Abstract
The human immunodeficiency virus type 1 (HIV-1) laboratory strains adapted to T-cell lines, as well as most syncytium-inducing primary isolates, replicate poorly in macrophages, which, beside CD4+ T lymphocytes, are major targets of HIV-1. In the present work, we used a semiquantitative PCR-based technique to study viral entry into cells, kinetics of reverse transcription, and translocation of the viral DNA into the nucleus of macrophages infected with different HIV-1 strains. Our results demonstrate that T-lymphotropic strains efficiently enter macrophages. Entry was inhibited by a monoclonal antibody against CD4 and by stromal cell-derived factor 1α, a natural ligand of CXCR4, suggesting that both CD4 and CXCR4 act as receptors on macrophages for HIV-1 T-lymphotropic strains. Analysis of the kinetics of reverse transcription and nuclear import revealed that the most pronounced differences between T-lymphotropic and macrophagetropic strains occurred at the level of nuclear translocation of viral DNA, although a delay in reverse transcription was also observed. These results suggest that postentry steps are critical for restricted replication of T-lymphotropic HIV-1 strains in macrophages.
Human immunodeficiency virus type 1 (HIV-1) is characterized by a high degree of genetic variability, resulting in differences in biological properties such as replicative rate, syncytium-inducing capacity, and preferential infection of specific target cells (3, 12, 28, 58). Beside CD4+ T lymphocytes, macrophages are the major targets of HIV-1. Although most primary isolates can infect cells of both types (59, 64), there is a clear strain-specific preference toward one or the other target, which correlates with the clinical outcome of HIV-1 infection (39, 64). Viruses isolated during primary infection have a predominantly macrophagetropic and non-syncytium-inducing phenotype (60). During the period between the initial infection and the full-blown disease, a shift from macrophage tropism to T-cell tropism, associated with the emergence of syncytium-inducing viruses, has been observed in serial peripheral blood virus isolates (17, 54, 64). A similar change in tropism can be seen during laboratory adaptation of primary isolates to transformed T-cell lines. Viruses adapted to T-cell lines can still infect primary T lymphocytes but lose the ability to replicate efficiently in macrophages. Since biological diversity plays an important role in the pathogenesis of HIV-1 infection, numerous genetic studies have been directed toward characterization of viral determinants responsible for selective tropism. A specific region of the envelope gp120 protein, the V3 loop, was demonstrated to be a main determinant of HIV-1 tropism (34, 44, 55), suggesting that the major block to HIV-1 replication in macrophages was at the step of virus entry. This hypothesis was further supported by demonstration of the correlation between the fusion capacity of the envelope glycoprotein and the tropism of different HIV-1 strains (6). However, other investigators arrived at different conclusions. A measurement of fluorescence dequenching of virus-cell fusion indicated that T-lymphotropic viruses fuse efficiently with primary macrophages, suggesting that a block at a postentry step in the viral life cycle was responsible for restricted replication of these strains in macrophages (49). Other reports supported this conclusion, demonstrating an efficient synthesis of HIV-1 DNA in macrophages infected with T-lymphotropic strains (33, 53).
The CD4 glycoprotein is the major receptor for HIV-1 on T lymphocytes and monocytes/macrophages (16, 18, 35). Several studies indicated that entry of HIV-1 into target cells requires additional cell cofactors besides CD4 (7, 14). Members of the seven-transmembrane-domain G-protein-coupled receptors have been recently identified as such cofactors. An α-chemokine receptor CXCR4 was shown to act as a coreceptor for T-lymphotropic strains (27). The natural ligand for this receptor was later identified as stromal cell-derived factor 1 (SDF1) (5, 46). Subsequently, several groups identified CCR5, a member of the β-chemokine receptor family, as a coreceptor for macrophage-tropic viruses (2, 13, 19, 21, 22). These findings suggested that cell tropism of HIV-1 may be determined by differential expression of chemokine receptors on target cells. However, this simple model was questioned in recent studies, where expression of CXCR4 mRNA was detected in primary macrophages refractory to infection by T-lymphotropic viruses (38, 41).
Here, we used a semiquantitative PCR-based technique to determine the critical step at which replication of HIV-1 T-lymphotropic strains is restricted in primary macrophages. Our results demonstrate that these viruses enter efficiently macrophages, using CD4 and CXCR4 as coreceptors. According to our results, replication is restricted at a postentry level, with the most pronounced defect observed at the level of nuclear import of viral DNA.
MATERIALS AND METHODS
Isolation and culture of human macrophages and lymphocytes.
Peripheral blood mononuclear cells from healthy donors undergoing leukopheresis were separated on a Ficoll-Hypaque (Pharmacia) gradient. Suspensions of 8 × 106 cells/ml, prepared in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated human serum, were allowed to adhere to plastic for 2 h at 37°C. Afterwards, T-lymphocyte and macrophage cultures were prepared as follows. For T-lymphocyte cultures, nonadherent cells were transferred to new flasks and subjected to a second round of adherence to eliminate contaminating monocytes. On the next day, nonadherent cells were collected by centrifugation, resuspended in RPMI 1600 medium supplemented with 10% heat-inactivated fetal calf serum, and stimulated with 5 μg of phytohemagglutinin per ml for 3 days. For macrophage cultures, adherent cells were washed extensively, and medium containing 2 ng of human macrophage colony-stimulating factor (MCSF; Sigma) per ml was added to the cultures. After 24 h, adherent monocytes were treated with 10 mM EDTA, washed, detached, and seeded into 24-well plates (PRIMARIA; Falcon) at the concentration of 106 cells/ml. Cells were allowed to differentiate for 7 days in the presence of MCSF. At this time, 99.5 to 99.8% of the cells were macrophages, as determined by cytochemical staining for nonspecific esterase (Alpha-Naphthyl Acetate Esterase kit; Sigma).
Viruses and infection.
