Summary
Generating cell types with properties of embryo cells with full developmental potential is of great biological importance. Here, we present a protocol for generating mouse morula-like cells (MLCs) resembling 8- to 16-cell stage embryo cells. We describe steps for induction, via increasing Stat3 activation, and the isolation of MLCs. We then detail procedures for segregating MLCs into blastocyst cell fates and how to create embryo-like structures from them. This system provides a stem-cell-based embryo model to study early embryo development.
For complete details on the use and execution of this protocol, please refer to Li et al.1
Subject areas: Developmental biology, Stem Cells, Organoids
Graphical abstract

Highlights
-
•
Protocol for generating mouse morula-like cells (MLCs) from ESC cells
-
•
Procedure to induce MLCs via pSTAT3 stimulation and isolate MLCs by sorting
-
•
Instructions for segregating MLCs and assembling post-implantation embryoids
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Generating cell types with properties of embryo cells with full developmental potential is of great biological importance. Here, we present a protocol for generating mouse morula-like cells (MLCs) resembling 8- to 16-cell stage embryo cells. We describe steps for induction, via increasing Stat3 activation, and the isolation of MLCs. We then detail procedures for segregating MLCs into blastocyst cell fates and how to create embryo-like structures from them. This system provides a stem-cell-based embryo model to study early embryo development.
Before you begin
Generation of Gata6+/H2BVenus mouse embryonic cell line with GY118F receptor transgene
Timing: 12 days
To induce increased levels of pSTAT3 in mESCs, chimeric GCSF-LifR Y118F (GY118F) transgene will be constructed and then transfected into Gata6+/H2BVenus mESCs. This transgene will activate STAT3 (pSTAT3) upon addition of granulocyte colony-stimulating factor (GCSF)2,3,4 to the medium. The GCSF-GY118F system guarantees stable and robust STAT3 activation by circumventing the negative feedback regulative mechanism of JAK/STAT3 signaling.2,4
-
1.Construction of PB.CAG.GY118F.IRES.Bsd plasmid (Figure 1).
-
a.PB.CAG.IRES.BSD plasmid (which provides constitutive expression to both the gene of interest and to the blasticidin resistance gene), pDONR221 plasmid and the synthesized GY118F sequence flanked by attB1/attB2 gateway cloning sequences are required.
-
b.Synthesize the GY118F receptor coding sequence, described in.3Note: The sequence of synthetic GY118F is provided in Table 1.
Table 1.
The sequence of synthetic GY118FCAAGTTTGTACAAAAAAGCAGGCTCACCATGGCAAGGCTGGGAAACTGCAGCCTGACTTGGGCTGCCCTGATCATCCTGCTGCTCCCCGGAAGT
CTGGAGGAGTGCGGGCACATCAGTGTCTCAGCCCCCATCGTCCACCTGGGGGATCCCATCACAGCCTCCTGCATCATCAAGCAGAACTGC
AGCCATCTGGACCCGGAGCCACAGATTCTGTGGAGACTGGGAGCAGAGCTTCAGCCCGGGGGCAGGCAGCAGCGTCTGTCTGATGGGACCC
AGGAATCTATCATCACCCTGCCCCACCTCAACCACACTCAGGCCTTTCTCTCCTGCTGCCTGAACTGGGGCAACAGCCTGCAGATCCTGGAC
CAGGTTGAGCTGCGCGCAGGCTACCCTCCAGCCATACCCCACAACCTCTCCTGCCTCATGAACCTCACAACCAGCAGCCTCATCTGCCA
GTGGGAGCCAGGACCTGAGACCCACCTACCCACCAGCTTCACTCTGAAGAGTTTCAAGAGCCGGGGCAACTGTCAGACCCAAGGGGACTCC
ATCCTGGACTGCGTGCCCAAGGACGGGCAGAGCCACTGCTGCATCCCACGCAAACACCTGCTGTTGTACCAGAATATGGGCATCTGGGTGCAGGCA
GAGAATGCGCTGGGGACCAGCATGTCCCCACAACTGTGTCTTGATCCCATGGATGTTGTGAAACTGGAGCCCCCCATGCTGCGGACCATGGACCCCA
GCCCTGAAGCGGCCCCTCCCCAGGCAGGCTGCCTACAGCTGTGCTGGGAGCCATGGCAGCCAGGCCTGCACATAAATCAGAAGTGTGAGCTG
