Abstract
Crotonylation is an importantly conserved post-translational modification, which is completely different from acetylation. In recent years, it has been confirmed that crotonylation occurs on histone and non-histone. Crotonylated Histone primarily affects gene expression through transcriptional regulation, while non-histone Crotonylation mainly regulates protein functions including protein activity, localization, and stability, as well as protein-protein interactions. The change in protein expression and function will affect the physiological process of cells and even cause disease. Reviewing previous studies, this article summarizes the mechanisms of histone and non-histone crotonylation in regulating diseases and cellular physiological processes to explore the possibility of precise regulation of crotonylation sites as potential targets for disease treatment.
Keywords: Post-translational modifications, Protein crotonylation, Cancer, Diseases
Highlights
-
•
We discussed that crotonylation in specific sites of proteins can cause functional changes in both histone and non-histone proteins.
-
•
Proteins crotonylation is closely related to the occurrence and development of numerous diseases, including cancer, kidney diseases, and others.
-
•
Crotonylation participates in a variety of processes in organisms
1. Introduction
With the development of human understanding on protein function and biological mechanisms, the importance of post-translational modifications (PTM) of proteins has increased greatly. Various modifications of different amino acid residues have been reported on histone, such as histone methylation [1], formylation, acetylation, butyrylation, lactylation [2], and crotonylation [3]. Histone crotonylation is conserved in eukaryotes such as yeast [4] and mammals, but there are few reports about crotonylation in rice [5], tea plants [6], and papaya [7]. Histone crotonylation usually occupies a similar position as acetylation, but it is mechanistically and functionally distinct from histone lysine acetylation (Kac) because of the extended hydrocarbon chains and C–C π-bond in Kcr chemical structures [8]. This dynamic process is regulated by crotonyltransferases (“writer”) and decrotonylases (“eraser”) together [8], and the function of proteins to recognize Kcr are identified by "reader". Crotonylation of histone occurs mainly on lysine residues, and affects gene expression. Studies have reported that histone crotonylation plays vital roles in human physiological developments such as gene expression [8,9], cell cycle [10], DNA damage [11], aging [12], and spermatogenesis [13,14]. In addition, in recent years, it has been found that crotonylation occurs on histone as well as on non-histone proteins [15]. This modification can affect protein functions including protein localization, activity [16], and stability, as well as protein-protein interactions. Furthermore, crotonylation affects protein activity by modifying the serine of proteins [17]. Protein crotonylation is very important for the growth and development of organisms, participating in embryonic cell differentiation and division [18,19], maintaining metabolic homeostasis [20], and other physiological processes. Crotonylation of protein could also affect the development and progress of diseases, such as tumors, kidney diseases, heart diseases, etc.
To better understand the biological function of crotonylation in physiological processes and its roles in disease development, this review will focus on the diseases and biological processes related to histone and non-histone crotonylation, and elaborate its mechanism in physiological processes and diseases, which may provide new insights into the epigenetic research of related diseases.
2. Regulation of crotonylation
Crotonylation is a histone post-translational modification first reported by high-sensitivity mass spectrometry in 2011 [8]. Protein crotonylation level in cells was precisely regulated by the activity of crotonyltransferases (writer) and decrotonylases (eraser), and can be influenced by the concentration of crotonyl-CoA in cells, therefore, enzymes that regulate crotonyl-COA metabolism are also involved in the regulation of crotonylation (Fig. 1).
Fig. 1.
Schematic representation of the chemical formula of crotonylation and the biosynthesis of crotonyl-CoA.
2.1. Crotonyl-CoA
Protein crotonylation, is derived from crotonyl-CoA, an intermediate metabolite during fatty acid oxidation or lysine and tryptophan metabolism. Intracellular crotonyl-CoA could directly regulate Kcr [21], and is crucial for mesoderm/endoderm differentiation [22]. Crotonyl-CoA can be maintained at a relatively suitable concentration through various metabolic pathways: (1). Crotonate can be effectively converted to crotonyl-CoA by ACSS2 (acyl-CoA synthetase short chain family member 2), which is related to the level of crotonyl-CoA and crotonylation in the body [23], knock down ACSS2 in cultured cells notably changed the level of histone crotonylation[9]. (2) In amino acid metabolism, lysine, hydroxylysine, and tryptophan are metabolized to glutaryl-CoA, then glutaryl-CoA is oxidized to crotonyl-CoA, and this process is catalyzed by GCDH (glutaryl-CoA dehydrogenase). (3) The short-chain fatty acids (SCFAs) produced by microbiota are transported into cells via membrane receptors MCT or SMCT, and then converted into butyryl-CoA through β-oxidation pathway, which is then transformed into crotonyl-CoA by BCDH, moreover, ACADS [24], ACOX3 [22] and ACOX1 involved in crotonyl-CoA metabolism synthesis. ACOX2 is a indirect regulator of lysine crotonylation through interacts with methyl crotonyl coA carboxylase (MCCC1/2) and inhibits its enzyme activity [20]. MCCC1/2 a heterodimer and catalyze the carboxylation of 3-methylcrotonyl-CoA to 3-methylglutaconyl-CoA [25]. (4) ECHS1 (Short-chain enoyl–coenzyme A hydratase) has the highest activity for hydrolyzing crotonyl-CoA, and reduce intracellular crotonyl-CoA [26]. The chromodomain protein CDYL converts crotonyl-CoA to β-hydroxybutyryl-CoA, which is a negative regulator of histone crotonylation [21].
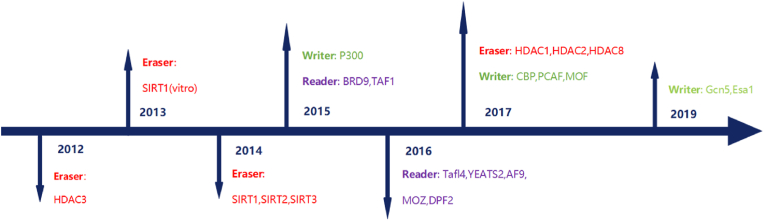
Several factors are associated with the process of crotonylation, namely writers, readers and erasers (Fig. 2).
Fig. 2.
Timeline depicting the discovery and characterization of crotonylation regulators.
2.2. Writer
Biochemically, histone acetyltransferase p300 induces histone crotonylation [27], which generates the crotonyllysine mark on histone H3 Lys18 (H3K18). Thus the connection between crotonylation and acetylation was clarified. One study reported that the acetyltransferases hMOF [28] PCAF [18] and CBP [27]act as crotonyltransferases for non-histone proteins [29]. Another study revealed that MOF is also the histone crotonyl transferase (HCT) that catalyzes the crotonylation of histones at multiple sites on H3 and H4, and MOF possesses potent HCT activity. KAT2B (PCAF) was also identified as an HCT [30]. Furthermore, the study also generated novel CBP/p300 mutants that were defective in histone acetyltransferase but maintained normal HCT activity. Using the CBP I1432G and p300 I1395G mutants, the study not only verified the HCT of mutants but also demonstrated that the mutants correlate with the recruitment of the promoter histone crotonylation and the crotonylation reader [30]. Recently, Gcn5 and Esa1 were found to possess crotonyltransferase activity due to the lysine of Gcn5-Ada2-Ada3 (ADA) and Esa1-YnG2-Epl1 (PiccoloNuA4) being crotonylated at the N-terminal tail of H4 and H3 [31].
