Abstract
The local cellular immune response to herpes simplex virus (HSV) is important in the control of recurrent HSV infection. The antiviral functions of infiltrating CD4-bearing T cells may include cytotoxicity, inhibition of viral growth, lymphokine secretion, and support of humoral and CD8 responses. The antigens recognized by many HSV-specific CD4 T cells localizing to genital HSV-2 lesions are unknown. T cells recognizing antigens encoded within map units 0.67 to 0.73 of HSV DNA are frequently recovered from herpetic lesions. Expression cloning with this region of DNA now shows that tegument protein VP22 and the viral dUTPase, encoded by genes UL49 and UL50, respectively, are T-cell antigens. Separate epitopes in VP22 were defined for T-cell clones from each of three patients. Reactivity with the tegument protein encoded by UL21 was identified for an additional patient. Three new epitopes were identified in VP16, a tegument protein associated with VP22. Some tegument-specific CD4 T-cell clones exhibited cytotoxic activity against HSV-infected cells. These results suggest that herpes simplex tegument proteins are processed for antigen presentation in vivo and are possible candidate compounds for herpes simplex vaccines.
Cellular immune responses are required to limit the severity of recurrent herpes simplex virus (HSV) infections in humans (38). HSV-specific CD4 and CD8 T cells infiltrate herpetic lesions (16, 17, 19). In vitro, some HSV-specific CD4 T cells are cytotoxic toward virus-infected cells (41, 42). HSV-specific CD4 T cells make large amounts of gamma interferon (7), which may overcome HSV-mediated HLA class I downregulation and permit lysis of HSV-infected cells by CD8 cytotoxic T lymphocytes (CTL) (35). Gamma interferon also upregulates HLA class II on HSV-infected keratinocytes to allow recognition by cytolytic CD4 T cells (26) and has direct antiviral effects (13).
The antigenic specificity of HSV-specific CD4 T cells in infected humans is incompletely defined. CD4 T-cell responses in peripheral blood mononuclear cells (PBMC) to HSV envelope glycoproteins B, C, D, and H have been detected by bulk culture, limiting dilution, or clonal analyses (22, 26, 36, 44). PBMC-derived CD4 T-cell clones (TCC) commonly recognize envelope glycoproteins (43, 44), and bulk cultures of PBMC-derived HSV-specific CD4 CTL efficiently recognize glycoprotein-expressing target cells (26). However, we determined that only a minority of lesion-infiltrating CD4 TCC recognize glycoprotein B, C, or D (17), and bulk cultures of lesion-infiltrating T cells show reactivity with glycoprotein B or D for only 50 to 70% of donors (19). We therefore undertook studies to define additional HSV T-cell antigens.
The region near 0.7 map units on the HSV type 2 (HSV-2) genome was identified as rich in HSV-2 type-specific T-cell antigens (17). The unique long (UL) 48 gene, encoding tegument protein VP16, contains at least three T-cell epitopes (14) but does not account for all the antigenicity in this region (17). Expression cloning with subgenomic DNA from this region initially identified VP22 (encoded by UL49) and dUTPase (encoded by UL50) as T-cell antigens. Expression cloning with full-length viral DNA independently showed recognition of VP22 by a lesion-derived TCC from a second donor, and local VP22-specific responses were also detected for a third donor at bulk and clonal levels. Three separate epitopes were identified in VP22. Reactivity with an additional tegument protein, UL21, was also detected. VP22 is tightly associated with VP16 in the virion tegument as well as within infected cells (10). We therefore extended our previous studies with VP16 and identified three additional epitopes recognized by lesion-infiltrating T cells.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 strain E115 (34), HSV-2 strain 333 (15), and intertypic recombinant viruses (IRV) RS1G31 (29), DX32 (31), and RP-2 (17) were grown and titered in Vero cells (20). Epstein-Barr virus-transformed lymphoblastoid cell lines (EBV-LCL) included both autologous cell lines created from donors with genital herpes as previously described (20) and the characterized lines AMAI (homozygous for HLA DPB1*0402), HOM2 (homozygous for HLA DQB1*0501), MAT (homozygous for HLA DQB1*0201), and ARENT (homozygous for HLA DPB1*2001) (4).
HSV-specific T cells were obtained after approval by the Institutional Review Board. Most clones were derived without secondary in vitro stimulation with antigen. Donors 1 through 4 are numbered as previously described (17), and donors 1, 2, and 4 were the sources of lesion-derived clones 1.L3D5.10.8, 2.3, and 4.2E1, respectively; clones 2.3 and 4.2E1 have been described previously (17). Additional lesion-derived clones came from donor 5, from whose second, third, and fourth lesion samples (each separated by 1 year) clones ESL2.20, ESL3.334, ESL4.34, and ESL4.9 were derived. Other clones, such as RH.13 and KM.7, were derived from donors 6 and 7. Clones 2.3, 4.2E1, ESL2.20, RH.13, and KM.17 were derived directly from herpetic vesicle fluid (16). To derive CD4 TCC ESL4.9, biopsy of a recurrent genital HSV-2 lesion (day 3 of symptoms) was performed and bulk lesion-infiltrating cells were expanded with phytohemagglutinin and interleukin 2 (Schiaperelli Biosystems, Columbia, Md.) in the presence of acyclovir as described elsewhere (19). After 16 days, cells were cloned at 1 cell/well (16). Previously described VP16-specific clones 1A.B.25, ESL3.334, and ESL4.34 (8, 14, 18) were similarly derived from bulk cultures.
Some clones were derived by using secondary in vitro stimulation with antigen. To derive additional TCC from donor 1 (17), phytohemagglutinin-driven bulk cultures were prepared from each of four 2-mm biopsy specimens (day 5 of symptoms) obtained 6 years after the occurrence of the lesion from which clone 1A.B.25 (mentioned above) was derived. After 16 days, 1.5 × 106 bulk lymphocytes from one biopsy culture were stimulated with 10 μg of HSV-2 VP22, amino acids 105 to 190 (VP22 105-190)/ml (see below) and an equal number of autologous irradiated (3,300 rads) PBMC in 2 ml of T-cell medium (16). Interleukin 2 (32 U/ml) was added starting on day 6. TCC 1.L3D5.10.8 was isolated from this line on day 12 as described elsewhere (16). To create PBMC-derived TCC SB.17 and BM.17, 1.5 × 106 PBMC of HSV-2-seropositive donors 8 and 9 were stimulated for 12 days with 4 μg of baculovirus-derived full-length VP16/ml in 24-well plates; responding cells were cloned at 1 cell/well. TCC and lines were used 10 to 14 days after the last stimulation.
All cell lines were negative for mycoplasmas except ARENT. ARENT was initially positive for mycoplasmas by DNA probe test (Genprobe, San Diego, Calif.) and was treated with ciprofloxacin at 10 μg/ml (12) for 2 weeks prior to utilization, with conversion of the test to negative.
