstrated for the antibody-FMDV complex (20).
The HRV14 capsid surfaces in all three virus-Fab complexes appear to be similar to that in HRV14 alone. Hence, the binding of Fab12 or Fab1 does not induce conformational changes that can be detected in the cryo-TEM reconstructions. This correlates with the results of the determination of the crystal structure of the HRV14-Fab17 complex at a 4-Å resolution, in which antibody-induced conformational changes in the viral capsid were not observed (49).
Fitting of atomic models into the cryo-TEM density maps.
Interpretation of each virus-Fab complex is based on docking experiments in which the atomic models of Fab17 (28) and Fab1 (6a) were fitted into the cryo-TEM density maps of the virus-Fab12 and virus-Fab1 complexes (Fig. 4), together with the atomic model of HRV14 (40). The 4-Å-resolution map of the Fab17-HRV14 crystal structure (49) guided the interpretation of these pseudo-atomic models. The model of Fab12 was built by starting with the Fab17-HRV14 crystal structure and then making adjustments to account for the 10-residue difference between the Fab12 and Fab17 sequences. Each Fab model was first fit manually into the cryo-TEM map and then refined as a rigid body by using X-PLOR.
FIG. 4.
Pseudo-atomic fitting of different Fab models into cryo-TEM density maps. The top panels and middle left panels show the C-α backbones of VP1, VP2, VP3, VP4, the Fab light chains (L), and the Fab heavy chains (H). The electron density is represented by the black lines. The RNA interior is towards the bottom of the panels, and the nearest fivefold axis is at the left. The middle right panel shows orientations of the bound Fab1 and Fab12 in the same view as the top row. The bottom two panels are a stereo diagram of the Fab1 and Fab12 models with the view direction from the nearest fivefold axis towards the nearest twofold axis. This figure was generated with the program MOLVIEW (48) (http://bilbo.bio.purdue.edu/~tom).
The 4-Å structure of the Fab17-HRV14 complex provided an important means to ascertain the effectiveness of X-PLOR rigid-body refinement. We first tested whether the radius of convergence of the rigid-body refinement was sufficiently large to be effective at the resolution of the cryo-TEM studies. The Fab17-HRV14 model was used to calculate structure factors (from 200- to 25-Å resolution) from which an electron density map was calculated. The Fab17 model, using the elbow angle as determined from the crystal structure, was then purposely displaced halfway (∼20 Å) out of the Fab density envelope. The rigid-body refinement process correctly moved the model back to its original position, with initial and final R factors of 56 and 0%, respectively. With the cryo-TEM-derived structure factors, refinement of a variety of starting models (R factor = 60%) led to final fits (R factor = 40%) that were consistent with each other but not identical to the crystal structure of the Fab17-HRV14 complex. Compared to the crystal structure, X-PLOR refinement reduced the average C-α error from 4.0 Å in the original model to 2.1 Å. No significant improvement was afforded when the constant domains were refined separately from the variable domains. Given the success of these tests, this X-PLOR rigid-body refinement procedure was then used to place the Fab1 structure into the cryo-TEM density map of the Fab1-HRV14 complex. All three Fab models gave good qualitative fits to the respective image reconstructions (Fig. 4, top and middle, left).
The results of the atomic model fitting experiments validate the proposed binding modes for weakly and strongly neutralizing antibodies. For the three NIm-IA antibodies we studied, the two strong neutralizers bound bivalently to the virion, whereas the weak neutralizer bound in an orientation that favors interparticle cross-linking (Fig. 4, bottom), although bidentate binding may occur to a small extent. Thus, these image reconstruction studies correlate well with the proposal that links the neutralizing efficacies of these antibodies with their binding modes. Previous studies of φX174 modified with 2,4-dinitrophenol showed that bivalently bound IgGs have a 1,000-fold-higher affinity to virions than monovalently bound Fab fragments (21). Therefore, the correlation between valency and neutralization efficacy may be due entirely to these avidity differences. If this hypothesis is correct, the intrinsic affinity of MAb23 (Fig. 2) should be equivalent to the avidity (bidentate binding) of MAB17 and MAb12.
Sequences of Fab variable domains.
All three of our modeling studies as well as the crystal structure determination for the Fab17 complex have indicated or shown that charged residues at the capsid-Fab17 interface are determinants of binding (28, 49, 52). Asp54H and Asp56H in the CDR2 loop of the Fab17 heavy chain contact R1094 and K1097 at the base of the NIm-IA loop in HRV14. At the top of the NIm-IA loop, E1091 and D1095 interact with Arg91L in the CDR3 loop of the light chain. To determine if similar coulombic interactions occur in the Fab12 and Fab1 virion contacts, the variable domains of these antibodies were cloned and sequenced (Fig. 5). Fab17 and Fab12 have nearly identical sequences: the respective heavy and light chains each differ by only five residues. Because only a few nucleotide differences distinguish these two monoclonal antibodies and because both hybridoma cell lines were isolated from the same mouse, the B cells probably originated from the same mother cell prior to somatic hypermutations. Hence, this sequence comparison cannot by itself prove that these charge interactions are conserved among all NIm-IA antibodies.
