Abstract
The chemokine receptors CCR5 and CXCR4 are used by human immunodeficiency virus type 1 (HIV-1) in conjunction with CD4 to infect cells. In addition, some virus strains can use alternative chemokine receptors, including CCR2b and CCR3, for infection. A polymorphism in CCR2 (CCR2-V64I) is associated with a 2- to 4-year delay in the progression to AIDS. To investigate the mechanism of this protective effect, we studied the expression of CCR2b and CCR2b-V64I, their chemokine and HIV-1 coreceptor activities, and their effects on the expression and receptor activities of the major HIV-1 coreceptors. CCR2b and CCR2b-V64I were expressed at similar levels, and neither molecule affected the expression or coreceptor activity of CCR3, CCR5, or CXCR4 in cotransfected cell lines. Peripheral blood mononuclear cells (PBMCs) from CCR2-V64I heterozygotes had normal levels of CCR2b and CCR5 but slightly reduced levels of CXCR4. CCR2b and CCR2b-V64I functioned equally well as HIV-1 coreceptors, and CCR2-V64I PBMCs were permissive for HIV-1 infection regardless of viral tropism. The MCP-1-induced calcium mobilization mediated by CCR2b signaling was unaffected by the polymorphism, but MCP-1 signaling mediated by either CCR2b- or CCR2-V64I-encoded receptors resulted in heterologous desensitization (i.e., limiting the signal response of other receptors) of both CCR5 and CXCR4. The heterologous desensitization of CCR5 and CXCR4 signaling by both CCR2 allele receptor types provides a mechanistic link that might help explain the in vivo effects of CCR2 gene variants on progression to AIDS as well as the reported antiviral activity of natural CCR2 ligands.
Infection of cells by human immunodeficiency virus type 1 (HIV-1) requires the presence of the viral receptor CD4 and an appropriate coreceptor on the cell surface. Direct interactions between the viral Env protein and the coreceptor are thought to trigger conformational changes in Env that lead to fusion between the viral and cellular membranes, allowing the viral genome to enter the host cell cytoplasm (reviewed in references 7, 10, 12, 23, and 43). The recent identification of certain chemokine receptors as coreceptors for HIV-1, HIV-2, and simian immunodeficiency virus (SIV) has provided tremendous insight into the mechanisms underlying viral entry and tropism. The virus strains responsible for transmission and which are the predominant virus type isolated from asymptomatic, HIV-positive individuals use CCR5 as a coreceptor, while viruses that emerge later during the course of infection use CXCR4 either in place of or in addition to CCR5 for cellular entry (2, 9, 16, 21, 26, 27, 31). By virtue of their differential use of the major HIV-1 coreceptors, it has been proposed that these viruses be referred to as R5, X4, and R5X4 strains, respectively (8, 22). In addition, a host of other chemokine and orphan seven-transmembrane domain receptors have been shown to support infection by one or more virus strains in vitro, including CCR2b and CCR3 (16, 22, 26, 50). Evolution of coreceptor use in vivo from CCR5 to additional coreceptors has been linked to disease progression and may help explain certain aspects of viral pathogenesis (18, 52).
The importance of CCR5 for viral transmission is shown by the fact that approximately 1% of Caucasians are CCR5 negative due to a naturally occurring polymorphism and that these individuals are very highly resistant to virus infection (20, 35, 39, 51). The critical role of CCR5 for viral transmission and the association between the emergence of viruses that use CXCR4 and accelerated disease progression has triggered a search for additional polymorphisms in chemokine receptor genes that may influence viral transmission and disease course. Recently, a polymorphism in CCR2 in which Val 64 is replaced by Ile (CCR2-V64I) has been reported (55). This polymorphism, which occurs at an allele frequency of 10 to 25%, depending on the ethnic population, is associated with a 2- to 4-year delay in the progression to AIDS. However, relatively few virus strains that can use CCR2b in conjunction with CD4 to infect cells have been reported (22). Therefore, the mechanism underlying this protective effect is not apparent.
To investigate the protective effect of the CCR2 polymorphism, we studied the expression as well as the chemokine receptor and HIV-1 coreceptor activities of the major CCR2 isoform, CCR2b, with and without the V64I mutation in peripheral blood mononuclear cells (PBMCs) and transfected cell lines. CCR2b-V64I was expressed at levels similar to CCR2b in cell lines and in PBMCs, and both functioned equally well as viral coreceptors. The CCR2b polymorphism did not affect CCR5 or CXCR4 coreceptor activity or expression levels in cell lines or in stimulated PBMCs, though unstimulated PBMCs from CCR2b-V64I heterozygotes had slightly reduced CXCR4 levels. The addition of the CCR2b ligand MCP-1 lead to reduction of (i.e., desensitized) subsequent signaling induced by addition of RANTES, a CCR5 ligand. Similarly, heterologous desensitization of CXCR4 by CCR2b was observed. By contrast, RANTES and SDF-1 only partially suppressed subsequent signaling mediated by MCP-1 and CCR2b. Together, these results suggest that the protective effects of CCR2-V64I are apt to be subtle and indirect. Since HIV-1 Env has been shown to induce intracellular signals mediated by either CXCR4 or CCR5 (19, 57), and receptor signaling may play a role in both receptor surface expression and postentry events in viral replication (13, 28), this mechanistic link between CCR2b and the major HIV-1 coreceptors provides a possible explanation for the antiviral effects reported for CCR2b ligands.
MATERIALS AND METHODS
Cells.
Heparinized whole blood was collected from healthy volunteers previously screened by PCR-restriction fragment length polymorphism (RFLP) for the CCR2b-V64I mutation (see below). Unstimulated PBMCs were isolated by Ficoll-Hypaque gradient centrifugation (Pharmacia Biotech, Uppsala, Sweden) and placed in RPMI 1640 (GibcoBRL, Grand Island, N.Y.) supplemented with 10% fetal calf serum (GibcoBRL) and allowed to recover in media at 37°C for 6 h before staining for fluorescence-activated cell sorting (FACS) analyses. We also stained PBMCs in whole blood followed by selective erythrocyte lysis using PharM Lyse according to the manufacturer’s directions (Pharmingen, San Diego, Calif.) to determine if the way in which PBMCs were isolated affected coreceptor expression levels. PBMCs were considered stimulated if they were first cultured in medium supplemented with phytohemagglutinin (PHA) for 2 days followed by 3 days in recombinant interleukin-2 (IL-2; 100 U/ml) (called PHA/IL-2 treatment) before FACS analysis was performed. 239T cells and quail QT6 cells were cultured in Dulbecco’s minimal essential medium (GibcoBRL) supplemented with 10% fetal calf serum, 2 mM glutamine, and 2 mM penicillin-streptomycin.