Flow cytometry.
Flow cytometry used a combination of murine monoclonal antibodies (MAb) to human CD4 (clone SFCI 12T4D11) and CD8 (clone SFCI 21Thy2D3, recognizing the α chain of human CD8) (Coulter, Hialeah, Fla.).
Immunoblotting.
Lysates of HSV-infected Vero cells were prepared, electrophoresed through sodium dodecyl sulfate (SDS)–10% polyacrylamide gels, and transferred to a nitrocellulose membrane as described elsewhere (1). Blots were blocked with phosphate-buffered saline (PBS)–0.05% Tween 20–1% nonfat dried milk. Antigen was detected by sequential incubation with a 1:100 dilution of MAb P43, specific for the UL49 gene product, VP22 (11), affinity-purified goat anti-mouse immunoglobulin M-peroxidase conjugate (Sigma, St. Louis, Mo.), and the TMB substrate system (Kirkegaard and Perry, Gaithersburg, Md.) with three washes (5 min each) in PBS-Tween between each step.
Viral DNA libraries and cloning.
For subgenomic DNA, the HSV-2 strain HG-52 BamHI w fragment was subcloned from the BglII i fragment and gel purified. Viral DNA was digested with SmaI, BamHI ends were blunted with Klenow DNA polymerase, and DNA fragments were purified by phenol extraction and alcohol precipitation. For whole-virus DNA, confluent Vero cells were infected with HSV-2 strain HG52. Total nucleic acids from three 150-cm2 cell cultures were prepared by proteinase K digestion, chloroform-phenol extraction, and isopropanol precipitation. The resultant material was treated with RNAse H and was reextracted and precipitated. Aliquots (1 μg) of HG52 DNA were digested with SmaI or AluI, and 80% of these digests were combined and recovered as described above for creation of expression libraries.
Expression cloning used pUEX vectors (5). pUEX1, -2, and -3 DNAs were linearized with SmaI, dephosphorylated with calf intestinal phosphatase, and gel purified. Approximately 100 ng of vector and 500 ng of DNA fragments were ligated, and 10% of ethanol-precipitated reaction mixtures were used to transform Escherichia coli DH10 Electromax (GIBCO) by electroporation (BTX, San Diego, Calif.) in 1-mm cuvettes. After 1 h of recovery in 1 ml of SOC medium, portions were frozen as glycerol stocks (100 μl each), titered on ampicillin plates at 30°C (250 μl), or used directly (250 μl) for protein induction to create fusion protein libraries. Several thousand ampicillin-resistant colonies were isolated per transformation. To amplify genomic libraries, glycerol stocks were grown overnight at 30°C in 2× YT-ampicillin medium and refrozen.
Confirmatory subcloning of VP22 105-190, UL50 118-312, and UL50 118-250 was performed by isolating the 262-bp SmaI-StuI fragment of UL49, the 583-bp SmaI fragment of UL50, and the 397-bp SmaI-StuI fragment of UL50, respectively. Fragments were cloned into the appropriate linearized, gel-purified pUEX vector, and protein was expressed in E. coli DH5α. Constructs were confirmed by sequencing.
Antigens.
Whole-virus preparations containing 108 to 109 PFU/ml were UV inactivated for 30 min (16) and used at a 1:100 final dilution. Peptides of VP22 were synthesized as described elsewhere (18) and used as stocks at 2 mg/ml in dimethyl sulfoxide. Peptides of UL48, 13 amino acids long and overlapping by 4 amino acids, and VP16 of HSV-2, amino acids 1 to 416, and full-length VP16, both expressed in baculovirus, were a kind gift of Rae L. Burke and Michael Tigges, Chiron Corporation, Emeryville, Calif.
Bacterium-derived protein antigen expression was induced for 2 h at 42°C in cells growing logarithmically (optical density at 600 nm, 0.4 to 0.6) in 2× YT-ampicillin broth at 30°C. Protein was purified as described elsewhere (27), omitting gel purification. Bacterial cultures of 50 ml (libraries) or 5- to 10-ml cultures (pools and clones) yielded fine particulate suspensions in 200 to 400 μl of PBS (Ca and Mg free). Protein concentrations were determined by bicinchoninic acid assay (Pierce, Rockford, Ill.) after proteins were solubilized in 1% SDS at 60°C for 10 min. In some experiments, heat-induced bacteria were washed with PBS and PBS–10 mM EDTA, heated to 56°C for 10 min, and washed in PBS prior to use as antigens.
After identification of an active library of viral DNA, antigen identification used 30 to 60 clones for subgenomic viral DNA fragments or 2,000 to 3,000 clones for full-length viral DNA. For the less complex library, 1-ml cultures of each clone were processed as pools of six to eight clones. Individual clones within the active pool, and confirmatory subclones containing known viral DNA fragments, were processed as 5-ml cultures. A combinatorial method (27) was used to screen libraries from whole-virus DNA. Glycerol stocks of amplified libraries of transformed bacteria were titered on ampicillin plates; 20 to 30 colonies/well were cultured overnight at 30°C in a 96-well plate in a rotating shaker. Cultures were diluted 1:100 into 1-ml cultures, and fusion protein synthesis was induced as described above. Portions (400 μl) of cultures were pooled row- and columnwise for protein purification and evaluation in lymphoproliferation assays. If more than one row and column were positive, wells at the intersections of positive rows and one positive column were used to prepare protein from 5- to 10-ml cultures to definitively identify a positive well. Cultures (n = 96 colonies) of bacteria were derived from ampicillin plates seeded with diluted broth from positive wells. These were evaluated as pools (of 12 bacterial colonies) and then individual clones.
Lymphocyte functional assays.
Triplicate proliferation assay wells contained 104 cloned T cells, 105 irradiated (3,300 rads) PBMC, or 2.5 × 104 irradiated (8,000 rads) EBV-LCL as antigen-presenting cells (APC) and antigen in 200 μl of T-cell medium (18) in 96-well U-bottom plates. When heat-killed bacteria were used as the antigen, the equivalent of 105 CFU/well (prior to inactivation) was added and gentamicin (20 μg/ml) was included. After 72 h, 1 μCi of [3H]thymidine/well was added for 18 h, cells were harvested, and incorporation of thymidine was evaluated by liquid scintillation counting. Standard deviations were less than 10% of the mean values. Results are reported as mean counts per minute or as change in counts per minute (Δcpm), equal to mean counts per minute for the experimental antigen minus mean counts per minute for the control antigen. The control antigen was mock-infected cell lysate for whole-virus antigens and pUEX2-derived β-galactosidase for recombinant protein preparations. To determine the reactivity of bulk-cultured lesion-derived T cells, fusion proteins or control β-galactosidase was used at 10 μg/ml. Glycoproteins B and D and VP16 of HSV-2 were used at 1 μg/ml, and assays were performed as described previously (19). To determine HLA-restricting loci, HLA-DR-specific MAb L243 (32), HLA-DP-specific MAb B7.21 (37), or HLA-DQ-specific MAb SPV-L3 (33) was used as described previously (17).