CGCCACAAGCCGCAGCGTGGAGAAGCCAGCTGGGCACTGGTGGGCCCCCTCCCCTTGGAGGCCCTTCAGTATGAGCTCTGCGGGCTCCTCCCAG
CCACGGCCTACACCCTGCAGATACGCTGCATCCGCTGGCCCCTGCCTGGCCACTGGAGCGACTGGAGCCCCAGCCTGGAGCTGAGAACTACCGAAC
GGGCCCCCACTGTCAGACTGGACACATGGTGGCGGCAGAGGCAGCTGGACCCCAGGACAGTGCAGCTGTTCTGGAAGCCAGTGCCCCTGG
AGGAAGACAGCGGACGGATCCAAGGTTATGTGGTTTCTTGGAGACCCTCAGGCCAGGCTGGGGCCATCCTGCCCCTCTGCAACACCACAGAGCTC
AGCTGCACCTTCCACCTGCCTTCAGAAGCCCAGGAGGTGGCCCTTGTGGCCTATAACTCAGCCGGGACCTCTCGCCCCACCCCGGTGGT
CTTCTCAGAAAGCAGAGGCCCAGCTCTGACCAGACTCCATGCCATGGCCCGAGACCCTCACAGCCTCTGGGTAGGCTGGGAGCCCCCCAATCCAT
GGCCTCAGGGCTATGTGATTGAGTGGGGCCTGGGCCCCCCCAGCGCGAGCAATAGCAACAAGACCTGGAGGATGGAACAGAATGGGAGA
GCCACGGGGTTTCTGCTGAAGGAGAACATCAGGCCCTTTCAGCTCTATGAGATCATCGTGACTCCCTTGTACCAGGACACCATGGGACCCTCCCA
GCATGTCTATGCCTACTCTCAAGAAATGGCTCCCTCCCATGCCCCAGAGCTGCATCTAAAGCACATTGGCAAGACCTGGGCACAGCTGGAGTG
GGTGCCTGAGCCCCCTGAGCTGGGGAAGAGCCCCCTTACCCACTACACCATCTTCTGGACCAACGCTCAGAACCAGTCCTTCTCCGCCATCCTG
AATGCCTCCTCCCGTGGCTTTGTCCTCCATGGCCTGGAGCCCGCCAGTCTGTATCACGAATTCACTTTTACAACACCAAAGTTCGCTCAAGGAG
AAATAGAAGCCATAGTCGTGCCTGTGTGCTTAGCCTTCCTCCTGACAACCCTGCTGGGCGTCTTGTTCTGCTTTAACAAACGAGAC
CTAATTAAAAAACACATCTGGCCTAATGTTCCTGATCCTTCCAAGAGTCATATTGCCCAGTGGTCACCTCACACCCCCCCAAGGCACAA
TTTTAACTCCAAAGATCAAATGTACTCGGACGGCAATTTCACTGATGTAAGCGTTGTGGAAATAGAAGCAAACAACAAGAAGCCTTGTCCAGATGAC
CTGAAGTCCGTGGACCTGTTCAAGAAGGAGAAAGTGAGTACAGAAGGGCACAGCAGTGGCATCGGGGGCTCTTCATGCATGTCCTCCTCCAGG
CCCAGCATCTCCAGCAACGAGGAGAATGAGTCTGCTCAGAGCACCGCCAGCACGGTGCAGTTCTCCACTGTGGTGCACAGCGGCTACAGGCAC
CAGGTCCCGTCCGTGCAAGTGTTCTCAAGGTCCGAGTCCACCCAGCCCCTGCTAGACTCGGAGGAGCGGCCAGAAGACCTGCAGCTGGTGGA
TAGTGTAGACGGTGGGGATGAGATCTTGCCCAGGCAACCGTATTTCAAGCAGAACTGCAGTCAGCCTGAAGCCTGTCCAGAGATTTCACATTTTGAA
AGGTCAAACCAGGTTTTGTCCGGCAATGAGGAGGATTTTGTCAGACTGAAGCAGCAGCAGGTTTCAGATCACATTTCTCAGCCCTATG
GATCCGAGCAACGGAGGCTGTTTCAGGAAGGCTCTACAGCGGATGCTCTTGGCACGGGGGCTGATGGACAGATGGAGAGATTTGAATCTGTTG
GAATGGAGACCACAATTGATGAAGAAATTCCCAAAAGTTACTTGCCACAGACTGTAAGACAAGGTGGCTACATGCCGCAGTGACTCGAGGTAGGACC
CAGCTTTCTTGTACAAAGTGG. -
c.Insert GY118F into the pDONR221 plasmid via Gateway re-combinatorial cloning, reagent from kit Gateway Technology with Clonase II (Invitrogen, 12535-029). Firstly, perform a BP reaction (recombining the attB and attP sites) between the GY118F synthetic sequence and pDONR221 plasmid, followed by kanamycin (50 μg/mL) selection.
-
d.Perform a LR reaction (recombining the attL and attR sites) between the GY118F pDONR221 plasmid and the PB.CAG.IRES.Bsd plasmid, followed by ampicillin (50 μg/mL) selection.
-
e.Pick the resistant bacteria clones and send for verification by Sanger sequencing using the following primers: CAG-fw: CTACAGCTCCTGGGCAACGT; BSD-fw: ATGGCCACAACCATGCCTTT.
-
a.
-
2.Thawing and maintaining Gata6+/H2BVenus mESCs.
-
a.Coat with 1.5 mL of 0.1% gelatin into each well of a 6-well plate and incubate in the incubator at 37°C for at least 30 min. Remove the gelatin solution before seeding the cells.
-
b.Prewarm 4–5 mL of 2iLIF medium at 15°C–25°C in a 15 mL centrifuge tube.
-
c.Carefully take out a tube of Gata6+/H2BVenus mESCs from liquid nitrogen tank. Immediately thaw the cryopreserved cells in 37°C water bath until only a small piece of ice is visible.
-
d.Quickly transfer the cells into the 15 mL centrifuge tube containing prewarmed culture medium.
-
e.Centrifuge at 300 × g at 15°C–25°C for 5 min.
-
f.Carefully remove the supernatant and resuspend the cell pellet with appropriate volume of 2iLIF medium.
