Abstract
Mus dunni endogenous virus (MDEV) can be activated from M. dunni cells by exposing the cells to hydrocortisone or 5-iodo-2′-deoxyuridine. Interference analysis has revealed that MDEV uses a receptor for cell entry that is different from those used by other murine retroviruses. The entire genome has now been sequenced, revealing a long terminal repeat (LTR)-gag-pol-env-LTR structure typical of simple retroviruses of the murine leukemia virus genus, with no additional open reading frames between env and the 3′ LTR. The LTRs and other noncoding regions of MDEV are most closely related to those of VL30 elements, while the majority of the coding sequences are most closely related to those of gibbon ape leukemia virus. MDEV represents the first example of a naturally occurring, replication-competent virus with sequences closely related to VL30 elements. The U3 region of MDEV contains six nearly perfect 80-bp repeats and the beginning of a seventh, and the region expected to contain the packaging sequence contains approximately four imperfect 33-bp repeats. The receptor specificity domains of the envelope are unique among retroviruses and show no apparent similarity to regions of known proteins.
Mus dunni endogenous virus (MDEV) is transcriptionally inactive in cultured M. dunni cells but can be activated by treatment of the cells with either hydrocortisone or 5-iodo-2′-deoxyuridine (24). Activation can be easily measured because MDEV can package Moloney murine leukemia virus (Mo-MLV)-based retrovirus vectors. Once activated, MDEV will continue to replicate in M. dunni cells and can infect many other cell types (4). MDEV is endogenous to M. dunni wild mice (also known as Mus terricolor) at a proviral copy number of one to two per genome, and there is at least one other element in dunni cells that hybridizes to MDEV probes, but it has a different restriction map (4). MDEV has not been found in the genomes of laboratory mice or in mammalian genera other than Mus (4). It is unknown whether MDEV causes pathology or is ever activated in M. dunni mice.
MDEV does not interfere with known MLVs, indicating that it uses a different receptor for cell entry (26). MDEV also does not interfere with some nonmurine retroviruses, such as gibbon ape leukemia virus (GALV) (4). The endogenous cat retrovirus RD114 was found to interfere at a low level with MDEV, but this interference was observed in only one cell line (G355 cat glial cells) (4). Furthermore, this interference was weak and varied from one experiment to another, making it unclear whether MDEV and RD114 share a receptor in G355 cells. Other members of the RD114 interference group, such as spleen necrosis virus and Mason-Pfizer monkey virus, have not been found to interfere with MDEV (4). The MDEV receptor is widely expressed among different species, as indicated by the ability of a retroviral vector pseudotyped by MDEV to transduce cells from many species (4).
Molecular clones of MDEV were obtained to study its genome and receptor usage (4). However, the clones were unable to produce infectious virus after transfection into permissive cells. Here we describe the correction of a clone that renders it infectious and show that the resulting virus is in the same interference group as the biological isolates. In addition, we have determined the entire sequence of MDEV and describe some unique features of the MDEV genome.
MATERIALS AND METHODS
Nomenclature.
Cells that contain a retroviral vector and/or contain and express a retrovirus are indicated by the cell line name followed by a slash and the names of the vector or virus, e.g., G355/LAPSN for G355 cells containing the retroviral vector LAPSN or dunni/N2+ MDEV for M. dunni cells that contain both the N2 vector and MDEV. LAPSN(PA317) refers to the viral form of the LAPSN retroviral vector packaged by PA317 cells, which express the amphotropic MLV (AM-MLV) envelope.
Cell culture.
G355 feline embryonic glial cells (7) were grown in McCoy’s medium with 15% fetal bovine serum. D17 dog cells (ATCC CCL 183), 293 human kidney cells (13), and M. dunni tail fibroblasts (dunni cells) (19) were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. There are two M. dunni cell strains available that originated from the same mouse, and they can be distinguished by determining whether the medium becomes viscous upon exposure to the cells (26). dunni-v cells (M. dunni cells that make the medium viscous, as opposed to dunni-nv cells), which can be activated to produce MDEV, were used in the experiments described here.
dunni/N2 cells infected with activated MDEV were prepared by exposing dunni/N2 cells to 90 μM hydrocortisone sodium succinate. dunni/N2 cells infected with virus generated from pMDEV were prepared by transfecting pMDEV into G355/LAPSN cells, passaging the cells in the presence of 4 μg of Polybrene per ml for 24 days to allow virus spread, and transferring limiting dilutions of harvested medium to dunni/N2 cells. After passage, the resulting dunni/N2+MDEV cells were stained for alkaline phosphatase-positive (AP+) foci to confirm the absence of cells transduced by contaminating LAPSN vector. In preparation of dunni/N2 cells infected with virus derived from pMDEV, the cells were not exposed to hydrocortisone or 5-iodo-2′-deoxyuridine and so were not expressing the endogenous MDEV.
Retroviral vectors.
LAPSN is an Mo-MLV-based retroviral vector that contains a human placental AP reporter gene and a neomycin phosphotransferase gene (27), and N2 is a similar vector that contains a neomycin phosphotransferase gene (2).
MDEV molecular clones.
A circular-DNA form of MDEV, with two adjacent long terminal repeats (LTRs), was cut with EcoRI in the pol gene and cloned into the EcoRI site of pBluescript II KS(+) (pBSII KS+) (Stratagene) to generate pMDEV9 (4), the prototype MDEV clone that was sequenced and from which other MDEV plasmids were derived. A frameshift mutation in the env gene of pMDEV9 was corrected by site-directed mutagenesis, using the QuikChange site-directed mutagenesis kit (Stratagene) and primers of the sequences 5′-CAGGGTCAGAAAGGAAAGCTGCAACAAGAATG-3′ and 5′-CATTCTTGTTGCAGCTTTCCTTTCTGACCCTG-3′. The frameshift correction was confirmed by sequencing. Finally, a 13-kb plasmid called pMDEV, with an intact (depermuted) MDEV genome, was constructed. MDEV sequences in this pUC19-based plasmid extend from a SalI site in env through a single 5′ LTR, the entire region between the LTRs, and a single 3′ LTR and continue to the SalI site in gag.
PCR.