Antibodies.
Monoclonal antibodies (MAbs) used in FACS analyses against CCR2b, CCR3, CCR5, and CXCR4 were R02 (biotinylated) (32), 7B11 (NIH AIDS Reference and Reagent Program), clone 45549 (R&D Systems, Minneapolis, Minn.), and 12G5 (29), respectively.
Plasmids and viruses.
Plasmids encoding the HIV-1 ADA, HxB2, and NL4-3 Envs for use in making luciferase virus were provided by John Moore (Aaron Diamond AIDS Research Center). The NL4-3 luciferase virus backbone (pNL-luc-E−R−) was provided by Ned Landau (Aaron Diamond AIDS Research Center) (15, 17). The plasmid expressing the CCR2b-V64I variant was cloned from an CCR2-V64I/V64I individual by using primers to amplify a 412-bp fragment from the 5′ end of the CCR2b gene encompassing the V64I mutation. The upstream primer introduces a HindIII site, while the downstream primer encompasses the unique ClaI site at position 407 (counting from the ATG start codon) of CCR2b. This PCR fragment was used to replace the identical HindIII-ClaI fragment save for the V64I mutation in pcDNA3-CCR2b, previously cloned from a wild-type CCR2b individual (26). The regenerated pcDNA3-CCR2b-V64I plasmid was confirmed to possess the V64I mutation by restriction mapping and direct sequencing. All other plasmids have been described by our lab previously (50). Vaccinia viruses encoding HIV-1 Envs included vSC60 (BH8), vCB51 (BK132; provided by Chris Broder), and vBD3 (89.6). We also used the recombinant virus vTF1.1, encoding the T7 RNA polymerase (1).
PCR-RFLP detection of CCR2b-V64I.
The G→A nucleotide substitution at position 190 (counting from the ATG start codon) resulting in the V64I mutation also introduces an additional FokI site into the open reading frame of the CCR2b gene. Genomic DNA was isolated from buccal swabs by incubating samples with DNA lysis buffer as previously described (47), subjected to 35 cycles of amplification by using Taq polymerase (95°C for 30 s, 62°C for 30 s, and 72°C for 30 s), separated on a 3% agarose gel, and stained with ethidium bromide. Primers were designed to amplify a 521-bp fragment from the Kozak sequence immediately upstream of the ATG codon to position 516 of the CCR2b gene. The amplified fragment comprises either one FokI site at position 379 in wild-type CCR2b or an additional FokI site at position 175 in CCR2b-V64I. Restriction digestion of this fragment with FokI results in either two fragments of 384 and 137 bp when amplified from a CCR2-+/+ individual or three fragments of 204, 180, and 137 bp when amplified from a CCR2-V64I/V64I individual (−/−). Restriction digestion of PCR fragments from heterozygous individuals (CCR2-+/V64I) results in four bands of 384, 204, 184, and 134 bp. The presence of at least one FokI site in the PCR fragment serves as an internal control for enzyme activity and digestion completion.
Cell-cell fusion and virus infection assays.
The efficiency of Env-mediated cell-cell fusion was determined by a gene reporter assay as described previously (45, 49). Briefly, 293T effector cells expressing vaccinia virus-driven Env and T7 RNA polymerase were mixed with quail QT6 cells transiently expressing CD4 and the indicated coreceptor with the reporter luciferase gene under control of the T7 promoter. Cytoplasmic mixing as a result of Env-mediated membrane fusion results in luciferase production which can be readily quantified with a luminometer.
Luciferase reporter viruses were prepared as previously described (15, 17) by cotransfecting 293T cells with the indicated Envs and the NL4-3 luciferase virus backbone (pNL-luc-E−R−). Target cells were prepared by transfecting U87-MG cells with CD4 and the appropriate coreceptors or mix of coreceptors as indicated. A constant amount of DNA was used for all target cell transfections by using pcDNA3 vector as a filler. Four days postinfection, cells were lysed with 0.5% Triton X-100 in phosphate-buffered saline (PBS), and an appropriate aliquot was analyzed for luciferase activity.
To measure viral entry, pcDNA3, pcDNA3-CCR5, pcDNA3-CXCR4, pcDNA3-CCR2b, and pcDNA3-V64I were transfected (2 μg of each plasmid) into 105 CD4-expressing QT6 (QT6/CD4) cells in 48-well tissue culture plates, using calcium phosphate precipitation. Four hours later, the cells were washed and placed in fresh medium; 48 h later, cells were infected with DNase-treated (50 U/ml for 30 min at room temperature) cell-free virus, using 50 ng of viral p24. After 2 days, cells were washed three times, suspended in 50 μl of lysis buffer (100 mM KCl, 20 mM Tris [pH 8.4], 0.1% Nonidet P-40, 0.5 mg of proteinase K per ml), incubated for 2 h at 60°C, and the boiled for 15 min. HIV-1-specific DNA sequences were detected by PCR using 35 cycles on 2.5 μl of cell lysate to amplify a 430-bp region of U3/U5 long terminal repeat (LTR) DNA sequences, using primers LTR-plus/LTR-minus (5′-ACAAGCTAGTACCAGTTGAGCC-3′/5′-CACACACTACTTGAAGCACTCA-3′). Fourfold serial dilutions of cell lysate were used under the same conditions. Products were resolved by electrophoresis on 2% agarose gels, transferred to Hybond N+ (Amersham), and detected by using a 3′-End Labeling Biotin kit (DuPont; probe 5′-ATCTACAAGGGACTTTCCCGC-3′), followed by exposure.
Calcium mobilization assays.
