Abstract
Hypertranscription and temporal expression from the Autographa californica nuclear polyhedrosis (AcNPV) baculovirus polyhedrin promoter involves an α-amanitin-resistant RNA polymerase and requires a trans-acting viral factor(s). We previously reported that a 30-kDa host factor, polyhedrin promoter binding protein (PPBP), binds with unusual affinity, specificity, and stability to the transcriptionally important motif AATAAATAAGTATT within the polyhedrin (polh) initiator promoter and also displays coding strand-specific single-stranded DNA (ssDNA)-binding activity (S. Burma, B. Mukherjee, A. Jain, S. Habib, and S. E. Hasnain, J. Biol. Chem. 269:2750–2757, 1994; B. Mukherjee, S. Burma, and S. E. Hasnain, J. Biol. Chem. 270:4405–4411, 1995). We now present evidence which indicates that an additional factor(s) is involved in stabilizing PPBP-duplex promoter and PPBP-ssDNA interactions. TBP (TATA box binding protein) present in Spodoptera frugiperda (Sf9) cells is characteristically distinct from PPBP and does not interact directly with the polh promoter. Replacement of PPBP cognate sequences within the polh promoter with random nucleotides abolished PPBP binding in vitro and also failed to express the luciferase reporter gene in vivo. Phosphocellulose fractions of total nuclear extract from virus-infected cells which support in vitro transcription from the polh promoter contain PPBP activity. When PPBP was sequestered by the presence of oligonucleotides containing PPBP cognate sequence motifs, in vitro transcription of a C-free reporter cassette was affected but was restored by the exogenous addition of nuclear extract containing PPBP. When PPBP was mopped out in vivo by a plasmid carrying PPBP cognate sequence present in trans, polh promoter-driven expression of the luciferase reporter was abolished, demonstrating that binding of PPBP to the polh promoter is essential for transcription.
The baculovirus expression vector system is very commonly used for foreign gene expression (20, 32). Its attraction lies in the high yields of foreign gene products and the eukaryotic environment for posttranslational modification provided by the insect host cell (29). Unfortunately, not much is known about factors regulating transcription from the very late polyhedrin (polh) and p10 gene promoters most commonly employed in this insect cell expression system.
Fine mapping of the Autographa californica nuclear polyhedrosis virus (AcNPV) polh gene promoter has been extensively done (39, 44, 46). Transcription from the polh promoter, which lacks a well-defined TATA or CAAT box, cannot proceed in the absence of viral infection. The minimal promoter elements capable of directing basal transcription from the promoter, albeit at low levels, are present within an 18-bp sequence surrounding the transcription start site (36). Transcription from the polyhedrin promoter is insensitive to α-amanitin (10, 11, 59). A number of viral late expression factor (lef) genes have been shown to support polyhedrin gene expression, probably in conjunction with cellular factors (55). Many of these lef genes were found to be involved in DNA replication (27), with the remainder likely functioning in late promoter recognition or stabilization of late transcripts or participating directly in transcription as subunits of the virus-induced RNA polymerase complex. A candidate for a very late expression factor gene (vlf-1) has been identified which regulates very late gene transcripts (34). Thus, none of the factors identified so far have been demonstrated to have a direct involvement in transcription regulation from the polh promoter.
We previously reported the isolation and characterization of an unusual 30-kDa host factor, PPBP (polyhedrin promoter binding protein), which binds to a hexamotif, AATAAA, and the octamotif TAAGTATT, encompassing the transcription start point (2). This factor has also been shown to bind to other baculovirus very late promoters, such as the p10 promoter (16). Interestingly, the minimal promoter element defined earlier (36) essentially consists of the PPBP-binding motifs within the polh promoter. A number of observations suggest that PPBP may be important in polyhedrin gene transcription. (i) Dephosphorylation of PPBP abolished binding to its cognate sequences within the promoter. (ii) Nuclear extracts prepared from five different insect cell lines expressing different levels of reporter protein displayed differences in the levels of PPBP binding to the promoter (38). (iii) Gel retardation assays with nuclear extract from a transcription-nonpermissive cell line generated a PPBP-promoter complex with decreased mobility, although the molecular mass of the factor in a UV cross-linking gel was found to be 30 kDa, suggesting the interplay of additional factors along with PPBP in the transcriptionally competent (virus-infected) cell lines. (iv) PPBP displayed both duplex promoter DNA binding and single-stranded DNA (ssDNA) binding restricted to the coding strand of the promoter (37). It is plausible that PPBP, after promoter recognition (double-stranded [dsDNA]-binding activity) and DNA melting, facilitates the availability of the template (via its ssDNA-binding activity) for transcription; this not only suggests the importance of PPBP in polh transcription but also provides an attractive model to explain repeated rounds of transcription.