To analyze the env frameshift mutation, PCR was performed with Taq polymerase (Promega) according to the manufacturer’s instructions with the primers 5′-TTGGTGGCCTGATCTCACACCTG-3′ and 5′-CTCTCCTATTTTGCAGTACTACCTC-3′. Thermocycling was done as follows: one cycle of 10 min at 95°C; 30 cycles of 1 min at 94°C, 1 min at 71°C, and 2 min at 72°C; and one cycle of 5 min at 72°C. Templates included genomic DNA from unactivated M. dunni cells and G355/LAPSN+MDEV cells, as well as negative controls such as DNA from uninfected G355 cells and no template. The PCR products were cloned into pT7Blue (Novagen) according to the manufacturer’s protocol. Several clones from separate PCR amplifications of various templates were sequenced.
Virus assays.
Marker rescue assays were employed to determine whether the MDEV molecular clones were capable of producing infectious virions. G355/LAPSN cells and 293/LAPSN cells were separately plated at a density of 2 × 105 per 3.5-cm-diameter well of six-well plates on day 1. On day 2, 5 μg of plasmid DNA was transfected into the cells by calcium phosphate precipitation. On day 3, the medium was replaced. On day 4, the medium was harvested, clarified by centrifugation at 4,000 × g for 5 min at 4°C, and stored at −80°C prior to assay for the LAPSN vector.
The LAPSN vector titer was determined by plating target cells at 1 × 105 or 2 × 105 cells per 3.5-cm-diameter well of a six-well dish on day 1. On day 2, the medium was replaced with medium containing 4 μg of Polybrene per ml and the virus stock was added. On day 4, the cells were stained for AP+ foci as described elsewhere (11), and virus titers were expressed as the number of AP+ foci per milliliter of test medium.
DNA sequencing.
Portions of the MDEV clone pMDEV9 were subcloned into pBSII KS+, and deletions were made with exonuclease III by the method of Clark and Henikoff (6). Sequencing was performed on the nested deletion constructs, using dye primer ABI PRISM sequencing kits, a model 373A DNA sequencer, and sequence analysis software (Applied Biosystems, Foster City, Calif.). Regions of poor sequencing were resequenced after the appropriate primers were made, using the ABI PRISM dye terminator kits. Both strands of MDEV were completely sequenced, and the sequences were assembled into contigs and further analyzed by using Sequencher 3.0 (Gene Codes Corporation, Inc., Ann Arbor, Mich.).
We have numbered the nucleotides of the MDEV genome beginning with the presumptive cap site, the first nucleotide of the R region. This putative cap site was identified by comparison of MDEV to the VL30 element VL3 (GenBank accession no. X03489), for which the cap site has been mapped (35).
Sequence analysis.
To elucidate the nature of the breakpoint regions, alignments of MDEV, VL30 BVL-1, and GALV sequences were performed with the MACAW program (36). To infer phylogenetic relationships between the MDEV sequences and those other retroelements and retroviruses, Clustal W was used to make sequence alignments, create phylogenetic trees, and evaluate the trees by bootstrap analysis (37). Clustal W creates alignments by a progressive pairwise method and creates phylogenetic trees by a neighbor-joining method. When sequences were of different lengths due to incomplete sequence data, as for the porcine endogenous retroviruses (PERVs) in the pol tree, positions with gaps were excluded from the phylogenetic analysis so that only those regions for which all sequences are represented were analyzed. TreeView (31) and Canvas (Deneba Software, Miami, Fla.), respectively, were used to view and print the phylogenetic trees. Some branches were rotated by using the Canvas program without changing the branch lengths. This does not alter the information contained in the tree, which is based only on branching order and branch length.
Nucleotide sequence accession number.
The complete nucleotide and polyprotein sequences of MDEV have been deposited in GenBank under accession no. AF053745.
RESULTS AND DISCUSSION
Correction of a mutation in the env gene renders an MDEV molecular clone infectious.
Six molecular clones of MDEV were isolated from a library of extrachromosomal DNA from G355 cat cells infected 1 day earlier with MDEV (4). To prepare the library, extrachromosomal circular DNA was cut with EcoRI and cloned into pBSII KS+. Sequencing demonstrated that the MDEV EcoRI site was in pol. Different orientations of the virus, the presence of one LTR versus two LTRs, and the presence of extraneous DNA fragments in the pBSII KS+ EcoRI cloning site demonstrated that the clones were independent. A vector rescue assay was employed to determine whether the molecular clones were competent to provide gag, pol, and env gene products in trans to a vector. The clones were digested with EcoRI, religated at a low concentration to rejoin the pol gene, and transfected into G355/LAPSN cells, but none of the clones was competent to rescue the LAPSN vector. However, three clones were competent to rescue LAPSN when cotransfected with pSX2 (25), a plasmid that expresses the 10A1 MLV envelope protein (data not shown). This indicated that these clones could express functional Gag and Pol proteins, but not a functional Env protein, after religation of the pol genes. Sequencing of one clone, pMDEV9, demonstrated that there was a +1 frameshift in the env gene. The other five clones have the same frameshift, indicating that they had all originated from a common defective virus.
To determine the exact the nature of the frameshift mutation, a portion of MDEV env was PCR amplified from DNA of unactivated M. dunni cells and of G355/LAPSN cells that had been infected with MDEV, using primers flanking the frameshift region as described in Materials and Methods. Amplification reactions performed on DNA from uninfected G355 cells or performed with no DNA did not yield a detectable product. Sequence analysis of cloned PCR products derived from both unactivated M. dunni cell DNA and G355/LAPSN+MDEV DNA demonstrated that pMDEV9 had an extra A residue at position 6168 within the first variable region (variable region A) of env, generating the +1 frameshift. We corrected the env frameshift by site-directed mutagenesis and constructed a plasmid containing an intact (depermuted) copy of MDEV, called pMDEV.
The clone pMDEV was then transfected into 293/LAPSN cells to assess the ability of the cloned virus to rescue the LAPSN vector. As a negative control, transfection of pBSII KS+ did not result in the rescue of the LAPSN vector, as determined by titration on D17 cells (LAPSN titer, <5 AP+ focus-forming units [FFU]/ml). Transfection of pMDEV did result in production of the LAPSN vector (average titer ± standard deviation, 2 × 102 ± 20 AP+ FFU/ml), although at a lower titer than achieved by transfection of pAMS, which encodes a replication-competent AM-MLV (3 × 105 ± 7 × 104 AP+ FFU/ml). It is unclear whether the LAPSN titers resulting from transfection of pMDEV were low because pMDEV represents a nonoptimal copy of MDEV (transfections had to be optimized before successful LAPSN rescue could be demonstrated) or whether this simply reflects the biology of the virus. Indeed, the biological isolates of MDEV also package LAPSN at a much (about 100-fold) lower titer than does AM-MLV.