Response to ligand was determined in transiently transfected human 293T cells. Cells were transfected with the desired coreceptor for 4 to 6 h, medium was replaced, and cells were allowed to express the coreceptor overnight. Cells were incubated in medium containing 2.5 μM Fura-2/AM (Molecular Probes) at 37°C in the dark for 1 h. Cell efflux was allowed to occur for 15 min in PBS before the cells were removed from the plate manually. The cells were centrifuged and resuspended in Dulbecco’s PBS containing calcium and magnesium (BioWhittaker) at 2 × 106 cells/ml and were warmed at 37°C for 10 min before measurement of ligand response. Ca2+ mobilization was measured in an Aminco-Bowman luminescence spectrometer in a constantly stirring cuvette and in a volume of 1.5 ml. Excitation of cells was monitored at 340 and 380 nm, and Ca2+ concentration was calculated as previously described, using an assumed molecular weight of 224 (34). MCP-1, RANTES, and SDF-1α were obtained from Peprotech and resuspended in PBS for use. All cells were also stimulated with 27 μM thrombin receptor agonist peptide (TAP), consisting of the amino acids SFLLRN, to confirm the integrity of cells by their ability to signal through the G-protein-coupled receptor PAR-1.
FACS analyses.
For analysis of coreceptor expression on PBMCs, fresh or stimulated PBMCs prepared as described above were stained with primary antibodies for CCR2 (R02), CCR5 (45549), or CXCR4 (12G5) at 10 μg/ml followed by secondary detection by either phycoerythrin-conjugated streptavidin (2.5 μg/ml; Pharmingen) for anti-CCR2b antibody or affinity-purified phycoerythrin-conjugated donkey anti-mouse antibody (1:100 dilution; Jackson ImmunoResearch Laboratories) for anti-CCR5 or anti-CXCR4 antibody. Analyses were performed only on the lymphocyte gate defined by broad forward scatter but low side scatter. The mean channel fluorescence (MCF) was used to compare the levels of coreceptor expression. For analysis of coreceptor expression on transfected cells, 293T cells were variously transfected (via CaPO4 precipitation) with CCR5, CCR3, and CXCR4 plasmids alone or in combination with either wild-type CCR2b or CCR2b-V64I plasmids, using a constant amount of DNA with pcDNA3 vector as a filler. Cells were allowed to express the coreceptor for 18 h and subsequently stained with the appropriate anticoreceptor antibodies followed by secondary detection as described above for the PBMCs. All staining and washing protocols have been described in detail elsewhere (50), and FACS analyses were performed on a Becton Dickinson FACScan using the CellQuest version 3 software (Becton Dickinson, San Jose, Calif.).
RESULTS
Effects of CCR2b-V64I on HIV-1 coreceptor expression.
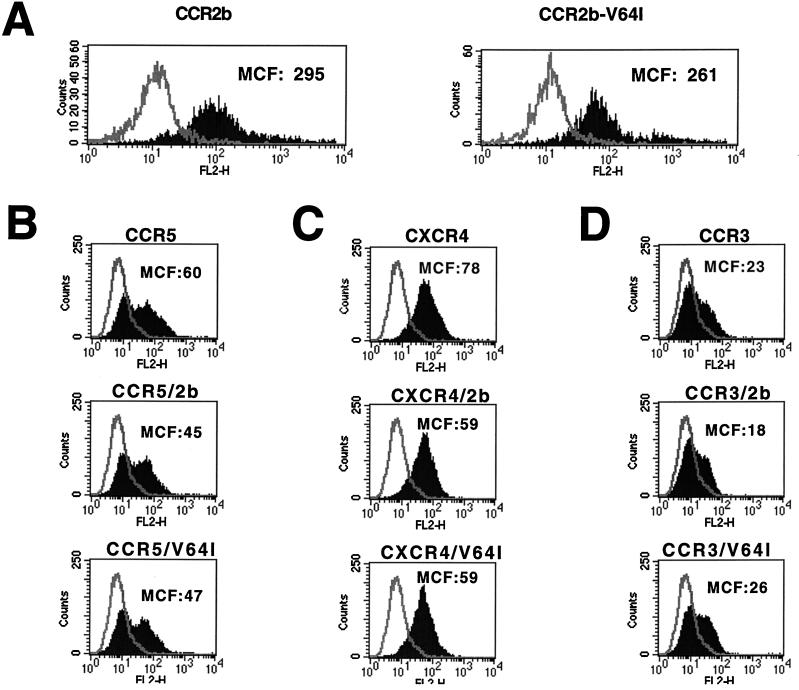
Before studying the chemokine receptor and HIV-1 coreceptor activities of CCR2-V64I, we determined whether the mutant receptor was expressed on the cell surface as efficiently as CCR2b since surface expression levels can strongly influence coreceptor function (38, 46, 50). Human 293T cells were transfected with plasmids encoding either CCR2b or CCR2b-V64I. CCR2b is the major CCR2 isoform derived by alternative RNA splicing (14). The plasmids were identical in both coding and noncoding regions save for the V64I alteration. Expression of both receptors on the cell surface was determined by FACS analysis using a previously described MAb to CCR2b (32). We found that the expression of CCR2b and CCR2b-V64I did not differ substantially when MCF intensities were compared (Fig. 1A). To determine if the V64I mutation could affect surface expression levels of other HIV-1 coreceptors, we coexpressed CCR2b and CCR2b-V64I with CCR5 (Fig. 1B), CXCR4 (Fig. 1C), or CCR3 (Fig. 1D) and measured their surface expression levels by using well-characterized MAbs. We found no differential effects on the surface expression levels of CCR3, CCR5, or CXCR4 as a consequence of either CCR2b or CCR2b-V64I coexpression. Thus, CCR2b-V64I is expressed as well as CCR2b and, relative to CCR2b, has no effect on the expression of other HIV-1 coreceptors in transfected 293T cells.
FIG. 1.
Effects of CCR2b or CCR2b-V64I on HIV-1 coreceptor expression. Equal amounts of CCR2b and the V64I variant were transfected into 293T cells, expression was allowed for 18 h, and subsequently staining was done with biotinylated anti-CCR2b antibody R02 followed by phycoerythrin-conjugated streptavidin. FACS analysis was used to determine surface expression levels, which are presented as the MCF (A). Similarly, CCR5 (B), CXCR4 (C), or CCR3 (D) was transfected into 293T cells alone or in combination with either CCR2b or V64I. Coreceptor expression levels were determined via FACS analysis by staining with MAbs 45549, 12G5, and 7B11 against CCR5, CXCR4 and CCR3, respectively (see Materials and Methods).
Effects of CCR2b-V64I on HIV-1 coreceptor activity.