2.3. Eraser
Eraser refers to the enzymes that can remove specific residues in proteins. In humans, histone deacetylases (HDACs) are classified into two families: the histone deacetylase family and the sirt regulator family [32]. The activity of histone deacetylase (HDCR) was found to be specific for class I histone deacetylation but not for class II and IV histone deacetylation by Rnai inactivation assay and in vitro histone deacetylation assay. For example, HDAC1 is active in decrotonylation of many proteins, such as H3K4, H3K9and H4K12. Moreover, it was confirmed to be an active HDCR enzyme [33,34]. Experiments found that HDAC2, HDAC3, and HDAC8 also could exhibit depyruvase activity in vitro [29,35]. Moreover, SIRT1, SIRT2, and SIRT3 were all confirmed that they have the ability of histone decrotonylases in vitro [36,37]. HDAC1/2 deficiency increases histone crotonylation expression while reduces total decrotonylase activity by 85% in embryonic stem cells [38]. These results suggest that "eraser" is vitally important in crotonylation, and the underlying mechanisms need to be further explored.
2.4. Reader
The reader is to identify the functions of Kcr modification in physiology and pathology. It can be recognized by three classes of domains: Double PHD finger (DPF), Bromodomain, and YEATS domain [39]. Bromodomains weakly with a crotonylated peptide, BRD9 and TAF1 bind more tightly to acetylated peptides [40,41]. Studies have verified that Bromodomains was capable of binding to acetylated lysine residues [42]. H3K9cr was shown to be a selective target of the Yeats domain of Taf14, which binds crotonyllysine through a unique π-π-π superposition mechanism. This implies that Taf14 is an H3K9 crotonylation reader [33,43]. Another study identified YEATS2 as a histone crotonylation resolving reader with H3k27 site specificity and revealed an aromatic sandwich pocket with open ends within YEATS2 for Kcr binding [44]. Further, AF9 was found to positively regulate gene expression in the YEATS domain and colocalize with the crotonylated H3 [45]. Additionally, MOZ and DPF2 with DPF domains are specific readers of H3K14cr [46].
3. Related diseases of histone crotonylation and non-histone crotonylation
Similar to other modifications, crotonylation is closely associated with many diseases and physiological processes. And a substantial body of research has revealed protein crotonylation modifications in numerous diseases through analysis. However, only a few diseases have established a clear association between crotonylation and disease progression, and in-depth investigations into the underlying mechanisms remain limited. In this section, we primarily summarize the confirmed crotonylation in diseases (Table 1, Table 2), and the importantly crotonylated proteins and their sites (Table 3). Although clinical research on the impact of crotonylation sites on diseases is lacking, multiple animal and cell experiments have demonstrated their significance.
Table 1.
Diseases associated with crotonylation.
| Diseases | Protein crotonylation | Regulatory mechanism | References |
|---|---|---|---|
| Colorectal cancer | ENO1 | ENO1 K420 crotonylation promoted the growth, migration, and invasion of colorectal cancer cells (CRC) in vitro by enhancing the activity of ENO1 and regulating the expression of tumor-associated genes. | [27] |
| Non-small-cell lung cancer cells | BEX2 | The crotonylation of BEX2 at the K59 site is found to be critical for mediating mitophagy in lung cancer cells. | [51] |
| Hepatocellular carcinoma | SEPT2 | Crotonylation facilitates cell invasion through the crotonylated SEPT2-K74-P85α-AKT pathway | [52] |
| Pancreatic cancer | MTHFD | The activation of MTHFD1 by decrotonylation at Lys354 and Lys553 promotes pancreatic cancer the development of by increasing resistance to ferroptosis | [53] |
| Acute kidney injury | – | Cell stress increases affecting the TWEAK, it then decreased PGC1a and Sirt-3 expression and with increased CCL2 expression. | [60,61] |
| Autosomal dominant polycystic kidney disease | H3K18 | CDYL increases the level of Kcr, and overexpression of CDYL decreased histone Kcr, inhibited the expression of cyst-related genes, and slowed the growth of cysts. | [64] |
| Depression | H3K27 | Increased H3K27 content and decreased the level of Kcr to inhibit the transcription of a group of genes such as neuronal VGF, then it leads to promote depression. | [85,86] |
| Alzheimer's disease | H3K27 | NEAT1 inhibition influences H3K27 acetylation (H3K27Ac) and H3K27 crotonylation (H3K27Cro) located nearby to the transcription start site of many genes, including endocytosis-related genes. | [81] |
| Hypertrophic cardiomyopathy | H3K18 and H2BK12 | Short-chain enoyl-CoA hydratase (ECHS1) downregulation was accompanied with the upregulation of H3K18cr and H2BK12cr in human hearts with hypertrophic cardiomyopathy. | [30] |
| HIV latency | H3K4 | Viral infection or addition of crotonyl-CoA induces the expression of the fatty acid metabolic enzyme ACSS2, and ACSS2 increased H3K4 crotonylation which leads to regulation of HIV latency/transcription. | [89] |
| Ischemic heart disease | IDH3a | IDH3a K199 and TPM1 L28/29 crotonyalation could not only protect cardiomyocytes but also preserve myocardial function after injury. | [72] |
| Immunoglobulin A nephropathy | – | Identified 353 crotonylated proteins | [68,119] |
| Chronic renal failure | – | Identified 1109 lysine modification sites | [69,120] |
| Hemodialysis | – | Identified total 1109 lysine crotonylation sites on 347 proteins | [57] |
| COPD Combined with Type II RF | – | Identified 32 sites of 23 proteins were upregulated and 914 sites of 295 proteins were downregulated | [121] |
Table 2.
Diseases associated with crotonylation metabolite.
| Diseases | Metabolite | Regulatory mechanism | References |
|---|---|---|---|
| Hepatocellular carcinoma | HDACs | By adding HDACs inhibitor, it founded that lysine crotonylation can affect the Hepatocellular carcinoma (HCC)cell migration, and decrease the proliferation ability of hepatoma cell. | [48] |
| Wilms tumor | YEATS domain of MLLT1 | Mutations in the YEATS domain of MLLT1 have been shown to be functionally relevant in Wilms tumor. | [122] |
| Hepatocellular carcinoma | ACOX2 | ACOX2 is a regulator of Kcr.Compared with normal mice, experiments on ACOX2 mice have uncovered that the level of non-histone Kcr was dowenregulated while the level of histone (H2b) K86cr was upregulated. | [54] |
| Ovarian cancer | HDACs | HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) could inhibit growth of cancer cells, inducing expression of tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. | [123] |
| inflammation | AF9 YEATS domain | AF9 YEATS domain has selectively higher binding affinity on crotonyllysine than acetyllysine, and AF9 YEATS Links the inflammatory genes' response to histone crotonylation. | [45] |
Table 3.