Cytolysis assays were performed in triplicate by using 4-h 51Cr release as described elsewhere (20). Target EBV-LCL were infected for 18 h with HSV-2 at a multiplicity of infection of 30 or pulsed with 1.0 μM peptide for 90 min prior to washing as described elsewhere (21). The effector-to-target ratio was 20:1. Spontaneous release was less than 28%.
DNA sequencing.
Viral inserts in plasmids in bacteria yielding active proteins were completely sequenced (Taq DyeDeoxy FS kit; Perkin-Elmer ABI, Foster City, Calif.) in both directions by starting with primers CATGGCTGAATATCGACGGT (5′ end of insert) and CTAGAGCCGGATCGATCCGGTC (3′ end of insert) and then using internal primers designed as required.
HLA typing.
HLA-DR and -DQ typing was performed at class II alleles by serologic methods or at the DNA level by reverse dot blot hybridization (25). HLA-DP typing was performed by sequencing (HLA-DP kit; Perkin-Elmer ABI).
RESULTS
Fine localization of T-cell epitopes.
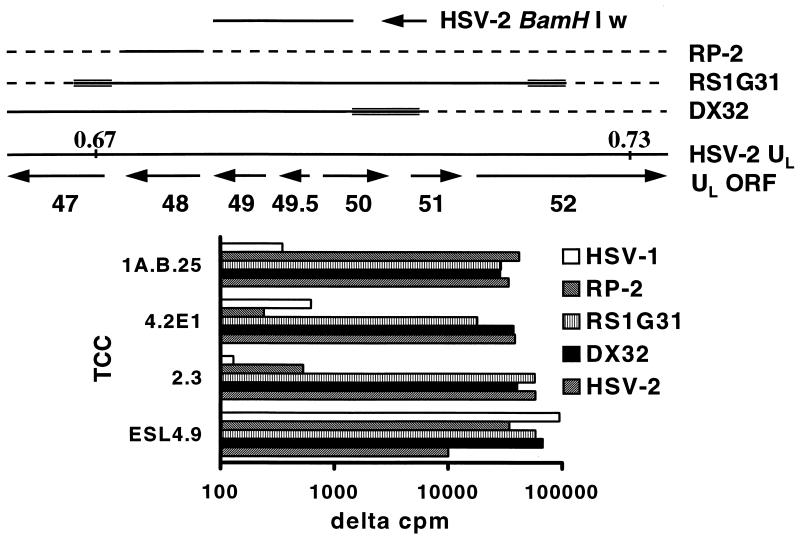
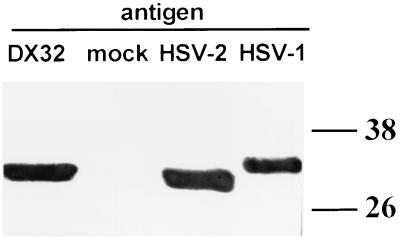
To reduce the complexity of libraries for expression cloning, we selected a TCC-recognizing antigen(s) partially mapped by using HSV-1 × HSV-2 IRV. We have previously determined that HSV DNA near 0.7 map units encodes T-cell antigens in addition to VP16. Epitope mapping for TCC 4.2E1 and 2.3 (17) was improved with IRV DX32 (Fig. 1). This HSV-2-based virus contains a block of HSV-1 DNA near 0.7 map units (31). The UL48 gene product has the HSV-2 phenotype, as shown by reactivity with the HSV-2 type-specific, VP16-specific (17) TCC 1A.B.25. The UL49 (Fig. 2) and UL50 gene products (39, 40) also have an HSV-2 phenotype. The HSV-2 DNA present in IRV DX32 therefore includes UL48, UL49, UL50, and most likely the intervening UL49.5. Since TCC 4.2E1 and 2.3 react with RS1G31 and DX32, but not with RP2 (Fig. 1), recognition of UL49, UL49.5, or UL50 is most likely.
FIG. 1.
(Top) Organization of the HSV genome in the region of map units 0.67 to 0.73. Boundaries are approximate. HSV-1 × HSV-2 IRV are also shown. HSV-2 DNA is indicated by a solid line, HSV-1 DNA by a dashed line, and indeterminate regions by multiple lines. The HSV-2 BamHI w fragment used for expression cloning is also shown. ORF, open reading frame. (Bottom) Proliferative responses of TCC to the indicated IRV. Data are Δcpm, expressing [3H]thymidine incorporation compared to that in the medium, which was less than 500 cpm in each case.
FIG. 2.
Determination of the HSV viral phenotype of the UL49 gene product (VP22) of IRV DX32. Lysates of mock-infected cells and cells infected with the indicated viral strains were separated by SDS-polyacrylamide gel electrophoresis, blotted, and probed with VP22-specific MAb. The molecular weights (in thousands) of marker proteins are shown at the right.
Expression cloning to determine T-cell antigens.
The BamHI w fragment of HSV-2 was selected for expression cloning, since it contains the UL49 and UL49.5 coding sequences and most of the UL50 coding sequence (6, 11, 24). Seventy to ninety percent of random colonies contained an insert; all were of viral origin. The most active libraries (Table 1) for each TCC (pUEX1 for TCC 4.2E1 and pUEX3 for TCC 2.3) were selected, and an individual reactive bacterial clone was detected by sequential testing of pools and individual colonies (Table 2). Clone 1.1.3 encodes a fusion protein eliciting proliferation by TCC 4.2E1. This clone contains a backwards 80-bp SmaI fragment of UL49, a 262-bp SmaI fragment of HSV-2 UL49 DNA predicted to encode amino acids 105 to 190, forward and in frame with regard to β-galactosidase, and a 246-bp SmaI fragment of UL49 forward but out of frame due to a deletion of a single C residue at the 262-bp SmaI fragment–242-bp SmaI fragment junction. Clone 3.19 contained a 583-bp SmaI fragment encoding amino acids 118 to 312 of UL50, followed by backwards 80- and 96-bp SmaI fragments of UL49.
TABLE 1.