FIG. 5.
Sequence comparisons between Fab17, Fab12, and Fab1 light-chain (A) and heavy-chain (B) variable domains. The numbering scheme (23) is shown above the amino acid sequences, and the CDR regions are noted below the sequences. Residues that are common to Fab17 are highlighted in gray, and residues in contact with HRV14 are outlined in boxes. The GenBank accession numbers for Fab12-VL, Fab12-VH, Fab1-VL, and Fab1-VH are AF045893, AF045892, AF045894, and AF045895, respectively.
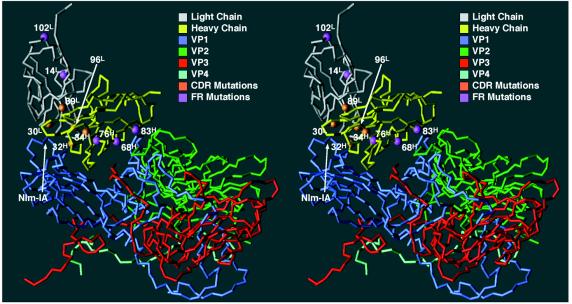
The crystal structure of the HRV14-Fab17 complex showed that several residues in the FR1 and FR3 framework regions of the Fab heavy chain contact the south wall of the canyon (49). Two of the five amino acid differences between the heavy chains of Fab17 and Fab12 (32H and 34H) lie within the CDR1 loop. The remaining three (68H, 82aH, and 76H) are in the FR3 region, with the first two making contact with the south wall of the canyon (Fig. 5 and 6). It seems unlikely that this cluster of mutations would occur by coincidence at the south wall interface. Thus, these FR3 residues may contribute to the higher neutralization efficacy of Fab12 compared to Fab17 (Fig. 1).
FIG. 6.
The C-α backbone of the HRV14-Fab17 complex, with the residues that differ from Fab12 in the framework (FR) and hypervariable (CDR) regions highlighted. The orientation and program used are the same as for Fig. 4.
Fab1, which was isolated from a different mouse than Fab17, differs from Fab17 by 42 residues in the heavy chain and 23 residues in the light chain. Fab1 binds in an orientation distinct from that of Fab17 (Fig. 4). This is also manifested in their footprints on the viral surface (Fig. 7), which indicate that Fab1 contact is more towards the west of the NIm-IA site. However, both Fabs contain what appear to be two key aspartic acid residues in the CDR2 loops of their heavy chains. In Fab1, these Asp residues (52H and 53H) lie upstream compared to those in Fab17 (54H and 56H). The crucial role of these Asp residues in the HRV14-Fab17 epitope-paratope interactions provides evidence that a pair of charged residues may likewise be important in the binding of other antibodies at the NIm-IA site. This interpretation is further corroborated by the effects of site-directed mutations (see below).
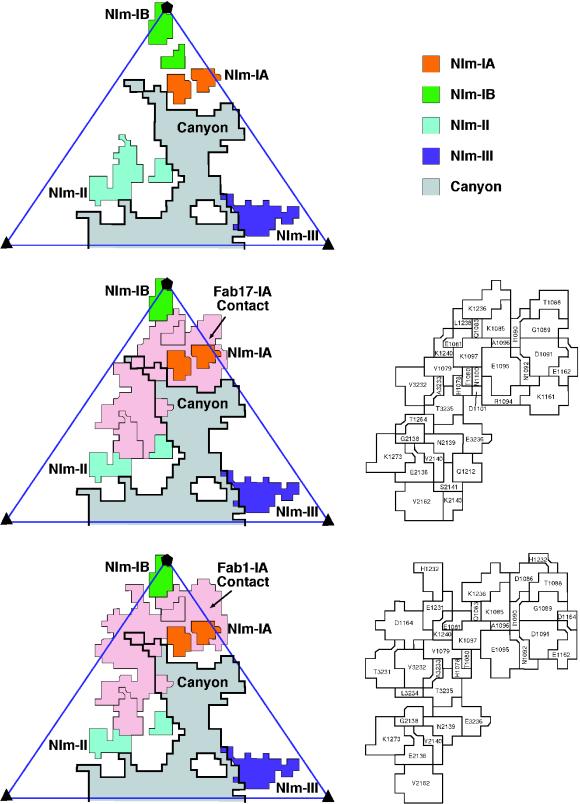
FIG. 7.
(Left) Surface diagrams of HRV14 denoting the Fab17 and Fab1 contact regions within an icosahedral asymmetric unit. The contact region for Fab17 was determined by using the Fab17-HRV14 crystal structure, whereas the Fab1 contact area was determined from the pseudo-atomic model derived from the cryo-TEM density. (Right) Stereo views of a portion of the HRV14 van der Waals surface, with the Fab17 (upper panels) and Fab1 (lower panels) contact areas in blue and the remainder of the viral surface in gray (made by using the program GRASP [35rsqb;).
