Abstract
The current naso-oropharyngeal swab for SARS-CoV-2 detection faces several problems, such as waste issues and its use for quantitative studies. This study aimed to evaluate the total RNA and viral loads from different upper respiratory tract swabs types and whether SARS-CoV-2 quantification can use the current internal control for normalization. This cross-sectional study collected positive specimens with single oropharyngeal or nasopharyngeal swabs and naso-oropharyngeal swabs. The samples were extracted, tested with qualitative RT‒PCR, and then tested with quantitative RT‒PCR. The RNA eluate was measured for the total RNA concentration. The total RNA concentration, viral load, and RNaseP Ct values were collected and analysed statistically. The positive results came from 41 oropharyngeal swabs, 34 nasopharyngeal swabs, and 36 naso-oropharyngeal swabs. The total RNA increased significantly from oropharyngeal swabs to nasopharyngeal swabs to naso-oropharyngeal swabs. Significant differences in RNaseP Ct values between groups and their correlations with total RNA were found. In addition, the increase in the total RNA and the RNaseP Ct values were unrelated to the viral load. The physical features in the naso-oropharyngeal area and the swabbing procedures could affect the total RNA but not the viral load. However, since the virus particles could present inside and outside human cells, the increase in collected human cells may not always be followed by the viral load increase. Normalization using the RNaseP Ct value became unnecessary due to the factors mentioned above. Therefore, a careful approach is needed in viral load studies of swab specimens.
Keywords: SARS-Cov-2, Quantitative PCR, Viral load, Total RNA
1. Introduction
According to World Health Organization interim guidance in 2020, one of the minimum respiratory materials needed for SARS-CoV-2 diagnosis is nasopharyngeal and oropharyngeal swabs [1]. The combination of nasopharyngeal and oropharyngeal swabs has been found to provide a wider diagnostic yield of respiratory viruses [2]. Therefore, combination swabs have been adapted as the ideal specimen for SARS-CoV-2 diagnosis in Indonesia [3]. However, single swabs (either oro- or nasopharyngeal swabs) have been widely used, and their decent sensitivity has been reported [4]. Hence, the use of a single type of swab could help to reduce medical waste and simplify the diagnostic protocol [5], especially considering the future of SARS-CoV-2 as an endemic disease.
In addition to qualitative real-time polymerase chain reaction (PCR), quantitative methods have been used in many studies to develop guidance for clinicians in patient management and as a baseline for infection control measures and policies [[6], [7], [8], [9]]. However, the current approach for SARS-CoV-2 quantification has raised awareness due to inconsistent standard curves and the variety of estimated viral loads [10]. Some quantitative studies reported their findings with the defined measurement (copies/ml [7] or Log10 copies/ml [11]), and the other studies used the cycle threshold (Ct) values from qualitative PCR methods to represent the viral load [6,[12], [13], [14]]. Since the amount of specimen within the swab could be influenced by the amount of the absorbed fluid [15] and the type of swab itself [16], several studies have explored the need for normalization of viral load [17,18].
This study aimed to observe the variety of total amounts of extracted RNA from different types of swabs, whether the increase in total RNA would affect virus quantification, and the potential of the current internal control (human RNaseP) as the component to normalize viral load from respiratory swab specimens. This report is important since, to our knowledge, current published studies rarely discussed the type of swab's performance for SARS-CoV-2 detection from this point of view.
2. Materials and methods
This study was conducted after obtaining ethical clearance from the Research Ethics Committee of Universitas Indonesia (protocol number 20-05-0516). Patients provided written consent after they were informed about the study details. All the patients involved in this study agreed to have their SARS-CoV-2-positive results and upper respiratory tract specimens used for analysis.