Three HIV-1 strains with different cell tropism were used in this study: macrophagetropic strain ADA (30); and two T-lymphotropic strains, LAI (4) and NDK (25). The stocks of HIV-1 LAI and HIV-1 NDK were prepared in primary lymphocytes, and the stock of HIV-1 ADA was prepared in primary macrophages. Aliquoted viruses were stored at −70°C. Before infection, viral stocks were treated with 200 U of RNase-free DNase per ml (1 h at room temperature) to eliminate contamination with viral DNA. Seven days after isolation, macrophages were infected for 2 h at 37°C with an amount of virus corresponding to 1.5 × 105 cpm of reverse transcriptase (RT) activity per 106 cells. After viral adsorption and washing, cells were cultivated in medium without MCSF.
RT and p24 antigen assays.
RT activity was measured by using a standard RT assay (51). The p24 antigen capture enzyme-linked immunosorbent assay (ELISA) was performed as instructed by the manufacturer (DuPont).
Detection of HIV-1-specific DNA by PCR.
Cells growing in 24-well plates (106 cells/well) were lysed with 200 μl of PCR buffer, consisting of 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.1 mg of gelatin per ml, 0.45% Nonidet P-40, 0.45% Tween 20, and 100 μg of proteinase K per ml (32). After protein digestion (2 h at 56°C) and inactivation of the proteinase (10 min at 95°C), 25 μl of cell lysate was subjected to 25 cycles (α-tubulin primers) or 35 cycles (HIV-1-specific primers) of PCR in a total volume of 50 μl containing 0.2 μM oligonucleotide primers, 200 μM each deoxynucleotide, 50 mM KCl, 10 mM Tris, pH 8.3, 2 mM MgCl2, and 1.25 U of Taq DNA polymerase (Perkin-Elmer). Each cycle comprised a 30-s denaturation step (94°C), a 30-s annealing step (Tm − 5°C for each pair of primers), and a 1-min extension (72°C). After agarose gel electrophoresis, amplified DNA was analyzed by Southern blot hybridization with the 32P-labeled probe. Amplified fragments of the correct size were quantified with an Instant Imager (Packard) and expressed as counts per minute. The following primers and probes were used in the study (numbering of nucleotide positions corresponds to that for the HIV-1 HXB-2 DNA sequence [50]): for LTR (long terminal repeat) R/U5, sense primer (5′-GGCTAACTAGGGAACCCACTG-3′; nucleotides 496 to 517), antisense primer (5′-CTGCTAGAGATTTTCCACACTGAC-3′; nucleotides 612 to 635), and probe (5′-TGTGTGCCCGTCTGTTGTGTG-3′; nucleotides 557 to 577); for LTR U3/R, sense primer (5′-CAGATATCCACTGACCTTTGG-3′; nucleotides 110 to 130), antisense primer (5′-GAGGCTTAAGCAGTGGGTTC-3′; nucleotides 507 to 526), and probe (5′-AAGCTAGTACCAGTTGAGCC-3′; nucleotides 141 to 160); for pol, sense primer (5′-TTCTTCAGAGCAGACCAG-3′; nucleotides 2131 to 2149), antisense primer (5′-ACTTTTGGGCCATCCATT-3′; nucleotides 2592 to 2610), and probe (5′-GGAAGCTCTATTAGATACAGG-3′; nucleotides 2311 to 2331); for LTR/gag, sense primer (LTR U3 as showed above), antisense primer (5′-GCTTAATACTGACGCTCTCGCA-3′; nucleotides 794 to 815), and probe (LTRU3/R antisense primer); for two-LTR (2LTR) DNA, sense primer (5′-GCCTCAATAAAGCTTGCCTTG-3′; nucleotides 522 to 542), antisense primer (5′-TCCCAGGCTCAGATCTGGTCTAAC-3′; nucleotides 465 to 488), and probe (330-bp MroI/NarI fragment of plasmid MJ2 containing HIV-1 NL4-3 nucleotide sequence [1]); for α-tubulin, sense primer (5′-GTTGGTCTGGAATTCTGTCAG-3′; cDNA nucleotides 489 to 507) and antisense primer (5′-AAGAAGTCCAAGCTGGAGTTC-3′; cDNA nucleotides 756 to 777).
PCR in situ hybridization analysis of HIV-1-specific DNA.
HIV-1 DNA was amplified and detected according to a previously published protocol (42, 43).
Flow cytometric analysis.
Seven-day-old macrophages were detached by treatment with cold 10 mM EDTA and washed, and suspensions of 5 × 105 cells were preincubated with 20% normal human serum in phosphate-buffered saline for 20 min at room temperature. Cells were then washed and colabeled with anti-CXCR4 monoclonal antibody (MAb) 12G5, directly labeled with phycoerythrin (PE; Pharmingen), and anti-CD14 MAbs, directly labeled with fluorescein isothiocyanate (FITC; Becton Dickinson), or with anti-CD3 MAbs, directly labeled with peridin chlorophyll protein (PerCP; Becton Dickinson), and anti-CD14 MAbs-FITC for 30 min at room temperature in a 100-μl volume containing 0.5 μg of each antibody. As a control, mouse isotype antibodies, immunoglobulin G2a (IgG2a)-PE, IgG1-PerCP, and IgG2b-FITC, were used. After washing, cells were resuspended in 2% formaldehyde in phosphate-buffered saline and analyzed on FACS 440 (Becton Dickinson).
Calcium mobilization assay.
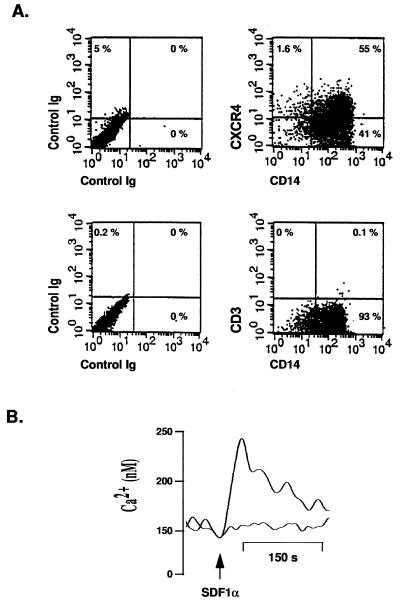
Detached macrophages were washed and incubated for 30 min at 37°C with 5 μM Fura-2/AM (Calbiochem) at the concentration of 107 cells/ml of Hanks’ balanced salt solution (HBSS; 2 mM CaCl2, 145 mM NaCl, 5 mM KCl, 1 mM MgSO4, 10 mM d-glucose, 10 mM HEPES) containing 1% fetal calf serum. After the initial loading, cells were diluted with HBSS without CaCl2 and incubated for additional 10 min at 37°C. Afterwards, cells were resuspended in serum-free HBSS prewarmed at 37°C for at least 20 min, and 3 × 106 cells were added to a stirred cuvette in a fluorimeter (LS50B; Perkin-Elmer) and stimulated with 1 μg of recombinant human SDF1α (PeproTech Inc.). Fluorescence was measured at 500-nm emission wavelength, following excitation at both 340 and 380 nm. Final intracellular Ca2+ levels were calculated from the 340/380-nm ratio according to the standard equation, with a dissociation constant of 224 nM for Fura-2.
