ABSTRACT
Background
Assessment of bone marrow involvement (BMI) in non‐Hodgkin lymphoma (NHL) is crucial for determining patient prognosis and treatment strategy. We assessed the prognostic value of next‐generation sequencing (NGS)–based immunoglobulin (Ig) gene clonality analysis as an ancillary test for BMI evaluation in NHL.
Methods
A retrospective cohort of 124 patients newly diagnosed with B‐cell NHL between 2019 and 2022 was included. NGS‐based Ig clonality analysis was conducted using LymphoTrak IGH FR1 Assay and IGK Assay (Invivoscribe Technologies, San Diego, CA, USA) on BM aspirate samples, and the results were compared with those of histopathological BMI (hBMI).
Results
Among the 124 patients, hBMI was detected in 16.9% (n = 21). The overall agreement of BMI between Ig clonality analyses and histopathological analysis for IGH, IGK, and either IGH or IGK was 86.3%, 92.7%, and 90.3%. The highest positive percent agreement was observed with clonal rearrangements of either IGH or IGK gene (90.5%), while the highest negative percent agreement was observed with clonal rearrangement of IGK gene (96.1%). For the prediction of hBMI, positive prediction value ranged between 59.1% and 80.0% and the negative prediction value ranged between 91.3% and 97.9%.
Conclusion
NGS‐based clonality analysis is an analytic platform with a substantial overall agreement with histopathological analysis. Assessment of both IGH and IGK genes for the clonal rearrangement analysis could be considered for the optimal diagnostic performance of BMI detection in B‐cell NHL.
Keywords: bone marrow involvement, immunoglobulin, lymphoma, NGS
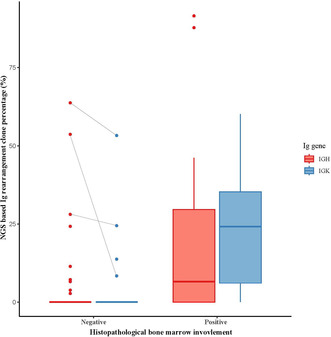
Distribution of next‐generation sequencing–based immunoglobulin rearrangement clone percentages of IGH and IGK genes among patients with positive and negative histopathological bone marrow involvement.
1. Introduction
In non‐Hodgkin lymphoma (NHL), bone marrow involvement (BMI) is classified as an extra‐nodal involvement with adverse prognostic implications and included as a component of the Ann Arbor staging and clinical risk‐stratification indexes, such as the International Prognostic Index (IPI) [1, 2]. BMI status is essential in treatment decisions, influencing choices such as local or systemic treatment for certain types of NHL, including follicular lymphoma (FL) and mucosa‐associated lymphoid tissue lymphoma [3, 4]. Therefore, accurate assessment of BMI is crucial for establishing patient prognosis and appropriate treatment strategy.
BMI is typically assessed by histologic and immunohistochemical (IHC) analysis of bone marrow biopsies and aspirations [5]; however, histopathological detection of BMI has limitations, notably the possibility of false‐negative results in cases with nontypical histomorphologic features, minimal BMI, or inadequate specimens. Immunophenotyping with flow cytometry and molecular studies using immunoglobulin (Ig) gene rearrangement can aid in determining the clonality of lymphoproliferative tissues [1, 6, 7, 8, 9, 10, 11]. Recent studies demonstrate that positron emission tomography (PET)/computed tomography (CT) may possess adequate sensitivity for BMI assessment in some diffuse large B‐cell lymphoma (DLBCL) cases [12, 13]. However, for most NHL histologic subtypes, the ability of PET/CT to accurately detect BMI is not well‐defined [14]. Furthermore, PET/CT has low sensitivity for diffuse, low‐volume BMI in DLBCL [15, 16].