Identification of protein libraries eliciting proliferation of HSV-specific TCCa
| TCC and libraryb | [3H]thymidine incorporation (mean cpm) with:
|
||
|---|---|---|---|
| Stimulus | Controlc
|
||
| Medium | HSV-2 | ||
| 4.2E1 | 286 | 21,591 | |
| pUEX1–BamHI w-SmaI | 10,105 | ||
| pUEX2–BamHI w-SmaI | 4,150 | ||
| pUEX3–BamHI w-SmaI | 1,903 | ||
| 2.3 | 102 | 11,014 | |
| pUEX1–BamHI w-SmaI | 418 | ||
| pUEX2–BamHI w-SmaI | 785 | ||
| pUEX3–BamHI w-SmaI | 2,279 | ||
| ESL4.9 | 146 | 66,013 | |
| pUEX1–HG52–SmaI-AluI | −52 | ||
| pUEX2–HG52–SmaI-AluI | −25 | ||
| pUEX3–HG52–SmaI-AluI | 16,235 | ||
| ESL2.20 | 123 | 13,359 | |
| pUEX1–HG52–SmaI-AluI | 1 | ||
| pUEX2–HG52–SmaI-AluI | 768 | ||
| pUEX3–HG52–SmaI-AluI | 5,427 | ||
Proliferation is expressed as [3H]thymidine incorporation. Autologous EBV-LCL (clones 4.2E1 and 2.3) or PBMC were used as APC, and library-derived fusion protein antigens were diluted 1:300.
Library designations consist of an expression vector, the HSV-2 restriction fragment or strain of full-length viral DNA, and the restriction enzyme(s) used to digest viral DNA.
Controls consisted of 105 autologous irradiated (3,300 rads) PBMC and either mock-infected cell lysate or UV-treated HSV-2 antigen.
TABLE 2.
Antigenic specificity of HSV-2-reactive TCC
| TCC and bacterial clonea (viral sequenceb) | [3H]thymidine incorporation (Δcpmc) with:
|
|||
|---|---|---|---|---|
| Recom- binant antigen | Control antigend
|
|||
| pUEX-derived β-Gal | HSV-1 | HSV-2 | ||
| 4.2E1 | 93 | ND | ND | |
| 1.1.3 (VP22 105-190) | 4,875 | |||
| 49.262.12e (VP22 105-190) | 6,898 | |||
| 2.3 | 231 | 543 | 53,032 | |
| 3.19 (UL50 118-312) | 43,971 | |||
| 50.583.44f (UL50 118-312) | 34,453 | |||
| 50.397f (UL50 118-250) | 66,501 | |||
| ESL4.9 and C11 (VP22 177-220) | 59,400 | 166 | 112,803 | 64,685 |
| ESL2.20 and C9D10 (UL21 148-181) | 23,543 | 173 | 0 | 37,989 |
Bacterially derived recombinant fusion protein antigens were used at a 1:900 dilution. Autologous EBV-LCL (clone 4.2E1) or PBMC were used as APC.
Amino acids predicted forward and in frame with β-galactosidase from sequence data.
Compared to [3H]thymidine incorporation in medium, which was less than 500 cpm in each case.
β-Gal, β-galactosidase. ND, not done.
Confirmatory subclone of 1.1.3, containing only a 262-bp SmaI fragment of UL49 DNA forward and in frame with pUEX3 (see text).
Confirmatory subclones of 3.19, containing a 583-bp SmaI fragment of UL50 or a 397-bp SmaI-StuI fragment of UL50 DNA forward and in frame with pUEX3 (see text).
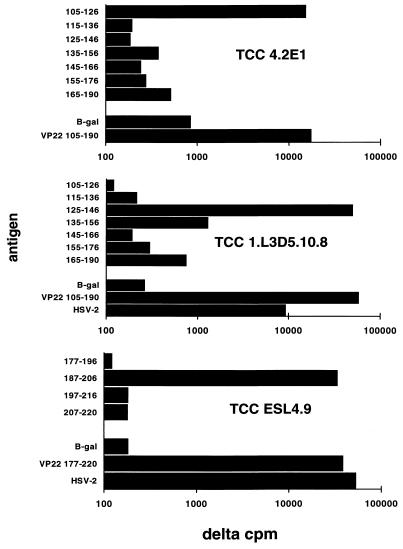
Identification of T-cell antigens was confirmed by targeted subcloning and overlapping peptides. The 262-bp SmaI fragment of UL49 of HSV-2 encoding amino acids 105 to 190 was subcloned into pUEX3 to yield plasmid 49.262.12. This protein stimulated TCC 4.2E1 (Table 2). Only peptide 105-126 of VP22 of HSV-2 (GGPVGAGGRSHAPPARTPKMTR) was stimulatory (Fig. 3). DNA fragments encoding UL50 118-312 and 118-250 were subcloned into pUEX3. Fusion proteins expressing these fragments were active (Table 2).
FIG. 3.
Proliferative responses of VP22-specific TCC to peptide epitopes in VP22 of HSV-2. APC were autologous EBV-LCL (for TCC 4.2E1) or autologous PBMC (for other TCC). Antigens included β-galactosidase and fusion proteins used at 10 μg/ml and peptides used at 3 μM (for TCC 4.2E1) or 1 μM (for other TCC). Data are Δcpm, expressing [3H]thymidine incorporation compared to that in the medium, which was less than 500 cpm in each case.
Evaluation of random colonies from full-length HSV-2 DNA libraries showed that 80 to 100% contained plasmids with an insert; 80 to 100% of inserts were of viral origin. For both TCC ESL4.9 and ESL2.20, only the pUEX3 protein library elicited lymphoproliferation (Table 1). Since the libraries were more complex than those made from the BamHI w fragment, 2,000 to 3,000 bacterial transformants were screened by a combinatorial method. In preliminary experiments, heat-killed, washed bacteria were found to substitute for inclusion body preparations of protein in lymphoproliferation assays at the pool (5 to 12 bacterial clones) and final-assay stages (data not shown).
Sequencing of plasmids in positive bacteria showed that TCC ESL4.9 recognized a 44-amino-acid fragment of UL49 gene product VP22 (amino acids 177 to 220), while TCC ESL2.20 recognized a 34-amino-acid fragment of the UL21 gene product (amino acids 148 to 181) (Table 2). In both cases, single AluI fragments of HSV-2 DNA were inserted in frame and forward. Peptide mapping revealed that amino acids 187 to 206 (Fig. 3) stimulated TCC ESL4.9.
Fusion proteins as probes of bulk lesion-infiltrating T cells.
Newly discovered T-cell antigens were added to the panel of HSV antigens used to probe bulk cultures of lesion-infiltrating T cells. The first available specimens in our ongoing studies (19) were a set of four biopsy specimens (2 mm each) obtained from a day-5 (virus culture positive) buttock lesion due to a recurrence of HSV-2 in patient 1 (17). All four biopsy specimens showed reactivity with VP22 105-190 but not with β-galactosidase, glycoprotein B or D, or VP16 (data not shown). TCC were derived after the original bulk culture was restimulated for one cycle with VP22 105-190 fusion protein. The proliferative responses of TCC 1.L3D5.10.8 (Fig. 3) to VP22 105-190 and constituent peptides document a third T-cell epitope in VP22, contained within amino acids 125 to 146.