The study design was cross-sectional, using positive specimens collected from a referral laboratory in Jakarta on November 13–23, 2020. We selected specimens that were sent to the lab within 24 h after collection from the health-care provider (both hospitals and public health centres), used the same volume and type of viral transport medium (BioVTM, Biofarma, Indonesia), used the same type of nasopharyngeal and oropharyngeal swab (Dacron swab, China), and were already positive for the N and ORF genes (Da An Gene, China). To ensure that the entire range of cycle threshold (Ct) values was covered, we collected specimens with Ct values under 20, 20 to 30, and above 30. Since many viral load studies have been widely reported, we decided to take each type of swab from different patients to emulate the field situation and observe the factors that could influence the results. We collected 111 positive specimens, with 41 oropharyngeal swabs, 34 nasopharyngeal swabs, and 36 naso-oropharyngeal swabs. All specimens were stored at −80 °C in an ultra freezer before being processed for this study.
The extraction was performed according to the manufacturer's instructions (PureLink™ Viral RNA Mini Kit, Thermo Fisher Scientific, USA). In brief, 200 μl of specimen was used for extraction. The total RNA extracted from every specimen was measured using a NanoDrop™ (Thermo Fisher Scientific, USA). The specimens with a total RNA 260/280 ratio >2.0 were included in this study. Before proceeding to quantitative RT‒PCR, positive specimens were retested by qualitative real-time RT‒PCR with N2 as the gene target. The cut-off cycle threshold (Ct) value was 40, and Ct values < 40 were defined as positive. rRT-PCR was performed within 30 min after extraction. The rRT-PCR mixture was made following the manufacturer's instructions (SensiFAST™, Meridian Bioscience, USA). The primer used was the N2 gene recommended by the CDC, with the forward primer TTA CAA ACA TTG GCC GCA AA and reverse primer GCG CGA CAT TCC GAA GAA; the probe was FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1. Confirmatory rRT-PCR was conducted if the first reaction resulted in a Ct value between 38 and 40. The RNaseP primer was used as the internal control. The RNAaseP Ct values were collected; the cut-off cycle threshold (Ct) value was 40, and Ct values < 40 were defined as positive.
The 20 μl qualitative RT‒PCR system contained 12.1 μl of reaction mixture and 7.9 μl of RNA template (SensiFAST™, Meridian Bioscience, USA). The qRT‒PCR protocol was 50 °C for 30 min of RT incubation, followed by 95 °C for 2 min as the cDNA denaturation step, and then 45 cycles consisting of 94 °C for 15 s and 55 °C for 45 s. The viral copy number was plotted against a standard curve constructed based on standard products (AMPLIRUN® SARS-CoV-2 RNA CONTROL, 20MBC137103-R1). All RT‒PCRs were conducted using the same instrument (MA6000, Shuzou Molarray Co. Ltd., China).
The total RNA concentration is provided as ng/μl, and the viral load is provided as copies/ml. The data were numerical and tested for normality and presented as the mean (CI 95%, lower–upper) or median (interquartile range (IQR), number), depending on the normality result. The statistical analysis was performed with an appropriate test.
3. Results
When grouped according to the N2 gene Ct value from the rRT-PCR, we found 18 specimens with a Ct value under 20, 54 specimens with a Ct value of 20–30, and 39 specimens with a Ct value above 30 (Supplementary 1). The specimen total RNA, viral load, and RNaseP Ct values are described in Table 1, including the p value of the comparison of mean/median between the three groups.
Table 1.
Total RNA concentration, viral load, and Ct value of RNaseP from oropharyngeal swabs, nasopharyngeal swabs, and naso-oropharyngeal swabs.