Among the ancillary tests, the detection of monoclonal Ig rearrangements using a PCR‐based analysis of Ig receptor is a useful supplementary tool for BMI assessment. The development and standardization of the multiplex PCR protocol by the BIOMED‐2 European Consortium have made high detection rates possible in most NHL cases [17]. A combination of immunoglobulin heavy chain (IGH) and Ig light chain analysis has been suggested to maximize clonality detection [18]. The diagnostic accuracy of PCR‐based Ig gene clonality showed positive rates ranging from 58% to 74% in histopathological BMI (hBMI)–positive cases [11]. A study evaluating a PCR‐based IGH rearrangement analysis for BMI detection reported a positivity rate of 66.7% and suggested that combining PET/CT assessment may be necessary for increasing positivity rates up to 85.2% [19].
With the advent of next‐generation sequencing (NGS) technology, its application to Ig gene clonality analysis has become prominent. NGS offers higher sensitivity and specificity than multiple PCR, making high‐resolution examinations of all rearrangement events in a sample possible. Ig clonality analysis using NGS has proven to be a sensitive tool for detecting clonality and minimal residual disease (MRD) in various lymphoid hematologic neoplasm [20, 21, 22, 23]. Although NGS‐based Ig testing for clonality detection and MRD monitoring has been previously studied, to the best of our knowledge, its clinical implications and use in BMI assessment have not been described. Therefore, the purpose of this study was to evaluate NGS‐based Ig gene clonality analysis as an ancillary test for BMI assessment in NHL.
2. Materials and Methods
2.1. Study Design and Patients
A retrospective cohort of 124 patients, newly diagnosed with B‐cell lymphoma between January 2019 and June 2022, was included. The study protocol was approved by the Institutional Review Board of Korea University Guro Hospital (2023GR0504). Clinical and demographic characteristics at diagnosis were collected, and the data were analyzed by June 2023. All patients were staged using the Ann Arbor staging system and IPI scores [2, 14].
2.2. Histopathological Evaluation of BMI
The B‐cell lymphoma diagnosis was made according to the 2017 WHO classification. Two hematopathologists reviewed the BM slides for the histopathological detection of BMI. Immunohistochemical staining was performed using monoclonal antibodies against the following antigens: CD3 (polyclonal, 1:200; Dako), CD5 (SP19, RTU; Roche), CD10 (56C6, 1:200; Novocastra), CD20 (L26, 1:4; Novocastra), CD79a (JCB117, 1:200; Dako), BCL2 (124, 1:200; Dako), BCL6 (GI191E/A8, RTU; Roche), cyclin D1 (SP4‐R, RTU; Roche), FMC7 (4C7, 1:100; Novocastra), Ki‐67 (MIB‐1, 1:100; Dako), and MUM‐1 (MUM1p, 1:400; Dako).
2.3. IGH and IGK Clonality Analysis Using NGS
IGH and IGK clonality analysis using NGS was conducted on DNA isolated from BM aspirate samples. The samples were collected in K2 EDTA tubes, and DNA extraction was performed using a QIAamp DNA blood mini kit (Qiagen, Venlo, the Netherlands). DNA was quantified using the Qubit 3.0 Fluorometer (Thermo Fisher Scientific, MA, USA). Ig clonality analysis using NGS was performed with the LymphoTrak IGH FR1 Assay and IGK Assay (Invivoscribe Technologies, San Diego, CA, USA), according to the manufacturer's instructions. Briefly, 200–400 ng of DNA underwent PCR amplification with master mixes containing primers for each target (IGH FR1 or IGK). After purification and quantification, the libraries were sequenced using the MiSeq platform (Illumina, San Diego, CA, USA). Data were analyzed using the LymphoTrack Dx Data Analysis software (version 2.4.3), with the cutoff for the presence of clonality for IGH and IGK determined according to the manufacturer's interpretation criteria.
2.4. Statistical Analysis
The Mann–Whitney U test was applied to compare continuous variables, and the Kruskal–Wallis test was used where appropriate. Categorical variables were compared using Pearson's chi‐squared test and Fisher's exact test. p < 0.05 was considered statistically significant. Data analyses and visualizations were performed using the R software (version 3.4.3).