Definition of additional T-cell epitopes in tegument protein VP16.
We previously found three epitopes within VP16 (Table 3), all HSV-2 type specific (14), and detected proliferative responses to full-length VP16 in bulk cultures of genital HSV-2 lesion-infiltrating lymphocytes from four of seven (57%) patients (19). We sought additional peptide epitopes within VP16 by two strategies. The first strategy was to screen panels of clones recovered from lesion vesicle fluid for reactivity with recombinant VP16 of HSV-2, followed by epitope mapping with peptides. Peptides containing amino acids 185 to 197 and the overlapping pair 209-221 and 213-225 were stimulatory for TCC RH.13 and KM.7, respectively (Table 3). All other VP16 peptides were negative (<500 cpm [data not shown]). The second strategy involved use of PBMC as the starting material and secondary in vitro stimulation with recombinant baculovirus-derived VP16. Clones (BM.17 and SB.17) from two individuals recognized the same peptide (amino acids 437 to 449), as well as a β-galactosidase–VP16 fusion protein and whole virus. All three newly defined VP16 epitopes were type common, shared by HSV-1 and HSV-2 whole-virus preparations, as expected from sequence data (6).
TABLE 3.
Epitopes within VP16 of HSV-2 recognized by lesion- and PBMC-derived CD4 TCC
| TCC (origin) | [3H]thymidine incorporation (Δcpma) with:
|
HSV-2 VP16 peptidec
|
||||
|---|---|---|---|---|---|---|
| Whole-virus antigen
|
Recombinant HSV-2 proteinb
|
|||||
| HSV-1 | HSV-2 | VP16 1-492 | β-Gal–VP16 180-492 | Amino acids | Δcpm | |
| Newly reported epitopes | ||||||
| RH.13 (lesion) | 3,340 | 3,407 | 32,991 | ND | 185–197 | 55,614 |
| KM.7 (lesion) | 6,093 | 5,847 | 5,627 | ND | 209–221 | 10,075 |
| BM.17 (PBMC) | 30,784 | 13,777 | ND | 45,958 | 437–449 | 79,723 |
| SB.17 (PBMC) | 2,207 | 4,187 | ND | 12,178 | 437–449 | 36,442 |
| Previously reported epitopes | ||||||
| ESL4.34 (lesion) | 256 | 8,780 | 17,302 | ND | 389–401 | 12,968 |
| 393–405 | 95,942 | |||||
| ESL3.334 (lesion) | 253 | 14,232 | 22,754 | 16,434 | 430–444 | 27,283 |
| 1A.B.25 (lesion) | 414 | 33,493 | 24,919 | 41,123 | 431–440 | 38,664 |
Compared to [3H]thymidine incorporation in the medium, which was less than 500 cpm in each case.
VP16 1-492 (baculovirus-derived) was used at 1 μg/ml. β-Gal–VP16 180-492 was used at a 1:1,000 dilution. β-Gal, β-galactosidase; ND, not done.
Peptides were used at 1 μM.
HLA restriction.
The HLA restriction of the TCC-recognizing antigens encoded near 0.7 map units was determined in detail. Proliferation of TCC 4.2E1, specific for VP22 105-126, is inhibited 84% by an anti-DP MAb but less than 20% by an anti-DR or anti-DQ MAb. TCC 4.2E1 is from a DPB1*2001/DPB1*0402 heterozygous donor. Allogeneic EBV-LCL bearing DPB1*2001 but not DPB1*0402 present antigen (Table 4), establishing restriction by DPB1*2001. Proliferation of TCC 2.3, specific for UL50, was inhibited by an anti-DR MAb but not by an anti-DP or anti-DQ MAb. This clone is from a DRB1*0301/BRB1*0701 heterozygous donor. Allogeneic PBMC from a DRB1*0301 donor presented antigen, consistent with binding of antigenic peptide to this allele. However, presentation by one of the linked DR gene products DRw52 and DRw53 cannot be ruled out. Additional HLA restriction studies are summarized in Table 5.
TABLE 4.
Determination of restricting HLA allele of lesion-derived CD4 TCC 4.2E1 and 2.3a
| TCC | Antigen | APC | HLA typeb | Δcpmc |
|---|---|---|---|---|
| 4.2E1 | 1.1.3 | Autologous EBV-LCL | DPB1*0402, 2001 | 30,719 |
| AMAI EBV-LCL | DPB1*0402 | 2,732 | ||
| ARENT EBV-LCL | DPB1*2001 | 26,218 | ||
| 2.3 | 50.583.44 | Autologous PBMC | DRB1*0301, 0701 | 8,964 |
| Allogeneic PBMC A | DRB1*0701, 1001 | 45 | ||
| Allogeneic PBMC B | DRB1*0301, 1301 | 19,223 |
Antigens were β-galactosidase fusion proteins (Table 2) at a 1:900 deletion.
HLA type at the HLA class II locus as determined by inhibition with MAb (data not shown).
In comparison to pUEX2 control protein (1:1,000 dilution) with the same APC, which caused less than 500 cpm of [3H]thymidine incorporation in each case.
TABLE 5.
Cytolytic activity of lesion-derived, tegument-specific CD4 TCC, with summary of fine specificity and HLA restriction
| TCC | Specificitya | HLA restrictionb | % Specific releasec with the following cytolysis assay target:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Auto HSV-2 | Auto peptide | Auto mock | Allo HSV-2 | Allo peptide | Allo mock | |||
| Newly reported epitopes | ||||||||
| 4.2E1 | VP22 105-126 | DPB1*2001 | 20.7 | 44.2 | −4.1 | −2.9 | −1.7 | 4.6 |
| ESL4.9 | VP22 187-206 | DRd | −0.6 | 20.2 | 1.3 | 0 | 0 | 0 |
| ESL2.20 | UL21 148-181 | DRd | 2.7 | NA | 0.9 | 0 | NA | 0 |
| 1.L3D5.10.8 | VP22 125-146 | DRe | 1.1 | 61.1 | −0.3 | −0.4 | −0.6 | −0.4 |
| 1.L3D5.10.12 | VP22 125-146 | DRe | 2.5 | 57.6 | 1.6 | −0.1 | −2.5 | −1.4 |
| RH.13 | VP16 185-197 | DR4 | 62.5 | 55.2 | −0.9 | 9.6 | 0.3 | 1.8 |
| KM.7 | VP16 209-221 | DR2 | 38.7 | 43.6 | 2.7 | −2.2 | 4.3 | −1.1 |
| BM.17 | VP16 437-449 | DQB1*0501 | 10.1 | 28.5 | −0.3 | ND | ND | ND |
| SB.17 | VP16 437-449 | DQB1*0501 | 48.7 | 60.6 | 5.4 | ND | ND | ND |
| 2.3 | UL50 118-250 | DRB1*0301 | 0.8 | NA | 0 | 1.1 | NA | 0 |
| Previously described epitopes | ||||||||
| ESL4.34 | VP16 393-405 | DRB1*0402 | 2.1 | 10.4 | 1.0 | 0.5 | 0.6 | 0.3 |
| ESL3.334 | VP16 430-444 | DQB1*0302 | 12.3 | 33.6 | 0.7 | 1.4 | 0.3 | 2.2 |
| 1A.B.25 | VP16 431-440 | DQB1*0201 | 24.3 | 42.2 | 1.9 | 1.7 | 2.1 | −0.4 |
Indicates peptide used (1 μM) to load targets in CTL assay for selected TCC.