-
g.Seed the cells (2–3×104 cells/ well) on the gelatin coated 6-well plate.
-
h.Refresh the culture medium daily.
-
i.Passage the cells every 2–3 days according to cell density. Add appropriate amount of Accutase (300 μL/ 6-well plate) to Petri dish and incubate the dish in incubator at 37°C for 4 min.
-
j.Collect the cells by centrifugation at 300 × g, 15°C–25°C for 5 min.
-
k.Seed 2×105 cells per 6-well plate.
-
l.Refresh the culture medium daily.
-
a.
-
3.Construction of GY118F Gata6+/H2BVenus mESCs by Electro transfection or Lipofectamine transfection (optional) and subsequent cell selection.
-
a.Aspirate culture medium and wash cells with DPBS once.
-
b.Digest the cells with Accutase as described before and add appropriate amount of N2B27 medium to resuspend.
-
c.Count resuspended cells and then centrifuge approximately 2×105 Gata6+/H2BVenus mESCs for 5 min at 300 × g at 15°C–25°C. Remove the supernatant with caution.
-
d.Set up Neon transfection system apparatus according to the manufacturer’s manual.
-
e.Add 2 mL of 2iLIF medium per well in the gelatin-coated 6-well plate and prewarm in the incubator for 5–10 min before electro transfection.
-
f.Resuspend the cells with 100 μL Resuspension Buffer R (Neon transfection system, Thermo Fischer) containing 2 μg piggyBac (PB) carrying the GY118F transgene and 2 μg non-integrating piggyBac transposase (PBase) expression vector (CAG.PBase).
-
g.Set up the electroporation parameters as following: 10 ms, 1200 V, 3 pulses. Conduct electroporation.
-
h.Seed the electroporated cells into 1 well of a 6-well plate containing the prewarmed culture medium. Place the Petri dish into the incubator at 37°C.
-
i.Observe the cells daily, start with 2iLIF medium only and then switch to 2iLIF medium containing blasticidin S-HCl at 20 μg/mL 2–3 days after transfection.
-
j.Transfected GY118F Gata6+/H2BVenus cells could be considered selected after three passages in 2iLIF + Bsd medium. These cells can be further expanded, under BSD selection, for follow up experiments.Optional: Construction of GY118F Gata6+/H2BVenus mESCs by Lipofectamine transfection.
-
k.Seed 5×104 GY118F Gata6+/H2BVenus mESCs in one 6-well the day before conducting Lipofectamine transfection (optimal time: 14–18 h).
-
l.Check the cell confluency at around 50%–70% at transfection. Fresh the cells with 2 mL of 2iLIF medium 30 min before the transfection.
-
m.Dilute 10 μL Lipofectamine 3000 reagent into 250 μL Opti-MEM medium and mix thoroughly.
-
n.Prepare master mix of 2 μg piggyBac (PB) carrying the GY118F transgene and of 2 μg non-integrating piggyBac transposase (PBase) expression vector (CAG.PBase) in 250 μL Opti-MEM medium, mix thoroughly.
-
o.Add diluted DNA to diluted Lipofectamine 3000 reagent at 1:1 ratio with a total volume of 500 μL.
-
p.Incubate at 15°C–25°C for 10–15 min.
-
q.Add the DNA-lipid complex to cells and incubate at 37°C for 12–14 h.
-
r.Remove the supernatant and replace with appropriate amount of fresh 2iLIF culture medium. Culture the cells for 2–3 days at 37°C incubator before switching to 2iLIF medium containing blasticidin S-HCl at 20 μg/mL.
-
s.Apply selection to the transfected GY118F Gata6+/H2BVenus cells in 2iLIF + Bsd medium for three passages. Expand the cells under continuous BSD selection, for follow up experiments.Note: Transfection of mouse pluripotent stem cells with the GY118F transgene does not affect pluripotent stem cell morphology or cell proliferation. To ensure the transgene response, GY118F Gata6+/H2BVenus mESCs are maintained under continuous blasticidin S-HCL selection at 20 μg/mL. It’s recommended to use this cell line with 8 passages to make sure the generation of MLCs.
-
a.
Figure 1.