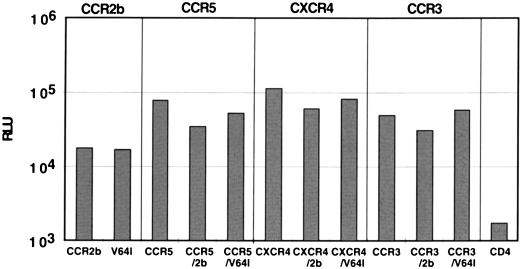
Since CCR2b-V64I was expressed normally, we next tested its HIV-1 coreceptor activity by using two different assays. Since only a handful of viruses that can use CCR2b as a coreceptor have been identified (22), we also asked if the V64I mutation affects the coreceptor activity of CCR3, CCR5, or CXCR4. To do this, we first used a quantitative cell-cell fusion assay in which HIV-1 Env and T7 polymerase are expressed in HeLa cells, while plasmids encoding CD4, the desired coreceptors, and luciferase under control of the T7 promoter are transfected into a target cell population (45, 49). The two cell populations are mixed, and if cell-cell fusion occurs, luciferase is produced as a consequence of cytoplasmic mixing. The results of a representative experiment are shown in Fig. 2. In all experiments, CCR2b-V64I and CCR2b supported cell-cell fusion by the HIV-1 89.6 Env protein equally well but at levels much lower than what was observed with CCR5. Further, coexpression of either CCR2b or CCR2b-V64I with CCR5, CXCR4, or CCR3 had no deleterious effects on the coreceptor activities of these molecules when tested against either 89.6 Env (Fig. 2) or other R5 (JR-FL) or X4 (BK132) Env proteins (data not shown). Thus, the V64I mutation did not affect the coreceptor activity of CCR2b, nor did it affect the activity of other HIV-1 coreceptors in transiently transfected cells.
FIG. 2.
Fusion activity of CCR2b and V64I. 293T cells expressing the R5X4 89.6 Env protein (which also utilizes CCR2b) and T7 polymerase were mixed with quail QT6 cells expressing CD4, the indicated coreceptor(s), and luciferase under control of the T7 promoter. Cell-cell fusion was assessed 8 h later by lysis of cells followed by quantitation of luciferase activity in relative light units (RLU). The results shown are from a typical experiment representative of four independent experiments. The trends observed were identical even when different effector cells (simian COS cells) and target cells (feline CCC cells) were used.
While the cell-cell fusion assay has been used to identify the coreceptors used by different virus strains, quantitative differences are sometimes observed between this and virus infection assays. Therefore, to determine whether CCR2b-64I was able to mediate entry by a virus that uses CCR2 for CD4-mediated fusion, QT6/CD4 cells were transfected with wild-type or mutant CCR2b and infected with HIV-1 89.6, and the cell lysate was analyzed 2 days later for the presence of specific viral DNA sequences, using a PCR-based assay (26). As shown in Fig. 3, both CCR2b-V64I and CCR2b supported 89.6 entry. Within each experiment, there was often some variability in the intensity of signal with the two CCR2b variants, but these differences were slight and not reproducible. However, the intensity of signal with CCR2b- and CCR2b-64I-mediated infection was always markedly less than that seen in parallel wells with CCR5 (Fig. 3) or CXCR-4 (data not shown), consistent with our earlier findings (26).
FIG. 3.
Viral entry and infection. QT6/CD4 cells were transfected with cofactor or with vector alone and the following day infected with DNase-treated HIV-1 89.6 virus stock. Two days later, the cells were lysed and viral entry was determined by PCR detection of viral DNA reverse transcription products, using LTR primers, followed by Southern blotting.
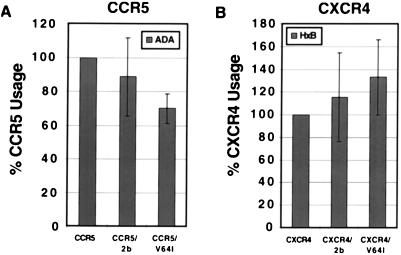
To determine if expression of CCR2b or CCR2b-V64I affected the coreceptor activity of CCR5 or CXCR4, cells were cotransfected with various combinations of coreceptors and then infected with luciferase reporter viruses bearing Env proteins known to use either CCR5 (HIV-1 ADA) or CXCR4 (HIV-1 HxB2) (15, 17). Different ratios of plasmids were used in order to obtain equivalent levels of CCR2b and CCR2b-V64I. As shown in Fig. 4, coexpression of either CCR2b or CCR2b-V64I with CCR5 reduced, on average, infection mediated by the R5 Env protein ADA. However, the reduction was slight and there were no statistically significant differences between the effects seen with CCR2b and CCR2b-V64I. Likewise, there were no statistically significant differential effects of either CCR2b or CCR2b-V64I coexpression on the use of CXCR4 by the X4 virus strain HxB2. Taken together, these results show that in transfected cells, the V64I mutation does not adversely affect the coreceptor activity of CCR2b, nor does it significantly affect the coreceptor activity of CCR5 or CXCR4 as measured by cell-cell fusion and virus infection assays.
FIG. 4.
Coexpression of CCR2b or CCR2b-V64I with CCR5 or CXCR4 does not affect infectability by R5 or X4 virus. CCR5 (A) or CXCR4 (B) was transfected alone or in combination with CCR2b or V64I and subsequently infected by luciferase reporter viruses pseudotyped with the R5 ADA or the X4 HxB2 Env as indicated. Luciferase activity was measured 4 days after infection. Results are normalized to the percentage of relative light units obtained by infection of cells transfected with CCR5 or CXCR4 alone. Data are presented as mean ± standard error of the mean of four independent experiments.
Expression of HIV-1 coreceptors on PBMCs.