| Oropharyngeal swabs (n = 41) | Nasopharyngeal swabs (n = 34) | Naso-oropharyngeal swabs (n = 36) | ||
|---|---|---|---|---|
| Specimens characteristic | ||||
| Total RNA concentration (ng/μl) | 3.20 (1.20) | 5.05 (6.20) | 8.15 (10.35) | p < 0.001 |
| Viral Load (copies/ml) | 9.8 × 105 (3.1 × 106) | 7.3 × 105 (4.6 × 106) | 10.4 × 105 (6.1 × 106) | p = 0.36 |
| Ct value of RNaseP |
28.87 (28.10–29.63) |
29.57 (28.85–30.27) |
25.83 (24.93–26.73) |
p < 0.001 |
| Correlation test | ||||
| Total RNA Concentration and Viral Load |
p = 0.024 r = - 0.35 |
p = 0.84 | p = 0.35 | |
| Total RNA Concentration and Ct value of RNaseP |
p = 0.001 r = - 0.5 |
p = 0.003 r = - 0.5 |
p < 0.001 r = - 0.8 |
|
| Viral Load and Ct value of RNaseP | p = 0.35 | p = 0.053 | p = 0.94 | |
We were expecting a significant difference in viral load between groups, but our statistical analysis showed otherwise because each swab type covered almost the same range of Ct values despite the number of specimens (Supplementary 1). Since our study was directed to evaluate the factors affecting the performance of a single swab to collect virus from each intended area, this situation benefitted our study, and we continued with the analysis.
The medians of the total RNA concentration and viral load between the three specimen groups were analysed with the Kruskal‒Wallis test (Table 1); the Mann‒Whitney test was used for the post hoc analysis. The median total RNA concentration extracted from the naso-oropharyngeal swabs was significantly different from that of the oropharyngeal swabs (p < 0.001) but not from that of the nasopharyngeal swabs (p = 0.2). However, there was a significant difference between oropharyngeal swabs and nasopharyngeal swabs (p < 0.001).
The mean RNaseP Ct values between groups were analysed by ANOVA and showed a significant difference. The Bonferroni correction was used as the post hoc analysis, and the differences were significant between naso-oropharyngeal and oropharyngeal swabs (p < 0.001; CI 95% −4.3 to −1.7) and between naso-oropharyngeal swabs and nasopharyngeal swabs (p < 0.001; CI 95% −5.1 to −2.3). No difference was found between oropharyngeal swabs and nasopharyngeal swabs.
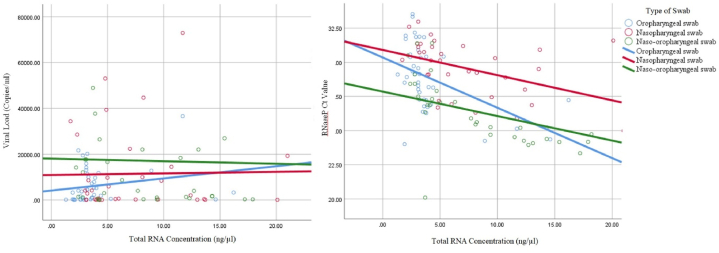
The correlation between the total RNA concentration and viral load was only statistically significant for the oropharyngeal swabs, with weak power (p = 0.02, r = 0.35). The correlation between the total RNA concentration and RNaseP Ct value was significant for every type of specimen, with the strongest correlation seen with the naso-oropharyngeal swabs (Table 1, Fig. 1). There was no correlation between the RNaseP Ct value and viral load (Table 1, Fig. 1).
Fig. 1.
Correlation between total RNA concentration with viral load and Ct value of RNaseP.
4. Discussion
From our observation, the increase in total RNA was not in parallel with the viral load, although there were differences in total RNA between the types of swabs. Both nasopharyngeal swabs and naso-oropharyngeal swabs had higher total RNA concentrations but showed no significant increase in viral load, and only oropharyngeal swabs showed a significant correlation between total RNA and viral load (Fig. 1). We consider several factors to have roles in these results, including the anatomy and histology of the nasopharynx and oropharynx and the swabbing method.