RESULTS
T-lymphotropic strains infect primary human macrophages.
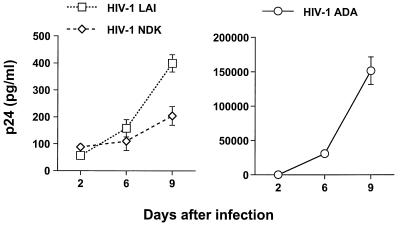
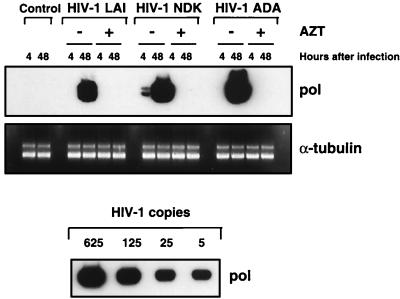
Infection of primary macrophages with HIV-1 T-lymphotropic viruses does not yield efficient viral replication. In cultures infected with T-lymphotropic strains, we never observed the RT production or formation of multinucleated giant cells, which are easily detected in the cells infected with macrophagetropic viruses. However, low levels of p24 antigen were detected in the supernatants of these cultures (Fig. 1). Since previous studies of HIV-1 macrophage tropism yielded conflicting results with regard to the ability of T-lymphotropic strains to enter and undergo reverse transcription in primary macrophages (6, 33, 53), we wished to confirm that T-lymphotropic viruses can infect primary macrophages. First, we analyzed the synthesis of HIV-1-specific DNA in macrophages infected with a macrophagetropic strain, HIV-1 ADA, or two T-lymphotropic viruses, HIV-1 LAI and HIV-1 NDK. To control for possible contamination of the viral inoculum with viral DNA, a parallel infection was carried out in the presence of 10 μM zidovudine (AZT), an inhibitor of reverse transcription. Infection with all three viruses yielded similar results, showing a positive PCR signal in samples collected 48 h after infection and cultivated without AZT (Fig. 2). This result suggests that T-lymphotropic viruses can enter and initiate reverse transcription in primary macrophages.
FIG. 1.
Replication of different HIV-1 strains in primary macrophages. Seven-day-old macrophages were infected with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA. At days 2, 6, and 9 after infection, supernatants were collected and assayed for p24 antigen, using an ELISA kit (DuPont). Results are expressed as means ± standard deviations.
FIG. 2.
De novo synthesis of HIV-1-specific DNA in primary macrophages. Macrophages were infected with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA in the presence or absence of 10 μM AZT for 2 h at 37°C. PCR lysates were prepared at 4 and 48 h after infection, and HIV-1-specific DNA was amplified by using pol gene primers. PCR analysis of the α-tubulin gene in cell lysates was used to standardize DNA recovery. Control amplification was performed in lysates prepared from uninfected macrophages. Different dilutions of 8E5/LAI cells, containing one HIV-1 genome per cell, were used as PCR standards.
T-lymphotropic strains enter macrophages via a CD4-dependent mechanism.
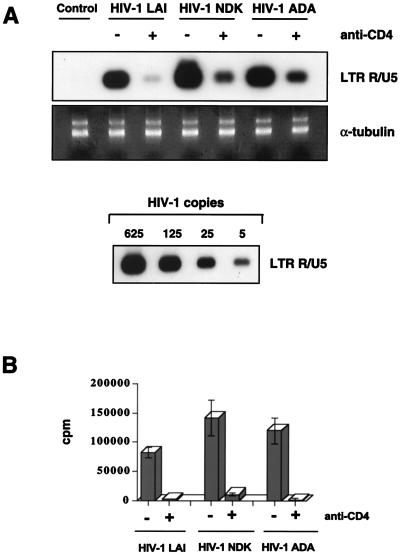
The CD4 glycoprotein is the major receptor for HIV-1 on T lymphocytes and macrophages: CD4-dependent entry into macrophages has been shown for a variety of macrophagetropic strains of HIV-1 (15, 16). Due to inefficient replication of HIV-1 T-lymphotropic viruses in macrophages, little is known about the dependence of their entry on specific surface receptors, although infection of macrophages by simian immunodeficiency virus (SIV) T-lymphotropic strains has been shown to depend on CD4 (40). To determine if CD4 acts as a receptor on macrophages for HIV-1 T-lymphotropic strains, we infected cultured macrophages with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA in the presence of a MAb against CD4 (Leu3a; Becton Dickinson). After infection, cells were lysed and analyzed for the presence of strong-stop DNA, the early product of reverse transcription synthesized shortly after viral entry (see Fig. 6). Anti-CD4 antibody inhibited production of strong-stop DNA in cultures infected with any of the three viruses (Fig. 3A). The magnitudes of inhibition were similar for all strains (Fig. 3B), indicating that the CD4 receptor plays the major role in the entry of T-lymphotropic viruses into primary macrophages.