Proteins and their Kcr sites associated with crotonylation.
| Protein Names | Enter Name | ID | Sites | Diseases and Biological Processes | References |
|---|---|---|---|---|---|
| Histone 2b | H2b | – | K12 | Hypertrophic cardiomyopathy (HCM) | [30] |
| Histone 2b | H2b | – | K86 | Metabolic homeostasis, Hepatocellular carcinoma | [54,100] |
| Histone H3 | H3 | Q6NXT2 | K18 | Autosomal dominant polycystic kidney disease (ADPKD), HCM | [30,64] |
| Histone H3 | H3 | Q6NXT2 | K27 | Depression, Alzheimer's disease (AD) | [81,85,86] |
| Histone H3 | H3 | Q6NXT2 | K4 | HIV | [89] |
| Histone H4 | H4 | P62805 | K77, K91 | Stem cell endoderm differentiation | [22] |
| Enolase 1, chloroplastic | ENO1 | Q9C9C4 | K40 | colorectal cancer | [27] |
| Protein ENL | ENL | Q03111 | S46 | Wilms tumor | [122] |
| Cellular tumor antigen P53 | P53 | P04637 | Render the tolerance of cancer cells to chemotherapeutics and drugs | [17] | |
| Platelet glycoprotein 4 | CD36 | P16671 | Chronic renal failure (CRF) | [69] | |
| Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | IDH3a | Q9D6R2 | K199 | Ischemic heart disease (IHD) | [72] |
| Microtubule-associated protein RP/EB family member 1 | EB1 | Q8WQ86 | K66 | Spindle positioning | [94] |
| Tropomyosin alpha-1 chain | TPM1 | P58771 | K28 | Ishemia-Reperfusion Injury | [72] |
| Brain expressed X-linked gene 2 | BEX2 | Q9BXY8 | K59 | Non-small lung cancer | [51] |
| Septin2 | SEPT2 | Q15019 | K74 | Hepatocellular carcinoma (HCC) | [52] |
| methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 | MTHFD1 | P11583 | K354, K553 | Pancreatic cancer | [53] |
3.1. Cancer
With the development of medical technology, modern medicine can cure many diseases, but cancer is still the most concerned public problem. Fortunately, many recent studies have indicated that lysine crotonylation has a positive effect in the treatment of cancer [47] Report found that the lysine crotonylation expression increases in the esophagus, thyroid, pancreas, colon, and lung cancer, but decreases in the stomach, liver, kidney cancer [48]. From this, it can be seen that there is a significant correlation between crotonylation modification and tumors. However, due to the lack of clinical research data, further in-depth studies are still needed in the future. Here, we will mainly introduce the potential crotonylation modification sites have been identified in tumors.
A recent study found that HDACs inhibitor increased the crotonylation level and lysine crotonylation can affect the migration of hepatocellular carcinoma (HCC) cells and reduce the proliferation ability of hepatoma cells [48]. Furthermore, histone crotonylation in paracancerous tissues was lower than PCa tissues and Kcr increased with the enhanced malignancy of PCa [49]. Crotonylation was also observed in small cell lung cancer (SCLC) tissues, providing a new angle to understand the mechanisms of SCLC malignancy, such as metastasis, immunosuppression, and chemoradiotherapy resistance [50].
Compared with histone crotonylation and its relationship with cancer, non-histone crotonylation is much under-explored. Recently, the study reported that K420 was the main Kcr site of ENO1 (α enolase), ENO1 K420 crotonylation can enhance ENO1 activity and regulate the expression of tumor-related genes, thus promoting the growth, invasion, and migration of colorectal cancer cells (CRC) in vitro [27]. And the crotonylation of BEX2 at the K59 site is found to be critical for mediating mitophagy in lung cancer cells [51]. In addition, by modifying the serine 46 sites of p53, crotonic acid (CA) can negatively regulate the transcriptional level and activity of p53, increasing resistance of cancer cells to chemotherapy drugs [17]. High levels of SEPT2-K74 crotonylation promoted HCC metastasis both in vitro and in vivo, and predicted poor prognosis and a high recurrence rate in HCC patients [52]. And the crotonylation of MTHFD1 at Lys354 and Lys553 could inhibit the development of pancreatic cancer [53].
One of the regulators of lysine crotonylation is Acyl-CoA oxidase 2 (Acox2), its main point of action is Kcr and its deletion leads to liver cancer in mice [54]. H4K77cr and H4K91cr are two Kcr sites and critical for endoderm differentiation [22]. And studies have found that the occurrence and development of the disease can be better controlled if the cancer stem cell program is activated [55]. Both studies indicated that crotonylation-meditated mechanism may be important for cancer related stem cell therapy.
Although seldom known about the field of non-histone crotonylation, non-histone proteins occupy important positions in numerous vital biological processes. Consequently, delving deeper into the study of crotonylation modifications on non-histone proteins holds immense prospects for future cancer treatments.
3.2. Kidney diseases
Kidney is one of the main organs of the human body, and many diseases will cause damage to kidney function. Commonly known kidney diseases include Acute kidney injury (AKI), Autosomal dominant polycystic kidney disease (ADPKD) and others. And studies have shown that crotonylation is closely related to these diseases [56]. By the liquid chromatography tandem mass spectrometry (LC-MS/MS) with highly sensitive immune-affinity purification, many studies have revealed the widespread presence of crotonylation modifications on proteins and sites in various kidney disease [57]. However, there is limited research on the specific protein targets and ways of action. Only few studies specified the accurate crotonylation affect kidney diseases’ mechanism. There is still need more basic research to elucidate the proteins and specific sites of crotonylation, providing new insights for future clinical studies.
3.2.1. Acute kidney injury
AKI has high morbidity and mortality and is a major public health burden [19]. Research found the expression of histone crotonylation in experimental nephrotoxic AKI. Further experiments indicate that this increase in histone crotonylation is associated with the activation of the inflammatory cytokine TWEAK [58,59], and TWEAK promotes the crotonylation of cultured tubular cells. Additionally, cellular stress affects TWEAK expression, leading to reduced expression of Sirt-3 and PGC1a, as well as increased expression of CCL2. Regarding this matter, adding exogenous crotonic or HDAC inhibitor, resulted in increased expression of kidney SIRT3 and PGC-1α in both in vivo and cultured cell settings, and decreased expression of CCL2, providing protection against AKI [60,61]. Overall, both cellular stress and crotonic promote histone crotonylation and thus the protective effect of crotonylation against AKI.
3.2.2. Autosomal dominant polycystic kidney disease
ADPKD is the most common hereditary kidney disease that can impair kidney function and eventually lead to kidney failure [62,63]. A recent study revealed that crotonylation might have a crucial role in ADPK. Animal experiments, and statistical analysis found that the expression of chromodomain Y-like protein (CDYL) [21], was inhibited in ADPKD, whereas the level of Kcr was increased. Overexpression of CDYL can decrease histone Kcr, suppress the expression of cyst-related genes, and slow down cysts growth. Furthermore, H3K18 has been identified as a CDYL targeted crotonylation site in ADPKD cells [64].
3.2.3. Other kidney diseases
Immunoglobulin type A nephropathy (IgAN) and chronic renal failure (CRF) are severe kidney diseases [[65], [66], [67]]. Study analyzed lysine crotonylation in patients with IgA nephropathy (IgAN), chronic renal failure (CRF), and healthy controls. The results identified numerous crotonylated proteins and modification sites [68,69]., associated with processes like inflammation, oxidative stress [68] and fibrosis [69]. Also, crotonylation levels were slightly lower in maintenance hemodialysis patient group [61]. Although the main mechanisms of crotonylation affected these diseases are still unknown, they open up new ways for the development of treatment strategies and interventions patients, and even identifies crotonylation as a potential therapeutic target.
3.3. Cardiac diseases
Ischemic heart disease (IHD) is a serious condition that causes complex and multifaceted pathophysiology [70,71]. New research shows that when IHD ischemia-reperfusion injury triggers cardiac muscle cell contraction, the required protein is first crotonylated by lysine. Specifically, mitochondrial protein IDH3a (isocitrate dehydrogenase 3 [NAD+] alpha) at K199 and cytoskeletal protein TPM1 (tropomyosin alpha-1 chain) at K28/29 were selected for the regulation of crotonylation, adding addition of exogenous sodium crotonate to enhance crotonylation, and the results indicated that the two proteins crotonylation could not only protect cardiomyocytes from apoptotic structural rearrangement through inhibiting mitophagy or cytoskeleton mediated by BNIP3 (Bcl-2 adenovirus E18 19 kDa interacting protein 3), but also maintain myocardial function after injury through inhibiting fibrosis and apoptosis [72].