3. Results
3.1. Clinical Characteristics of Patients
Table 1 presents the baseline characteristics of the 124 patients with B‐cell lymphoma included in the study. Histopathological BMI was detected in 16.9% (n = 21) patients. Among the 21 patients with positive BMI, a relatively higher diagnostic frequency of follicular cell lymphoma (FL) and mantle cell lymphoma was observed, whereas a lower frequency of DLBCL was noted compared with the negative group, as previously reported [1]. As expected, the IPI profile was significantly different between BMI‐positive and BMI‐negative groups (p < 0.001), with BMI‐positive patients being classified as low (19.0%), low intermediate (14.3%), high intermediate (42.9%), and high (23.8%) risk. There were no statistically significant differences in the percentage of patients > 60 years (p = 0.196), sex distribution (p = 0.522), and levels of lactate dehydrogenase (LDH) (p = 0.205) and hemoglobin (Hb) (p = 0.069) between BMI‐positive and BMI‐negative patients.
TABLE 1.
Baseline characteristics of patients with B‐cell non‐Hodgkin lymphoma included in this study.
| Total patients (N = 124) | Histologic and immunohistochemical analyses | |||
|---|---|---|---|---|
| Negative BM involvement | Positive BM involvement | p | ||
| (N = 103) | (N = 21) | |||
| Sex (%) | 0.522 | |||
| Men | 78 (62.9%) | 63 (61.2%) | 15 (71.4%) | |
| Women | 46 (37.1%) | 40 (38.8%) | 6 (28.6%) | |
| Age | 0.196 | |||
| < 60 years | 48 (38.7%) | 43 (41.7%) | 5 (23.8%) | |
| ≥ 60 years | 76 (61.3%) | 60 (58.3%) | 16 (76.2%) | |
| Diagnosis (%) | < 0.001 | |||
| Diffuse large B‐cell lymphoma | 72 (58.1%) | 65 (63.1%) | 7 (33.3%) | |
| Follicular lymphoma | 18 (14.5%) | 11 (10.7%) | 7 (33.3%) | |
| Mantle cell lymphoma | 3 (2.4%) | 1 (1.0%) | 2 (9.5%) | |
| Marginal zone lymphoma | 24 (19.4%) | 21 (20.4%) | 3 (14.3%) | |
| Other B‐cell lymphoma a | 7 (5.6%) | 5 (4.9%) | 2 (9.5%) | |
| Ann Arbor stage (%) | < 0.001 | |||
| I | 33 (26.6%) | 33 (32.0%) | 0 (0.0%) | |
| II | 34 (27.4%) | 34 (33.0%) | 0 (0.0%) | |
| III | 12 (9.7%) | 12 (11.7%) | 0 (0.0%) | |
| IV | 45 (36.3%) | 24 (23.3%) | 21 (100.0%) | |
| International Prognostic Index (%) | < 0.001 | |||
| Low (0–1) | 63 (50.8%) | 59 (57.3%) | 4 (19.0%) | |
| Low intermediate (2) | 27 (21.8%) | 24 (23.3%) | 3 (14.3%) | |
| High intermediate (3) | 22 (17.7%) | 13 (12.6%) | 9 (42.9%) | |
| High (4–5) | 12 (9.7%) | 7 (6.8%) | 5 (23.8%) | |
| Lactate dehydrogenase concentration (g/dL) | 0.205 | |||
| Mean ± SD | 573.6 ± 756.9 | 495.6 ± 387.4 | 956.4 ± 1604.1 | |
| Hb (g/dL) | 0.069 | |||
| Mean ± SD | 12.9 ± 2.0 | 13.0 ± 2.0 | 12.2 ± 1.8 | |
Abbreviations: BM, bone marrow; Hb, hemoglobin; SD, standard deviation.
Corresponds to ALK‐positive large B‐cell lymphoma, lymphoplasmacytic lymphoma, primary mediastinal large B‐cell lymphoma, and T‐cell/histiocyte‐rich large B‐cell lymphoma.