The surface size of the oropharynx is larger than that of the nasopharynx [19]. However, tongue size and variations in the gag reflex between patients could affect the medical worker's manoeuvre while swabbing the intended oropharynx area, therefore reducing the obtained amount of fluid and cells. The size variation of nasopharyngeal tonsils might affect the nasopharyngeal swab process, but the lack of an expulsion reflex gives medical workers enough time to place the flocked swab inside the nasopharynx cavity, let the flocked swab absorb any extracellular fluid, and brush the mucosa, which is impossible to perform with an oropharyngeal swab [20]. In addition, 60% of the total epithelial surface of the nasopharyngeal mucosa is lined by a stratified squamous epithelium, with alternating patches of squamous and ciliated epithelia [21]. Similar to the nasopharynx, the soft palate and posterior oropharynx are lined with a stratified squamous epithelium [22]. A previous study on SARS-CoV-2 showed viral shedding from ciliated epithelia [23]. The report suggests that virus entry and shedding take place on ciliated epithelia found in the nasopharyngeal mucosa. Stratified squamous epithelium also demonstrates ACE protein expression [24]. Therefore, the oropharynx had the same opportunity to be the location for virus entry and shedding. We assume that the total RNA difference between each type of swab was influenced by anatomy and the swabbing method rather than by real differences in the amount of viral shedding in the oropharynx.
We also examined the Ct values of the RNaseP gene as the internal control. There were significant differences in the mean RNaseP Ct value, especially between naso- or oropharyngeal swabs and naso-oropharyngeal swabs, and a significant correlation was observed between total RNA and the RNaseP Ct value in all types of swabs. This result was in concordance with the assumption that the more swabs obtained, the higher the number of human cells collected [25]. Nonetheless, the correlation between the internal control and the total RNA did not guarantee an increase in viral load. This was supported by the independence of our RNaseP Ct value from the viral load, in line with the report from Van Wesenbeeck et al.’s study [17]. Our findings showed that the number of host cells on the swab was near the optimum and that the number of secreted viral particles was sufficient, in concordance with Akmatov et al.’s study [26]. Therefore, similar to the Piralla et al. study [18], normalization using internal controls seemed unnecessary for upper respiratory swab specimens.
In addition, RNaseP only came from the intracellular compartment, but the viral load came from the intracellular compartment (the infected cells) and extracellular compartment (the shedding virus particles). The different disease stages showed different viral loads [27]. When the virus was at its highest shedding period, the virus particles obtained from the extracellular compartment would increase, regardless of the number of collected human cells. This circumstance could explain the insignificant correlation of viral load with total RNA within the single and combination swabs, despite the obvious correlation between the Ct value of the internal control and the total RNA concentration. Hence, more rigorous sampling might not increase viral detection in our study, similar to the report by Akmatov et al. [26].
As described above, each type of swab is sufficient to obtain viral particles for diagnosis, in concordance with a report by Zhang et al. on similar yields of SARS-CoV-2 detection in nasopharyngeal swabs and oropharyngeal swabs [28]. Calame et al. also reported comparable sensitivity from both swabs [4]. When the skill of the medical worker is reliable, either a single nasopharyngeal swab or single oropharyngeal swab for SARS-CoV-2 diagnosis is equivalent to a naso-oropharyngeal swab.
5. Study limitations
The limitation of our study was our inability to evaluate the medical worker's technique in performing oropharyngeal swabs, the small number of specimens, and the unknown stage of the patient's disease. We could also not obtain the control plasmid to generate the RNaseP standard curve; therefore, we only used the Ct value. The positivity rate of each type of swab was also not mentioned to avoid ascertainment bias because the specimens were sent from various health-care centres with different rates of COVID-19 cases.
6. Conclusions
In conclusion, we found that the nasopharynx had anatomical and histological advantages in determining the total RNA obtained from nasopharyngeal and combination (naso- and oropharyngeal) swabs. We also found that the increase in total RNA concentration did not indicate a higher viral load, and the RNaseP Ct value was not suitable for normalizing the SARS-CoV-2 viral load from upper respiratory swabs. Hence, we suggest a careful approach using upper respiratory swabs for future SARS-CoV-2 viral load research.