Hypertrophic cardiomyopathy (HCM) is a prevalent inherited cardiovascular disease with unknown etiology. Recent studies found that HCM patients exhibit a downregulation of short-chain enyl-coA hydrase (ECHS1) along with an up-regulation of H3K18cr and H2BK12cr [73]. Additionally, the hydratase ECHS1 could maintain the maturity and homeostasis of cardiomyocytes, as well as regulating the intracellular crotonyl-CoA through histone crotonylation and other ways [26]. It mentions the association between hemodialysis (HD) and crotonylation, suggesting that HD-induced stress and injury to the cardiovascular system may be linked to crotonylation changes [61,74].
Vascular smooth muscle cells (VSMCs) are essential components of tissue structure constituting to the maintenance of vascular tone [75,76]. And the LC-MS/MS assay identifies 2138 lysine crotonylation sites among 534 proteins. these non-histone crotonylated proteins involve in multiple essential biological processes, including glycolysis, cellular skeleton modulation, and VSMC contraction [77].
3.4. Neuropsychiatric diseases
Alzheimer's disease (AD) is characterized by severe cytoskeletal alterations in only a few neuronal types in the human central nervous system [78]. A study revealed that the repression of Nuclear Paraspeckle Assembly Transcript 1 (NEAT1) [79,80] in early AD mediates the clearance of Aβ by inhibiting the expression of endocytosis-related genes [81]. NEAT1 has a complex relationship with P300/CBP, and its inhibition influence the H3K27 crotonylation (H3K27Cr) and H3K27 acetylation (H3K27Ac) of various genes, including toxic-related genes [81]. Whether NEAT1 could influence H3K27Cr to meditate Aβ still need further research.
Depression is a major mental illness, affecting a vast number of individuals. [82]. Despite its prevalence, the pathogenesis of depression remains elusive, and our understanding of the disease is limited. Compared with B6 mice, BTBR mice had higher levels of lysine crotonylation, which could contribute to neuropsychiatric disorders, like depressive disorder and Alzheimer's disease [83]. But this study did not identify the exact histone or non-histone lysine crotonylation [84]. By the dual effects of increased H3K27 and decreased level of Kcr, CDYL inhibits the transcription of a group of genes such as neuronal VGF, leading to the promotion of depression [85,86].
In addition to the aforementioned neuropsychiatric disorders, Kcr is widely present in macrophages, sensory neurons, astrocytes, and microglia of the medulla oblongata. Furthermore, histone crotonylation plays a crucial role in regulating neuroinflammation and neuralgia pain, although the precise mechanism remains unclear [87]. Although the findings suggested that the Kcr may greatly affect the progress of the neuropsychiatric diseases, the crotonylation modifications in neuro-psychiatric disorders is not yet widely explored, and more foundational research data is needed to validate this.
3.5. Acquired immunodeficiency syndrom (AIDS)
AIDS is caused by Human immunodeficiency virus (HIV) infection. Approximately a decade ago, HDACs inhibitors trohostatin and trapoxin were found to reactive latent HIV transcriptionally [88]. Recently, a study uncovered that induced by crotonyl-CoA or viral infection, ACSS2 could increase crotonylation of H3K4 as well as acetylation of H3K4 and acetylation of H3K18 (H3K18Ac), leading HIV latency/transcription [89]. More importantly, combined with PKC agonists PEP005, vorinostat, or JQ1 in T cell cultures in vitro and/or CD4+T cells, histone crotonylation was found to significantly enhance latent HIV reactivation in HIV-infected patients, suggesting a synergistic effect of histone crotonylation with PKC agonists [89]. These findings highlight the potential for investigating the combination of histone crotonylation and HIV latency therapy to improve treatment outcomes.
3.6. Inflammation
Inflammation serves as an adaptive response to harmful stimuli such as infection and tissue damage [[90], [91], [92]]. A study found that the AF9 YEATS structure domain has higher binding compatibility on lysine crotonylation and AF9 YEATS junction inflammatory gene response to histone crotonylation. AF9 was also found to be recruited to LPS-stimulated genes, and its recruitment can be further augmented through crotonate pre-treatment in a YEATS-dependent manner [45]. Additionally, inflammation in osteoarthritis chondrocytes could lead to increased HDACs expression and activity [93]. Although studies have indicated the presence of crotonylation in inflammation, no research has provided a specific mechanistic explanation to elucidate the connection between the two.
4. Physiological processes related to crotonylation
In addition to the research on crotonylated diseases, multiple studies have revealed the link between crotonylated diseases and physiological processes. Specifically, histone crotonylation have been shown to impact spindle positioning [94], metabolic homestasis [54], and stem cell endoderm differentiation [22]. Furthermore, non-histone crotonylation has been linked to subcellular organelles [72].
4.1. Spindle positioning
The accurate spindle localization is crucial physiological functions, including cell passage and development, as well as a significant prerequisite for proper mitotic and meiotic cell divisions [95]. Previous work has shown that P300/CBP-associated factor-(PCAF) regulates EB1K220 acetylation to make sure dynamic interaction between kinetochore and microtubule in early mitosis [96,97]. A recent study revealed that dynamic crotonylation of EB1 during mitosis, mediated by TIP60, plays a crucial role in ensuring precise spindle localization [94]. Specifically, TIP60 primarily catalyzes the crotonylation of EB1 over Lys66, rather than acetylation. The synergistic effect of HDAC3 enhances TIP60-catalyzed crotonylation. Further analysis showed that tip60 mediated crotonylation of EB1 by regulating the positive end dynamics of astral microtubules to ensure the orientation of mitotic spindles [94]. The investigation of the relationship between crotonylation and spindle positioning provides new avenues for the studying physiological functions both in vivo or ex vivo.
4.2. Metabolic homeostasis
ACOX2 (acyl-CoA oxidase 2) is an important peroxisomal enzyme that could impact the oxidation of branched-chain fatty acids and the metabolism of bile acid, and various physiological processes [98,99]. Importantly, ACOX2 has been implicated in the development of hepatocellular carcinoma [100] and primary malignant cardiac tumors (PMCTs) [101,102]. Recently, ACOX2 is identified as a regulator of Kcr and contributes to metabolic homeostasis via crotonylation. And ACOX2 mice have down-regulation of non-histone crotonylation levels and up-regulation of histone 2B K86cr levels. Together, these results underscore the significance of ACOX2 crotonylation in metabolic homeostasis and liver cancer [54].
4.3. Stem cell endoderm differentiation
Acetyl-coenzyme A (CoA) is an important cofactor for post-translational modifications, particularly in Kcr [26]. A recent study found that the crotonylation of human embryonic stem cells (hESCs) promotes mesodermal differentiation both in vitro and in mice embryos. The only sites detected in differentiated endodermal cells were H4K77cr and H4K91cr [22]. Endothelial differentiation is associated with increased expression of the crotonyl-CoA-producing enzyme in vivo and cultured cells (Fig. 3). Crotonate significantly enhances hESCs’ endoderm differentiation efficiency [103]. Embryonic stem cells (ESCs) were enriched in histone crotonylation and the lack of HDAC1/2 affected the activity of the decrotonylase [34,38]. These findings have important physiological relevance and clinical implications. For example, short-chain acyl-CoA dehydrogenase deficiency (SCADD) is a disease mainly characterized by elevating butyryl-COA and its by-products in muscle and liver [[104], [105], [106]]. Combing these results, investigating histone crotonylation and other possible short-chain fat acylation in patients could help manage SCADD. Furthermore, as many organs including the pancreas, liver, digestive tract, and lung originate from the endoderm [107], these findings could potentially lead to new treatments for these organ diseases.