Maximum extent of definition of HLA restricting locus and/or allele. Subjects 6 and 7 were typed serologically; others were typed at the DNA level.
At an effector-to-target ratio of 20:1, except for ESL4.34 (10:1). Auto, autologous EBV-LCL as target cells; allo, allogeneic EBV-LCL mismatched at the relevant HLA locus (if known) or mismatched at HLA-DR and -DQ. NA, not available, since epitope mapping was not done and synthetic antigenic peptide was not made; ND, not done.
Subject is heterozygous for HLA DRB1*0402 and DRB1*1301, and restricting allele has not been determined.
Subject is heterozygous for HLA DRB1*0301 and DRB1*1102, and restricting allele has not been determined.
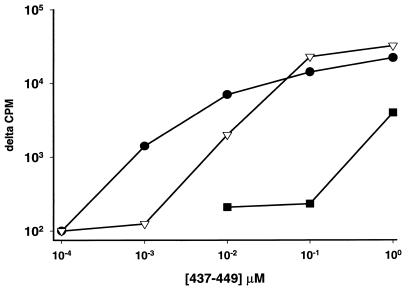
The HLA restriction of TCC BM.17 was studied in detail, since the antigenic peptide, VP16 437-449, overlaps two previously described (18, 21) VP16 epitopes (Table 3). Proliferation of TCC BM.17 and the similar clone SB.17 was inhibited 90% by an anti-DQ MAb but less than 25% by an anti-DR or anti-DP MAb. Donors 8 and 9 are heterozygous for HLA DQB1*0201/0501. At high concentrations of peptide, both DQB1*0201 and DQB1*0501 homozygous EBV-LCL appeared to present antigen to TCC BM.17. However, DQB1*0501 homozygous cells presented peptide much more efficiently than DQB1*0201 homozygous cells (Fig. 4). Thus, three different but overlapping epitopes in VP16 431-449 are presented by HLA DQB1*0302, DQB1*0201, and DQB1*0501.
FIG. 4.
HLA restriction element for TCC BM.17 response to peptide 437-449 of VP16 of HSV-2. Proliferative responses are plotted against concentrations of viral peptide. APC are EBV-LCL that are either autologous (solid circles), homozygous for HLA DQB1*0501 (open inverted triangles), or homozygous for HLA DQB1*0201 (solid squares).
CTL activity of tegument-specific CD4 TCC.
Cytotoxic activities of the CD4 TCC with newly and previously identified specificities were tested by using EBV-LCL target cells (Table 5). All clones tested displayed cytolytic activity towards peptide-loaded target cells. Cytolytic activity against target cells infected with HSV-2 showed greater variability. VP22-specific TCC 4.2E1 was active, while VP22-specific TCC from other donors were not. Among the seven VP16-specific TCC tested, six displayed greater than 10% cytotoxicity towards HSV-2-infected target cells. The single UL21- and UL50-specific TCC were not active against virus-infected target cells.
DISCUSSION
HSV-specific T cells selectively infiltrate recurrent genital HSV-2 lesions (16). Local CTL activity, with CD4- and CD8-mediated components, is correlated with viral clearance (19). The antigens recognized by local HSV-specific T cells are diverse and in many cases unknown (17). In this report, we document recognition of tegument proteins VP22 and UL21 and the viral dUTPase, and we extend our previous observations on tegument protein VP16.
HSV has several characteristics which facilitate the expression cloning system described in this report. Genomic double-stranded DNA can be used directly, since introns are rare in the HSV genome. The same HSV-2 strain, HG52 (9), was used to screen candidate lesion-derived TCC and make protein libraries. The relatively low degree of strain variability (28) between HSV-2 strains in the donors and HG52 might rarely lead to omission of an epitope(s) recognized in vivo; application to viruses with more strain variation would benefit from the use of autologous isolates.
Notably, reactivity with VP22 was detected in two independent expression cloning experiments with lesion-infiltrating TCC from two donors. VP22 reactivity was also detected during screening of the first available set of bulk lesion-infiltrating lymphocyte cultures. We plan assessment of the reactivity of bulk and cloned T cells from the herpetic lesions of additional subjects after expression of full-length antigens. Thus far, 10 additional clones from three patients have been negative with fragments of UL49, UL21, and UL50 discussed in this report; assessment of the relative immunodominance of tegument-specific responses in comparison to HSV glycoproteins is not possible at this time.
Tegument antigens may be suitable targets for lesion-infiltrating CD4 T cells because of their abundance. VP16 and VP22 are present in large amounts: on the order of 1.6 × 103 molecules of VP16 (45) and 2.5 × 103 to 2.8 × 103 molecules of VP22 (23) are incorporated into each virion in HSV-1. Less information is available for UL21 (2, 3). The viral dUTPase is the first nonvirion component documented to be a target of the HSV-specific CD4 T-cell response; no information is available concerning its relative abundance. This enzyme, like VP16 and VP22, localizes to the nuclei of cells infected with HSV-2 (but not HSV-1) (40). Antigen presentation in vivo may occur after endogenous synthesis of dUTPase in infected cells or scavenging of dUTPase antigen from infected-cell debris. Lysis of HSV-infected cells by dUTPase-specific TCC 4.2E1 indicates that, at least in vitro, presentation of endogenous antigen can occur.
Immune responses to VP22 may be an obstacle to its use for delivery of exogenous materials (30) in vivo. Conversely, since polypeptides expressed as C-terminal fusions to VP22 can be cotransported into cells, expression of proteins as VP22 fusions may be of interest as a type of adjuvant preparation. This hypothesis can be tested by expression of heterologous epitopes in VP22. VP16 and VP22 of HSV-1 are strongly, noncovalently associated in infected cells as shown by coimmunoprecipitation. These proteins colocalize in the perinuclear areas of cells (10, 11). Possibly, this association plays a role in stimulating the apparently high level of CD4 T-cell response to VP16.