Data availability
All data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
Fera Ibrahim: Writing – review & editing, Writing – original draft, Supervision, Resources, Methodology, Investigation, Formal analysis, Conceptualization. Augustine Natasha: Writing – review & editing, Writing – original draft, Project administration, Investigation, Formal analysis, Data curation, Conceptualization. Andi Yasmon: Writing – review & editing, Visualization, Validation, Supervision, Resources, Methodology, Formal analysis, Conceptualization. Chairunnisa Tawadhu Rizal: Writing – review & editing, Resources, Project administration, Methodology, Data curation. Fithriyah: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Methodology, Formal analysis, Data curation, Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Methodology, Formal analysis, Data curation. Anis Karuniawati: Writing – review & editing, Validation, Supervision, Resources, Project administration, Formal analysis. Yulia Rosa Saharman: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Investigation, Formal analysis. Pratiwi Sudarmono: Writing – review & editing, Supervision, Resources, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to thank Universitas Indonesia, through PUTI Grant with contract number NKB-1493/UN2.RST/HKP.05.00.2020.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28647.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization . World Health Organization; 2020. Laboratory Testing for Coronavirus Disease 2019 (Covid-19) in Suspected Human Cases: Interim Guidance, 2 March 2020. [Google Scholar]
- 2.Ek P., Böttiger B., Dahlman D., Hansen K.B., Nyman M., Nilsson A.C. A combination of naso- and oropharyngeal swabs improves the diagnostic yield of respiratory viruses in adult emergency department patients. Infectious diseases (London, England) 2019;51(4):241–248. doi: 10.1080/23744235.2018.1546055. [DOI] [PubMed] [Google Scholar]
- 3.Indonesia Ministry of Health Guidelines for prevention and control of coronavirus diseases (covid-19) 2020. https://covid19kemkesgoid/document/pedoman-pencegahan-dan-pengendalian-covid-19/view (in Bahasa)
- 4.Calame A., Mazza L., Renzoni A., Kaiser L., Schibler M. Sensitivity of nasopharyngeal, oropharyngeal, and nasal wash specimens for sars-cov-2 detection in the setting of sampling device shortage. Eur. J. Clin. Microbiol. Infect. Dis. 2020:1–5. doi: 10.1007/s10096-020-04039-8. official publication of the European Society of Clinical Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni B.N., Anantharama V. Repercussions of covid-19 pandemic on municipal solid waste management: challenges and opportunities. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M.J. Quantifying sars-cov-2 viral load: current status and future prospects. Expert Rev. Mol. Diagn. 2021;21(10):1017–1023. doi: 10.1080/14737159.2021.1962709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han MS, Seong MW, Heo EY, et al. Sequential Analysis of Viral Load in a Neonate and Her Mother Infected With Severe Acute Respiratory Syndrome Coronavirus 2. Clin. Infect. Dis. 2020;71(16):2236–2239. doi: 10.1093/cid/ciaa447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation Between Relative Nasopharyngeal Virus RNA Load and Lymphocyte Count Disease Severity in Patients with COVID-19. Viral. Immunol. 2021;34(5):330–335. doi: 10.1089/vim.2020.0062. [DOI] [PubMed] [Google Scholar]
- 9.Xia X.Y., Wu J., Liu H.L., Xia H., Jia B., Huang W.X. Epidemiological and initial clinical characteristics of patients with family aggregation of covid-19. J. Clin. Virol. : the official publication of the Pan American Society for Clinical Virology. 2020;127 doi: 10.1016/j.jcv.2020.104360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M.S., Byun J.-H., Cho Y., Rim J.H. Rt-pcr for sars-cov-2: quantitative versus qualitative. Lancet Infect. Dis. 2020;S1473–3099(20) doi: 10.1016/S1473-3099(20)30424-2. 