Transfection of pMDEV into G355/LAPSN cells, followed by passaging of the cells for 24 days, resulted in production of the LAPSN vector at levels equivalent to those achieved with a biological isolate of MDEV (9 × 104 AP+ FFU/ml), whereas transfection of pBSII KS+ again resulted in undetectable production of LAPSN. The virus resulting from transfection of pMDEV was successfully passaged into naive G355/LAPSN cells (data not shown), showing that the virus was fully replication competent.
Virus derived from the molecular clone uses the same receptor as the MDEV biological isolates.
Northern and Southern blot analyses indicated that the MDEV molecular clones represent the activated MDEV virus (4). Additional evidence demonstrating that virus derived from the molecular clone is the same as the activated viruses can be provided by interference analysis to show that both viruses use the same receptor. Viral interference experiments rely on the observation that cells that have been productively infected with a replication-competent retrovirus are resistant to superinfection by a virus or vector that uses the same receptor for cell entry. As seen in Table 1, the entry of LAPSN pseudotyped by MDEV from either an activated or a molecular source was strongly impeded by the presence of MDEV from either source. The LAPSN(PA317) control had equivalent titers on all cells, showing that the block is not due to a general resistance to retroviral infection. Similar experiments have been done with chimeric virion containing the MDEV Env protein and the Mo-MLV Gag and Pol proteins, demonstrating that the block is due to Env protein rather than Gag and Pol proteins (data not shown). This interference demonstrates that the virus derived from the MDEV molecular clone uses the same receptor as that used by hydrocortisone-activated MDEV. Because the MDEV receptor is not known to be used by any other retrovirus, these experiments strongly indicate that the molecular clone represents MDEV. The reciprocal nature of the interference also shows that the virus derived from activation of M. dunni cells with hydrocortisone does not contain a mixture of viruses that use different receptors for cell entry.
TABLE 1.
MDEV derived from the molecular clone displays reciprocal interference with a biological isolatea
| LAPSN pseudotype | LAPSN titer (AP+ FFU/ml) on cell type:
|
||
|---|---|---|---|
| dunni/N2 | dunni/N2 + activated MDEV | dunni/N2 + cloned MDEV | |
| PA317 | 106 | 106 | 106 |
| Activated MDEV | 8 × 104 | <10 | <10 |
| Cloned MDEV | 4 × 104 | <10 | <10 |
Uninfected dunni/N2 cells and dunni/N2 cells that had been previously infected with either MDEV activated from M. dunni cells or MDEV derived from the pMDEV molecular clone were plated at a density of 2 × 105 cells per 3.5-cm-diameter well on day 1. The cells were exposed to LAPSN with the indicated pseudotypes on day 2 and were stained for AP+ foci on day 4. Values are expressed as the means of duplicates which varied by no more than 20% from the mean. The experiment was repeated by plating 105 cells per 3.5-cm-diameter well, with nearly identical results being achieved.
Genomic structure and ORFs of MDEV.
MDEV has the genomic structure of a simple retrovirus of the MLV group, with three major open reading frames (ORFs), corresponding to gag, pol, and env, and no significant ORFs between the env gene and the 3′ LTR (Fig. 1A). MDEV has a UAG (amber) stop codon separating the gag and pol genes and two stop codons each at the ends of the pol and env genes. The MDEV provirus is predicted to be 9,480 bp after the loss of 4 bp on integration. The genomic and spliced env RNAs are predicted to be 8,655 and 3,383 bases long, respectively, not including the poly(A) tails. These predicted sizes and the restriction map of the clone are consistent with the results of previous Northern and Southern blot analyses (4) (data not shown).
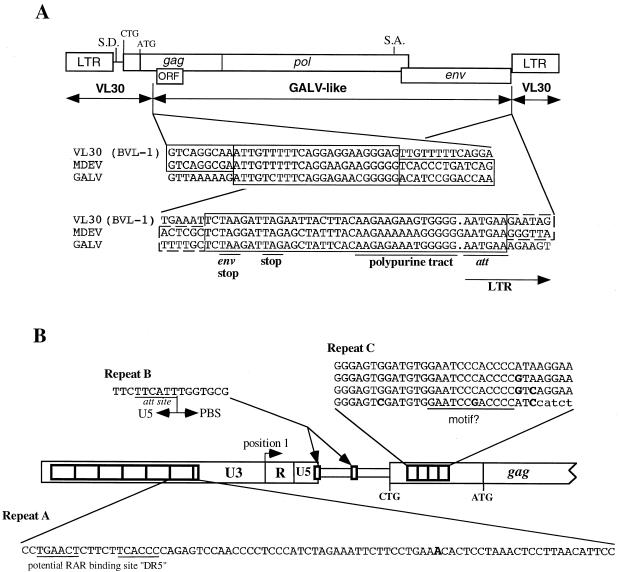
FIG. 1.
Genomic structure of the proviral form of MDEV. (A) Recombinant structure of MDEV. The sequences of a VL30 element and of the MDEV and GALV genomes were aligned by using the MACAW program; the alignment at the putative breakpoint regions is shown. The recombinations are presumed to have occurred in the homologous regions shown in the shaded boxes. S.D., putative splice donor; S.A., putative splice acceptor; CTG, start codon of the extended glycosylated Gag; ATG, start codon of the nonglycosylated Gag. (B) Repeated regions of MDEV. Repeats A are nearly perfect, so only one copy is shown; the nucleotide A indicated in boldface is a C in the sixth repeat. Repeat A has a consensus retinoic acid binding site DR5, consisting of the underlined imperfect direct repeats separated by five spaces. Repeats B are perfect repeats, so only one copy is shown. The first copy of repeat B overlaps the U5-PBS boundary. Repeat C is imperfect, so all four copies are shown, with differences indicated in boldface; these 33-bp repeats are within the coding region of the extended glycosylated Gag. Repeat C has an underlined sequence similar to a sequence found in corresponding repeats of VL30 elements. CTG indicates the start codon of the glycosylated Gag, and ATG indicates the start codon of the nonglycosylated Gag.