PB.CAG.GY118F.IRES.Bsd expression plasmid
Primers used, and their location within the plasmid, for the verification of plasmid are indicated.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SOX2 monoclonal antibody (Btjce) (IF, 1:100) | Invitrogen eBioscience | Cat#14-9811-82; RRID: AB_11219471 |
| Anti-CDX-2 (CDX2-88) (IF, 1:50) | BioGenex | Cat#MU392A-UC; RRID:AB_2923402 |
| Oct-4A (D6C8T) rabbit mAb (IF, 1:200) | Cell Signaling Technology | Cat#83932; RRID: AB_2721046 |
| Anti-TBR2/Eomes antibody (IF, 1:100) | Abcam | Cat#ab23345; RRID: AB_778267 |
| Recombinant anti-Brachyury/Bry antibody (IF, 1:1,000) | Abcam | Cat#ab209665; RRID: AB_2750925 |
| Tox3 polyclonal antibody (IF, 1:100) | Invitrogen eBioscience | Cat# PA5-112771; RRID: AB_2867506 |
| Cerberus antibody (C-1) (IF, 1:100) | Santa Cruz | Cat#; sc-515324: RRID: AB_2923406 |
| Experimental models: Cell lines | ||
| Gata6+/H2BVenus mESCs | Christian Schröter lab, Max Planck Institute of Molecular Physiology; Anna-Katerina Hadjantonakis, Sloan Kettering Institute | N/A |
| GY118F Gata6+/H2BVenus mESCs | This study | N/A |
| Recombinant DNA | ||
| PB.CAG.GY118F.IRES.bsd plasmid | This study | N/A |
| PB.CAG.IRES.BSD plasmid | Addgene | Cat#48760 |
| pDONR221 | Invitrogen eBioscience | Cat#12536017 |
| pBase | SBI | PB210PA-1 |
| Other | ||
| Blasticidin-S-HCl, 20 μg/mL | Thermo Fisher Scientific | Cat#A1113903 |
| DMEM/F-12 | Gibco | Cat#C11330500CP |
| Neurobasal | Gibco | Cat#21103049 |
| GlutaMAX | Gibco | Cat#35050061 |
| MEM NEAA | Gibco | Cat#11140050 |
| 2-Mercaptoethanol | Gibco | Cat#21985023 |
| B27 supplement, serum free, 50x | Gibco | Cat#17504044 |
| N2 supplement, 100x | Gibco | Cat#17502048 |
| Murine LIF, 200 μg/mL in H2O | QKine | Cat#Qk018 |
| CHIR99021, 10 mM in DMSO | Selleck | Cat# S2924 |
| PD0325901, 10 mM in DMSO | Selleck | Cat# S1036 |
| Accutase | STEMCELL | Cat#7920 |
| Paraformaldehyde | Beyotime | Cat#P0099 |
| Triton X-100 | Sigma-Aldrich | Cat#X100 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Neon transfection system | Thermo Fisher Scientific | Cat#MPK5000 |
| 96-well U-bottom suspension plate | Greiner | Cat#650185 |
| Human GCSF, 30 μg/mL | PeproTech | Cat#300-23-10 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | Cat#15140122 |
| Primocin, 50 mg/mL | InvivoGen | Cat#ant-pm-05 |
| DMEM | Thermo Fisher Scientific | Cat#A1443001 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat#11360070 |
| FBS | Capricorn Scientific | Cat#FBS-52A |
| 40 μm cell strainer | BD | Cat#352340 |
| ART wide bore filtered pipette tips | Thermo Fisher Scientific | Cat#2079GPK |
| Advanced DMEM/F-12 | Thermo Fisher Scientific | Cat# 12634010 |
| ITS-X | Thermo Fisher Scientific | Cat#51500056 |
| β-Estradiol, 80 μM in DMSO | Sigma-Aldrich | Cat#E8875-1G |
| Progesterone, 2 mg/mL in DMSO | Sigma-Aldrich | Cat#P0130 |
| N-acetyl-L-cysteine, 250 mM in H2O | Sigma-Aldrich | Cat#C7880-100G |
| 3,3′,5-Triiodo-L-thyronine sodium salt, 1 mM in DMSO | Sigma-Aldrich | Cat#T6397 |
| DPBS | Thermo Fisher Scientific | Cat#14190144 |
| BSA, 10% in H2O | Absin | Cat#abs9156 |
| Anti-adherence rinsing solution | STEMCELL | Cat#7010 |
| AggreWell 400 | STEMCELL | Cat#34415 |
| Tween 20 | Thermo Fisher Scientific | Cat#85113 |
| Triton X-100 | Thermo Fisher Scientific | Cat#28314 |
| ART wide bore filtered pipette tips | Thermo Fisher Scientific | Cat#269G |
| Ampicillin | Thermo Fisher Scientific | Cat#11593027 |
| Kanamycin | Thermo Fisher Scientific | Cat#11815024 |
| Gateway Technology with Clonase II | Invitrogen | Cat#12535-029 |
Materials and equipment
N2B27 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12 | N/A | 240 mL |
| Neurobasal | N/A | 240 mL |
| N2 (50×) | 0.5× | 2.5 mL |
| B27 (100×) | 1× | 5 mL |
| GlutaMAX (100×) | 1× | 5 mL |
| MEM Non-Essential Amino Acids (100×) | 1× | 5 mL |
| 2-mercaptoethanol (55 mM) | 0.1 mM | 0.91 mL |
| BSA (10%), optional | 0.1% | 5 mL |
| Total | N/A | 500 mL |
Store basal medium at 4°C and use within 2 weeks.
2iLIF medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 medium | N/A | 50 mL |
| PD0325901 (10 mM) | 1 μM | 5 μL |
| murine LIF (200 mg/ mL) | 20 ng/mL | 5 μL |
| CHIR99021 (10 mM) | 3 μM | 15 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
2iLIF + Bsd medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 medium | N/A | 50 mL |
| PD0325901 (10 mM) | 1 μM | 5 μL |
| murine LIF (200 mg/ mL) | 20 ng/mL | 5 μL |
| CHIR99021 (10 mM) | 3 μM | 15 μL |
| Blasticidin-S-HCL (10 mg/mL) | 20 μg/mL | 100 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
CL medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 medium | N/A | 50 mL |
| murine LIF (200 mg/ mL) | 20 ng/mL | 5 μL |
| CHIR99021 (10 mM) | 3 μM | 15 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
CLG medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 medium | N/A | 50 mL |
| murine LIF (200 mg/ mL) | 20 ng/mL | 5 μL |
| CHIR99021 (10 mM) | 3 μM | 15 μL |
| GCSF (30 μg/mL) | 30 ng/mL | 50 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
CG medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 medium | N/A | 50 mL |
| CHIR99021 (10 mM) | 3 μM | 15 μL |
| GCSF (30 μg/mL) | 30 ng/mL | 50 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 medium | N/A | 10 mL |
| Penicillin-Streptomycin (10,000 U/mL) | 100 U/mL | 100 μL |
| Primocin (50 mg/mL) | 50 μg/mL | 10 μL |
| Total | N/A | 10 mL |
Make fresh medium immediately before experiment. Do not store FACS buffer.