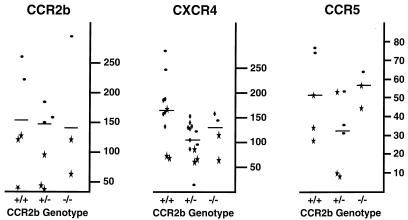
While the V64I mutation did not affect CCR2b, CCR5, or CXCR4 expression in transfected cells, it is possible that the effects of the mutation are evident only in primary cells. Therefore, we screened 84 individuals for the V64I mutation by PCR-RFLP analysis and identified 13 heterozygotes (+/V64I) and 2 homozygotes (V64I/V64I) for allelic frequencies of 0.083 for Caucasians and 0.225 for Asians, closely matching the published frequencies from large HIV-infected cohorts (55). PBMCs were isolated from race-matched volunteers with either CCR2b +/+, +/V64I, or V64I/V64I genotypes and stimulated with PHA and IL-2. FACS analysis using antibodies to CCR2, CCR5, and CXCR4 revealed no genotype-associated differences in expression levels on stimulated PBMCs (data not shown). However, stimulation of PBMCs in vitro is known to elevate CCR5 receptor expression levels (59) and may also influence the expression of other chemokine receptors (40). Therefore, to control for any effects that in vitro stimulation of PBMCs might have, we also tested unstimulated cells. Since Ficoll purification of PBMCs has been found to result in transient down regulation of CXCR4 (56a), cells were analyzed either by direct staining of whole blood followed by selective erythrocyte lysis or after a 6-h recovery period in medium not supplemented with either mitogen or IL-2 (see Materials and Methods). Similar results were obtained with both approaches. While there were no differences in CCR2b expression levels among the three genotypes, we found a mean decrease of approximately 37% in cell surface expression levels of CXCR4 between +/+ and +/V64I individuals (Fig. 5). While this difference was statistically significant (P < 0.025), PBMCs from +/V64I individuals were fully permissive to infection by X4 virus strains (see below). A slight decrease in CCR5 expression levels was also observed in +/V64I individuals, but this difference was not statistically significant. Finally, as only two homozygotes were identified, we can draw no conclusions about the effects of the V64I/V64I genotype on receptor expression levels. Examination of a larger number of individuals who bear the CCR2b-V64I mutation will have to be performed to more accurately determine the expression levels of the major HIV-1 coreceptors and to determine the effects of different culture conditions and purification protocols on the cell surface expression of these molecules.
FIG. 5.
Comparison of HIV-1 coreceptor expression on PBMCs from individuals with or without the V64I variant. Unstimulated PBMCs isolated as described in Materials and Methods were stained and FACS analyzed for CCR2b, CXCR4, and CCR5 expression levels. Due to the logistic difficulties of staining fresh PBMCs for multiple coreceptors on multiple volunteers, data are presented as compiled from several different FACS experiments. Data points with similar symbols indicate data from a single FACS experiment performed on the same day, with each data point representing the results obtained with cells from a different individual. Expression levels are presented as MCF, and only positive data points defined as being at least threefold above the MCF obtained for the isotype-matched negative controls are presented. Solid bars indicate the mean for each data spread. All non-Asians used for comparison studies of coreceptor expression were screened for the Δ32-ccr5 allele in order to control for the contribution of this allele to CCR5 expression levels. The Asian population in this study was not screened because the 32-ccr5 allele is nonexistent in the Asian population (4).
Infection of PBMCs from individuals with the CCR2b-V64I polymorphism.
To determine if the CCR2b-V64I mutation could have any unforeseen effects on HIV-1 entry or replication in primary cells, PHA/IL-2-stimulated PBMCs from +/+, +/V64I, and V64I/V64I individuals were infected with equal amounts of the X4 virus strain IIIB or the R5 virus strain SF162 or BR-2. In each experiment, cells were harvested from two to four donors in each category and analyzed in duplicate experiments. Viral p24 values were measured 7 and 14 days after infection, and the results were averaged. Results from three independent experiments are shown in Table 1. As differences were not observed between +/V64I and V64I/V64I samples, these were considered as a single group. While there was some variability, we observed no consistent differences between the abilities of +/V64I and V64I/V64I PBMCs to support virus infection compared to +/+ PBMCs. Thus, the CCR2b-V64I polymorphism did not affect the ability of HIV-1 to productively infect PBMCs, at least for the virus strains examined and under the tissue culture conditions used.
TABLE 1.
HIV-1 replication in CCR2b wild-type or CCR2b-V64I PBMCsa
| Expt | Mean supernatant p24 antigen level (ng/ml) ± SEM
|
|||||
|---|---|---|---|---|---|---|
| IIIB
|
SF162
|
BR-2
|
||||
| Day 7 | Day 14 | Day 7 | Day 14 | Day 7 | Day 14 | |
| 1 | ||||||
| Wild type | 9.5 ± 3.0 | 26.5 ± 5.5 | 183 ± 8.4 | 103 ± 5.0 | 164 ± 12.0 | 71.4 ± 7.8 |
| CCR2b-V64I | 13.8 ± 5.5 | 69.2 ± 10.3 | 114 ± 9.3 | 68.3 ± 6.9 | 111 ± 12.4 | 51.0 ± 8.2 |
| 2 | ||||||
| Wild type | 811 ± 21.6 | 850 ± 18.5 | 613 ± 10.3 | 79.9 ± 8.9 | 373 ± 5.3 | 107 ± 4.5 |
| CCR2b-V64I | 664 ± 16.6 | 490 ± 17.5 | 384 ± 12.8 | 238 ± 10.3 | 161 ± 8.0 | 67.0 ± 4.2 |
| 3 | ||||||
| Wild type | 30.7 ± 3.4 | 351 ± 8.6 | 257 ± 13.8 | 480 ± 15.8 | 129 ± 7.6 | 269 ± 10.9 |
| CCR2b-V64I | 23.1 ± 5.1 | 174 ± 10.6 | 381 ± 9.9 | 328 ± 5.2 | 98.2 ± 8.4 | 120 ± 6.8 |
Equal amounts of all viruses (20 ng of p24 antigen) were used to infect PHA/IL-2-stimulated PBMCs, and supernatant p24 antigen levels were measured by enzyme-linked immunosorbent assay. CCR2b-V64I heterozygous and homozygous donors were considered as a single group. In each experiment, cells were harvested from two to four donors in each category and analyzed in duplicate infections.
Asymmetric, heterologous desensitization of CCR5 and CXCR4 by CCR2b.