In addition to the Gag protein whose translation is initiated at an ATG in the gag ORF, many MLVs synthesize a glycosylated Gag protein whose translation is initiated upstream of the gag ATG start codon. Translation of this protein in Mo-MLV has been shown to initiate at an upstream CTG start codon (33). The gag ORF of MDEV also extends upstream of the ATG start codon and contains a CTG (underlined) embedded in a sequence identical to that of Mo-MLV (ACC CTG GGA GAC GTC CCA GG, positions 419 to 438), indicating that MDEV makes a similar glycosylated Gag protein. The high level of sequence identity between these two distantly related retroviruses (there is 44% amino acid identity between the two in the gag ORF, starting at the putative CTG start codon) is suggestive of an important role for this region.
There is an additional ORF within the gag gene, in the +1 reading frame with respect to gag. This ORF has at its beginning an ATG codon (underlined below), making it potentially capable of encoding a protein of 163 amino acids, but the sequence surrounding this codon (CCGATGC, positions 1059 to 1065) does not fit the consensus for efficient translation (18). GALV has a similar ORF, potentially capable of encoding a protein of 117 amino acids, and an overlapping stretch of 66 amino acids shows similarity to that encoded by the MDEV ORF. However, the similar regions of these ORFs coincide with a highly conserved region of gag; thus, their similarity is likely to be due to sequence constraints on the gag gene. The potential MDEV ORF protein shares no similarity with those predicted from sequences in the GenBank database. We have not determined whether this ORF is translated into a protein.
Several areas of repeats exist in the MDEV genome (Fig. 1B). The first is a series of more than six nearly perfect tandem 80-bp repeats in the beginning of the U3 region (repeat A, at positions 7,838 to 7,917, 7,918 to 7,997, 7,998 to 8,077, 8,078 to 8,157, 8,158 to 8,237, 8,238 to 8,317; and a partial repeat at 8,318 to 8,329). The second consists of two identical but separate copies of a 16-bp repeat (repeat B, at positions 170 to 185 and 304 to 319), and the third consists of more than three tandem imperfect 33-bp repeats (repeat C, at positions 485 to 517, 518 to 550, and 551 to 583; and a partial repeat at 584 to 611) located between the LTR and the start of gag. The repeats will be discussed below.
MDEV appears to be a chimera between a VL30 element and a virus similar to GALV.
VL30 elements are retroelements, found in mice, rats, and potentially other species, that are similar to retroviruses in their structure and replication (12). They have LTRs that are structurally similar to those of retroviruses but are not similar in sequence. VL30s have gag and pol genes that have domains with recognizable similarity to retroviral gag and pol genes, but of the VL30s characterized thus far, none would be expected to encode a functional protein, due to the presence of numerous stop codons and frameshift mutations (1, 15). Furthermore, no VL30 that has been characterized has an env gene. Indeed, the elements were originally defined as having genomic RNAs of 30S, a size that would not be predicted to have a complete env gene. VL30 elements have been called retrotransposons because their genomes are indicative of an intracellular replication cycle.
Many cis-acting sequences of VL30 elements have been found to be intact, including promoters and sequences necessary for packaging, reverse transcription, and integration. Thus, VL30 elements can be propagated by parasitizing an MLV-type virus (14). Copackaging of VL30 and retrovirus genomes is thought to facilitate recombination between the elements.
Sequence analysis demonstrated that MDEV has a recombinant structure, with the LTRs and untranslated region (UTR) being most similar to VL30 sequences and the majority of the coding sequences being most similar to those of GALV (Fig. 1A). Thus, MDEV has cis-acting sequences derived from a VL30 element(s), and the majority of its trans-acting sequences were derived from a virus similar to GALV. Inspection of both putative recombination breakpoint regions indicates that homologous recombination events generated MDEV. The putative 5′ recombination breakpoint appears to have occurred in an area that is conserved between VL30s and MLV-type viruses in a region of the gag gene between nucleotides 919 and 944; similarly, the 3′ recombination appears to have occurred in a region of conservation encompassing the env termination codon and the polypurine tract between nucleotides 7770 and 7813. This homologous recombination may have occurred during first-strand DNA synthesis by jumping of the elongating chain to the corresponding region of the other copackaged RNA template. GALV is a primate virus, but it belongs to the MLV group of retroviruses due to its genomic sequence, and is thought to have originated from a xenotropic mouse retrovirus (xenotropic here referring to host range, not the MLV interference group) (5, 21). It is therefore, likely that the recombination event that generated MDEV occurred in a mouse.
Other examples of MLV-VL30 recombinants exist, but apparently none has been replication competent. From a BALB/c genomic library, Itin and Keshet (17) successfully selected clones that hybridized to both a VL30 clone and an Mo-MLV clone. Hybridization analysis demonstrated that the clone VM-1 has VL30 LTRs and gag and pol sequences similar to those of Mo-MLV (17). This clone did not hybridize to the Mo-MLV env, however, and its size indicates that it could not contain a full complement of genes. The structure of the genomes of Harvey, Kirsten, and Rasheed murine sarcoma viruses, isolated from rats after inoculation with MLVs, is the inverse of that of MDEV. They have MLV LTRs and some interior sequences related to rat VL30s, which incidentally do not have sequences closely similar to murine VL30s. In this case, it has been proposed that recombination with rat VL30s facilitated acquisition of the ras oncogene (23). VL30-MLV recombinants have also been selected by using an integration-defective MLV in which recombination with VL30 sequences provides intact att sites (28). MDEV differs from these examples in that it is naturally occurring and replication competent.
The MDEV LTR defines a new VL30 class.