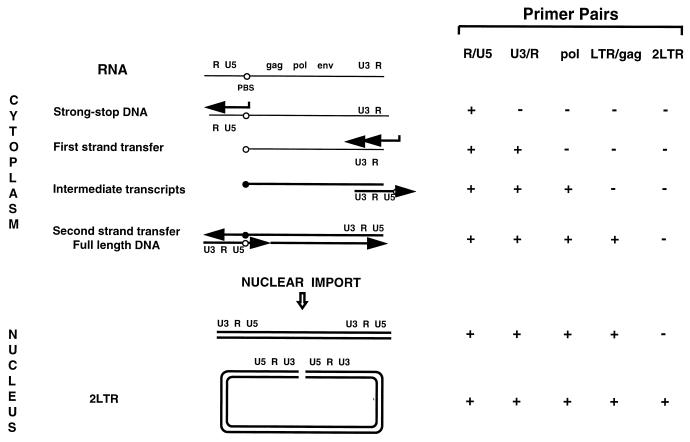
FIG. 6.
Schematic model of retroviral reverse transcription, adapted from reference 8. (A) Steps in viral DNA (thick lines) synthesis following entry and liberation of viral RNA (thin lines) into the cytoplasm of target cells. (B) Reactivity (+) of oligonucleotide primers used in this study. Reverse transcription is initiated by binding of the tRNA primer to the primer binding site (PBS; open circle) and synthesis of a short transcript through the U5/R region at the 5′ end of the RNA. The R/U5 transcript, known also as strong-stop DNA, undergoes the first template switch and anneals to the R sequence at the 3′ end of the viral RNA. Synthesis of minus-strand DNA continues through the PBS region and forms complementary PBS (closed circle). After the second template switch, two complementary PBSs hybridize and the full-length DNA is synthesized. After import of the synthesized viral DNA into the nucleus, some linear DNA undergoes circularization, yielding 2LTR circle forms.
FIG. 3.
Infection of macrophages is CD4 dependent. (A) Cells were pretreated with 5 μg of anti-CD4 MAb (Leu3a) per ml for 30 min at room temperature. Treated and control cultures were infected with macrophagetropic HIV-1 ADA and T-lymphotropic HIV-1 LAI and HIV-1 NDK for 2 h at 37°C. In treated cultures, anti-CD4 antibody was present also during infection. Strong-stop DNA was amplified by using HIV-1-specific primers from the LTR R/U5 region. Amplification of the α-tubulin gene was used to control the amount of DNA in each sample. Dilutions of 8E5/LAI cells were used as standards. (B) Quantification of PCR products. After hybridization with the 32P-labeled probe, the PCR bands were quantified by using an Instant Imager (Packard). Data show results of one representative experiment of three, each performed in duplicate. Standard deviations are shown as vertical bars.
Expression of CXCR4 on primary macrophages.
Expression of mRNA for CXCR4, the main coreceptor for HIV-1 T-lymphotropic strains on T lymphocytes, has been demonstrated in primary macrophages (38, 41). Since surface expression of this molecule can be influenced by macrophage cultivation conditions (20), we assayed expression of CXCR4 in our macrophage cultures by flow cytometry. Results presented in the Fig. 4A demonstrate that CXCR4 is expressed on CD14-positive cells. More than 50% of macrophages expressed this coreceptor for T-lymphotropic viruses. Also, colabeling with a anti-CD3 antibody did not show contamination of these cultures with T lymphocytes (Fig. 4A). To examine the function of CXCR4 on macrophages, we assayed the ability of SDF1α, a natural ligand of CXCR4, to induce Ca2+ mobilization in primary macrophages. Increased intracellular Ca2+ levels in SDF1α-treated macrophages (Fig. 4B) indicated that CXCR4 initiated signal transduction. Preincubation of cells with MAb 12G5, which specifically binds to CXCR4 (26), inhibited the SDF1-mediated increase in intracellular calcium (Fig. 4B). We conclude from these results that CXCR4 is expressed on macrophages in a functional form.
FIG. 4.
Expression of CXCR4 on primary macrophages. (A) Seven-day-old macrophages were stained with 12G5-PE and CD14-FITC antibodies (upper right) or CD14-FITC and CD3-PerCP antibodies (lower right) and analyzed by a two-color flow cytometry. PE-, FITC-, and PerCP-conjugated isotypes were used as controls (left). (B) At day 7 after isolation, macrophages loaded with Fura-2/AM were stimulated with 1 μg of SDF1α at the time indicated by the arrow. Ca2+-dependent fluorescence changes were recorded, and final intracellular Ca2+ levels were calculated as described in Materials and Methods. The lower curve represents results obtained after SDF1α stimulation of cells preincubated with MAb 12G5 for 15 min before stimulation; the upper curve represents results obtained after stimulation of untreated cells.
Entry of T-lymphotropic strains into macrophages is inhibited by SDF1α, a natural ligand of the CXCR4 receptor.
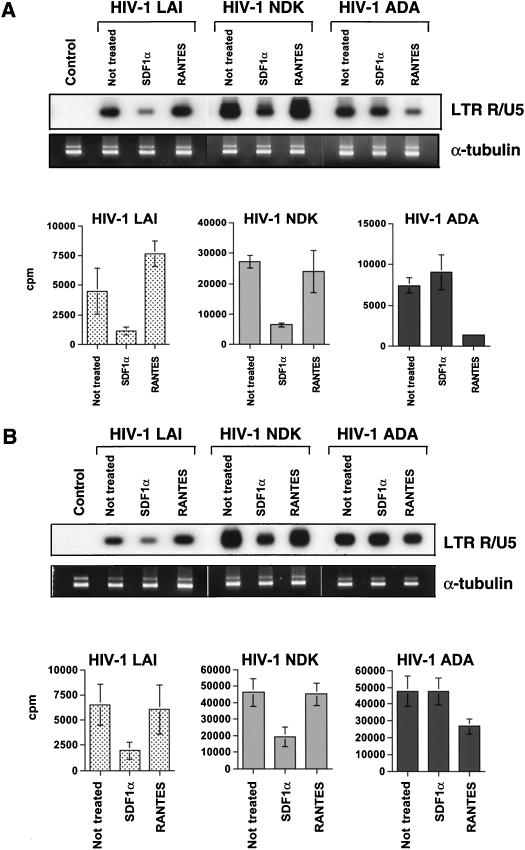
Next, we wished to know if CXCR4 acts as a coreceptor for T-lymphotropic viruses on macrophages. To address this question, we pretreated cells either with SDF1α or with RANTES (PeproTech Inc.), a natural ligand of CCR5. Treated and untreated cells were then infected with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA. Two hours after infection, cells were lysed, and viral DNA was amplified by using primers specific for the LTR R/U5 region (see Fig. 6). As expected, RANTES did not inhibit strong-stop DNA synthesis in lymphocytes (Fig. 5A) or macrophages (Fig. 5B) infected with T-lymphotropic strains. In contrast, SDF1α inhibited entry of HIV-1 LAI and HIV-1 NDK into both lymphocytes and macrophages, indicating that CXCR4 is used as a coreceptor for these viruses in both cell types. The same degree of macrophage infection inhibition by SDF1α was observed with another T-lymphotropic strain, HIV-1 RF (data not shown). RANTES strongly inhibited entry of HIV-1 ADA into lymphocytes but was much less effective against entry into primary macrophages (Fig. 5). In addition, in long-term macrophage cultures, the partial inhibitory effect of RANTES on HIV-1 ADA entry was overcome: 14 days after infection, RT levels in the treated cultures were even slightly higher than those in controls (data not shown and reference 52).