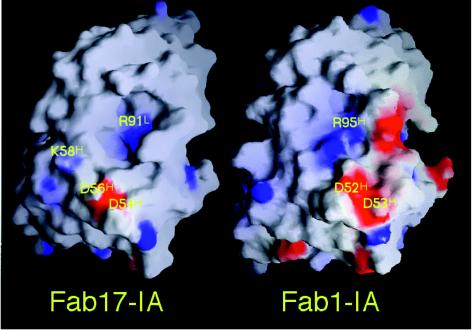
The structure-based alignment of Fab17 and Fab1 reveals other regions of paratope conservation. The electrostatic characters of both paratope surfaces are quite similar (Fig. 8). In Fab17, Arg91L (which interacts with D1091 and E1095) contributes to a positively charged patch located in the cleft between the heavy and light chains. The two aspartic acid residues (Asp54H and Asp56H) in the CDR2 loop of the Fab17 heavy chain form a negatively charged patch juxtaposed with viral residues R1094 and K1097. Fab1 also has a positively charged patch, but it is comprised of heavy-chain residues Arg50H and Arg95H. The negatively charged patch is formed by the two conserved aspartic acid residues Asp52H and Asp53H. Interestingly, the two charged patches are aligned in Fab1 in an orientation that is rotated counterclockwise by ∼25° compared to that in Fab17, and this directly correlates with the differences in the Fab1 and Fab12 binding orientations. This agrees with the previous suggestions that electrostatic field interactions may be important in NIm-IA antibody binding (52) and with recent calculations of electrostatic field complementarity at protein-protein interfaces (32).
FIG. 8.
Electrostatic character of the Fab17 and Fab1 paratope surfaces. The positive charge (blue) and the negative charge (red) are mapped onto the van der Waals surface. The heavy-chain hypervariable region is towards the bottom of the image and the light chain is towards the top. This figure was made by using the program GRASP (35).
Interactions between HRV14 and NIm-IA antibodies.
Despite differences in the orientation of bound Fab1 compared to those of Fab17 and Fab12, the light-chain CDR2 loops of all three antibodies contact very little of the viral surface (Fig. 5). As the light chains of these antibodies contact a steep surface east of the NIm-IA site, it is difficult for them to make extensive contact. In contrast, the heavy chain fits quite well into the canyon, thereby allowing all three heavy-chain CDRs to contact the viral surface (Figs. 5 to 7). Such dominance of antigen contact via the antibody heavy-chain contact has been observed in other Fab-antigen complexes (see, e.g., reference 10) and is not unexpected in view of the inherent genetic diversity of heavy chains compared to light chains (e.g., the D genetic cassettes and the activation of terminal dideoxynucleotide transferase during heavy-chain somatic recombination). In addition, recent results with camelid antibodies have shown that antibodies comprised of only heavy chains do occur in vivo and bind antigens (13).
Electrostatic interactions dominate the interface between the NIm-IA loop region and Fab1 (Table 1; Fig. 5). E1095, one of the two residues that define the NIm-IA site, is clamped by the positively charged cleft between the heavy- and light-chain hypervariable regions (Fig. 8). The Fab1 heavy-chain arginines (Arg50H and Arg95H) have direct interactions with E1095. The other NIm-IA residue, D1091, lies outside this region of positive charge. The side chain of D1091 in the crystal structure of Fab17-HRV14 rotates to form interactions with the corresponding bases in Fab17. K1097 interacts with only one aspartic acid residue (Asp52H) in Fab1 but with two (Asp54H and Asp56H) in Fab17. The other aspartic acid of Fab1 (Asp53H) forms a salt bridge with K1240, which lies near the NIm-IA loop. R1094 is located close to the negatively charged region. It is quite possible that its side chain would move into this region, as was observed in the Fab17-HRV14 crystal structure.
TABLE 1.