Fig. 3.
The function of crotonylation in stem cell endoderm differentiation (Left). crotonylation was observed during the differentiation of hESCs into mesondodermal and endodermal cells. Two crotonylation sites were identified on endodermal: H4K77cr and H4K91cr. Function of non-histone crotonylation in subcellular organelles (Right). crotonylation at the K28/29 site of TMP1 protein was found to rearrange the cytoskeleton structure, and crotonylation at the IDH3a K99 was observed to reduce myocardial fibrosis and protein BNIPS, thereby significantly preserving cardiac function.
Furthermore, histone crotonylation can activate Zscan4, leading to a decrease in telomere damage and maintenance of telomere length in chemically induced pluripotent stem cells (CiPSCs) [12], Combined with great potential of CiPSCs in stem cell-based therapy [108]. These results highlight the potential significance of histone crotonylation in shaping the future of stem cell research.
4.4. Autophagy and the MTORC1 pathway
Autophagy usually refers to the process that involves lysosomes invading and degrading the substrates directly. Previous studies have suggested that leucine can affect this process, but the precise mechanism remains unclear. However, a recent study identified the relationship between lysine crotonylation levels and autophagy induced by leucine-deprivation [109]. Combined with previous findings on the interaction between HDAC7 and 14-3-3 proteins [[110], [111], [112]], leucine deprivation inhibits HDAC7 increases the 14-3-3ε crotonylation level, and releases the protein phosphatase 1B (PPM1B) from its interaction with 14-3-3ε. This dephosphorylates ULK1 and activate autophagy. Furthermore, quantitative crotonylomic profiling identified two lysine crotonylation sites, K73 and K78, on 14-3-3ε, which have a significant effect on leucine-deprivation-induced autophagy.
Regarding MTORC1 pathway, CANX (calnexin) is an essential regulator for the leucine-stimulated MTORC1 (mechanistic target of rapamycin kinase complex 1) pathway [113]. Specifically, in response to leucine deprivation, CANX lysine undergoes 525 crotonylation, which is mediate by KAT7(lysine acetyltransferase 7). The combination of CANX and LAMP2 (lysosomal associated membrane protein 2) further decreased the MTORC1 pathway activity. Together, these components playing a role in the interplay between lysine deprivation and the MTORC1 pathway [114].
4.5. Non-histone crotonylation in subcellular organelles
Previous studies have investigated the relationship between PTMs and myofilament proteins, such as short-term phosphorylation at multiple sites in the myosin light chain (MLC) [115]. Lysine crotonylation has been observed in myofilm and ribosomal proteins in zebrafish embryos. Biological process analysis revealed that Kcr was mainly involved in metabolic processes (22%) and cellular processes (28%), and the detected Kcr proteins were primarily localized in the cytoplasm (58%) and mitochondria (11%). These findings suggest the possible Kcr sites and modification in human [116]. Furthermore, proteins containing Kcr sites have been detected and predicted to localize in mitochondria [4].
Crotonylation also exists in subcellular organelles and is closely related to cellular physiological functions. Multiple cytoskeletal proteins have Kcr sites, K28/29 site of the TMP1 protein was found to preserve cardiac function and reduce myocardial fibrosis [117]. To detect Kcr in mitochondrial, the K199 site of IDH3a protein was selected because it participates in multiple physiological processes, and regulate mitochondrial function [118] (Fig. 3). The findings elucidate the role of non-histone crotonylation in suborganelles and cell physiological processes, greatly expanding the research scope of crotonylation [72].
5. Conclusion and perspectives
In conclusion, this review has provided a comprehensive overview of the recent research of Kcr, focusing on the role of histone and non-histone crotonylation in various diseases and physiological processes, and the potential for Kcr-targeted treatment. The review also summarizes the subcellular locations of Kcr in several diseases and future directions for the development of novel therapeutic approaches to combat diseases.
Kcr had an important impact on gene expression, aging, DNA damage and repair [[8], [9], [10], [11], [12], [13], [14]]. Despite the significant progress made in the study of Kcr, much remains to be understood about its precise and complete mechanism of Kcr in diverse diseases. Consequently, there is a pressing need for further research in this area, with a particular emphasis on identifying the targeted sites of Kcr, and fully elucidating the physiology of this post-translational modification. Additionally, in-depth research on readers, writers, and erasers is essential to advance our understanding of this critical regulatory mechanism.
The clarification of the Kcr mechanism has the potential to yield practical therapeutic modalities for the treatment of several diseases, including those that are difficult to cure, such as cancer and kidney diseases. Furthermore, the identification of Kcr sites in cytoskeletal proteins and mitochondria indicates that Kcr sites may be the target for the treatment of muscle and mitochondrial diseases. Moreover, Kcr play a crucial role in spindle positioning and stem cell endodermal differentiation, which are the basis of multiple physiological processes and have broad implications for the treatment of many diseases. Therefore, future studies are warranted for developing drugs that can specifically target protein crotonylation to maximize the therapeutic potential of Kcr.
Overall, this review underscores the pivotal role of Kcr in disease and highlights the need for continued research in this field. By deepening our understanding of the mechanisms underlying Kcr, more effective treatments can be developed to combat diseases.
Findings
This work was supported by the National Natural Science Foundation of China under Grant 82003046, the Natural Science Foundation of Chongqing Municipal Science and Technology Bureau under Grant CSTB2022NSCQ-MSX0561 and CSTB2022NSCQ-JQX0010 , the Chongqing Municipal Health Commission under Grant 2024GDRC011, and Chongqing University under Grant 2023CDJYGRH-YB10.
CRediT authorship contribution statement
Dongling Li: Writing – original draft, Visualization. Ling Lin: Writing – original draft. Fan Xu: Data curation. Tianlin Feng: Data curation. Yang Tao: Supervision. Hongming Miao: Funding acquisition. Fan Yang: Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Yang Tao, Email: 36117392@qq.com.
Hongming Miao, Email: hongmingmiao@sina.com.
Fan Yang, Email: yangfanbio@cqu.edu.cn.
Data availability
No data was used for the research described in the article.