The 928-bp LTR is composed of U3, R, and U5 regions with predicted lengths of 750, 99, and 79 bp, respectively. The beginning of the U3 region was identified by the consensus att site AATGAA (positions 7807 to 7812), which is immediately downstream of the polypurine tract AAGAAAAAAGGGGGG (positions 7792 to 7806) (Fig. 1A). The U3-R junction was identified by comparison to the VL30 element VL3 (GenBank accession no. X03489), for which the cap site has been mapped (35). The R-U5 boundary of the LTR was identified by alignment with several VL30s that were sequenced from cDNA clones in which the beginning of the poly(A) tail indicates the end of the R region (29). The end of the U5 region was identified by the presence of the inverted att site TTCATT (positions 173 to 178), which matches the U3 att site and is adjacent to the primer binding site (PBS) (Fig. 1B, repeat B).
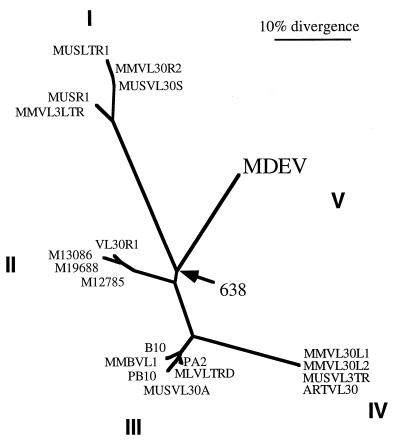
The U3 region is the least-conserved region of VL30 LTRs, and it has been used to divide VL30 elements among four families, each with multiple representatives (Fig. 2) (29). The MDEV LTR represents a fifth family and is about twice as long as a standard VL30 LTR due to the presence of multiple nearly perfect 80-bp repeats (repeats A) in the beginning of the U3 region (Fig. 1B). Because the U3 region contains the promoter and enhancers, we examined the repeat for the presence of consensus enhancer elements. At the beginning of each repeat is a consensus VL30 retinoic acid receptor binding domain of the sequence TGAACTCTTCTTCACCC, the underlined sequences being the imperfect direct repeats that are characteristic of such binding site (16). Retinoic acid receptor binding domains consist of short direct repeats separated by a spacer, and the MDEV sequence differs from the VL30 sequences in that it has a 5-nucleotide spacer instead of a 2-nucleotide spacer. In other systems, however, a spacing of 5 nucleotides shows a more vigorous response to retinoic acid than does a 2-nucleotide spacing (32). Our attempts to activate MDEV from M. dunni cells with retinoic acid were unsuccessful, but these experiments may have been complicated by the lack of appropriate retinoic acid receptor expression in the dunni cells. Other consensus binding sites in the MDEV U3 are those for NF-1 (positions 8461 to 8474) and NF-κB (positions 8489 to 8499) (29). A putative TATA box with the sequence TATATAA is present (positions 8528 to 8534), but a CAAT box is not apparent. A putative polyadenylation signal of the sequence ATTAAA is located in each R region (positions 83 to 88 and 8639 to 8644). MDEV can be activated by exposing dunni cells to hydrocortisone, but no glucocorticoid response elements were observed in the U3 region.
FIG. 2.
The MDEV U3 region defines a fifth VL30 group. The phylogenetic tree is based on the nucleotide sequences of the U3 regions of VL30s and MDEV. The sequences are indicated by GenBank locus names. The VL30 BVL-1, which was compared to MDEV in Fig. 1A, is indicated here as MMBVL1 in group III. The four VL30 groups defined by Nilsson and Bohm (29) are indicated, along with some additional VL30 sequences, and the MDEV U3 defines a fifth group. Five of the perfect 80-bp repeats of the MDEV U3 region were eliminated prior to the alignment to make the U3 regions approximately the same size. The alignment, phylogenetic tree, and bootstrap analysis were performed with the Clustal W program, and the tree was drawn with the TreeView program. All major branchpoints between VL30 groups have confidence values of >98% except for the branchpoint that indicates that MDEV is most closely related to group 1, which was observed in 638 of 1,000 trials.
The MDEV UTR is similar to those of VL30 elements.
The UTR between the LTR and gag is most similar to the UTRs of VL30 elements. This region contains the PBS, repeats B, the putative splice donor, and the putative packaging signal, which may extend into gag.
The 18-nucleotide PBS (positions 179 to 196) is similar to the 3′ end of the glycine tRNA, which is characteristic of VL30 elements. However, the MDEV PBS has a single-base mismatch (TGGTGCGTTGGCCGGGAA, with the mismatch underlined) for the 3′ end of murine tRNAGly (TGGTGCATTGGCCGGGAA), while those sequenced from other VL30s are a perfect match. It is not known whether this mismatch is a mutation in our molecular clone, whether MDEV is tolerant for polymorphism at this site, or whether the molecularly cloned PBS matches the 3′ end of the cat tRNAGly perfectly. The latter is possible since MDEV was last replicated in G355 cat cells during the cloning process; the 3′ end of the tRNA is copied during reverse transcription, and so the PBS could be altered to match the tRNA of the cells in which it was last propagated.
Repeats B are located at the end of the LTR and upstream of the presumed glycosylated Gag CTG start codon (Fig. 1B). The first copy (TTCTTCATTTGGTGCG, with the beginning of the PBS underlined; positions 170 to 185) overlaps the U5-PBS border, and the second occurs downstream of the putative splice donor (position 233 to 239, with the second repeat being at positions 304 to 319). The VL30 BVL-1 sequence aligns well with that of MDEV throughout this region, and the regions that correspond to the MDEV repeats can be readily identified in the VL30s. Although the two repeats within MDEV are identical, the repeats within VL30 BVL-1 exhibit only 60% identity and are 81 and 75% similar to the MDEV repeats. The original copy of the MDEV repeat is likely the one that overlaps the U5-PBS boundary, since this is a critical region that would be sensitive to mutation.
The putative MDEV splice donor (AG · GTAAG; positions 233 to 239) is located in a region highly similar to those of VL30s, while the putative splice acceptor sequence (CCCTCCTCTTTGCTTATTCATTTAAAG · G; positions 5480 to 5507) is located in the pol gene in a sequence similar to that of GALV. The consensus splice donor sequence exactly matches the corresponding sequence of VL30s BVL-1 and NVL-3 (30). However, no VL30 with an env gene has been identified, leaving the requirement of a splice donor questionable. Although it is possible that a short consensus sequence is coincidentally present at the proper location in a VL30, it is also possible that VL30s were originally derived from infectious retroviruses that had env genes and, therefore, a splicing requirement. Perhaps a VL30 with an env gene will eventually be identified. Indeed, an intracisternal A-type particle, a member of a class that had also been previously considered to be void of env genes, that appears to have an env sequence has been cloned (34).