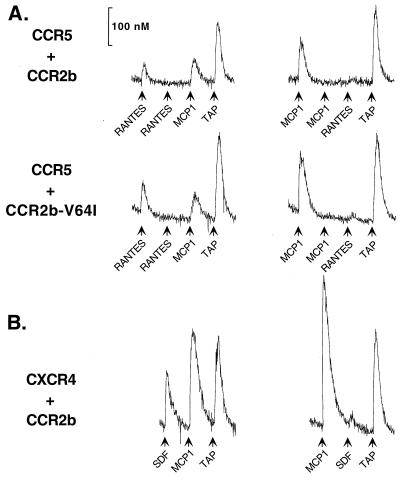
Our results indicate that the V64I mutation does not directly affect CCR2b expression or its already limited HIV-1 coreceptor capability. In addition, CCR2b-V64I did not affect the coreceptor activity of CCR3, CCR5, or CXCR4. These data argue that if the V64I polymorphism affects CCR3, CCR5, or CXCR4 coreceptor function in vivo, it is unlikely to be due to mere expression of the CCR2b-V64I receptor. However, G-protein-mediated signaling events can induce both intracellular changes that affect cellular activation and functional changes that alter the number and state of receptors on the surface of a cell. Peptide chemoattractant receptors have previously been reported to be capable of heterologous desensitization (limiting the signaling response of other receptors), suggesting a cross-communication between receptors that could affect surface expression (11, 14, 36, 56). Moreover, the recent demonstration that HIV-1 Env can signal through CCR5 and CXCR4 (19, 57) suggests that if this signal is important for HIV-1 replication and if other receptors have the ability to abrogate this signal, then factors that desensitize CCR5 or CXCR4 may protect against HIV infection. In fact, heterologous desensitization of a RANTES response by MCP-1 has been reported for the human monocytic leukemia MonoMac 6 cell line (14). We therefore determined whether the V64I mutation affected CCR2b-dependent signaling and whether activation of CCR2b desensitized the major HIV-1 coreceptors. Addition of MCP-1 to 293T cells expressing either CCR2b or CCR2b-V64I resulted in similar calcium mobilization profiles (Fig. 6). In addition, we found that addition of MCP-1 to either +/+, +/V64I, or V64I/V64I PBMCs resulted in similar calcium mobilization responses (data not shown). Thus, the V64I mutation has no obvious effects on the capability of CCR2b to signal upon binding MCP-1.
FIG. 6.
Signaling capability of CCR2b and CCR2b-V64I and effects on CCR5 and CXCR4. (A) 293T cells cotransfected with CCR5 and either CCR2b (top) or CCR2b-V64I (bottom). In the experiment shown, RANTES activated CCR5, MCP-1 activated CCR2b and CCR2b-V64I, and TAP is a control agonist for cell viability. 293T cells exhibited no background activation in response to either RANTES or MCP-1 (data not shown). Surface expression of receptors was monitored by FACS of parallel sets of cells and was equivalent for both sets of cells. Cells in the right- and left-hand columns differ only in the order of chemokine stimulation. CCR2b and CCR2b-V64I responded to MCP-1 nearly identically in the experiment shown and when transfected into cells without additional coreceptors (data not shown). (B) 293T cells cotransfected with CXCR4 and CCR2b. Cells in the right- and left-hand columns differ only in their order of chemokine stimulation. Similar results were obtained when CCR2b-V64I was used (data not shown).
To determine if CCR2b signaling could affect either of the major HIV-1 coreceptors, 293T cells were transfected with various combinations of CCR2b, CCR2b-V64I, CCR5, and CXCR4. Chemokines were then added sequentially, and receptor signaling was determined by measuring calcium mobilization. Equivalent levels of chemokine receptors were ensured in these experiments by transfecting limiting amounts of CCR2b DNA and subsequently measuring surface expression and Ca2+ mobilization in parallel sets of cells. We found that addition of MCP-1 to cells expressing either CCR2b or CCR2b-V64I completely desensitized subsequent signaling via either CCR5 or CXCR4 following addition of RANTES or SDF-1, respectively (Fig. 6). CCR2b and CCR2b-V64I functioned equally well in this regard. By contrast, prior addition of RANTES or SDF-1 did not abrogate a subsequent signal mediated by either CCR2b or CCR2b-V64I resulting from the addition of MCP-1. None of the chemokines tested desensitized PAR-1 (the thrombin receptor), a G-protein-coupled receptor unrelated to the chemokine receptors, which was used as an internal control in all experiments for cell viability. Additionally, prior signaling mediated by PAR-1 had only minor effects on RANTES/CCR5 and SDF-1/CXCR4 signaling (data not shown). Thus, addition of MCP-1 to cells expressing either CCR2b or CCR2b-V64I results in the asymmetric heterologous desensitization of both major HIV-1 coreceptors.
DISCUSSION
Infection of cells by HIV-1 requires the presence of a coreceptor such as CCR5 or CXCR4 (reviewed in references 7, 10, 12, 23, and 43). Three naturally occurring polymorphisms that demonstrate the importance of chemokines and chemokine receptors for HIV-1 infection in vivo have been described. Elimination of surface expression of CCR5, caused by a 32-bp deletion in the CCR5 open reading frame (CCR5-Δ32), renders homozygotes highly resistant to viral infection and delays progression of disease in heterozygotes (20, 39, 41, 51). A recently identified polymorphism in the 3′ untranslated region of the SDF-1β gene suggests that expression levels of this chemokine may affect progression to disease (58). In the case of CCR2-V64I, there is no protection against virus transmission, but individuals with a single copy of this allele progress to AIDS 2 to 4 years more slowly than individuals without this polymorphism (55). A subsequent study, using a seroprevalent cohort rather than a cohort that followed individuals from the time of seroconversion, disputed this finding (42). However, the protective effect of the CCR2-V64I polymorphism has now been confirmed by several groups using cohorts followed from the time of seroconversion (37, 48, 54). Understanding how this polymorphism results in delayed disease progression may provide additional insight into the interplay between HIV-1, chemokines, and chemokine receptors and may also suggest new therapeutic strategies.
Our results indicate that the effects of the CCR2b-V64I polymorphism are unlikely to be directly linked to use of CCR2b as an HIV-1 coreceptor. Besides the fact that few viruses appear to use CCR2b as a coreceptor (22), we found that the polymorphism had no effects on CCR2b coreceptor function for a virus which does use this receptor (HIV-1 89.6) at the level of either cell-cell fusion or virus infection. For these reasons, we considered the possibility that the effects of CCR2b-V64I would be exerted in trans on the major HIV-1 coreceptors, CCR5 and CXCR4. However, we did not detect any differential effect of CCR2b or CCR2b-V64I expression on CCR3, CCR5, or CXCR4 coreceptor function, as judged by both cell-cell fusion and virus infection assays, nor did coexpression of CCR2-V64I with CCR5 or CXCR4 alter their surface expression. Thus, in transfected cell lines, CCR2b and CCR2b-V64I were indistinguishable in their influences on the expression and activity of the major HIV-1 coreceptors.