Percentages of surface contact according to residue type
| Residue type | Contact with:
|
|||
|---|---|---|---|---|
| Fab17
|
Fab1
|
|||
| Area (Å2) | % of total | Area (Å2) | % of total | |
| Acidic (D, E) | 96 | 11 | 95 | 14 |
| Basic (H, K, R) | 142 | 17 | 84 | 12 |
| Polar (N, Q, S, T) | 187 | 22 | 191 | 28 |
| Small (A, G) | 61 | 7 | 76 | 11 |
| Hydrophobic (C, I, L, M, P, V) | 23 | 3 | 84 | 12 |
| Aromatic Polar (W, Y) | 336 | 38 | 158 | 23 |
| Total | 845 | 100 | 685 | 100 |
Further study of HRV14 site-directed mutants, constructed previously for our Fab17-virus complex work (52), supports our interpretations of the two new Fab-virus complexes and demonstrates the importance of electrostatic interactions in Fab binding to the NIm-IA site. Of the residues tested, mutation of K1097, which makes extensive interactions with Fab17 (49), had the greatest effect on the neutralization of all NIm-IA antibodies (Table 2). The K1097E mutation reduced NIm-IA antibody neutralization by 102- to 104-fold. This mutation is just as effective as a naturally occurring escape mutation in blocking neutralization. When an uncharged residue (Gln) was substituted for K1097, little to no effect was observed except for MAb17. The K1085E mutation affected almost all NIm-IA antibodies by about 10-fold, whereas the K1236E mutation had little to no effect on any of these antibodies. This result agrees with the crystal structure study of the Fab17-HRV14 complex, in which Fab17 was observed to contact K1085 but made very little contact with K1236. These mutagenesis results demonstrate that coulombic interactions with these viral surface lysine residues (not identified in the initial selection of naturally occuring escape mutations) are very important to both weakly and strongly neutralizing antibodies.
TABLE 2.
Residual infectivity of HRV14 wild-type (WT) and mutant viruses after treatment with NIm-IA antibodies
| Antibody | % of residual plaques
|
||||
|---|---|---|---|---|---|
| LP1 (WT) | K1085E | K1236E | K1097Q | K1097E | |
| MAb1 | 0.0021 | 0.032 | 0.0088 | 0.0054 | 0.27 |
| MAb3 | 0.0064 | 0.060 | 0.014 | 0.011 | 83 |
| MAb4 | 0.0031 | 0.045 | 0.0082 | 0.0059 | 0.44 |
| MAb6 | 0.0045 | 0.050 | 0.014 | 0.012 | 71 |
| MAb7 | 0.0064 | 0.063 | 0.014 | 0.015 | 106 |
| MAb14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.19 |
| MAb17 | 0.0190 | 0.052 | 0.016 | 3.1 | 87 |
| MAb20 | 0.0000 | 0.003 | 0.0013 | 0.00066 | 0.018 |
Antibody-mediated stabilization of HRV14 against acid inactivation.
The crystal structure of the Fab17-HRV14 complex demonstrated that large conformational changes in the virion are not required for antibodies to mediate neutralization. Thus, antibodies may achieve their effect by stabilizing virions without necessitating large conformational changes. To test this hypothesis, HRV14 was complexed with several antibodies, incubated in buffers at various pHs, and examined for residual infectivity (Table 3). Interestingly, all NIm-IA antibodies stabilized virions against acid inactivation, whereas antibodies to the other antigenic sites did not. For NIm-IA antibodies, stabilization was independent of binding valancy, since both aggregating MAb1, -14, and -23) and nonaggregating (MAb3, -4, -6, -7, -17, and -20) antibodies protected HRV14. Although binding orientation and valency differ among these antibodies, Fab1, Fab12, and Fab17 make extensive contact with the bottom and north side of the canyon. The NIm-IB, NIm-II, and NIm-III sites are further from the canyon than NIm-IA, and antibodies to these sites are less likely to make extensive contact with the canyon. Therefore, the stabilization of virions by antibodies is probably mediated by direct contact with the canyon region, but such stabilization is not necessary for neutralization.
TABLE 3.
Effects of antibodies on pH inactivation of HRV14 with strong neutralizers (SN), intermediate neutralizers (IN), weak neutralizers (WN), precipitating antibodies (P), and nonprecipitating antibodies (NP) to all four antigenic sites
| MAb | Nim site | Type | % Residual infectivity after incubation at pHa:
|
||
|---|---|---|---|---|---|
| 7.0 | 5.0 | 4.5 | |||
| None | 100 | 0.0 | 0.0 | ||
| 3 | IA | SN, NP | 101 | 99 | 0.14 |
| 4 | IA | SN, NP | 84 | 89 | 49 |
| 6 | IA | SN, NP | 116 | 91 | 61 |
| 7 | IA | SN, NP | 127 | 104 | 35 |
| 14 | IA | SN, P | 65 | 67 | 5.6 |
| 23 | IA | SN, P | 82 | 87 | 0.0 |
| 17 | IA | IN, NP | 92 | 91 | 0.0 |
| 20 | IA | IN, NP | 80 | 82 | 0.29 |
| 1 | IA | WN, P | 52 | 50 | 0.30 |
| 26 | IB | WN, P | 81 | 0.0012 | 0.0 |
| 18 | II | WN, P | 44 | 0.0047 | 0.0 |
| 8 | III | IN, NP | 104 | 0.0 | 0.0 |
| 9 | III | IN, P | 106 | 0.0 | 0.0 |
Residual infectivity after incubation at the indicated pH and removal of antibody by freezing.
DISCUSSION
Mechanism of antibody-mediated neutralization of HRV14.
Several mechanisms for antibody-mediated neutralization of picornaviruses have been proposed. These include aggregation, induction of conformational changes, virion stabilization, and abrogation of cellular attachment.