Gag.
MDEV appears to encode both a nonglycosylated Gag protein with an ATG translational start codon and an extended, glycosylated Gag protein with an upstream CTG translational start codon. The amino terminus of MDEV Gag, corresponding to approximately 70 amino acids of nonglycosylated Gag and 170 amino acids of glycosylated Gag, is encoded by sequence derived from a VL30 element. This region of a VL30 gag gene is one of the few regions of VL30s that show strong sequence similarity to retroviruses (50% identity among VL30 BVL-1, MDEV, GALV, and Mo-MLV in the first 70 amino acids of nonglycosylated Gag).
The MDEV Gag polyprotein is predicted to be 522 amino acids and 59 kDa, and the glycosylated Gag polyprotein is predicted to be 622 amino acids and 69 kDa prior to glycosylation. The nucleocapsid portion of Gag has a single Cys-His box that matches the consensus spacing of CX2CX4HX4C. Between the first two cysteines is usually found an aromatic amino acid; while this is tyrosine in most MLVs and GALV, it is phenylalanine in MDEV.
MDEV has approximately four imperfect ∼33-bp repeats (repeats C) between the CTG start codon of the glycosylated Gag and the ATG start codon of the nonglycosylated Gag (Fig. 1B). The repeats contain 33 bp and so would not disrupt the reading frame of gag. BVL-1 and NVL-3, two VL30s whose entire sequences are known, have five and six imperfect repeats, respectively, of about the same size in the same region. These repeats, while similar for these two VL30s, are generally of a different sequence than the MDEV repeat, with the exception of a potential motif of the sequence GAATCC(C/G)ACCCC in MDEV and GAGTCCCACCTC in the two VL30s. The significance of these repeats and putative motifs remains unknown, but they may play a role at the nucleotide level in the packaging signal or at the protein level in the extended glycosylated Gag.
Pol.
The molecular MDEV clones were originally obtained by cloning EcoRI-cut extrachromosomal, circular DNA into pBSII KS+. Sequence analysis showed that EcoRI had cut MDEV within the pol gene. To be sure that a portion of the pol gene was not lost due to the presence of two EcoRI sites in close proximity, we aligned the MDEV protein sequence that spans the EcoRI site with the corresponding sequence of GALV. No gaps in the MDEV sequence were observed. This evidence, coupled with data that demonstrated that the molecular clones encode functional Pol proteins, indicates that none of the MDEV pol gene was lost during cloning.
Pol proteins of MLV-type viruses are translated by misreading of the stop codon UAG as CAG to create a Gag-Pol fusion protein with glutamine occupying the position of the stop codon. The resulting MDEV Gag-glutamine-Pol fusion protein would be expected to contain 1,726 amino acids and have a molecular mass of 192 kDa.
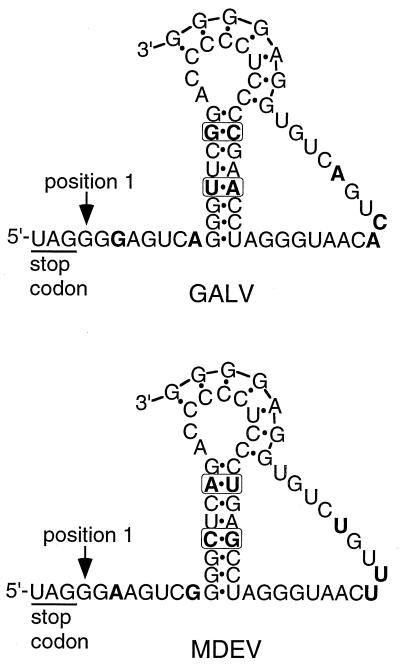
The readthrough needed to express pol is theorized to be due to effects of RNA secondary structure on translation rather than editing of the RNA (38, 39). The RNA has been predicted to form a pseudoknot just downstream of the UAG codon for MLV-type retroviruses, and support for the existence of hairpins in Mo-MLV has been obtained through complementary mutation analysis (10, 38, 39). A pseudoknot structure for GALV has been proposed (39), but mutational analysis of the predicted hairpins has not been conducted. A corresponding structure can be drawn with the related but different MDEV sequence so that the sequences can be compared to find evidence for the existence of the hairpin (Fig. 3). Comparison of the two hypothetical pseudoknots shows that differences in one strand of stem 1 are accompanied by changes in the other strand of the stem, consistent with the existence of stem 1 in GALV and in MDEV. The residues U6, C7, G35, G36, U38, A39, U40, U44, and U50 have previously been found to be absolutely conserved in MLV-type retroviruses and again are conserved in MDEV.
FIG. 3.
Differences between the MDEV and GALV sequences are consistent with a proposed model of a GALV pseudoknot. The sequences start at the pol stop codon. In this figure and in the text referring to this figure, position 1 is defined as the nucleotide immediately downstream of the stop codon. Hydrogen bonding is indicated by dots, and sequence differences are shown in boldface, with boxes drawn around differences in the proposed hairpins. The GALV pseudoknot is redrawn from reference 39, and the MDEV pseudoknot has been drawn by analogy.