FC medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | N/A | 37.909 mL |
| FBS | 20% | 10 mL |
| GlutaMAX (100 ×) | 1× | 500 μL |
| MEM Non-Essential Amino Acids (100 mM) | 1× | 500 μL |
| Sodium pyruvate (100 mM) | 1 mM | 500 μL |
| 2-mercaptoethanol (55 mM) | 0.1 mM | 91 μL |
| Penicillin-Streptomycin (10,000 U/mL) | 100 U/mL | 500 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
IVC medium (Amadei, G. et al.)5
| Reagent | Final concentration | Amount |
|---|---|---|
| advanced DMEM/F12 | N/A | 33.485 mL |
| FBS | 30% | 15 mL |
| GlutaMAX (100 ×) | 1× | 500 μL |
| ITS-X (100 ×) | 1× | 500 μL |
| β-estradiol (80 μM) | 8 nM | 5 μL |
| Progesterone (2 mg/mL) | 200 ng/mL | 5 μL |
| N-acetyl-L-cysteine (250 mM) | 25 μM | 5 μL |
| 3,3′,5-Triiodo-L-thyronine sodium salt (1 mM) | 100 nM | 5 μL |
| Penicillin-Streptomycin (10,000 U/mL) | 100 U/mL | 500 μL |
| Total | N/A | 50 mL |
Store medium at 4°C and use within 1 week.
Blocking buffer for immunofluorescence
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 45 mL |
| FBS | 5% | 2.5 mL |
| BSA (10%) | 2% | 1 mL |
| Tween-20 (10%) | 0.5% | 250 μL |
| Triton X-100 (10%) | 0.3% | 1.5 mL |
| Total | N/A | 50 mL |
Store medium at −20°C and use within 3 months.
Step-by-step method details
Induction and capture of MLCs
Timing: 3 days
GCSF mediated pSTAT3 induction in GY118F Gata6+/H2BVenus mESCs and capture of MLCs with fluorescence-activated cell sorting (FACS).
-
1.Switch the culture medium of GY118F Gata6+/H2BVenus mESCs from 2iLIF + Bsd medium to CL medium for 24 h to remove MEK/ERK signaling inhibition. Treat the cells with GCSF (G) in either CLG medium or CG medium for GCSF-GY118F induced increased pSTAT3 and MLC generation.
-
a.Dissociate the GY118F Gata6+/H2BVenus mESCs with Accutase as mentioned above.
-
b.Seed 5×105 dissociated GY118F Gata6+/H2BVenus mESCs into a 100 mm Petri dish containing 10 mL of CL medium for 24 h.
-
c.Dissociate the pretreated cells with Accutase, wash with 5 mL N2B27 medium, transfer the cells/Accutase/wash suspension to a falcon tube and collect the cells by centrifugation.
-
d.Seed 9×105 dissociated pretreated cells into a 100 mm Petri dish containing 10 mL of CLG or CG medium.
-
e.Refresh the CLG or CG medium the next day.
-
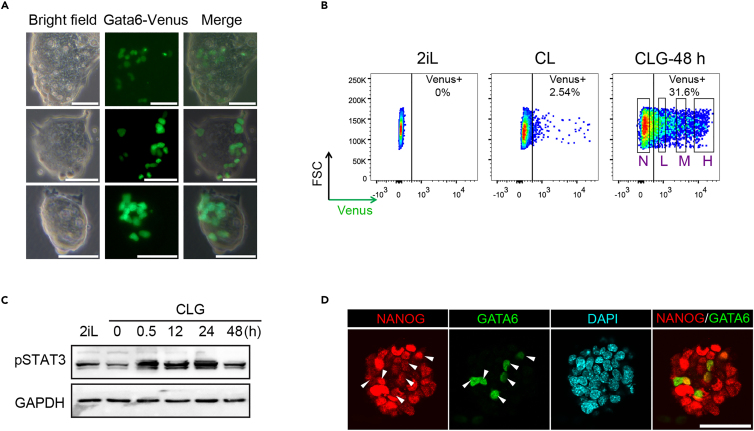
f.Induce the ESCs with CLG or CG medium for 48 h. Check under a microscope whether green fluorescence can be easily observed (Figure 2A).
-
a.
Note: CG induction is equally effective, as compared to CLG, in the generation of MLCs.
CRITICAL: The cell density of seeded cells will influence the induction efficiency according to our experience. When conducting GCSF induction, the seeded cell density should be no less than 8×105 cells per 100 mm Petri dish to ensure induction efficiency. Adjust the cell density according to the dish surface area.
-
2.Capture of MLCs by FACS.
-
a.Dissociate GCSF induced GY118F Gata6+/H2BVenus cells into single cells with Accutase at the 48h-time point.