FIG. 5.
Effects of chemokines on HIV-1 entry. Primary lymphocytes (A) and macrophages (B) were pretreated with 200 ng of SDF1α or RANTES per ml for 1 h at 37°C. Treated and control cultures were infected with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA and analyzed by PCR using primers from the LTR R/U5 region. DNA recovery was controlled by PCR with α-tubulin-specific primers. The control lane shows PCR amplification of lysates prepared from uninfected cells. Bottom panels show quantification of LTR R/U5-amplified products. Data represent averages of four experimental samples (obtained in two independent experiments, performed with cells from the same donor) ± standard deviations.
Analysis of reverse transcription in primary macrophages infected with T-lymphotropic and macrophagetropic strains of HIV-1.
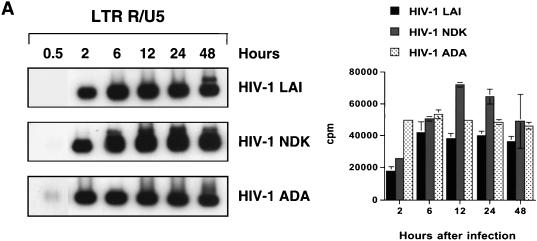
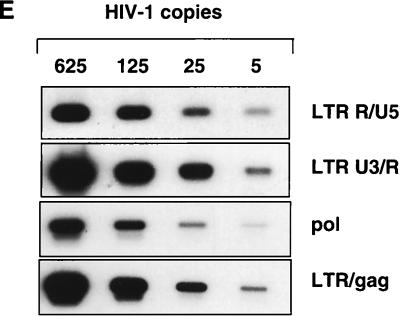
We next analyzed the sequential steps of reverse transcription in macrophages infected with different HIV-1 strains. Since the accumulation of full-length viral DNA in HIV-1-infected macrophages reaches a peak between 36 and 48 h after infection (45), we collected samples at 0.5, 2, 6, 12, 24, and 48 h after inoculation. Each cell lysate was analyzed by PCR, using primers (Fig. 6) designed according to a generally accepted model of retroviral reverse transcription (61). To evaluate the sensitivity of the chosen primer pairs, different dilutions of lysate prepared from 8E5/LAI cells, containing one proviral copy per cell, were amplified in parallel (Fig. 7E). The detection limit of all oligonucleotide primers ranged from two to five HIV-1 copies. The amount of total DNA was controlled by amplifying the cellular α-tubulin gene (data not shown).
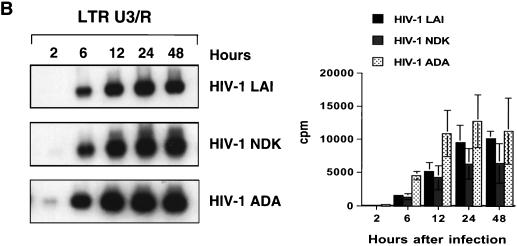
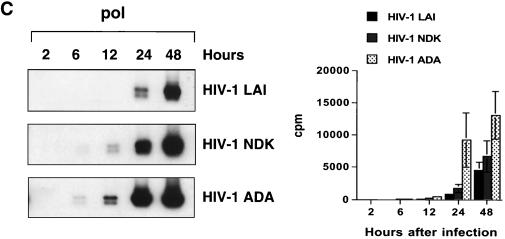
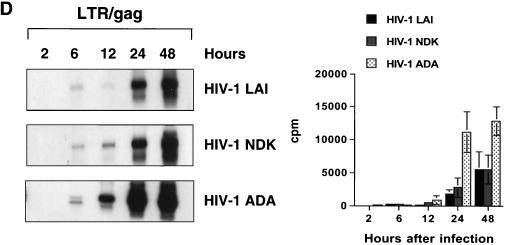
FIG. 7.
Kinetics of HIV-1 reverse transcription in primary macrophages. Macrophages were infected with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA. PCR lysates were prepared 0.5, 2, 6, 12, 24, and 48 h after infection. Each lysate was subjected to PCR amplification using HIV-1-specific primers from the LTR R/U5 region (A), LTR U3/R region (B), pol gene (C), and LTR/gag region (D). Amplified products were quantified after hybridization with the 32P-labeled probe by an Instant Imager (Packard), and results were expressed as counts per minute. Panels on the left show representative results from one of two independent experiments. Panels on the right represent data averaged from duplicates obtained after quantification of the same experiment ± standard deviation. Dilutions of PCR lysates prepared from 8E5/LAI cells, containing one HIV-1 provirus per cell, were amplified with the same set of primers (E).
Strong-stop DNA (amplified with LTR R/U5 primers) was synthesized rapidly in infected macrophages. The peak of synthesis was reached by 2 h after infection with HIV-1 ADA (Fig. 7A). Infection with HIV-1 LAI or HIV-1 NDK resulted in a slower accumulation of R/U5 DNA, with the peak reached at 6 h after infection. At that time, the levels of strong-stop DNA were similar in macrophages infected with any of the three viruses. In experiments presented in Fig. 3 and 5B, the levels of strong-stop DNA in cells infected with HIV-1 ADA and HIV-1 NDK were the same at 2 h after infection. These slight differences in kinetics of reverse transcription can be explained by the well-documented phenomenon of donor variability (47).