References
- 1.Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D., Tang Z., Huang H., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shvedunova M., Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022;23(5):329–349. doi: 10.1038/s41580-021-00441-y. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q., Li Y., Apaliya M.T., et al. The response of Rhodotorula mucilaginosa to patulin cased on lysine crotonylation. Front. Microbiol. 2018;9:2025. doi: 10.3389/fmicb.2018.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y., Xu Q., Liu Y., et al. Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence. Genome Biol. 2018;19(1):144. doi: 10.1186/s13059-018-1533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun J., Qiu C., Qian W., et al. Ammonium triggered the response mechanism of lysine crotonylome in tea plants. BMC Genom. 2019;20(1):340. doi: 10.1186/s12864-019-5716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu K., Yuan C., Li H., et al. A qualitative proteome-wide lysine crotonylation profiling of papaya (Carica papaya L.) Sci. Rep. 2018;8(1):8230. doi: 10.1038/s41598-018-26676-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M., Luo H., Lee S., et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabari B.R., Tang Z., Huang H., et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell. 2015;58(2):203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W., Mao A., Tang B., et al. Large-Scale identification of protein crotonylation reveals its role in multiple cellular functions. J. Proteome Res. 2017;16(4):1743–1752. doi: 10.1021/acs.jproteome.7b00012. [DOI] [PubMed] [Google Scholar]
- 11.Abu-Zhayia E.R., Machour F.E., Ayoub N. HDAC-dependent decrease in histone crotonylation during DNA damage. J. Mol. Cell Biol. 2019;11(9):804–806. doi: 10.1093/jmcb/mjz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu H., Tian C.L., Ye X., et al. Dynamics of telomere rejuvenation during chemical induction to pluripotent stem cells. Stem Cell Rep. 2018;11(1):70–87. doi: 10.1016/j.stemcr.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montellier E., Rousseaux S., Zhao Y., Khochbin S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays. 2012;34(3):187–193. doi: 10.1002/bies.201100141. [DOI] [PubMed] [Google Scholar]
- 14.Sin H.S., Barski A., Zhang F., et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012;26(24):2737–2748. doi: 10.1101/gad.202713.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J.F., Wu S.F., Liu S., et al. Global lysine crotonylation profiling of mouse liver. Proteomics. 2020;20(19–20) doi: 10.1002/pmic.202000049. [DOI] [PubMed] [Google Scholar]
- 16.Hou J.Y., Cao J., Gao L.J., et al. Upregulation of α enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem. Biophys. Res. Commun. 2021;578:77–83. doi: 10.1016/j.bbrc.2021.09.027. [DOI] [PubMed] [Google Scholar]
- 17.Liao P., Bhattarai N., Cao B., et al. Crotonylation at serine 46 impairs p53 activity. Biochem. Biophys. Res. Commun. 2020;524(3):730–735. doi: 10.1016/j.bbrc.2020.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., Li H., Chen S., et al. P300/CBP-associated factor (PCAF) attenuated M1 macrophage inflammatory responses possibly through KLF2 and KLF4. Immunol. Cell Biol. 2021;99(7):724–736. doi: 10.1111/imcb.12455. [DOI] [PubMed] [Google Scholar]
- 19.Sun C.Y., Chang S.C., Wu M.S. Suppression of Klotho expression by protein-bound uremic toxins is associated with increased DNA methyltransferase expression and DNA hypermethylation. Kidney Int. 2012;81(7):640–650. doi: 10.1038/ki.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Chen Y., Zhang Z., et al. Acox2 is a regulator of lysine crotonylation that mediates hepatic metabolic homeostasis in mice. Cell Death Dis. 2022;13(3):279. doi: 10.1038/s41419-022-04725-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H., Bu C., Liu Y., et al. Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair. Sci. Adv. 2020;6(11):eaay4697. doi: 10.1126/sciadv.aay4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Y., Xu X., Ding J., et al. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell. 2021;28(4):748–763 e7. doi: 10.1016/j.stem.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Zeng Y., He X., et al. Folate-deficiency induced acyl-CoA synthetase short-chain family member 2 increases lysine crotonylome involved in neural tube defects. Front. Mol. Neurosci. 2022;15 doi: 10.3389/fnmol.2022.1064509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X., Zhang T., Zeng Y., et al. Sodium butyrate mediates histone crotonylation and alleviated neonatal rats hypoxic-ischemic brain injury through gut-brain axis. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.993146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumgartner M.R., Dantas M.F., Suormala T., et al. Isolated 3-methylcrotonyl-CoA carboxylase deficiency: evidence for an allele-specific dominant negative effect and responsiveness to biotin therapy. Am. J. Hum. Genet. 2004;75(5):790–800. doi: 10.1086/425181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X., Chen X.F., Sun X., et al. Short-chain enoyl-CoA hydratase mediates histone crotonylation and contributes to cardiac homeostasis. Circulation. 2021;143(10):1066–1069. doi: 10.1161/circulationaha.120.049438. [DOI] [PubMed] [Google Scholar]
- 27.Hou J.Y., Cao J., Gao L.J., et al. Upregulation of alpha enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem. Biophys. Res. Commun. 2021;578:77–83. doi: 10.1016/j.bbrc.2021.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Pote N., Cros J., Laouirem S., et al. The histone acetyltransferase hMOF promotes vascular invasion in hepatocellular carcinoma. Liver Int. 2020;40(4):956–967. doi: 10.1111/liv.14381. [DOI] [PubMed] [Google Scholar]
- 29.Xu W., Wan J., Zhan J., et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27(7):946–949. doi: 10.1038/cr.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S., Yu H., Liu Y., et al. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol. Cell. 2017;67(5):853–866 e5. doi: 10.1016/j.molcel.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Kollenstart L., de Groot A.J.L., Janssen G.M.C., et al. Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J. Biol. Chem. 2019;294(52):20122–20134. doi: 10.1074/jbc.RA119.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seto E., Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor Perspect. Biol. 2014;6(4):a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews F.H., Shinsky S.A., Shanle E.K., et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 2016;12(6):396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei W., Liu X., Chen J., et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27(7):898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madsen A.S., Olsen C.A. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew Chem. Int. Ed. Engl. 2012;51(36):9083–9087. doi: 10.1002/anie.201203754. [DOI] [PubMed] [Google Scholar]
- 36.Bao X., Wang Y., Li X., et al. Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3 doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman J.L., Baeza J., Denu J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013;288(43):31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly R.D.W., Chandru A., Watson P.J., et al. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-32927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 40.Flynn E.M., Huang O.W., Poy F., et al. A subset of human Bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure. 2015;23(10):1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Filippakopoulos P., Picaud S., Mangos M., et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhalluin C., Carlson J.E., Zeng L., et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 43.Gowans G.J., Bridgers J.B., Zhang J., et al. Recognition of histone crotonylation by Taf14 links metabolic state to gene expression. Mol. Cell. 2019;76(6):909–921 e3. doi: 10.1016/j.molcel.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao D., Guan H., Zhao S., et al. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016;26(5):629–632. doi: 10.1038/cr.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., Sabari B.R., Panchenko T., et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell. 2016;62(2):181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong X., Panchenko T., Yang S., et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016;12(12):1111–1118. doi: 10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. Ca - Cancer J. Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 48.Wan J., Liu H., Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed. Pharmacother. 2019;111:976–982. doi: 10.1016/j.biopha.2018.12.148. [DOI] [PubMed] [Google Scholar]
- 49.Xu X., Zhu X., Liu F., et al. The effects of histone crotonylation and bromodomain protein 4 on prostate cancer cell lines. Transl. Androl. Urol. 2021;10(2):900–914. doi: 10.21037/tau-21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo, Z., M. Gu, J. Huang, P.-k. Zhou, and T. Ma, 2020. doi:10.1101/2020.06.29.175877.