3.2. Detection of Ig Clonal Sequence
Ig gene clonality analysis using NGS identified clonal rearrangements in either IGH or IGK gene in 23.4% (n = 29) patients; clonal IGH and IGK rearrangements were detected in 17.7% (n = 22) and 16.1% (n = 20) patients, respectively. Among the 21 hBMI‐positive cases, clonal IGH and IGK rearrangements were identified in 61.9% (n = 13) and 76.2% (n = 16), respectively, with either rearrangement identified in 90.5% (n = 19). Compared with the hBMI‐negative group, the BMI‐positive group showed a higher rearrangement clone percentage in IGH and IGK (both p < 0.001, Figure 1). A relatively small tumor burden was noted in two hBMI‐positive cases with negative Ig gene clonality results. One of these was a DLBCL case with an estimated tumor volume of about 1%, whereas the other was an FL case with a < 1% estimated tumor volume, both with suspected lymphomatous involvement.
FIGURE 1.
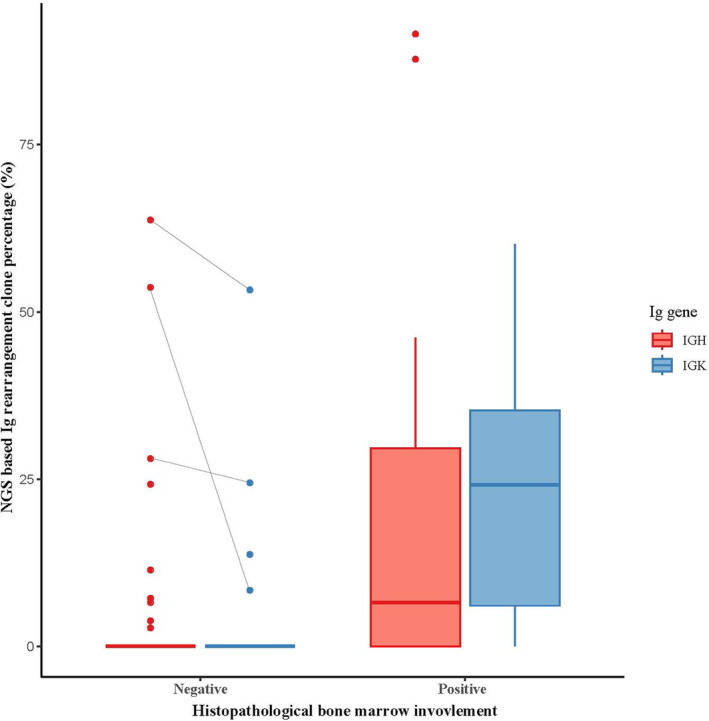
Distribution of next‐generation sequencing–based immunoglobulin rearrangement clone percentages of IGH and IGK genes among patients with positive and negative histopathological bone marrow involvement.
Among the 103 hBMI‐negative cases, clonal IGH and IGK rearrangements were identified in 8.7% (n = 9) and 3.9% (n = 4), respectively, with either IGH or IGK gene rearrangement identified in 9.7% (n = 10). Among these 10 hBMI‐negative patients, three showed possibility of BMI in other ancillary test results, such as flow cytometry (n = 2) and cytogenetic analysis (n = 1). All three cases showed a hypocellular marrow and BM cellularity ranging from 5% to 30% with some scattered lymphocytes, suggesting variability in involved tumor volume among the BM aspirate and section. Interestingly, clonal rearrangement was detected in IGH and IGK in these three cases, with a relatively high clone percentage (IGH, 28.1%–63.7%; IGK, 8.4%–53.3%) (Figure 1).
3.3. Diagnostic Performance of Ig Gene Clonality Analysis Using NGS
The overall agreement between Ig clonality analyses and histopathological analysis for IGH, IGK, and either IGH or IGK was 86.3%, 92.7%, and 90.3%, respectively. The highest positive percent agreement (PPA) was observed for clonal rearrangements of either IGH or IGK gene (90.5%), while the highest negative percent agreement (NPA) was observed with clonal rearrangement of IGK gene (96.1%) (Table 2). For the prediction of hBMI, positive prediction value ranged between 59.1% and 80.0% and the negative prediction value ranged between 91.3% and 97.9% (Table 2).
TABLE 2.