Oligonucleotide primers from the U3/R region of the LTR can detect the reverse transcription product synthesized after the first strand transfer (Fig. 6). Using this primer pair, we observed a delay in reverse transcription of HIV-1 LAI and HIV-1 NDK compared to HIV-1 ADA, but in samples collected 24 h after infection, the levels were similar to those in HIV-1 ADA-infected cultures (Fig. 7B). The first significant differences between the T-lymphotropic viruses and the macrophagetropic ADA strain were detected by using oligonucleotide primers amplifying the pol gene (Fig. 7C) and LTR/gag primers that amplify DNA formed after the second template switch (Fig. 7D). The levels of these transcripts in cells infected with HIV-1 LAI and HIV-1 NDK were significantly lower than in cells infected with HIV-1 ADA.
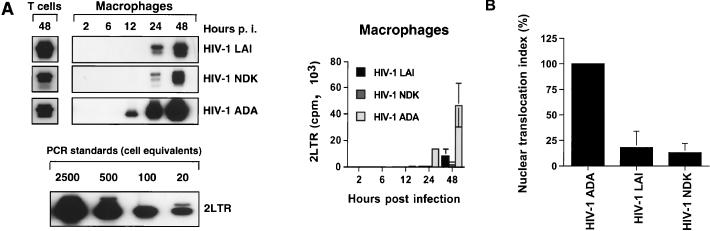
Inefficient nuclear translocation is an important factor contributing to the restricted replication of T-lymphotropic strains in macrophages.
2LTR DNA circles are formed exclusively in the nucleus (8, 9) and provide a useful marker for successful nuclear translocation of the HIV-1 DNA. Thus, to analyze nuclear import of the HIV-1 DNA, we measured production of viral 2LTR circular DNA in the samples collected 2, 6, 12, 24, and 48 h after infection with HIV-1 macrophagetropic or T-lymphotropic strains. The differences observed between T-lymphotropic and macrophagetropic strains during the intermediate and later phases of reverse transcription (Fig. 7C and D) increased markedly at the step of nuclear import (Fig. 8A). To control for efficiency of 2LTR circle formation, we measured production of viral 2LTR circular DNA in samples prepared 48 h after infection of primary T lymphocytes (Fig. 8A). Obtained results demonstrated that levels of 2LTR DNA in HIV-1 LAI-infected cultures were higher than those in HIV-1 ADA-infected T lymphocytes (Fig. 8A), correlating with the replication pattern of these viruses in primary T cells (data not shown). To quantitate the differences obtained in primary macrophages at the step of nuclear import, we calculated the nuclear translocation index (23), which reflects the efficiency of nuclear import of reverse-transcribed viral DNA. This index was calculated as the ratio of 2LTR circular DNA to total viral DNA in each culture infected with HIV-1 LAI or HIV-1 NDK, normalized to the ratio in HIV-1 ADA-infected culture. The nuclear translocation index, calculated and averaged from three independent experiments (performed with cells isolated from different blood donors), each done in duplicate, clearly demonstrated inefficient nuclear import in cultures infected with T-lymphotropic viruses (Fig. 8B). Consistent with this result, PCR in situ hybridization demonstrated a nuclear as well as cytoplasmic HIV-1 DNA-specific signal in macrophages infected with T-lymphotropic viruses, while only nuclear signal was detected in cells infected with macrophagetropic strain (data not shown).
FIG. 8.
Nuclear translocation of HIV-1-specific DNA in primary macrophages. (A) PCR lysates were prepared at 48 h after infection of primary T lymphocytes (T cells) and 2, 6, 12, 24, and 48 h after infection of macrophages with HIV-1 LAI, HIV-1 NDK, or HIV-1 ADA. Cell lysates were subjected to PCR amplification using a primer pair amplifying 2LTR DNA circles (upper left). The lower left panel represents PCR standards obtained by dilution of extrachromosomal DNA extracted from HIV-1-infected H9 cells to the equivalent of 20, 100, 500, and 2,500 infected cells and amplified with primers specific for 2LTR circular forms of viral DNA. The right panel shows quantification of this experiment. Data are shown as means ± standard deviations. p.i., postinfection. (B) Macrophages isolated from three different blood donors were infected as described above. After a 48-h incubation, cells were lysed and subjected to PCR amplification using LTR/gag primers, detecting complete reverse transcripts, and primers amplifying viral 2LTR circles. Efficiency of nuclear translocation for HIV-1 T-lymphotropic strains was calculated from the amount of 2LTR circle DNA (N2LTR) and total viral DNA (Ntot) in each culture and indexed to the same ratio of HIV-1 ADA cultures (C2LTR against Ctot), using the formula [(N2LTR/Ntot)/(C2LTR/Ctot)] × 100. Standard deviations are shown as vertical bars.
DISCUSSION
In this study, we investigated the determinants of HIV-1 replication in primary macrophages. Several lines of evidence suggest that the critical step at which replication of T-lymphotropic strains in macrophages is aborted occurs after entry and initiation of reverse transcription, most likely at the step of nuclear import of the viral preintegration complexes (PIC). First, we show that CXCR4, the major coreceptor for T-lymphotropic strains, is expressed on macrophages in a functional form (Fig. 4) and acts as a coreceptor for T-lymphotropic viruses (Fig. 5B). Second, only a slight delay in the synthesis of strong-stop DNA, an early product of reverse transcription, was found in macrophages infected with T-lymphotropic strains compared to HIV-1 ADA-infected cultures (Fig. 7A). Third, major differences were detected at the step of nuclear import (Fig. 8).