- 51.Mu N., Wang Y., Li X., et al. Crotonylated BEX2 interacts with NDP52 and enhances mitophagy to modulate chemotherapeutic agent-induced apoptosis in non-small-cell lung cancer cells. Cell Death Dis. 2023;14(9):645. doi: 10.1038/s41419-023-06164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X.Y., Liu Z.X., Zhang Y.F., et al. SEPT2 crotonylation promotes metastasis and recurrence in hepatocellular carcinoma and is associated with poor survival. Cell Biosci. 2023;13(1):63. doi: 10.1186/s13578-023-00996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y., Zhu L., Qin Z.Y., et al. Modulation of cellular metabolism by protein crotonylation regulates pancreatic cancer progression. Cell Rep. 2023;42(7) doi: 10.1016/j.celrep.2023.112666. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., Chen Y., Zhang Z., et al. Acox2 is a regulator of lysine crotonylation that mediates hepatic metabolic homeostasis in mice. Cell Death Dis. 2022;13(3) doi: 10.1038/s41419-022-04725-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lytle N.K., Barber A.G., Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer. 2018;18(11):669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fontecha-Barriuso M., Martin-Sanchez D., Ruiz-Andres O., et al. Targeting epigenetic DNA and histone modifications to treat kidney disease. Nephrol. Dial. Transplant. 2018;33(11):1875–1886. doi: 10.1093/ndt/gfy009. [DOI] [PubMed] [Google Scholar]
- 57.Chen W., Tang D., Xu Y., et al. Comprehensive analysis of lysine crotonylation in proteome of maintenance hemodialysis patients. Medicine. 2018;97(37) doi: 10.1097/md.0000000000012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz A.B., Sanchez-Nino M.D., Ramos A.M., et al. NF-kappaB in renal inflammation. J. Am. Soc. Nephrol. 2010;21(8):1254–1262. doi: 10.1681/ASN.2010020218. [DOI] [PubMed] [Google Scholar]
- 59.Sanz A.B., Sanchez-Nino M.D., Ortiz A. TWEAK, a multifunctional cytokine in kidney injury. Kidney Int. 2011;80(7):708–718. doi: 10.1038/ki.2011.180. [DOI] [PubMed] [Google Scholar]
- 60.Martinez-Moreno J.M., Fontecha-Barriuso M., Martin-Sanchez D., et al. Epigenetic modifiers as potential therapeutic targets in diabetic kidney disease. Int. J. Mol. Sci. 2020;21(11) doi: 10.3390/ijms21114113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz-Andres O., Sanchez-Nino M.D., Cannata-Ortiz P., et al. Histone lysine crotonylation during acute kidney injury in mice. Dis Model Mech. 2016;9(6):633–645. doi: 10.1242/dmm.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cornec-Le Gall E., Alam A., Perrone R.D. Autosomal dominant polycystic kidney disease. Lancet. 2019;393(10174):919–935. doi: 10.1016/s0140-6736(18)32782-x. [DOI] [PubMed] [Google Scholar]
- 63.Lanktree M.B., Haghighi A., Guiard E., et al. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J. Am. Soc. Nephrol. 2018;29(10):2593–2600. doi: 10.1681/ASN.2018050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dang L., Cao X., Zhang T., et al. Nuclear condensation of CDYL links histone crotonylation and cystogenesis in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2022 doi: 10.1681/ASN.2021111425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu Z., Zheng K., Zhang H., et al. Physical exercise and patients with chronic renal failure: a meta-analysis. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/7191826. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Ene-Iordache B., Perico N., Bikbov B., et al. Chronic kidney disease and cardiovascular risk in six regions of the world (ISN-KDDC): a cross-sectional study. Lancet Global Health. 2016;4(5):e307–e319. doi: 10.1016/S2214-109X(16)00071-1. [DOI] [PubMed] [Google Scholar]
- 67.Hassler J.R. IgA nephropathy: a brief review. Semin. Diagn. Pathol. 2020;37(3):143–147. doi: 10.1053/j.semdp.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Lin H., Tang D., Xu Y., et al. Quantitative analysis of protein crotonylation identifies its association with immunoglobulin A nephropathy. Mol. Med. Rep. 2020;21(3):1242–1250. doi: 10.3892/mmr.2020.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J., Tang D., Zheng F., Xu H., Dai Y. Comprehensive analysis of lysine crotonylation modification in patients with chronic renal failure. BMC Nephrol. 2021;22(1):310. doi: 10.1186/s12882-021-02445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Virani S.S., Alonso A., Benjamin E.J., et al. Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. 2020;141(9):e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 71.Severino P., D'Amato A., Pucci M., et al. Ischemic heart disease pathophysiology paradigms overview: from plaque activation to microvascular dysfunction. Int. J. Mol. Sci. 2020;21(21) doi: 10.3390/ijms21218118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai W., Xu D., Zeng C., et al. Modulating lysine crotonylation in cardiomyocytes improves myocardial outcomes. Circ. Res. 2022 doi: 10.1161/CIRCRESAHA.122.321054. [DOI] [PubMed] [Google Scholar]
- 73.Ganetzky R., Stojinski C. In: GeneReviews((R)) Adam M.P., et al., editors. 1993. Mitochondrial short-chain enoyl-CoA hydratase 1 deficiency. Seattle (WA) [PubMed] [Google Scholar]
- 74.Ahmadmehrabi S., Tang W.H.W. Hemodialysis-induced cardiovascular disease. Semin. Dial. 2018;31(3):258–267. doi: 10.1111/sdi.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lacolley P., Regnault V., Segers P., Laurent S. Vascular smooth muscle cells and arterial stiffening: relevance in development, aging, and disease. Physiol. Rev. 2017;97(4):1555–1617. doi: 10.1152/physrev.00003.2017. [DOI] [PubMed] [Google Scholar]
- 76.Petsophonsakul P., Furmanik M., Forsythe R., et al. Role of vascular smooth muscle cell phenotypic switching and calcification in aortic aneurysm formation. Arterioscler. Thromb. Vasc. Biol. 2019;39(7):1351–1368. doi: 10.1161/ATVBAHA.119.312787. [DOI] [PubMed] [Google Scholar]
- 77.Cao S.H., Chen Z.H., Ma R.Y., et al. Dynamics and functional interplay of nonhistone lysine crotonylome and ubiquitylome in vascular smooth muscle cell phenotypic remodeling. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.783739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braak E., Griffing K., Arai K., et al. Neuropathology of Alzheimer's disease: what is new since A. Alzheimer? Eur. Arch. Psychiatr. Clin. Neurosci. 1999;249(Suppl 3):14–22. doi: 10.1007/pl00014168. [DOI] [PubMed] [Google Scholar]
- 79.Yamazaki T., Souquere S., Chujo T., et al. Functional domains of NEAT1 architectural lncRNA induce Paraspeckle assembly through phase separation. Mol. Cell. 2018;70(6):1038–1053 e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 80.Clemson C.M., Hutchinson J.N., Sara S.A., et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z., Zhao Y., Xu N., et al. NEAT1 regulates neuroglial cell mediating Abeta clearance via the epigenetic regulation of endocytosis-related genes expression. Cell. Mol. Life Sci. 2019;76(15):3005–3018. doi: 10.1007/s00018-019-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith K. Mental health: a world of depression. Nature. 2014;515(7526):181. doi: 10.1038/515180a. [DOI] [PubMed] [Google Scholar]
- 83.Wang M., Chang Q., Yang H., et al. Elevated lysine crotonylation and succinylation in the brains of BTBR mice. Int. J. Dev. Neurosci. 2019;76:61–64. doi: 10.1016/j.ijdevneu.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 84.Wang M., Chang Q., Yang H., et al. Elevated lysine crotonylation and succinylation in the brains of BTBR mice. Int. J. Dev. Neurosci. 2019;76(1):61–64. doi: 10.1016/j.ijdevneu.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 85.Liu Y., Li M., Fan M., et al. Chromodomain Y-like protein-mediated histone crotonylation regulates stress-induced depressive behaviors. Biol. Psychiatr. 2019;85(8):635–649. doi: 10.1016/j.biopsych.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 86.Jiang C., Lin W.J., Sadahiro M., et al. VGF function in depression and antidepressant efficacy. Mol. Psychiatr. 2018;23(7):1632–1642. doi: 10.1038/mp.2017.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zou Y., Bai X.H., Kong L.C., et al. Involvement of histone lysine crotonylation in the regulation of nerve-injury-induced neuropathic pain. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.885685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verma M. Epigenetic regulation of HIV, AIDS, and AIDS-related malignancies. Cancer Epigenetics. 2015:381–403. doi: 10.1007/978-1-4939-1804-1_21. [DOI] [PubMed] [Google Scholar]