(i) Aggregation.
It has been suggested that aggregation occurs concomitantly with neutralization and that virus/antibody ratios in vivo are conducive to aggregation (4, 5, 54). However, our data strongly suggest that aggregation is not a major contributor to neutralization of HRV. First, antibodies that bind bivalently to virions do not aggregate them over a wide range of antibody/virus ratios, yet such antibodies are strong neutralizers (Fig. 1). Second, even antibodies that are strong aggregators neutralize virus at antibody/virus concentration ratios that do not favor aggregation. The neutralization profile for aggregating antibodies sometimes displays a dip that is often coincident with aggregation. Hence, in such circumstances, neutralization may be enhanced in a narrow range of antibody/virus ratios that favors precipitation. This enhancement may result from a decrease of independent infectious particles or from avidity effects caused by antibodies bound bivalently, in an interparticle manner, to the large immunocomplexes. Although aggregation probably does not play a significant role in vitro, it may facilitate innate immunological responses via opsonization in vivo.
(ii) Stabilization.
It has also been suggested that antibodies might neutralize virions by stabilizing the capsid (34), which might then prevent uncoating or receptor-induced conformational changes. All antibodies that bind to NIm-IA (aggregating and nonaggregating) stabilize virions against acidic pH to various extents. However, none of the non-NIm-IA antibodies that we tested cause such stabilization, although some are efficacious neutralizers. Therefore, these stabilization effects do not correlate well to neutralization efficacy or binding valency. In addition, antibodies to all four sites have been shown to block cellular attachment (8), and this would precede any stabilization effects. Notably, all known escape mutations map only to residues around the epitope. An escape mutation which does not affect antibody binding but prevents neutralization has not yet been observed. If capsid stabilization-destabilization was a major determinant of neutralization, at least some escape mutations that abrogated these effects might be expected to arise. Analogous distal-site resistance mutations have been found when poliovirus and rhinovirus are grown in the presence of capsid-stabilizing antiviral agents (19).
(iii) Conformational changes.
Antibodies and Fab fragments cause an apparent decrease in the pI of the viral capsid concomitant with neutralization (8, 29). This fact has been cited as evidence that antibodies neutralize by distorting the capsid. The crystal structure of the Fab17-HRV14 complex clearly demonstrated that efficacious neutralization occurs in the absence of large conformational changes. Instead, Fab17 undergoes large conformational changes to better accommodate the epitope without inducing structural changes in the virion (49). Even though all antibodies to the four different antigenic sites that were tested (MAb13, -17, -21, -28, -29, -33, -34, and -35) caused apparent changes in the pI of the capsid (8), it seems unlikely that dissimilar antibodies, which bind to distinct epitopes, would all cause the same effect on the capsid. Antibodies might cause conformational changes in protein structure upon binding, but such changes would be expected to occur on flexible portions of the viral structure. Antibody-induced conformational changes on less flexible regions would cost significant Gibbs free energy and would greatly affect antibody affinity. Therefore, it seems unlikely that induction of conformational changes contributes significantly to antibody neutralization.
(iv) Abrogation of cellular attachment.
Previous studies have clearly demonstrated that antibodies to all four HRV14 antigenic sites block cellular attachment (8). The three NIm-IA antibodies that we have studied clearly bind in a manner that overlaps the ICAM footprint as determined by cryo-TEM (37). Steric hindrance effects can be used to explain the competition between receptor and antibody binding. NIm-II is immediately adjacent to the ICAM binding region, and the ∼600- to 900-Å2 contact region of these antibodies possibly overlaps the ICAM site as well. However, NIm-III is quite distal (∼40 Å away) to the receptor binding region, yet NIm-III antibodies also compete with receptor binding (8). Perhaps, then, antibody competition with the receptor is merely a result of the sheer bulk of an antibody molecule (an IgG is ∼140 Å long and approximately equal to the radius of HRV14) and does not require direct overlap with the ICAM binding region. This steric model would also explain why cellular attachment is inhibited at a nonsaturating stoichiometry of 7 to 10 antibodies per virion (25, 52).
Summary.
Consideration of all of these results suggests that the mechanism of HRV neutralization in vitro may be much simpler than previously envisioned: antibodies bound to the surface of HRV14 are sufficient to block attachment of virus receptors. For poliovirus and rhinovirus, interactions with their receptors appear to be essential for the proper release of RNA into the cytoplasm of the host cell. Indeed, antibody-poliovirus complexes were shown to enter cells, but this import mode led to digested viral RNA (29). In contrast, the picornavirus FMDV needs its receptor only to enter the cell and not for RNA release (31), yet this virus is also effectively neutralized by antibodies. Therefore, a simple steric effect in which antibody binds to the virion surface and blocks receptor attachment is sufficient to explain the neutralization behavior of many antibodies. Although some antibodies might induce secondary effects (e.g., causing conformational changes in the viral capsid) upon binding (12, 27, 30, 60), such effects are not required for neutralization. In addition, antibodies that induce such secondary effects could not be exclusively selected for during B-cell clonal expansion.