A number of studies characterizing viral factors responsible for selective tropism of HIV-1 strains identified envelope glycoprotein gp120 as a major determinant of efficient viral replication in macrophages (34, 44, 55). Together with studies that reported a close correspondence between the fusion specificities of Env proteins and the tropism of isolates from which they were derived (6), these earlier results suggested that fusion between the viral and cellular membranes is the step at which the outcome of HIV-1 infection of macrophages is determined. However, other groups documented that replication of T-lymphotropic viruses in macrophages was restricted at a step following viral entry (33, 49, 53). Results presented in this study confirm those earlier observations that T-lymphotropic HIV-1 strains efficiently enter macrophages. In addition, we demonstrate that the entry is mediated by the CD4 and CXCR4 molecules on macrophages, the same receptors that mediate entry of these viruses into T cells (27). It was shown previously (57) that some dualtropic primary syncytium-inducing viruses infected CXCR4+ cells but failed to infect CCR5+ cells, and the authors suggested that these viruses may use an alternative coreceptor for infection of macrophages. Recently, CXCR4 was shown to mediate entry of dualtropic but not T-lymphotropic viruses into primary macrophages in a CCR5-independent fashion (63). Our results show that CXCR4 can act as a coreceptor for HIV-1 T-lymphotropic viruses on these cells. The discrepancies between our results and those obtained previously (63) could be due to differences in viral strains used. Studies with chimeric chemokine receptors (37, 48) have shown that individual HIV-1 strains interact differently with CXCR4, and the possibility raised in the previous work (63) that CXCR4 is expressed on macrophages in a form that is functional as a cofactor for entry of some but not other HIV-1 isolates can possibly explain the observed differences. Also, utilization of CXCR4 by T-lymphotropic isolates may depend on expression of CD4 (36), and since macrophages express lower levels of CD4 than T lymphocytes, the level of CXCR4 expression can be critical for efficient entry of certain HIV-1 strains. Thus, different culture conditions of primary macrophages, resulting in differences in expression of coreceptor molecules on the cell surface, can also account for observed differences.
It is puzzling why macrophages, which express both CD4 and CXCR4 molecules, do not support efficient replication of T-lymphotropic viruses. One possibility is that the CXCR4 coreceptor expressed on macrophages, while functional in the way of receptor-mediated signaling (Fig. 4), does not support fusion with the Env of T-lymphotropic viruses. This hypothesis would be in agreement with the results of Broder and Berger (6), describing inability of Env derived from T-lymphotropic viruses to fuse with macrophages in a cell-cell fusion assay. However, in contrast to these results, Simmons et al. (56) showed efficient fusion of T-cell lines, persistently infected with T-lymphotropic viruses, with primary macrophages, while the same T-lymphotropic viruses did not infect macrophages. These results, taken together with our observations, suggest that CXCR4-mediated virus entry can differ in macrophages and T lymphocytes.
Results presented here show that the replication block for T-lymphotropic viruses in macrophages occurs at a postentry level, and predominantly at the step of nuclear importation of the viral PICs. Although some delay in the reverse transcription and lower levels of the late viral transcripts were also observed, we believe that they reflect a lower efficiency of reverse transcription within the cytoplasmic compartment than in the nucleus, where late products of reverse transcription are synthesized more efficiently during productive infection (10). Thus, inefficient nuclear import of the PIC of T-lymphotropic strains in macrophages can account for lower levels of intermediate and late transcripts in these cultures. A fivefold difference (Fig. 8) between macrophagetropic and T-lymphotropic viruses in the level of nuclear import (nuclear translocation index) does not seem sufficient to account for the observed differences in viral replication. However, differences in the p24 antigen levels measured at the same time intervals were in the same range (data not shown). Amplification of these differences during several rounds of reinfection can explain observed phenomenon. However, we cannot exclude completely that restriction for T-lymphotropic viruses occurs also at the level of reverse transcription.
At the first glance, our finding that replication of T-lymphotropic HIV-1 strains in macrophages is restricted at the postentry steps, and predominantly at the level of nuclear import, is inconsistent with the demonstrated role of the Env protein in determining the replication fate of such viruses (34, 44, 55). Indeed, the viral envelope is not part of the PIC, which comprises viral nucleic acids and several viral proteins, including integrase, matrix antigen, Vpr, and RT (10, 29, 31, 62). Therefore, the steps in the viral life cycle preceding formation of the PIC can somehow affect the efficiency of nuclear import. A precedent for this phenomenon has been documented in a recent study with SIV, where differential use of the CCR5 coreceptor by different SIV strains determined the fate of viral infection of macrophages at a postentry step (24). Also, T-lymphotropic SIVmac239 enters macrophages as efficiently as macrophagetropic SIV isolates yet fails to replicate (40). Primary HIV-1 isolates can efficiently enter macaque cells expressing human CD4, presumably by using a simian coreceptor; however, expression of the human coreceptor is required for productive virus infection in these cells (11). Thus, coreceptors not only participate in viral entry but also influence later stages of virus replication, either through signaling or, more likely, by targeting viral PICs to a subcellular compartment, where postentry events take place. Therefore, differences in utilization of coreceptors on primary macrophages may result in targeting the viral PIC to a different compartment which does not support efficient nuclear import. Further work using a chimeras of macrophagetropic and T-lymphotropic viruses will be necessary to test this hypothesis. Thus, it remains to be seen whether the mode of viral entry or coreceptor usage determines the intracellular compartment to which the virus is targeted. Such studies would be useful for elucidating the mechanisms of HIV-1 replication in macrophages and a better understanding of AIDS pathogenesis.
ACKNOWLEDGMENTS
We thank Ivan Hirsch for helpful discussion, Michael Yamin for critical comments on the manuscript, and Tang Hao for performing p24 ELISA.