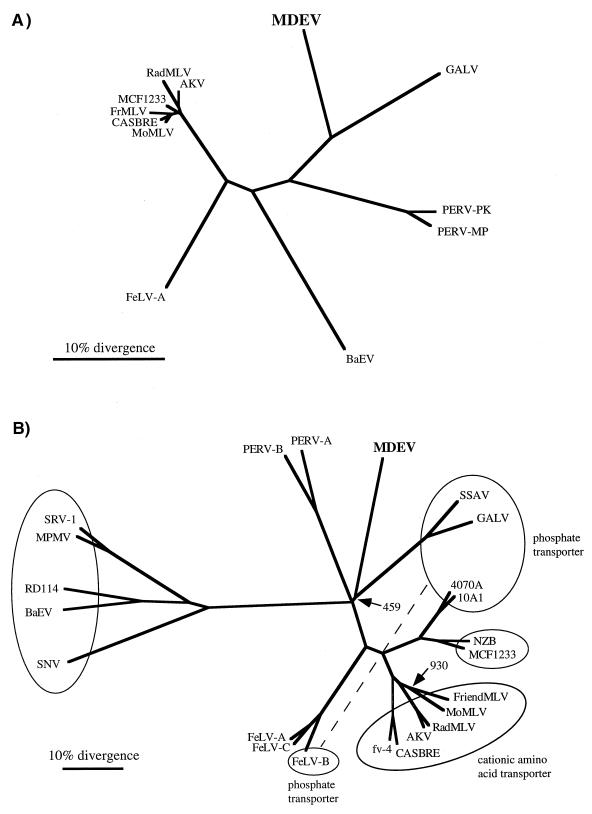
An unrooted tree based on the protease-reverse transcriptase (PR-RT) regions of Pol protein sequences of MLV-type retroviruses has been generated (Fig. 4A). The MDEV sequence is distinct but most closely resembles that of GALV. The PERVs, which have also recently been reported to resemble GALV (20), have been included in the phylogenetic analysis. It is apparent that MDEV is more closely related to GALV than are the PERVs. A tree based on Gag proteins has a similar structure (data not shown), but the PERVs could not be included because their Gag sequences have not been reported.
FIG. 4.
Phylogenetic trees based on Pol (A) and Env (B) proteins. The protein alignments, phylogenetic trees, and bootstrap analyses were performed with the Clustal W program, and the trees were viewed by using the TreeView program. (A) Tree based on PR-RT regions of Pol proteins. Only those positions of the alignment with no gaps were used to create the phylogenetic tree. All major branchpoints had confidence values of >98%, as measured by bootstrap analysis, except for branchpoints among the closely related members of the mouse virus group at the left. (B) Tree based on entire Env proteins. Viruses observed to share receptors in at least one cell type are grouped together in ovals, and the receptor is indicated in the cases where it is known. Note that feline leukemia virus type B (FeLV-B) belongs to the phosphate transporter group but cannot be included in that oval in a two-dimensional tree. MDEV and RD114 were not grouped due to the inconsistent nature of the interference observed in G355 cells. A phylogenetic tree created by excluding positions with a gap for any sequence was nearly identical. All major branchpoints had confidence values of >97%, as measured by bootstrap analysis, except for branchpoints indicated by the number of trials of the 1,000 that produced the displayed branchpoint. Abbreviations: AKV, AKR mouse ecotropic virus; BaEV, baboon endogenous virus; CASBRE, Casitas brain ecotropic virus; FrMLV, Friend MLV; NZB, New Zealand black xenotropic virus; RadMLV, radiation MLV; SNV, spleen necrosis virus; SRV-1, simian retrovirus type 1; SSAV, simian sarcoma-associated virus.
Env.
The env ORF overlaps the pol ORF by 146 nucleotides (∼49 amino acids), which is very similar to the overlap in GALV, PERV-A, and PERV-B (∼46, ∼41, and ∼42 amino acids, respectively). The immature MDEV Env protein is predicted to consist of a total of 673 amino acids prior to protease cleavage. The signal sequence of the Env protein is not sufficiently similar to those of other retroviruses to predict a cleavage site, but the regions surrounding the other presumed cleavage sites can be aligned with those of other retroviruses. Based on such comparisons, we predict that cleavage between the surface domain (SU) and transmembrane domain (TM) occurs immediately downstream of the sequence RWKR and that cleavage of the R peptide from the cytoplasmic tail occurs approximately 16 amino acids from the carboxy terminus. Such cleavage would produce an SU domain of 479 amino acids (including the signal peptide) and a TM domain of 178 amino acids (lacking the R peptide). The envelope is predicted to have a molecular mass of approximately 75 kDa prior to glycosylation.
There are six putative N-linked glycosylation sites of the structure N-X-S/T in the MDEV SU. All but the first, which is in variable region B, are well conserved among MLV-type viruses. The MDEV envelope has 19 cysteines in SU, 16 of which are conserved among various MLVs and are known to participate in disulfide bonding (8, 22).
To evaluate the relationship of the MDEV Env to those of other retroviruses, an alignment with Env proteins of MLV-type viruses as well as Mason-Pfizer monkey virus and simian retrovirus type 1, both of which have receptors identical to those of several MLV-type viruses, was performed. The alignment revealed that the MDEV Env has variable regions that correspond in position to the variable receptor specificity domains identified in other MLVs, with alignable scaffold regions surrounding the variable regions (3, 8). The TM region of the MDEV Env aligns very well with those of other retroviruses (9). Figure 4B shows an unrooted tree based on this alignment, with Env proteins that recognize common receptors grouped in ovals.
Retroviral interference experiments have shown that MDEV uses a receptor that is novel among murine retroviruses but may be identical to that of the endogenous cat virus RD114 in G355 cat glial cells. Figure 4B shows that the Env protein of MDEV is not closely related to that of any particular retrovirus, including RD114; furthermore, there exists no significant similarity in the variable regions that are responsible for receptor specificity. This does not rule out the possibility of common-receptor usage; while retroviruses that use common receptors tend to have closely related envelopes, there are exceptions. For example, receptor interference can be observed among 10A1, GALV, and feline leukemia virus type B, which also do not have closely related envelope sequences. Because the MDEV pol gene and the majority of gag are most similar to those of GALV, it is likely that the MDEV env gene was also derived from a virus similar to GALV. It is therefore interesting that the receptor-binding regions of the Env proteins of MDEV and GALV do not exhibit significant similarity and that the two Env proteins have been found to use distinct receptors in all cells tested thus far.
The mechanisms by which retroviruses evolve to use different receptors are poorly understood. One possibility is that a virus can dramatically change receptor usage by undergoing recombination with cellular sequences that encode a protein that binds to the new receptor. This type of event could produce a virus capable of infecting new cells, giving it a replicative advantage. Similar acquisition of cellular sequences by retroviruses is well documented in the case of oncogenes. A prediction of this envelope cassette-swapping hypothesis is that the variable regions of retroviral envelopes resemble cellular proteins. Therefore, the databases were searched for sequences similar to the amino acid sequences of the variable regions of MDEV and other MLV-type retroviruses, but none of statistical significance was found. Perhaps the rapid mutation rate of retroviruses obliterates the primary sequence while the envelope is fine tuned to mediate efficient entry via the new receptor.