The magnitude of the protective effect of CCR2b-V64I is similar to that observed with a single allele of CCR5- 32. PBMCs from individuals with a single copy of CCR5- 32 are readily infectable by HIV-1 in vitro (47), but they do exhibit reduced levels of CCR5 on the cell surface (59). Therefore, we reasoned that CCR2b-V64I PBMCs might exhibit similar properties. We found that PHA/IL-2-stimulated PBMCs from individuals with the V64I polymorphism had normal surface expression levels of CCR2, CCR5, and CXCR4. Since stimulation of PBMCs in culture can alter chemokine receptor expression, we also analyzed unstimulated PBMCs from CCR2-+/+, CCR2-+/64I, and CCR2-64I/64I individuals. Once again, equivalent surface expression levels of CCR2 and CCR5 were observed, though PBMCs from individuals with a single copy of the CCR2b-V64I allele had approximately one-third less CXCR4 on the cell surface. However, these cells were fully permissive to infection by X4 virus strains.
Another possible explanation for the protective effect of the CCR2b-V64I allele is that it is tracking a linked mutation through linkage disequilibrium (55). Most C-C chemokine receptor genes are clustered together on chromosome 3, with the CCR5 and CCR2 loci approximately 14 kb apart. While we have not detected a mutation in the CCR5 open reading frame that cosegregates with CCR2b-V64I, the possibility remains that a mutation in the noncoding region of CCR5 or some other mutation might be directly responsible for the protective effect. Indeed, Kostrikis et al. have found that the V64I polymorphism is linked to a single-base change in the CCR5 promoter, though they did not observe differences in CCR5 expression (37). Consistent with this, our experiments with +/V64I and V64I/V64I PBMCs indicate that differences in CCR5 expression levels are not apparent, nor are there obvious differences in the infectability of V64I PBMCs by HIV-1 in vitro.
While the coreceptors play a critical role in supporting entry of HIV-1 into cells, there is some evidence that coreceptors may also influence postentry events in virus replication (13, 28). For example, T-cell-tropic (T-tropic) strains of SIV use CCR5 as a coreceptor which enables them to enter macrophages, yet they fail to replicate in an Env-dependent manner (44). Differences in how macrophagetropic (M-tropic) and T-tropic SIV strains interact with CCR5 have been reported and may help account for this difference (28). More recently, several soluble HIV-1 and SIV Env proteins have been shown to interact with CCR5 or CXCR4 in a manner that results in receptor signaling (19, 57). While coreceptor signaling is not required for Env-mediated membrane fusion or virus infection of transformed cell lines (3, 5, 24, 25, 30, 33), receptor signaling could influence postentry events of virus replication in primary cells such as macrophages. In addition, the ability of chemokine receptors to desensitize each other suggests complex mechanisms of regulation that could potentially alter chemokine receptor signaling capability and surface expression. Therefore, we investigated the signaling capability of CCR2b-V64I and determined if it could influence the ability of other HIV-1 coreceptors to signal.
We found that activation of both CCR2b and CCR2b-V64I by MCP-1 resulted in the heterologous desensitization of both CCR5 and CXCR4. Desensitization of a RANTES response by MCP-1 was reported previously (14). Desensitization was asymmetric, since activation of either CCR5 or CXCR4 only minimally reduced subsequent CCR2b activation. Heterologous desensitization of other peptide chemoattractant receptors has been reported, though the mechanism by which this occurs is not well understood (11, 14, 36, 56). It is important to note that both CCR2b and CCR2b-V64I signaled equally well in response to MCP-1 and that both desensitized CXCR4 and CCR5. Thus, the ability of the CCR2b-V64I polymorphism to desensitize the major HIV-1 coreceptors is not unique to the mutant allele and is therefore not likely to explain its protective effect, although we cannot rule out the possibility that it can differentially desensitize the major HIV-1 coreceptors in vivo. In addition, any effects on other splice variants of CCR2 cannot be excluded. However, the CCR2b ligands MCP-1 and MCP-3 have been reported to inhibit the productive infection of PBMCs by both R5 and X4 strains of HIV-1 (32, 53). An anti-CCR2b antibody which possesses agonist activity comparable in potency to that of MCP-1 has also been shown to inhibit infection of PBMCs by some virus strains, although it is interesting that other MAbs to CCR2 that do not possess such agonist properties do not inhibit HIV (32). MCP-1 does not block direct infection of blood dendritic cells, which express mRNA for CCR2, CCR3, CCR5, and CXCR4, but does block virus spread to activated lymphocytes when cocultured with HIV-1-pulsed dendritic cells (6). Thus, the putative HIV-1-suppressive effect of CCR2b ligands seems not to be at the level of viral entry via CCR2b but appears to be exerted through its agonist activity. The means by which these CCR2b ligands exert their antiviral activity is not known, but our findings provide a mechanistic link between CCR2 and the major HIV-1 coreceptors that offers a potential explanation for the HIV-1-suppressive effect of various CCR2b ligands.
Under the conditions used, our studies failed to reveal functional differences between CCR2b and CCR2b-V64I. The mutant protein exhibited no significant differences from its wild-type CCR2 parent in either coreceptor function, chemokine receptor function, or effect on CCR3, CCR5, or CXCR4 functions. The importance of the minor difference associated with CXCR4 expression levels on +/V64I unstimulated PBMCs remains to be determined. Therefore, we consider it likely that the mutant CCR2-V64I protein is only indirectly related to the true protective effect associated with the polymorphism. We cannot rule out the possibility that the mutation exerts a subtle effect on CCR2b function that was not detected by our in vitro assays, or that the V64I polymorphism is linked to a second, more significant mutation in the CCR5 gene or elsewhere. In our investigation of this polymorphism, however, we have clearly established the ability of CCR2b, when signaled, to cross-regulate the major HIV coreceptors CCR5 and CXCR4. The significance of this cross-regulation for CCR5 and CXCR4, in terms of receptor signaling, receptor expression, and coreceptor usage, and whether these events can possibly help explain the protective effects of the V64I polymorphism remain to be determined.
ACKNOWLEDGMENTS
We thank Monica Tsang at R&D Systems for generously providing anti-CCR5 antibodies. We also thank Lawrence Brass, Mike Orsini, and Jeff Benovic for technical advice and support, and we especially thank Israel Charo for advice and helpful discussions about chemokine receptor desensitization. A number of reagents used in these experiments were provided by the NIH AIDS Research and Reference Reagent Program.
This work was supported by NIH grants R01 AI-40880 to R.W.D. and R01 AI-35502 to R.G.C. and by a Howard Hughes Medical Institute predoctoral fellowship to B.J.D. B.L. was supported by the Measey Foundation Fellowship for Clinicians (Wistar Institute).