-
b.Resuspend the centrifuged cells with ice cold FACS buffer at a density of 107 cell per mL.
-
c.Filter the resuspended cells through 40 μm cell strainer to eliminate cell clumps.
-
d.To ensure cell viability of sorted cells, from this point on until seeding, keep the cells on ice from this step until seeding.
-
e.Flow sort Gata6 positive cells. Since GY118F Gata6+/H2BVenus cells carry the Venus fluorescent protein, 488-FITC laser is required for successful sorting.
-
f.Sort the High Gata6 expressing cell population as indicated. This fraction is enriched for MLCs (Figure 2B).
-
a.
Figure 2.
Generation and isolation of MLCs
(A) Representative phase and fluorescent images of GY118F Gata6::Venus ESCs undergoing GCSF-GY118F mediated STAT3 activation in CLG medium for 48 h. Scale bar: 50 μm.
(B) Flow cytometry analysis of GY118F Gata6::Venus ESCs cultured in either: 2iLIF (MEK/ERK and GSK3β signaling inhibitors (2i) and LIF (L)) or CL medium or after 24 h in CL medium followed by 48 h in CLG medium. GCSF-GY118F STAT3 stimulated Gata6::Venus ESCs (CLG) were single cell sorted based on the GATA6::Venus reporter activity level as indicated; negative (N), low (L), medium (M), and high (H).
(C) Western blot time-course analysis for GCSF-GY118F mediated activation of STAT3 (Phospho(p)- STAT3) by CLG at 0.5, 12, 24 and 48 h compared to 0 h in CL. 2iLIF culture condition was introduced as an additional control.
(D) Representative immunofluorescence of GATA6 and NANOG. Scale bar, 50 μm. Figure reprinted with permission from Li et al.1
Generation of MLC-derived embryoids
Timing: 5 days
Sorted MLCs are immediately seeded into AggreWell400 for the generation of MLC-derived embryoids.
-
3.Preparation of AggreWell400 plate.
-
a.Add 500 μL anti-adherence solution per well and incubate at 15°C–25°C for 30 min.
-
b.Spin down at 1000 × g for 1 min at 15°C–25°C and observe under the microscope that no bubbles are present in the microwells.
-
c.Remove solution and rinse with DMEM medium twice.
-
a.
-
4.Seeding of captured MLCs into the AggreWell400.
-
a.Resuspend sorted MLCs with FC medium at a final density of 4.8×104 cells/mL.
-
b.Seed MLCs suspension at a density of 2.4×104 per well of AggreWell400 (20 MLCs per microwell) plate.
-
a.
-
5.Culture of MLC-embryoids.
-
a.On day 1 and day 2, aspirate medium gently and add 1 mL of fresh FC medium.
-
b.At day 3, aspirate the FC medium gently and add 1.5 mL of freshly prepared IVC medium.
-
c.At day 4, gently transfer the MLC-embryoids to a suspension 6-well plate, containing 4 mL of fresh IVC medium per well, and culture in a shaking incubator at 60–80 rpm, 37°C.
-
a.
-
6.
Collection of MLC-embryoids.
Collect MLC-embryoids at day 3, day 4 and day 5 for further examination.-
a.For day 3 and day 4 MLC-embryoids, remove the culture medium, and pipet gently up and down to harvest the MLC-embryoids with the wide bore pipette tips. Wash the harvest MLC-embryoids with PBS once.
-
b.To harvest the Day 5 MLC-embryoids, aspirate the medium gently and wash with PBS once.
-
a.
-
7.Staining of MLC-embryoids.
-
a.Fix the MLC-embryoids for 30 min at 15°C–25°C with 4% paraformaldehyde.
-
b.Wash the MLC-embryoids once with PBS and permeabilize with 0.5% Triton X-100 in PBS for 1 h at 15°C–25°C.
-
c.Block the MLC-embryoids with blocking buffer at 15°C–25°C for 1 h.
-
d.Incubate the MLC-embryoids with appropriate primary antibodies in blocking buffer at 4°C 12–14 h.
-
e.Wash the MLC-embryoids three times with PBST (PBS containing 0.05% Tween-20).
-
f.Incubate the MLC-embryoids with Alexa Fluor tagged secondary antibodies of choice at 15°C–25°C for 1 h.
-
g.Directly counterstain cell nuclei with DAPI for 20 min.
-
h.Wash the MLC-embryoids three times with PBST and store in PBS.
-
a.
Expected outcomes
PB.CAG.GY118F.IRES.Bsd expression plasmid
Constitutive PB.CAG.GY118F.IRES.Bsd plasmid should be verified by two primers; one designed to sequence the GY118F transgene and the other one to sequence the selection locus. CAG-fw: CTACAGCTCCTGGGCAACGT; BSD-fw: ATGGCCACAACCATGCCTTT (Figure 1).
MLC induction in GY118F Gata6+/H2BVenus ESCs
Since GY118F Gata6+/H2BVenus cells carry a Gata6 reporter, successful activation of this can be examined under the microscope. The green fluorescence can be observed at random position within the cell colonies at the 16 h-time point. At the 48 h-time point, Gata6 reporter green fluorescence is easily observed and is present in a significant proportion of the cells (Figure 2A). To further examine successful GCSF-GY118F mediated STAT3 activation, compare pSTAT3 levels of GCSF treated cells with parental CL ESCs. pSTAT3 levels should be detected at higher levels in GCSF treated cells (Figures 2C–2D).