Results of NGS‐based Ig rearrangement and histopathologic analyses of patients.
| Ig rearrangement analysis using NGS | Positive percent agreement (95% CI) | Negative percent agreement (95% CI) | Positive predictive value (95% CI) | Negative predictive value (95% CI) |
|---|---|---|---|---|
| Clonal IGH gene rearrangement | 61.9 (38.4–81.9) | 90.3 (82.4–95.5) | 59.1 (36.4–79.3) | 91.3 (83.6–96.2) |
| Clonal IGK gene rearrangement | 76.2 (52.8–91.8) | 96.1 (90.4–98.9) | 80.0 (56.3–94.3) | 95.2 (89.1–98.4) |
| Either IGH or IGK rearrangement | 90.5 (69.6–98.8) | 90.3 (82.9–95.3) | 65.5 (45.7–82.1) | 97.9 (92.6–99.7) |
Abbreviations: Ig, immunoglobulin; NGS, next‐generation sequencing.
4. Discussion
In this study, we assessed the utility of NGS‐based Ig gene clonality analysis as an ancillary test for BMI assessment in NHL. Our findings reveal a substantial overall agreement between NGS‐based clonality analysis and histopathological analysis. Specifically, in hBMI‐positive cases, NGS‐based clonality analysis, combining IGH and IGK, identified clonality in 90.5% cases, surpassing the clonal detection rates of previously reported PCR‐based methods. Furthermore, our results indicate a higher agreement in the identification of negative hBMI cases compared with PCR‐based methods, underscoring the enhanced specificity of clonal sequence detection using NGS.
Among the hBMI‐positive cases, two discrepant results were identified in two cases with suspected tumor burden approximately ≤1%. Retesting using alternative BM aspirate samples from the same patient was hindered by insufficient samples, and the possibility that the discrepant results stemmed from variations in tumor burden between samples could not be eliminated. Given that the intended use of the NGS‐based Ig gene clonality analysis in this study was to identify a disease‐associated clone for future monitoring, the direct application of clonality calling criteria as per the manufacturer's instruction may not have been optimal for detecting the clonality in samples with low tumor burden. Further evaluation and optimization of settings may be necessary to detect clonal reads amidst a high polyclonal background for BMI detection.
Clonal rearrangements can also be observed in nonneoplastic conditions, which may exhibit oligoclonality or monoclonality due to the predominance of specific antigen‐based subclones or reduced diversity in the B‐cell repertoire [24]. Although the detection of Ig clonal rearrangement does not necessarily indicate the presence of lymphoma, relatively high NPAs were observed between hBMI and NGS‐based Ig gene clonality assays in the current study. Furthermore, three of the 10 hBMI‐negative and NGS‐based Ig gene clonality‐positive cases showed possibility of BMI in other ancillary test results, particularly in cases with hypocellular samples. NGS‐based Ig clonality analysis may collectively show the presence of lymphoma in BM aspirate and section with variability in the tumor volume involved. However, the relatively high clone percentage observed in these samples may be due to the technical aspect that the NGS‐based Ig clonality assay is an amplicon‐based approach that selectively amplifies DNA derived from B‐cell lineage.
This study has certain limitations, including the small sample size focused on several NHL types, limiting our ability to draw definitive conclusions. Concurrent evaluation with other ancillary tests, such as flow cytometry or PCR‐based Ig clonality test or PET/CT, could have enhanced our understanding of the utility and effect of combining these tests. Future studies with larger populations and comparisons with other ancillary tests will offer a more comprehensive understanding of the diagnostic and prognostic significance of NGS Ig clonality testing for BMI detection.
In conclusion, our results indicate that the NGS‐based clonality analysis for the assessment of BMI exhibits a substantial overall agreement with histopathological analysis with enhanced clonal detection rates of previously reported PCR‐based methods. As the highest PPA and NPA were observed for clonal rearrangements of either IGH or IGK gene and IGK gene, respectively, the assessment of both IGH and IGK genes for clonal rearrangement analysis could be considered for the optimal diagnostic performance of BMI detection in B‐cell NHL.