All of the lesion-derived tegument-specific CD4 TCC studied had cytolytic potential against maximally sensitized (peptide-loaded) target cells. Why do only some clones kill virus-infected cells? Possibly, some epitopes are not presented in infected APC, and some may be processed at very low levels, below a threshold needed to trigger a cytolytic response. Thus far, with our small number of clones, we have not observed a consistent relationship between antigenic specificity or HLA restricting locus and sensitization, by viral infection, to lysis by CD4 T cells.
In summary, expression cloning has allowed the discovery of novel HSV T-cell antigens. The in situ enrichment of antigen-specific CD4 T cells in lesions allows study of the antigenic repertoire unbiased by secondary in vitro stimulation with antigen. The favorable characteristics of the HSV genome allow direct use of libraries of whole-virus DNA. In the future, a more complete description of the diversity and targets of HSV-specific T cells and correlation of these factors with the duration, clinical severity, or complications of HSV infections, such as HSV keratitis, may be assisted by application of the methods in this report. Tegument proteins are candidates, together with membrane glycoproteins, for evaluation as possible HSV vaccines in humans.
ACKNOWLEDGMENTS
This study was supported by NIH grants AI34616 and CA70017 (to D.M.K.).
HLA typing was performed by John Hansen and Effie Petersdorf. HSV-1 × HSV-2 IRV were graciously provided by Bernard Roizman (RS1G31), Howard Marsden (DX32), and Steve Triezenberg (RP-2). The HSV-2 HG52 BglII i fragment was provided by Steve Triezenberg and originally by Chris Preston. MAb p43, specific for VP22, was provided by David Meredith. John C. Hutton provided pUEX vectors, primer sequences, and valuable advice. Peptides and recombinant VP16, gB2, and gD2 were kindly provided by Rae L. Burke and Michael Tigges, Chiron Corporation. Lawrence Corey provided invaluable support and advice over several years. We thank Jeffrey Vieira for valuable advice and Mike Remington, Gail R. Barnum, and Mary Shaughnessy for assistance with specimen collection.
REFERENCES
- 1.Ashley R A, Militoni J, Lee F, Nahmias A, Corey L. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunoblot for detecting antibodies to herpes simplex types 1 and 2 in human sera. J Clin Microbiol. 1988;26:662–667. doi: 10.1128/jcm.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Koyama A H, Huang T, Roizman B. The UL21 gene products of herpes simplex virus 1 are dispensable for growth in cultured cells. J Virol. 1994;68:2929–2936. doi: 10.1128/jvi.68.5.2929-2936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 α regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 4.Bodmer J G, Marsh S G, Albert E D, Bodmer W F, Bonthrop R E, Charron D, Dupont B, Erlich H A, Fauchet R, Strominger J L, Svejgaard A, Terasaki P I. Nomenclature for factor of the HLA system, 1996. Tissue Antigens. 1997;49:297–321. doi: 10.1111/j.1399-0039.1997.tb02759.x. [DOI] [PubMed] [Google Scholar]
- 5.Bressan G M, Stanley K K. pUEX, a bacterial expression vector related to pEX with universal host specificity. Nucleic Acids Res. 1987;15:10056. doi: 10.1093/nar/15.23.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cress A, Triezenberg S J. Nucleotide and deduced amino acid sequence of the gene encoding virion protein 16 of herpes simplex virus type 2. Gene. 1991;103:235–238. doi: 10.1016/0378-1119(91)90278-j. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham A L, Merigan T C. Leu-3+ T cells produce gamma-interferon in patients with recurrent herpes labialis. J Immunol. 1984;132:197–202. [PubMed] [Google Scholar]
- 8.Doherty D G, Koelle D M, Kwok W W, Masewicz S, Domeier M E, Nepom G T. Allelic variants of MHC class II molecules can act as partial agonists of antigen specific T cell responses. Hum Immunol. 1996;47:149. [Google Scholar]
- 9.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott G, Mouzakitis G, O’Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol. 1992;73:723–736. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 12.Gignac S M, Brauer S, Hane B, Quentmeier H, Drexler H G. Elimination of mycoplasma from infected leukemia cell lines. Leukemia. 1991;5:162–165. [PubMed] [Google Scholar]
- 13.Ho M. Interferon as an agent against herpes simplex virus. J Investig Dermatol. 1990;95:158S–160S. doi: 10.1111/1523-1747.ep12875164. [DOI] [PubMed] [Google Scholar]
- 14.Jerome K R, Tait J F, Koelle D M, Corey L. Herpes simplex virus type 1 renders infected cells resistant to cytotoxic T-lymphocyte-induced apoptosis. J Virol. 1998;72:436–441. doi: 10.1128/jvi.72.1.436-441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kit S, Kit M, Qavi H, Trkula D, Otsuka H. Nucleotide sequence of the herpes simplex virus type 2 (HSV-2) thymidine kinase gene and predicted amino acid sequence of thymidine kinase polypeptide and its comparison with the HSV-1 thymidine kinase gene. Biochem Biophys Acta. 1983;741:158–170. doi: 10.1016/0167-4781(83)90056-8. [DOI] [PubMed] [Google Scholar]
- 16.Koelle D M, Abbo H, Peck A, Ziegweid K, Corey L. Direct recovery of HSV-specific T lymphocyte clones from human recurrent HSV-2 lesions. J Infect Dis. 1994;169:956–961. doi: 10.1093/infdis/169.5.956. [DOI] [PubMed] [Google Scholar]
- 17.Koelle D M, Corey L, Burke R L, Eisenberg R J, Cohen G H, Pichyangkura R, Triezenberg S J. Antigenic specificity of human CD4+ T-cell clones recovered from recurrent genital HSV-2 lesions. J Virol. 1994;68:2803–2810. doi: 10.1128/jvi.68.5.2803-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koelle D M, Johnson M L, Ekstrom A N, Byers P, Kwok W W. Preferential presentation of herpes simplex virus T-cell antigen by HLA DQA1*0501/DQB1*0201 in comparison to HLA DQA1*0201/DQB1*0201. Hum Immunol. 1997;53:195–205. doi: 10.1016/S0198-8859(97)00034-7. [DOI] [PubMed] [Google Scholar]