MLCs adopt cell identity and potency of natural morula embryo cell
MLCs show a molecular identity comparable to in vivo morula embryo cells on transcriptomic level (Figures 3A and 3B). MLCs develop rapidly and specifically into one of the three early embryo cell fates according to the culture conditions used, that is, into an ESC/EPI fate, using 2iL medium6 (Figure 4A), into a PrE fate using XEN medium (FBS plus Retinoic Acid plus Activin A)7 (Figure 4B) and into a TE fate using TE medium (FBS plus Fgf4 plus Heparin)8 (Figure 4C). These results show that the fate of MLCs is impacted by the signaling cues used in the medium and this response draws parallels to early embryo cells.
Figure 3.
Morula-like cells are molecularly similar to embryo Morula cells
(A) UMAP embeddings of single cells transcriptome of GY118F Gata6::Venus ESCs cultured in 2iLIF (Venus 2iL, blue), or in CL (Venus CL, yellow) or in CLG medium for 48 h (Venus H, green) . These were integrated with published datasets from early mouse embryo stages from zygote till E6.5.9,10,11,12,13
(B) Violin plots showing the expression levels of indicated genes in Gata6::Venus ESCs in 2iLIF medium or in CL medium or upon GCSF-GY118F mediated increased STAT3 activation (N, L, M or H cells upon ipSTAT3) in CLG medium for 48 h.10,11 Published datasets of early embryo developmental stages were also included (2-cell (C), 4C, 8C, 16C and E3.5 ICM). Data was generated by Smart-seq2 scRNA-seq. Figure reprinted with permission from Li et al.1
Figure 4.
MLCs respond to medium cues similarly to embryo cells
(A–C) Representative immunofluorescence for EPI markers (NANOG, OCT4, SOX2), TE marker (CDX2), PrE markers (GATA4, SOX17) in MLCs cultured in either ESC, TSC or XEN medium. Scale bar, 50 μm. Figure reprinted with permission from Li et al.1
Progression of MLC-derived embryoids
In the process of generating MLC-derived embryoids, the day we seed MLCs is defined as day 0 (Figure 5). At day 1, we can already start to observe cells forming compact aggregates. Starting from day 3, most MLC-derived aggregates appear elongated and form a cavitated structure. Embryoid formation rate per well reaches over 90% efficiency at day 3 according to our observations (Figure 5). Of these, approximately 35% adopt a peri-implantation embryo-like structure at around day 3–4. By day 5, MLC-embryoids are significantly increased in size and adopt gastrulating morphology.
Note: At day 1, if the cells appear not to have formed compact aggregates, keep cells un-interrupted and observe again on the following day. If cells still cannot aggregate, consider the possibility that cells may have died during the culture process.
Figure 5.
MLC-embryoid formation
Representative phase images of emerging embryo-like structures over a time course of 5 days. Scale bar, 100 μm. Figure reprinted with permission from Li et al.1
Staining of MLC-derived embryoids
We stain these MLC-derived embryoids for SOX2 and CDX2 to examine the lineage specification and correct positioning of each cell lineage. At day 4, MLC-embryoids should have morphology resembling natural embryo at post-implantation stage. We usually stain these with OCT4 and CDX2 to monitor lineage specification and the positioning of each cell lineage. At day 5, MLC-embryoids start to show AVE (anterior visceral endoderm) and primitive streak formation, as indicated by TOX3 and CER1 and by T staining respectively (Figure 6).
Figure 6.
Immunofluorescence staining of MLC-embryoid at day 3, day 4 and day 5
(A–C) Representative immunofluorescence images of lineage markers in day 3, day 4 and day 5. Scale bar, 50 μm. Figure reprinted with permission from Li et al.1
Limitations
In this protocol, robust generation of MLCs relies on a transgene (Chimeric GCSF-LifR Y118F) to elevate pSTAT3 levels and also on a reporter (Gata6) to isolate MLCs. MLCs are also transient, which means that they cannot be maintained in self-renewal. For this reason, MLCs must be evaluated for their molecular signature and employed in functional assays straight after being generated.
Troubleshooting
Problem 1
GY118F Gata6+/H2BVenus ESC colonies are poorly attached onto gelatin-coated plates and float (related to Step 1).
Potential solution
After spinning cell suspension with Accutase and wash buffer, make sure no residual liquid remains. Increase gelatin concentration to 0.2% and add 0.1% BSA to the culture medium.
Problem 2
GY118F Gata6+/H2BVenus ESCs are differentiated (related to Step 1).
Potential solution
In our experience, the starting condition of the GY118F Gata6+/H2BVenus cells is of critical importance.
Make sure you are using freshly made and high-quality medium. Also, avoid overgrowing culture. If the latter occurs thaw a new vial of cells.
Problem 3
No green fluorescence can be observed after CLG or CG induction (related to Step 1f)
Potential solution
Make sure you pretreat the cells with CL medium for no less than 18 h and no more than 24 h.
Problem 4
Only low green fluorescence can be observed after CLG or CG induction due to lack of fresh/high quality medium (related to Step 1f)
Potential solution
Make sure you are using high quality N2 and B27 and store your growth factors properly.