This work has demonstrated that a virus derived from an MDEV molecular clone uses the same receptor as do the biological isolates. Sequence analysis has demonstrated that MDEV is a chimera between a VL30 element(s) and a virus similar to GALV, making MDEV the first example of a naturally occurring, replication-competent retrovirus with major functional sequences similar to VL30 elements. Current work is directed at creating packaging cells that express the unique MDEV envelope for gene transfer applications, evaluating the recombinant nature of the native provirus in M. dunni cells, and exploring the significance of the LTR repeats.
ACKNOWLEDGMENTS
We thank Rebecca Gottschalk for technical assistance, B. Chesebro for the M. dunni tail fibroblasts, D. Blair for the G355 cells, and V. KewalRamani for the D17 cells. We thank Elizabeth Greene and the Biocomputing Shared Resource for assistance with sequence analysis.
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL36444 and HL54881) and the National Institute of Diabetes and Digestive and Kidney Diseases (DK47754).
REFERENCES
- 1.Adams S E, Rathjen P D, Stanway C A, Fulton S M, Malim M H, Wilson W, Ogden J, King L, Kingsman S M, Kingsman A J. Complete nucleotide sequence of a mouse VL30 retro-element. Mol Cell Biol. 1988;8:2989–2998. doi: 10.1128/mcb.8.8.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armentano D, Yu S-F, Kantoff P W, von Ruden T, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J-L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoprotein of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonham L, Wolgamot G, Miller A D. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J Virol. 1997;71:4663–4670. doi: 10.1128/jvi.71.6.4663-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan R, Meade C, Todaro G J. Isolation of an endogenous type C virus related to the infectious primate type C viruses from the Asian rodent Vandeleuria oleracea. J Virol. 1979;30:124–131. doi: 10.1128/jvi.30.1.124-131.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark D, Henikoff S. Ordered deletions using exonuclease III. Methods in Mol Biol. 1996;31:349–357. doi: 10.1385/0-89603-402-X:349. [DOI] [PubMed] [Google Scholar]
- 7.Dunn K J, Yuan C C, Blair D G. A phenotypic host range alteration determines RD114 virus restriction in feline embryonic cells. J Virol. 1993;67:4704–4711. doi: 10.1128/jvi.67.8.4704-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Burger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 9.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y-X, Yuan H, Rein A, Levin J G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields-Berry S C, Halliday A L, Cepko C L. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French N S, Norton J D. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 13.Graham F L, Smiley J. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 14.Hatzoglou M, Hodgson C P, Mularo F, Hanson R W. Efficient packaging of a specific VL30 retroelement by ψ2 cells which produce MoMLV recombinant retroviruses. Hum Gene Ther. 1990;1:385–397. doi: 10.1089/hum.1990.1.4-385. [DOI] [PubMed] [Google Scholar]
- 15.Hodgson C P, Fisk R Z, Arora P, Chotani M. Nucleotide sequence of mouse virus-like (VL30) retrotransposon BVL-1. Nucleic Acids Res. 1990;18:673. doi: 10.1093/nar/18.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam T C, Bugge T H, Bohm S. The long terminal repeat of VL30 retrotransposons contains sequences that determine retinoic acid-induced transcription in cultured keratinocytes. J Biol Chem. 1993;268:3251–3259. [PubMed] [Google Scholar]
- 17.Itin A, Keshet E. Apparent recombinants between virus-like (VL30) and murine leukemia virus-related sequences in mouse DNA. J Virol. 1983;47:178–184. doi: 10.1128/jvi.47.1.178-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander M R, Chattopadhyay S K. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Tissier P, Stoye J P, Takeuchi Y, Patience C, Weiss R A. Two sets of human-tropic pig retrovirus. Nature. 1997;389:681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- 21.Lieber M M, Sherr C J, Todaro G J, Benveniste R E, Callahan R, Coon H G. Isolation from the Asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc Natl Acad Sci USA. 1975;72:2315–2319. doi: 10.1073/pnas.72.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder M, Linder D, Hahnen J, Schott H-H, Stirm S. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur J Biochem. 1992;203:65–73. doi: 10.1111/j.1432-1033.1992.tb19828.x. [DOI] [PubMed] [Google Scholar]
- 23.Makris A, Patriotis C, Bear S E, Tsichlis P N. Structure of a Moloney murine leukemia virus–virus-like 30 recombinant: implications for transduction of the c-Ha-ras proto-oncogene. J Virol. 1993;67:1286–1291. doi: 10.1128/jvi.67.3.1286-1291.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller A D, Bonham L, Alfano J, Kiem H-P, Reynolds T, Wolgamot G. A novel murine retrovirus identified during testing for helper virus in human gene transfer trials. J Virol. 1996;70:1804–1809. doi: 10.1128/jvi.70.3.1804-1809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller A D, Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller A D, Wolgamot G. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy J, Goff S P. Forced integration of a Moloney murine leukemia virus DNA with a mutant integration site occurs through recombination with VL30 DNA. Virology. 1994;204:458–461. doi: 10.1006/viro.1994.1554. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson M, Bohm S. Inducible and cell type-specific expression of VL30 U3 subgroups correlate with their enhancer design. J Virol. 1994;68:276–288. doi: 10.1128/jvi.68.1.276-288.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norton J D, Connor J, Avery R J. Unusual long terminal repeats of a retrovirus transmissible mouse (VL30) genetic element: identification of functional domains. Nucleic Acids Res. 1984;12:3445–3460. doi: 10.1093/nar/12.8.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page R D M. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 32.Perlmann T, Rangarajan P N, Umesono K, Evans R M. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 33.Prats A-C, De Billy G, Wang P, Darlix J-L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 34.Reuss F U, Schaller H C. cDNA sequence and genomic characterization of intracisternal A-particle-related retroviral elements containing an envelope gene. J Virol. 1991;65:5702–5709. doi: 10.1128/jvi.65.11.5702-5709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotman G, Itin A, Keshet E. Promoter and enhancer activities of long terminal repeats associated with cellular retrovirus-like (VL30) elements. Nucleic Acids Res. 1986;14:645–656. doi: 10.1093/nar/14.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wills N M, Gesteland R F, Atkins J F. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wills N M, Gesteland R F, Atkins J F. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]