- 19.Koelle D M, Posavad C M, Barnum G R, Johnson M L, Frank J M, Corey L. Clearance of HSV-2 from recurrent genital lesions correlates with infiltration of HSV-specific cytotoxic T lymphocytes. J Clin Invest. 1998;101:1500–1509. doi: 10.1172/JCI1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koelle D M, Tigges M A, Burke R L, Symington F W, Riddell S R, Abbo H, Corey L. Herpes simplex virus infection of human fibroblasts and keratinocytes inhibits recognition by cloned CD8+ cytotoxic T lymphocytes. J Clin Invest. 1993;91:961–968. doi: 10.1172/JCI116317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok W W, Domeier M E, Johnson M L, Nepom G T, Koelle D M. HLA-DQB codon 57 is a critical determinant of peptide binding and recognition. J Exp Med. 1996;183:1253–1258. doi: 10.1084/jem.183.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langenberg A G, Burke R L, Adair S F, Sekulovich R, Tigges M, Dekker C L, Corey L. A recombinant glycoprotein vaccine for herpes simplex virus type 2: safety and immunogenicity. Ann Intern Med. 1995;122:889–898. doi: 10.7326/0003-4819-122-12-199506150-00001. [DOI] [PubMed] [Google Scholar]
- 23.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 24.Maitland N J, Smith I W, Peutherer J F, Robertson D H H, Jones K W. Restriction endonuclease analysis of DNA from genital isolates of herpes simplex virus type 2. Infect Immun. 1982;38:834–842. doi: 10.1128/iai.38.3.834-842.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mickelson E, Smith A, McKinney S, Anderson G, Hansen J A. A comparative study of HLA-DRB1 typing by standard serology and hybridization of non-radioactive sequence-specific oligonucleotide probes to PCR-amplified DNA. Tissue Antigens. 1993;41:86–93. doi: 10.1111/j.1399-0039.1993.tb01984.x. [DOI] [PubMed] [Google Scholar]
- 26.Mikloska A, Cunningham A L. Herpes simplex virus type 1 glycoproteins gB, gC, and gD are major targets for CD4 T-lymphocyte cytotoxicity in HLA-DR expressing human epidermal keratinocytes. J Gen Virol. 1998;79:353–361. doi: 10.1099/0022-1317-79-2-353. [DOI] [PubMed] [Google Scholar]
- 27.Neophytou P I, Roep B O, Arden S D, Muir E M, Duinkerken G, Kallan A, de Vries R R, Hutton J C. T-cell epitope analysis using subtracted expression libraries (TEASEL): application to a 38 kDa autoantigen recognized by T cells from an insulin-dependent diabetic patient. Proc Natl Acad Sci USA. 1996;93:2014–2018. doi: 10.1073/pnas.93.5.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novotny M J, Parish M L, Spear P G. Variability of herpes simplex virus 1 gL and anti-gL antibodies that inhibit cell fusion but not viral infectivity. Virology. 1996;221:1–13. doi: 10.1006/viro.1996.0347. [DOI] [PubMed] [Google Scholar]
- 29.Para M F, Zezulak K M, Conley A J, Weinberger M, Snitzer K, Spear P G. Use of monoclonal antibodies against two 75,000-molecular-weight glycoproteins specified by herpes simplex virus type 2 in glycoprotein identification and gene mapping. J Virol. 1983;45:1223–1227. doi: 10.1128/jvi.45.3.1223-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phelan A, Elliott G, O’Hare P. Intercellular delivery of the complete P53 protein by VP22. Natl Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 31.Preston V G, Davison A J, Marsden H S, Timbury M C, Subak-Sharpe J H, Wilkie N M. Recombinants between herpes simplex virus types 1 and 2: analyses of genome structures and expression of immediate-early polypeptides. J Virol. 1978;28:499–517. doi: 10.1128/jvi.28.2.499-517.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith L M, Petty H R, Parham P, McConnell H M. Cell surface properties of HLA antigens in Epstein-Barr virus-transformed cell lines. Proc Natl Acad Sci USA. 1982;79:608–612. doi: 10.1073/pnas.79.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spits H, Borst J, Giphart M, Cougan J, Terhorst C, De Vries J E. HLA-DC antigens can serve as recognition elements for human cytotoxic T lymphocytes. Eur J Immunol. 1984;14:299–304. doi: 10.1002/eji.1830140404. [DOI] [PubMed] [Google Scholar]
- 34.Spruance S L, Chow F S. Pathogenesis of herpes simplex virus in cultures of epidermal cells from subjects with frequent recurrences. J Infect Dis. 1980;142:671–675. doi: 10.1093/infdis/142.5.671. [DOI] [PubMed] [Google Scholar]
- 35.Tigges M A, Leng S, Johnson D C, Burke R L. Human herpes simplex (HSV)-specific CD8+ CTL clones recognize HSV-2 infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J Immunol. 1996;156:3901–3910. [PubMed] [Google Scholar]
- 36.Torseth J W, Cohen G H, Eisenberg R J, Berman P W, Lasky L A, Cerini C P, Heilman C J, Kerwar S, Merigan T C. Native and recombinant herpes simplex virus type 1 envelope proteins induce human immune T-lymphocyte responses. J Virol. 1987;61:1532–1539. doi: 10.1128/jvi.61.5.1532-1539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson A J, DeMars R, Trowbridge I S, Bach F H. Detection of a novel human class II HLA antigen. Nature. 1983;304:358–360. doi: 10.1038/304358a0. [DOI] [PubMed] [Google Scholar]
- 38.Whitley R J. Herpes simplex virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2297–2342. [Google Scholar]
- 39.Williams M V, Parris D S. Characterization of a herpes simplex virus type 2 deoxyuridine triphosphate nucleotidohydrolase and mapping of a gene conferring type specificity for the enzyme. Virology. 1987;156:282–292. doi: 10.1016/0042-6822(87)90408-9. [DOI] [PubMed] [Google Scholar]
- 40.Wohlrab F, Garrett B K, Francke B. Control of expression of the herpes simplex virus-induced deoxypyrimidine triphosphatase in cells infected with mutants of herpes simplex virus types 1 and 2 and intertypic recombinants. J Virol. 1982;43:935–942. doi: 10.1128/jvi.43.3.935-942.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasukawa M, Inatsuki A, Horiuchi T, Kobayashi Y. Functional heterogeneity among herpes simplex virus-specific human CD4+ T cells. J Immunol. 1991;146:1341–1347. [PubMed] [Google Scholar]
- 42.Yasukawa M, Zarling J M. Human cytotoxic T cell clones directed against herpes simplex virus-infected cells. II. Bifunctional clones with cytotoxic and virus induced proliferative activities exhibit herpes simplex virus type 1 and type 2 specific, or type common reactivities. J Immunol. 1984;133:2736–2742. [PubMed] [Google Scholar]
- 43.Yasukawa M, Zarling J M. Human cytotoxic T cell clones directed against herpes simplex virus-infected cells. III. Analysis of viral glycoproteins recognized by CTL clones by using recombinant herpes simplex viruses. J Immunol. 1985;134:2679–2687. [PubMed] [Google Scholar]
- 44.Zarling J M, Moran P A, Burke R L, Pachl C, Berman P W, Lasky L A. Human cytotoxic T cell clones directed against herpes simplex virus-infected cells. IV. Recognition and activation by cloned glycoproteins gB and gD-1. J Immunol. 1986;136:4669–4673. [PubMed] [Google Scholar]
- 45.Zhang Y, McKnight J L C. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]