Our results clearly do not rule out the existence of antibodies that induce changes in virion structure upon binding, just as they do not imply that all antibodies neutralize by abrogating cellular attachment. For example, the details of antibody interactions with FMDV and poliovirus might be expected to be quite different. The receptor binding region of FMDV is located on the end of a highly mobile, immunodominant loop. Receptor or antibodies binding to this loop are unlikely to transmit conformational changes to the virion but will most certainly affect cell attachment. In contrast, the top of the canyon region of poliovirus is involved in both receptor and antibody binding (18, 59). For poliovirus, therefore, some antibodies might bind to this region and either mimic receptor binding and cause conformational changes or inhibit changes in this region. Indeed, FMDV-antibody complexes can infect cells that have Fc receptors, whereas poliovirus-antibody complexes cannot (31). Antibodies to human immunodeficiency virus apparently neutralize by blocking attachment or events after uncoating, depending on which viral protein is targeted by the antibody (56). Antibodies to the hemagglutinin of influenza virus can prevent attachment and replication, but antibodies to the neuraminidase only interfere with virus release (24). Therefore, the effects of antibodies on viruses can be as diverse as the viruses.
Notably, in vivo studies have clearly shown that the types of in vitro mechanisms that we have described may be of limited consequence in protecting animals from viral infections. For example, antibodies that are not efficacious in vitro against Sindbis virus (43) and FMDV (33) are still capable of protecting animals from viral challenge. Although antibodies against neuraminidase from influenza virus are not neutralizing, they do affect disease progression in vivo (44). Therefore, the primary role of antibodies in vivo may be to act synergistically with other components of the immune system. This further implies that the design of vaccines should focus on the production of high-affinity antibodies rather than on a particular in vitro neutralization property. This has been recently shown to be true in the case of human immunodeficiency virus type 1, where the occupancy of binding sites on the virus is the major factor in neutralization efficacy irrespective of the epitope specificity (39). This goal of eliciting high-affinity antibodies is clearly more straightforward than having to create vaccines that yield antibodies which neutralize by a particular mechanism.
ACKNOWLEDGMENTS
We thank E. S. Chase for growing the hybridoma cells for MAb12 and MAb1, T. J. Schmidt for helpful suggestions in the processing of the Fab1 crystallographic data set, and Z. Zhu for help and advice in the sequence determinations of Fab1 and Fab12.
This work was supported by grants from the National Institutes of health (GM10704 to T.J.S. and GM33050 to T.S.B.).
REFERENCES
- 1.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 2.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 3.Baker T S, Newcomb W W, Olson N H, Cowsert L M, Olson C, Brown J C. Structures of bovine and human papilloma viruses: analysis by cryoelectron microscopy and three-dimensional image reconstruction. Cell. 1991;60:1007–1015. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brioen P, Dekegel D, Boeyé A. Neutralization of poliovirus by antibody-mediated polymerization. Virology. 1983;127:463–468. doi: 10.1016/0042-6822(83)90159-9. [DOI] [PubMed] [Google Scholar]
- 5.Brioen P, Thomas A A M, Boeyé A. Lack of quantitative correlation between the neutralization of poliovirus and the antibody-mediated pI shift of the virions. J Gen Virol. 1985;66:609–613. doi: 10.1099/0022-1317-66-3-609. [DOI] [PubMed] [Google Scholar]
- 6.Brünger A T. X-plor (version 3.1) user’s guide. New Haven, Conn: Yale University; 1992. [Google Scholar]
- 6a.Che, Z., and T. J. Smith. Unpublished data.