- 89.Jiang G., Nguyen D., Archin N.M., et al. HIV latency is reversed by ACSS2-driven histone crotonylation. J. Clin. Invest. 2018;128(3):1190–1198. doi: 10.1172/JCI98071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 91.Kuprash D.V., Nedospasov S.A. Molecular and cellular mechanisms of inflammation. Biochemistry (Mosc.) 2016;81(11):1237–1239. doi: 10.1134/S0006297916110018. [DOI] [PubMed] [Google Scholar]
- 92.Stewart A.G., Beart P.M. Inflammation: maladies, models, mechanisms and molecules. Br. J. Pharmacol. 2016;173(4):631–634. doi: 10.1111/bph.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen J., Abu-Amer Y., O'Keefe R.J., McAlinden A. Inflammation and epigenetic regulation in osteoarthritis. Connect. Tissue Res. 2017;58(1):49–63. doi: 10.1080/03008207.2016.1208655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song X., Yang F., Liu X., et al. Dynamic crotonylation of EB1 by TIP60 ensures accurate spindle positioning in mitosis. Nat. Chem. Biol. 2021;17(12):1314–1323. doi: 10.1038/s41589-021-00875-7. [DOI] [PubMed] [Google Scholar]
- 95.McNally F.J. Mechanisms of spindle positioning. J. Cell Biol. 2013;200(2):131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheeseman I.M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9(1):33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 97.Yao X., Abrieu A., Zheng Y., Sullivan K.F., Cleveland D.W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2(8):484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 98.Vilarinho S., Sari S., Mazzacuva F., et al. ACOX2 deficiency: a disorder of bile acid synthesis with transaminase elevation, liver fibrosis, ataxia, and cognitive impairment. Proc. Natl. Acad. Sci. U.S.A. 2016;113(40):11289–11293. doi: 10.1073/pnas.1613228113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferdinandusse S., Denis S., van Roermund C.W.T., et al. A novel case of ACOX2 deficiency leads to recognition of a third human peroxisomal acyl-CoA oxidase. Biochim. Biophys. Acta, Mol. Basis Dis. 2018;1864(3):952–958. doi: 10.1016/j.bbadis.2017.12.032. [DOI] [PubMed] [Google Scholar]
- 100.Zhang Q., Zhang Y., Sun S., et al. ACOX2 is a prognostic marker and impedes the progression of hepatocellular carcinoma via PPARalpha pathway. Cell Death Dis. 2021;12(1):15. doi: 10.1038/s41419-020-03291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou X., Wang H. ACOX2 deficiency in primary malignant cardiac tumors. Proc. Natl. Acad. Sci. U.S.A. 2017;114(18):E3590–E3591. doi: 10.1073/pnas.1701212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou X., Xu M., Zeng W., et al. Combined effects of FH (E404D) and ACOX2 (R409H) cause metabolic defects in primary cardiac malignant tumor. Cell Death Dis. 2018;4:18. doi: 10.1038/s41420-018-0072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fang Y., Li X. A simple, efficient, and reliable endoderm differentiation protocol for human embryonic stem cells using crotonate. STAR Protoc. 2021;2(3) doi: 10.1016/j.xpro.2021.100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhala A., Willi S.M., Rinaldo P., et al. Clinical and biochemical characterization of short-chain acyl-coenzyme A dehydrogenase deficiency. J. Pediatr. 1995;126(6):910–915. doi: 10.1016/s0022-3476(95)70207-5. [DOI] [PubMed] [Google Scholar]
- 105.Corydon M.J., Vockley J., Rinaldo P., et al. Role of common gene variations in the molecular pathogenesis of short-chain acyl-CoA dehydrogenase deficiency. Pediatr. Res. 2001;49(1):18–23. doi: 10.1203/00006450-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 106.Kim S.H., Park H.D., Sohn Y.B., et al. Mutations of ACADS gene associated with short-chain acyl-coenzyme A dehydrogenase deficiency. Ann. Clin. Lab. Sci. 2011;41(1):84–88. [PubMed] [Google Scholar]
- 107.Yiangou L., Ross A.D.B., Goh K.J., Vallier L. Human pluripotent stem cell-derived endoderm for modeling development and clinical applications. Cell Stem Cell. 2018;22(4):485–499. doi: 10.1016/j.stem.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 108.Masuda S., Wu J., Hishida T., et al. Chemically induced pluripotent stem cells (CiPSCs): a transgene-free approach. J. Mol. Cell Biol. 2013;5(5):354–355. doi: 10.1093/jmcb/mjt034. [DOI] [PubMed] [Google Scholar]
- 109.Zheng Z., Yan G., Li X., et al. Lysine crotonylation regulates leucine-deprivation-induced autophagy by a 14-3-3epsilon-PPM1B axis. Cell Rep. 2022;41(12) doi: 10.1016/j.celrep.2022.111850. [DOI] [PubMed] [Google Scholar]
- 110.Kao H.Y., Verdel A., Tsai C.C., et al. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 2001;276(50):47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 111.Li X., Song S., Liu Y., Ko S.H., Kao H.Y. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 2004;279(33):34201–34208. doi: 10.1074/jbc.M405179200. [DOI] [PubMed] [Google Scholar]
- 112.Dequiedt F., Martin M., Von Blume J., et al. New role for hPar-1 kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases. Mol. Cell Biol. 2006;26(19):7086–7102. doi: 10.1128/MCB.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Szwed A., Kim E., Jacinto E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021;101(3):1371–1426. doi: 10.1152/physrev.00026.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan G., Li X., Zheng Z., et al. KAT7-mediated CANX (calnexin) crotonylation regulates leucine-stimulated MTORC1 activity. Autophagy. 2022;18(12):2799–2816. doi: 10.1080/15548627.2022.2047481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schaub M.C., Hefti M.A., Zuellig R.A., Morano I. Modulation of contractility in human cardiac hypertrophy by myosin essential light chain isoforms. Cardiovasc. Res. 1998;37(2):381–404. doi: 10.1016/s0008-6363(97)00258-7. [DOI] [PubMed] [Google Scholar]
- 116.Kwon O.K., Kim S.J., Lee S. First profiling of lysine crotonylation of myofilament proteins and ribosomal proteins in zebrafish embryos. Sci. Rep. 2018;8(1):3652. doi: 10.1038/s41598-018-22069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma S., Xu Q., Bai R., et al. Generation of a TPM1 homozygous knockout embryonic stem cell line by CRISPR/Cas9 editing. Stem Cell Res. 2021;55 doi: 10.1016/j.scr.2021.102470. [DOI] [PubMed] [Google Scholar]
- 118.Liu X., Si W., He L., et al. The existence of a nonclassical TCA cycle in the nucleus that wires the metabolic-epigenetic circuitry. Signal Transduct. Targeted Ther. 2021;6(1):375. doi: 10.1038/s41392-021-00774-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kelly A., Trowsdale J. Genetics of antigen processing and presentation. Immunogenetics. 2019;71(3):161–170. doi: 10.1007/s00251-018-1082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Silverstein R.L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009;2(72):re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gan Q., Tang D., Yan Q., et al. Differential expression study of lysine crotonylation and proteome for chronic obstructive pulmonary disease combined with type II respiratory failure. Cancer Res. J. 2021;2021 doi: 10.1155/2021/6652297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wan L., Chong S., Xuan F., et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature. 2020;577(7788):121–126. doi: 10.1038/s41586-019-1842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen M.Y., Liao W.S., Lu Z., et al. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer. 2011;117(19):4424–4438. doi: 10.1002/cncr.26073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.