Problem 5
MLCs are not able to form compact aggregates at day 1 and day 2 (related to step 5a).
Potential solution
If compact aggregate cannot be observed at day 1, leave the plate undisturbed and observe the next day. If no compact aggregates are observed by day 2, this may indicate that seeded MLCs have low viability due to a technical issue.
To ensure good viability of MLCs.
-
•
use low flow rate when FACS sort for MLCs.
-
•
place sorted MLCs on cold environment until the point of seeding.
Resource availability
Lead contact
Further information and requests for resources or reagents should be directed to and will be fulfilled by the lead contact, José C. R. Silva (jose_silva@gzlab.ac.cn).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be answered by the technical contact, Huanhuan Li (li_huanhuan@gzlab.ac.cn).
Materials availability
The mouse lines and other research reagents generated in this study will be distributed upon request to other research investigators under a Material Transfer Agreement.
Data and code availability
This study did not generate new datasets.
Acknowledgments
We thank Drs. Christian Schröter and Anna-Katerina Hadjantonakis for kindly providing Gata6+/H2B-Venus (Gata6::Venus) mouse ESCs. We thank the flow cytometry core and imaging core in Guangzhou National Laboratory for technical help. This study was supported by grants from Department of Science and Technology of Guangdong Province, China (2021ZT09Y233 to J.C.R.S.), National Natural Science Foundation of China, China (32100653 to H.L.), Guangzhou National Laboratory, China (Major Project: GZNL2023A02005 and GZNL2023A02006, to J.C.R.S. and H.L.), and Guangzhou Municipal Science and Technology Bureau, China (202201010891 to H.L.). The J.C.R.S. laboratory is supported by Guangzhou National Laboratory.
Author contributions
H.L. wrote the manuscript as well as developed the experimental methods, performed the experiments, and supervised the project. L.C. and J.H. wrote the manuscript and performed the experiments. J.C.R.S. wrote the manuscript and supervised the project.
Declaration of interests
J.C.R.S. and H.L. submitted a patent application (202210437013.X) related to this work (filed by Guangzhou National Laboratory).
Contributor Information
Huanhuan Li, Email: li_huanhuan@gzlab.ac.cn.
José C.R. Silva, Email: jose_silva@gzlab.ac.cn.
References
- 1.Li H., Chang L., Wu J., Huang J., Guan W., Bates L.E., Stuart H.T., Guo M., Zhang P., Huang B., et al. In Vitro Generation of Mouse Morula-like Cells. Dev. Cell. 2023;58:2510–2527.e7. doi: 10.1016/j.devcel.2023.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Stuart H.T., van Oosten A.L., Radzisheuskaya A., Martello G., Miller A., Dietmann S., Nichols J., Silva J.C.R. NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr. Biol. 2014;24:340–346. doi: 10.1016/j.cub.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdon T., Stracey C., Chambers I., Nichols J., Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 4.Stuart H.T., Stirparo G.G., Lohoff T., Bates L.E., Kinoshita M., Lim C.Y., Sousa E.J., Maskalenka K., Radzisheuskaya A., Malcolm A.A., et al. Distinct Molecular Trajectories Converge to Induce Naive Pluripotency. Cell Stem Cell. 2019;25:388–406.e8. doi: 10.1016/j.stem.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amadei G., Handford C.E., Qiu C., De Jonghe J., Greenfeld H., Tran M., Martin B.K., Chen D.Y., Aguilera-Castrejon A., Hanna J.H., et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature. 2022;610:142–153. doi: 10.1038/s41586-022-05246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niakan K.K., Schrode N., Cho L.T.Y., Hadjantonakis A.K. Derivation of extraembryonic endoderm stem (XEN) cells from mouse embryos and embryonic stem cells. Nat. Protoc. 2013;8:1028–1041. doi: 10.1038/nprot.2013.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubaczka C., Senner C., Araúzo-Bravo M.J., Sharma N., Kuckenberg P., Becker A., Zimmer A., Brüstle O., Peitz M., Hemberger M., Schorle H. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Rep. 2014;2:232–242. doi: 10.1016/j.stemcr.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng S., Pei Y., He L., Peng G., Reinius B., Tam P.P.L., Jing N., Deng Q. Single-Cell RNA-Seq Reveals Cellular Heterogeneity of Pluripotency Transition and X Chromosome Dynamics during Early Mouse Development. Cell Rep. 2019;26:2593–2607.e3. doi: 10.1016/j.celrep.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 10.Deng Q., Ramsköld D., Reinius B., Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 11.Mohammed H., Hernando-Herraez I., Savino A., Scialdone A., Macaulay I., Mulas C., Chandra T., Voet T., Dean W., Nichols J., et al. Single-Cell Landscape of Transcriptional Heterogeneity and Cell Fate Decisions during Mouse Early Gastrulation. Cell Rep. 2017;20:1215–1228. doi: 10.1016/j.celrep.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowotschin S., Setty M., Kuo Y.Y., Liu V., Garg V., Sharma R., Simon C.S., Saiz N., Gardner R., Boutet S.C., et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature. 2019;569:361–367. doi: 10.1038/s41586-019-1127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posfai E., Petropoulos S., de Barros F.R.O., Schell J.P., Jurisica I., Sandberg R., Lanner F., Rossant J. Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. Elife. 2017;6 doi: 10.7554/eLife.22906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new datasets.