This work was supported in part by NIH grants R01 AI 38245 and R29 AI 33776 to M.B.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrvirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;2:660–662. [PubMed] [Google Scholar]
- 4.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 5.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 6.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broder C C, Dimitrov D S, Blumenthal R, Berger E A. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, La Rosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Clapham P R, Blanc D, Weiss R A. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by simian immunodeficiency virus. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collin M, Herbein G, Montaner L, Gordon S. PCR analysis of HIV1 infection of macrophages: virus entry is CD4-dependent. Res Virol. 1993;144:13–19. doi: 10.1016/s0923-2516(06)80006-3. [DOI] [PubMed] [Google Scholar]
- 16.Collman R, Godfrey B, Cutilli J, Rhodes A, Hassan N F, Sweet R, Douglas S D, Friedman H, Nathanson N, Gonzalez-Scarano F. Macrophage-tropic strains of human immunodeficiency virus type 1 utilize the CD4 receptor. J Virol. 1990;64:4468–4476. doi: 10.1128/jvi.64.9.4468-4476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor R I, Ho D D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalgleish A G, Beverly P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 19.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 20.DiMarzio P, Tse J, Landau N R. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res Hum Retroviruses. 1998;14:129–138. doi: 10.1089/aid.1998.14.129. [DOI] [PubMed] [Google Scholar]
- 21.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 22.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 23.Dubrovsky L, Ulrich P, Nuovo G J, Manogue K R, Cerami A, Bukrinsky M. Nuclear localization signal of HIV-1 as a novel target for therapeutic intervention. Mol Med. 1995;1:217–230. [PMC free article] [PubMed] [Google Scholar]
- 24.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellrodt A, Barre-Sinoussi F, Le Bras P, Nugeyre M T, Palazzo L, Rey F, Brun-Vezinet F, Rouzioux C, Segond P, Caquet R, et al. Isolation of human T-lymphotropic retrovirus (LAV) from Zairian married couple, one with AIDS, one with prodromes. Lancet. 1984;1:1383–1385. doi: 10.1016/s0140-6736(84)91877-4. [DOI] [PubMed] [Google Scholar]
- 26.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 27.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 28.Fenyo E M, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 30.Gendelman H E, Orenstein J M, Martin M A, Ferrua C, Mitra R, Phipps T, Wahl L A, Lane H C, Fauci A S, Burke D S, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167:1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higuchi R. Simple and rapid preparation of samples for PCR. In: Erlich H A, editor. PCR technology. Principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 34–38. [Google Scholar]
- 33.Huang Z B, Potash M J, Simm M, Shahabuddin M, Chao W, Gendelman H E, Eden E, Volsky D J. Infection of macrophages with lymphotropic human immunodeficiency virus type 1 can be arrested after viral DNA synthesis. J Virol. 1993;67:6893–6896. doi: 10.1128/jvi.67.11.6893-6896.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 35.Klatzmann K, Champagne E E, Chamaret K S, Gruest J, Guetard D, Hercent T, Gluckman J, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 36.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriuchi H, Moriuchi M, Combadiere C, Murphy P M, Fauci A S. CD8+ T-cell-derived soluble factor(s), but not beta-chemokines RANTES, MIP-1 alpha, and MIP-1 beta, suppress HIV-1 replication in monocyte/macrophages. Proc Natl Acad Sci USA. 1996;93:15341–15345. doi: 10.1073/pnas.93.26.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuovo G J, Becker J, Burk M W, Margiotta M, Fuhrer J, Steigbigel R T. In situ detection of PCR-amplified HIV-1 nucleic acids in lymph nodes and peripheral blood in patients with asymptomatic HIV-1 infection and advanced-stage AIDS. J Acquired Immune Defic Syndr. 1994;7:916–923. [PubMed] [Google Scholar]
- 43.Nuovo G J, Gallery F, MacConnell P, Braun A. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acids and tumor necrosis factor-alpha RNA in the central nervous system. Am J Pathol. 1994;144:659–666. [PMC free article] [PubMed] [Google Scholar]
- 44.O’Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 45.O’Brien W A, Namazi A, Kalhor H, Mao S H, Zack J A, Chen I S. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 47.Olafsson K, Smith M S, Marshburn P, Carter S G, Haskill S. Variation of HIV infectibility of macrophages as a function of donor, stage of differentiation, and site of origin. J Acquired Immune Defic Syndr. 1991;4:154–164. [PubMed] [Google Scholar]
- 48.Picard L, Wilkinson D A, McKnight A, Gray P W, Hoxie J A, Clapham P R, Weiss R A. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 49.Potash M J, Zeira M, Huang Z B, Pearce T E, Eden E, Gendelman H E, Volsky D J. Virus-cell membrane fusion does not predict efficient infection of alveolar macrophages by human immunodeficiency virus type 1 (HIV-1) Virology. 1992;188:864–868. doi: 10.1016/0042-6822(92)90543-x. [DOI] [PubMed] [Google Scholar]
- 50.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 51.Rey M A, Spire B, Dormont D, Barre-Sinoussi F, Montagnier L, Chermann J C. Characterization of the RNA dependent DNA polymerase of a new human T-lymphotropic retrovirus (lymphadenopathy associated virus) Biochem Biophys Res Commun. 1984;121:126–133. doi: 10.1016/0006-291x(84)90696-x. [DOI] [PubMed] [Google Scholar]
- 52.Schmidtmayerova H, Sherry B, Bukrinsky M. Chemokines and HIV replication. Nature. 1996;382:767. doi: 10.1038/382767a0. [DOI] [PubMed] [Google Scholar]
- 53.Schmidtmayerova H, Bolmont C, Baghdiguian S, Hirsch I, Chermann J C. Distinctive pattern of infection and replication of HIV1 strains in blood-derived macrophages. Virology. 1992;190:124–133. doi: 10.1016/0042-6822(92)91198-4. [DOI] [PubMed] [Google Scholar]
- 54.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, De Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 56.Simmons G, McKnight A, Takeuchi Y, Hoshino H, Clapham P R. Cell-to-cell fusion, but not virus entry in macrophage by T-cell line tropic HIV-1 strains: a V3 loop-determined restriction. Virology. 1995;209:696–700. doi: 10.1006/viro.1995.1307. [DOI] [PubMed] [Google Scholar]
- 57.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tersmette M, De Goede R E, Al B J, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valentin A, Albert J, Fenyo E M, Asjo B. Dual tropism for macrophages and lymphocytes is a common feature of primary human immunodeficiency virus type 1 and 2 isolates. J Virol. 1994;68:6684–6689. doi: 10.1128/jvi.68.10.6684-6689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varmus H E, Swanstrom R. Replication of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 369–512. [Google Scholar]
- 62.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]