30424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of sars-cov-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Chen S, Yang Z, et al. SARS-CoV-2 Viral Load in Clinical Samples from Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2020;201(11):1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., et al. Clinical and biochemical indexes from 2019-ncov infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with sars-cov-2 infection in a community treatment center in the Republic of Korea. JAMA Intern. Med. 2020;180(11):1447–1452. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angel D.E., Lloyd P., Carville K., Santamaria N. The clinical efficacy of two semi-quantitative wound-swabbing techniques in identifying the causative organism(s) in infected cutaneous wounds. Int. Wound J. 2011;8(2):176–185. doi: 10.1111/j.1742-481X.2010.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wise N.M., Wagner S.J., Worst T.J., Sprague J.E., Oechsle C.M. Comparison of swab types for collection and analysis of microorganisms. Microbiol. 2021;10(6) doi: 10.1002/mbo3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Wesenbeeck L., Meeuws H., D'Haese D., Ispas G., Houspie L., Van Ranst M., et al. Sampling variability between two mid-turbinate swabs of the same patient has implications for influenza viral load monitoring. Virol. J. 2014;11:233. doi: 10.1186/s12985-014-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piralla A., Giardina F., Rovida F., Campanini G., Baldanti F. Cellular DNA quantification in respiratory samples for the normalization of viral load: a real need? J. Clin. Virol. : the official publication of the Pan American Society for Clinical Virology. 2018;107:6–10. doi: 10.1016/j.jcv.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceylan I., Oktay H. A study on the pharyngeal size in different skeletal patterns. Am. J. Orthod. Dentofacial Orthop. 1995;108(1):69–75. doi: 10.1016/s0889-5406(95)70068-4. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention . 2021. Interim Guidelines for Collecting and Handling of Clinical Specimens for Covid-19 Testing.https://wwwcdcgov/coronavirus/2019-ncov/lab/guidelines-clinical-specimenshtml [Google Scholar]
- 21.Ali M. Histology of the human nasopharyngeal mucosa. J. Anat. 1965;99(Pt 3):657. [PMC free article] [PubMed] [Google Scholar]
- 22.Fossum C.C., Chintakuntlawar A.V., Price D.L., Garcia J.J. Characterization of the oropharynx: anatomy, histology, immunology, squamous cell carcinoma and surgical resection. Histopathology. 2017;70(7):1021–1029. doi: 10.1111/his.13140. [DOI] [PubMed] [Google Scholar]
- 23.Lee IT, Nakayama T, Wu CT, et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun. 2020;11(1):5453. doi: 10.1038/s41467-020-19145-6. Published 2020 Oct 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Descamps G., Verset L., Trelcat A., Hopkins C., Lechien J.R., Journe F., et al. Ace2 protein landscape in the head and neck region: the conundrum of sars-cov-2 infection. Biology. 2020;9(8) doi: 10.3390/biology9080235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mawaddah A., Gendeh H.S., Lum S.G., Marina M.B. Upper respiratory tract sampling in covid-19. Malays. J. Pathol. 2020;42(1):23–35. [PubMed] [Google Scholar]
- 26.Akmatov M.K., Gatzemeier A., Schughart K., Pessler F. Equivalence of self- and staff-collected nasal swabs for the detection of viral respiratory pathogens. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0048508. -e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J.Y., Ko J.H., Kim Y., Kim Y.J., Kim J.M., Chung Y.S., et al. Viral load kinetics of sars-cov-2 infection in first two patients in korea. J. Kor. Med. Sci. 2020;35(7) doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabbris C., Cestaro W., Menegaldo A., Spinato G., Frezza D., Vijendren A., et al. Is oro/nasopharyngeal swab for sars-cov-2 detection a safe procedure? Complications observed among a case series of 4876 consecutive swabs. Am. J. Otolaryngol. 2021;42(1) doi: 10.1016/j.amjoto.2020.102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in article/supp. material/referenced in article.