Author Contributions
Min Ji Jeon, Jung Ah Kwon, and Jung Yoon contributed to the conception and design of the study; Eun Sang Yu, Dae Sik Kim, and Chul Won Choi were involved in clinical evaluation; Ha Nui Kim, Jung Ah Kwon, and Soo‐Young Yoon interpreted the results; Jung Yoon performed the statistical analysis; Min Ji Jeon and Jung Yoon drafted the manuscript; and Jung Yoon supervised the study. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors have nothing to report.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Park Y., Park B. B., Jeong J. Y., et al., “Assessment of Bone Marrow Involvement in Patients with Lymphoma: Report on a Consensus Meeting of the Korean Society of Hematology Lymphoma Working Party,” The Korean Journal of Internal Medicine 31, no. 6 (2016): 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The International Non‐Hodgkin's Lymphoma Prognostic Factors Project , “A Predictive Model for Aggressive Non‐Hodgkin's Lymphoma,” The New England Journal of Medicine 329, no. 14 (1993): 987–994. [DOI] [PubMed] [Google Scholar]
- 3. Zucca E., Arcaini L., Buske C., et al., “Marginal Zone Lymphomas: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow‐Up,” Annals of Oncology 31, no. 1 (2020): 17–29. [DOI] [PubMed] [Google Scholar]
- 4. Dreyling M., Ghielmini M., Rule S., et al., “Newly Diagnosed and Relapsed Follicular Lymphoma: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow‐Up,” Annals of Oncology 32, no. 3 (2021): 298–308. [DOI] [PubMed] [Google Scholar]
- 5. Arber D. A. and George T. I., “Bone Marrow Biopsy Involvement by Non‐Hodgkin's Lymphoma: Frequency of Lymphoma Types, Patterns, Blood Involvement, and Discordance with Other Sites in 450 Specimens,” The American Journal of Surgical Pathology 29, no. 12 (2005): 1549–1557. [DOI] [PubMed] [Google Scholar]
- 6. Kim B., Lee S. T., Kim H. J., and Kim S. H., “Bone Marrow Flow Cytometry in Staging of Patients with B‐Cell Non‐Hodgkin Lymphoma,” Annals of Laboratory Medicine 35, no. 2 (2015): 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martín‐Moro F., Piris‐Villaespesa M., Marquet‐Palomanes J., et al., “Bone Marrow Infiltration by Flow Cytometry at Diffuse Large B‐Cell Lymphoma NOS Diagnosis Implies Worse Prognosis without Considering Bone Marrow Histology,” Cytometry. Part B, Clinical Cytometry 98, no. 6 (2020): 525–528. [DOI] [PubMed] [Google Scholar]
- 8. Aren M., Marce S., Jurado R., et al., “Flow Cytometry to Detect Bone Marrow Involvement by Follicular Lymphoma,” Cytometry. Part B, Clinical Cytometry 102, no. 6 (2022): 427–439. [DOI] [PubMed] [Google Scholar]
- 9. Cho Y. A., Yang W. I., Song J. W., Min Y. H., and Yoon S. O., “The Prognostic Significance of Monoclonal Immunoglobulin Gene Rearrangement in Conjunction with Histologic B‐Cell Aggregates in the Bone Marrow of Patients with Diffuse Large B‐Cell Lymphoma,” Cancer Medicine 5, no. 6 (2016): 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berget E., Helgeland L., Liseth K., Løkeland T., Molven A., and Vintermyr O. K., “Prognostic Value of Bone Marrow Involvement by Clonal Immunoglobulin Gene Rearrangements in Follicular Lymphoma,” Journal of Clinical Pathology 67, no. 12 (2014): 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Talaulikar D. and Dahlstrom J. E., “Staging Bone Marrow in Diffuse Large B‐Cell Lymphoma: The Role of Ancillary Investigations,” Pathology 41, no. 3 (2009): 214–222. [DOI] [PubMed] [Google Scholar]
- 12. Adams H. J., Kwee T. C., de Keizer B., Fijnheer R., de Klerk J. M., and Nievelstein R. A., “FDG PET/CT for the Detection of Bone Marrow Involvement in Diffuse Large B‐Cell Lymphoma: Systematic Review and Meta‐Analysis,” European Journal of Nuclear Medicine and Molecular Imaging 41, no. 3 (2014): 565–574. [DOI] [PubMed] [Google Scholar]