- 7.Cheng R H, Olson N H, Baker T S. Cauliflower mosaic virus: a 420 subunit (T=7), multilayer structure. Virology. 1992;186:655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colonno R J, Callahan P L, Leippe D M, Rueckert R R. Inhibition of rhinovirus attachment by neutralizing monoclonal antibodies and their Fab fragments. J Virol. 1989;63:36–42. doi: 10.1128/jvi.63.1.36-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly M L. Analytical molecular surface calculation. J Appl Crystallogr. 1983;16:548. [Google Scholar]
- 10.Corper A L, Sohi M K, Bonagura V R, Steinitz M, Jefferis R, Feinstein A, Beale D, Taussig M J, Sutton B J. Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody-antigen interaction. Nat Struct Biol. 1997;4:374–381. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]
- 11.Crowther R A. Procedure for the three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc London B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 12.Delaet I, Boeye A. Monoclonal antibodies that disrupt poliovirus only at fever temperatures. J Virol. 1993;67:5299–5302. doi: 10.1128/jvi.67.9.5299-5302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desmyter A, Transue T R, Ghahroudi M A, Thi M H, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 14.Emini E A, Kao S, Lewis A J, Crainic R, Wimmer E. Functional basis of poliovirus neutralization determined with monospecific neutralizing antibodies. J Virol. 1983;46:466–474. doi: 10.1128/jvi.46.2.466-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emini E A, Ostapchuk P, Wimmer E. Bivalent attachment of antibody onto poliovirus leads to conformational alteration and neutralization. J Virol. 1983;48:547–550. doi: 10.1128/jvi.48.2.547-550.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson J W, Frankenberger E A, Rossmann M G, Fout G S, Medappa K C, Rueckert R R. Crystallization of a common cold virus, human rhinovirus 14: “isomorphism” with poliovirus crystals. Proc Natl Acad Sci USA. 1983;80:931–934. doi: 10.1073/pnas.80.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 18.Harber J, Bernhardt G, Lu H H, Sgro J Y, Wimmer E. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology. 1995;214:559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- 19.Heinz B A, Rueckert R R, Shepard D A, Dutko F J, McKinlay M A, Francher M, Rossmann M G, Badger J, Smith T J. Genetic and molecular analysis of spontaneous mutants of human rhinovirus 14 resistant to an antiviral compound. J Virol. 1989;63:2476–2485. doi: 10.1128/jvi.63.6.2476-2485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewat E A, Verdaguer N, Fita I, Blakemore W, Brookes S, King A, Newman J, Domingo E, Mateau M G, Stuart D I. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 1997;16:1492–1500. doi: 10.1093/emboj/16.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornick C L, Karush F. Antibody affinity III. The role of multivalence. Immunochemistry. 1972;9:325–340. doi: 10.1016/0019-2791(72)90096-1. [DOI] [PubMed] [Google Scholar]
- 22.Jones T A, Zou J-Y, Cowan S W. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Kabat E A, Wu T T, Reid-Miller M, Perry H M, Gottesman K S. Sequences of proteins of immunological interests. Bethesda, Md: National Institutes of Health; 1987. [Google Scholar]
- 24.Kilbourne E D, Laver W G, Shulman J L, Webster R G. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J Virol. 1968;2:281–288. doi: 10.1128/jvi.2.4.281-288.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Kolatchar, P., et al. Unpublished data.
- 25.Lee W M. Human rhinovirus 14: synthesis and characterization of a molecular cDNA clone which makes highly infectious transcripts. University of Wisconsin, Madison; 1992. [Google Scholar]
- 26.Leippe D M. Stoichiometry of picornavirus neutralization by murine monoclonal antibodies. University of Wisconsin, Madison; 1991. [Google Scholar]
- 27.Li Q, Yafal A G, Lee Y M H, Hogle J, Chow M. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of the sequences at physiological temperatures. J Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Smith T J, Lee W M, Mosser A, Rueckert R R, Olson N H, Cheng R H, Baker T S. Structure determination of an fab fragment that neutralizes human rhinovirus 14 and analysis of the fab-virus complex. J Mol Biol. 1994;240:127–137. doi: 10.1006/jmbi.1994.1427. [DOI] [PubMed] [Google Scholar]
- 29.Mandel B. The interaction of neutralized poliovirus with HeLa cells. II. Elution, penetration, uncoating. Virology. 1967;31:247–259. doi: 10.1016/0042-6822(67)90168-7. [DOI] [PubMed] [Google Scholar]
- 30.Mandel B. Neutralization of poliovirus: a hypothesis to explain the mechanism and the one-hit character of the neutralization reaction. Virology. 1976;69:500–510. doi: 10.1016/0042-6822(76)90480-3. [DOI] [PubMed] [Google Scholar]
- 31.Mason P W, Baxt B, Brown F, Harber J, Murdin A, Wimmer E. Antibody-complexed foot-and-mouth disease virus, but not poliovirus, can infect normally insusceptible cells via the Fc receptor. Virology. 1993;192:568–577. doi: 10.1006/viro.1993.1073. [DOI] [PubMed] [Google Scholar]
- 32.McCoy A J, Chandana Epa V, Coleman P M. Electrostatic complementarity at protein/protein interfaces. J Mol Biol. 1997;268:570–584. doi: 10.1006/jmbi.1997.0987. [DOI] [PubMed] [Google Scholar]