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Locati M, Kennedy P E, Murphy P M, Berger E A. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 4.Ansari-Lari M A, Liu X M, Metzker M L, Rut A R, Gibbs R A. The extent of genetic variation in the CCR5 gene. Nat Genet. 1997;16:221–222. doi: 10.1038/ng0797-221. [DOI] [PubMed] [Google Scholar]
- 5.Aramori I, Ferguson S S G, Bieniasz P D, Cullen B R, Caron M G. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 1997;16:4606–4616. doi: 10.1093/emboj/16.15.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayehunie S, Garcis-Zepeda E A, Hoxie J A, Horuk R, Kupper T S, Luster A D, Ruprecht R M. Human immunodeficiency virus-1 entry into purified blood dendritic cells through CC and CXC chemokine receptors. Blood. 1997;90:1379–1386. [PubMed] [Google Scholar]
- 7.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 8.Berger E A, Doms R W, Fenyö E-M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. HIV-1 phenotypes classified by co-receptor usage. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 9.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieniasz P D, Cullen B R. Chemokine receptors and human immunodeficiency virus infection. Front Biosci. 1998;3:d44–d58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 11.Blackwood R A, Hartiala K T, Kwoh E E, Transue A T, Bower R C. Unidirectional heterologous receptor desensitization between both the fMLP and C5a receptor and the IL-8 receptor. J Leukocyte Biol. 1996;60:88–93. doi: 10.1002/jlb.60.1.88. [DOI] [PubMed] [Google Scholar]
- 12.Broder C C, Collman R G. Chemokine receptors and HIV. J Leukocyte Biol. 1997;62:20–29. doi: 10.1002/jlb.62.1.20. [DOI] [PubMed] [Google Scholar]
- 13.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charo I F, Meyers S J, Herman A, Franci C, Connolly A J, Coughlin S R. Molecular cloning and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxyl-terminal tails. Proc Natl Acad Sci USA. 1994;91:2752–2756. doi: 10.1073/pnas.91.7.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B K, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 17.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 18.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE Study. Genetic resistance of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 21.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 22.Doms R W, Moore J P. HIV-1 coreceptor use: a molecular window into viral tropism. In: Korber B, Foley B, Leitner T, Meyers G, Hahn B, McCutchan F, Mellors J, Kuuiken C, editors. Human retroviruses and AIDS 1997. Theoretical biology and biophysics, part III. Los Alamos, N.Mex: Los Alamos National Laboratory; 1998. pp. 1–12. [Google Scholar]
- 23.Doms R W, Peiper S C. Unwelcome guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 24.Doranz B J, Lu Z, Rucker J, Zhang T, Sharron M, Cen Y, Wang Z, Guo H, Du J, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doranz, B. J., M. J. Orsini, J. D. Turner, T. D. Hoffman, J. F. Berson, Z.-H. Lu, J. A. Hoxie, S. C. Peiper, L. F. Brass, and R. W. Doms. Separation of SDF-1 binding, SDF-1 activation, and HIV-coreceptor utilization of CXCR4. Submitted for publication.
- 26.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 27.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 28.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T-cell tropic SIV strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 30.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein 1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 31.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane domain, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 32.Frade J M R, Liorente M, Mellado M, Alcami J, Gutierrez-Ramos J C, Zaballos A, del Real G, Martinez-A C. The amino-terminal domain of the CCR2 chemokine receptor acts as coreceptor for HIV-1 infection. J Clin Invest. 1997;100:497–502. doi: 10.1172/JCI119558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grynkiewicz G, Poenie M, Tsien R Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 35.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 36.Kitayama J, Carr M W, Roth S J, Buccola J, Springer T A. Contrasting responses to multiple chemotactic stimuli in transendothelial migration. J Immunol. 1997;158:2340–2349. [PubMed] [Google Scholar]
- 37.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 38.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. CD4, CXCR-4, and CCR5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 40.Loetscher P, Seitz M, Baggiolini M, Moser B. Interleukin-2 regulates CC chemokine receptor expression and chemotactic responsiveness in T lymphocytes. J Exp Med. 1996;184:569–577. doi: 10.1084/jem.184.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 42.Michael N L, Louis L G, Rohrbaugh A, Schultz K A, Dayhoff D E, Wang C E, Sheppard H W. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat Med. 1997;3:1160–1162. doi: 10.1038/nm1097-1160. [DOI] [PubMed] [Google Scholar]
- 43.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 44.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by Env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platt E J, Wehrly K, Kuhnman S E, Chesbro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J, Guo H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by M-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the ccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizzardi G P, Morawetz R A, Vicenzi E, Ghezzi S, Poli G, Lazzarin A, Pantaleo G. CCR2 polymorphism and HIV disease. Nat Med. 1998;4:252–253. doi: 10.1038/nm0398-252. [DOI] [PubMed] [Google Scholar]
- 49.Rucker J, Doranz B J, Edinger A E, Long D, Berson J F, Doms R W. Use of a cell-cell fusion assay to study the role of chemokine receptors in human immunodeficiency virus type 1 (HIV-1) entry. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 50.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoumèroulie C, Cogniaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection of Caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 52.Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyö E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 53.Schols D, Proost P, Damme J V, Clercq E D. RANTES and MCP-3 inhibit the replication of T-cell-tropic human immunodeficiency virus type 1 strains (SF-2, MN, and HE) J Virol. 1997;71:7300–7304. doi: 10.1128/jvi.71.10.7300-7304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith M W, Carrington M, Winkler C, Lomb D, Dean M, Huttley G, O’Brien S J. CCR2 chemokine receptor and AIDS progression. Nat Med. 1997;3:1052–1053. doi: 10.1038/nm1097-1052c. [DOI] [PubMed] [Google Scholar]
- 55.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J Hemophilia Growth and Development Study (HGDS); Multicenter AIDS Cohort Study (MACS); Multicenter Hemophilia Cohort Study (MHCS); San Francisco City Cohort (SFCC); ALIVE Study. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 56.Tomhave E D, Richardson R M, Didsbury J R, Menard L, Snyderman R, Ali H. Cross-desensitization of receptors for peptide chemoattractants. J Immunol. 1994;153:3267–3275. [PubMed] [Google Scholar]
- 56a.Weissman, D. Personal communication.
- 57.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 58.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 59.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]