- 13. Shah H. J., Keraliya A. R., Jagannathan J. P., Tirumani S. H., Lele V. R., and DiPiro P. J., “Diffuse Large B‐Cell Lymphoma in the Era of Precision Oncology: How Imaging is Helpful,” Korean Journal of Radiology 18, no. 1 (2017): 54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheson B. D., Fisher R. I., Barrington S. F., et al., “Recommendations for Initial Evaluation, Staging, and Response Assessment of Hodgkin and Non‐Hodgkin Lymphoma: The Lugano Classification,” Journal of Clinical Oncology 32, no. 27 (2014): 3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen‐Liang T. H., Martín‐Santos T., Jerez A., et al., “Bone Marrow Biopsy Superiority Over PET/CT in Predicting Progression‐Free Survival in a Homogeneously‐Treated Cohort of Diffuse Large B‐Cell Lymphoma,” Cancer Medicine 6, no. 11 (2017): 2507–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adams H. J., Kwee T. C., Fijnheer R., Dubois S. V., Nievelstein R. A., and de Klerk J. M., “Bone Marrow 18F‐Fluoro‐2‐Deoxy‐D‐Glucose Positron Emission Tomography/Computed Tomography Cannot Replace Bone Marrow Biopsy in Diffuse Large B‐Cell Lymphoma,” American Journal of Hematology 89, no. 7 (2014): 726–731. [DOI] [PubMed] [Google Scholar]
- 17. van Krieken J. H., Langerak A. W., Macintyre E. A., et al., “Improved Reliability of Lymphoma Diagnostics via PCR‐Based Clonality Testing: Report of the BIOMED‐2 Concerted Action BHM4‐CT98‐3936,” Leukemia 21, no. 2 (2007): 201–206. [DOI] [PubMed] [Google Scholar]
- 18. Liu H., Bench A. J., Bacon C. M., et al., “A Practical Strategy for the Routine Use of BIOMED‐2 PCR Assays for Detection of B‐ and T‐Cell Clonality in Diagnostic Haematopathology,” British Journal of Haematology 138, no. 1 (2007): 31–43. [DOI] [PubMed] [Google Scholar]
- 19. Kim M., Ahn S. Y., Ahn J. S., et al., “Diagnostic Accuracy and Prognostic Relevance of Immunoglobulin Heavy Chain Rearrangement and 18F‐FDG‐PET/CT Compared With Unilateral Bone Marrow Trephination for Detecting Bone Marrow Involvement in Patients With Diffuse Large B‐Cell Lymphoma,” Journal of Korean Medical Science 37, no. 1 (2022): e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arcila M. E., Yu W., Syed M., et al., “Establishment of Immunoglobulin Heavy (IGH) Chain Clonality Testing by Next‐Generation Sequencing for Routine Characterization of B‐Cell and Plasma Cell Neoplasms,” The Journal of Molecular Diagnostics 21, no. 2 (2019): 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ha J., Lee H., Shin S., et al., “Ig Gene Clonality Analysis Using Next‐Generation Sequencing for Improved Minimal Residual Disease Detection with Significant Prognostic Value in Multiple Myeloma Patients,” The Journal of Molecular Diagnostics 24, no. 1 (2022): 48–56. [DOI] [PubMed] [Google Scholar]
- 22. Lee J. W., Kim Y., Ahn A., et al., “Clinical Implication of Minimal Residual Disease Assessment by Next‐Generation Sequencing‐Based Immunoglobulin Clonality Assay in Pediatric B‐Acute Lymphoblastic Leukemia,” Frontiers in Oncology 12 (2022): 957743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Svaton M., Skotnicova A., Reznickova L., et al., “NGS Better Discriminates True MRD Positivity for the Risk Stratification of Childhood ALL Treated on an MRD‐Based Protocol,” Blood 141, no. 5 (2023): 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeon Y. K., Yoon S. O., Paik J. H., et al., “Molecular Testing of Lymphoproliferative Disorders: Current Status and Perspectives,” Journal of Pathology and Translational Medicine 51, no. 3 (2017): 224–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