- 33.McCullough K C, De Simone F, Brocchi E, Capucci L, Crowther J R, Kihm U. Protective immune response against foot-and-mouth disease. J Virol. 1992;66:1835–1840. doi: 10.1128/jvi.66.4.1835-1840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosser A G, Leippe D M, Rueckert R R. Neutralization of picornaviruses: support for the pentamer bridging hypothesis. In: Semler B L, Ehrenfeld E, editors. Molecular aspects of picornavirus infection and detection. Washington, D.C: American Society for Microbiology; 1989. pp. 155–167. [Google Scholar]
- 34a.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 35.Nicholls A. GRASP: graphical representation and analysis of surface properties. New York, N.Y: Columbia University; 1993. [Google Scholar]
- 36.Olson N H, Baker T S, Willingmann P, Incardona N L. The three-dimensional structure of frozen-hydrated bacteriophage φX174. J Struct Biol. 1992;108:168–175. doi: 10.1016/1047-8477(92)90016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinoski Z. DENZO. In: Sawyer L, Isaacs N, Bailey S, editors. Data collection and processing. Warrington, United Kingdom: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 39.Parren P W H I, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 41.Rossmann M G, McKenna R, Tong L, Xia D, Wu H, Choi H. Molecular replacement real-space averaging. J Appl Cryst. 1992;25:166–180. [Google Scholar]
- 42.Rueckert R R. Picornaviridae and their replication. New York, N.Y: Raven Press; 1996. [Google Scholar]
- 43.Schmaljohn A L, Johnson E D, Dalrymple J M, Cole G A. Nonneutralizing monoclonal antibodies can prevent lethal alphavirus encephalitis. Nature. 1982;297:70–72. doi: 10.1038/297070a0. [DOI] [PubMed] [Google Scholar]
- 44.Schulman J L. Immunology of influenza. In: Kilbourne E D, editor. The influenza viruses and influenza. New York, N.Y: Academic Press; 1975. pp. 373–393. [Google Scholar]
- 45.Sheriff S, Hendrickson W A, Stenkamp R E, Sieker L C, Jensen L H. Influence of solvent accessibility and intermolecular contacts on atomic mobilities in hemerythrins. Proc Natl Acad Sci USA. 1985;82:1104–1107. doi: 10.1073/pnas.82.4.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherry B, Mosser A G, Colonno R J, Rueckert R R. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J Virol. 1986;57:246–257. doi: 10.1128/jvi.57.1.246-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherry B, Rueckert R R. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J Virol. 1985;53:137–143. doi: 10.1128/jvi.53.1.137-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith T J. MolView: a program to analyze and display atomic structures on the Macintosh personal computer. J Mol Graphics. 1995;13:122–125. doi: 10.1016/0263-7855(94)00019-o. [DOI] [PubMed] [Google Scholar]
- 49.Smith T J, Chase E S, Schmidt T J, Olson N H, Baker T S. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith T J, Mosser A G, Baker T S. Structural studies on the mechanisms of antibody-mediated neutralization of human rhinovirus. Semin Virol. 1995;6:233–242. [Google Scholar]
- 51.Smith T J, Olson N H, Cheng R H, Chase E S, Baker T S. Structure of a human rhinovirus-bivalently bound antibody complex: implications for virus neutralization and antibody flexibility. Proc Natl Acad Sci USA. 1993;90:7015–7018. doi: 10.1073/pnas.90.15.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith T J, Olson N H, Cheng R H, Liu H, Chase E, Lee W M, Leippe D M, Mosser A G, Ruekert R R, Baker T S. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J Virol. 1993;67:1148–1158. doi: 10.1128/jvi.67.3.1148-1158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas A A, Vrijsen R, Boeye A. Relationship between poliovirus neutralization and aggregation. J Virol. 1986;59:479–485. doi: 10.1128/jvi.59.2.479-485.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas A A M, Brioen P, Boeyé A. A monoclonal antibody that neutralizes poliovirus by cross-linking virions. J Virol. 1985;54:7–13. doi: 10.1128/jvi.54.1.7-13.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thouvenin E, Laurent S, Madelaine M-F, Rasschaert D, Vautherot J-F, Hewat E A. Bivalent binding of a neutralizing antibody to a calicivirus involves the torsional flexibility of the antibody hinge. J Mol Biol. 1997;270:238–246. doi: 10.1006/jmbi.1997.1095. [DOI] [PubMed] [Google Scholar]
- 56.Ugolini S, Mondor I, Parren P W, Burton D R, Tilley S A, Klasse P J, Sattentau Q J. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vrijsen R, Mosser A, Boeye A. Postabsorption neutralization of poliovirus. J Virol. 1993;67:3126–3133. doi: 10.1128/jvi.67.6.3126-3133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wade R H, Taveau J C, Lamy J N. Concerning the axial rotational flexibility of the Fab regions of immunoglobulin G. J Mol Biol. 1989;206:349–356. doi: 10.1016/0022-2836(89)90484-1. [DOI] [PubMed] [Google Scholar]
- 59.Wien M W, Curry S, Filman D J, Hogle J M. Structural studies of poliovirus mutants that overcome receptor defects. Nat Struct Biol. 1997;4:666–674. doi: 10.1038/nsb0897-666. [DOI] [PubMed] [Google Scholar]
- 60.Wien M W, Filman D J, Stura E A, Guillot S, Delpeyroux F, Crainic R, Hogle J M. Structure of the complex between the Fab fragment of a neutralizing antibody for type 1 poliovirus and its viral epitope. Nat Struct Biol. 1995;2:232–243. doi: 10.1038/nsb0395-232. [DOI] [PubMed] [Google Scholar]