This indirect evidence correlating a host factor with transcription from the polh promoter represents an enigma, given the definitive role of a virus-specific trans-acting factor(s) in this process. We now address the critical question of the involvement of PPBP per se in transcription from the polh promoter and provide experimental evidence which, while categorizing the host factor as a transcription factor, unequivocally documents the involvement of this initiator-binding protein in transcription from the polh promoter.
MATERIALS AND METHODS
Gel mobility shift assays.
Spodoptera frugiperda cells were maintained in TNMFH medium in the presence of 10% fetal calf serum (40). Crude nuclear protein extracts were prepared as described earlier (14). Complementary synthetic oligonucleotides were synthesized, annealed, and labeled with T4 polynucleotide kinase (Boehringer Mannheim GmbH, Mannheim, Germany) by using [γ-32P]ATP (DuPont, NEN, Boston, Mass.). The binding reaction mixture consisted of 1 μg of nuclear extract with 1 ng of labeled annealed oligonucleotide (∼104 cpm) and was carried out as described previously (2). The DNA-protein complex was resolved at 4°C on a 5% (acrylamide/bisacrylamide ratio, 29:1) nondenaturing polyacrylamide gel in TAE buffer (7 mM Tris-HCl [pH 7.5], 3 mM sodium acetate, 1 mM EDTA). The gel was then covered with plastic wrap, dried, and exposed overnight to Hyperfilm MP (Amersham, Bucks, United Kingdom) at −70°C. For competition analyses, an excess of the appropriate unlabeled, dsDNA was added along with the labeled DNA in the binding reaction.
For determination of the DNA major and minor groove binding activities of PPBP, 1 ng of 5′-end-labeled oligonucleotide was preincubated with different concentrations of actinomycin D or distamycin A (Sigma, St. Louis, Mo.) or Hoechst 33258 or methyl green (Polysciences Inc., Warrington, Pa.) for 30 min at room temperature in a 10-μl reaction volume, followed by a 15-min incubation at room temperature with 1 μg of crude insect cell nuclear extract. The protein-DNA complexes obtained were then resolved on a polyacrylamide gel as described above.
For estimation of the half-life of the PPBP-DNA complex, a preformed complex of protein and labeled probe was challenged with excess unlabeled probe and the reactions were loaded onto a running gel over a period ranging from 0 to 60 min, as described earlier (37). The decay of radioactivity in the original complexes was quantitated with a phosphorimager (GS-250 molecular imager; Bio-Rad), and the percent maximal binding was plotted against time.
For TFIID binding studies, 1 μg of crude extract (HeLa or Sf9 cell) or 20 ng of purified human TATA box binding protein (hTBP) was incubated for 15 min at room temperature with 1 ng of end-labeled oligonucleotide in the presence of TFIID binding buffer (10% glycerol, 20 mM Tris-HCl [pH 7.9], 80 mM KCl, 10 mM MgCl2, 2 mM dithiothreitol [DTT]). The reactions were electrophoresed at 4°C for 90 min in 0.5× TBE buffer (45 mM Tris-borate, 10 mM EDTA, pH 8.0) containing 5 mM MgCl2 and 0.05% Nonidet P-40 on a 6% nondenaturing gel containing 0.05% Nonidet P-40, which was prerun at 250 V for 20 min.
Western blot analysis.
One hundred micrograms of Sf9 or HeLa cell nuclear extracts or 60 ng of commercially available TBP or Sp1 (Promega, Madison, Wis.) was electrophoresed on a sodium dodecyl sulfate–15% polyacrylamide gel (23) and electrophoretically transferred to a nylon membrane (Hybond C-extra; Amersham) at 300 mA for 2 h at 4°C in transfer buffer (25 mM Tris, 190 mM glycine, 20% methanol). The membrane was blocked for 1 h in phosphate-buffered saline (PBS) (49) containing 1% nonfat dry milk and then incubated in a 1:3,000 dilution of anti-TBP antiserum in PBS for 1 h. Goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (procured from the National Institute of Immunology reagent bank) was used as the second antibody. H2O2 (0.03%) and 50 μg of diaminobenzidine (Sigma)/ml in PBS were added to develop the bands.
Phosphocellulose fractionation.
Nuclear proteins from Sf9 cells were extracted as described by Xu et al. (58) with minor modifications. All operations were carried out at 4°C. The AcNPV-infected cells (∼700 million) were collected at 36 h postinfection (p.i.), pelleted at 2,500 rpm in a Sorvall GS3 rotor for 10 min, and washed twice with chilled PBS. The cells were lysed in 5 ml of lysis buffer, and the nuclei were pelleted as described previously (2). Further treatment of the nuclei was carried out as described by Xu et al. (58). The pelleted nuclei were resuspended and lysed, and globular nuclear proteins were precipitated by the dropwise addition of ammonium sulfate (0.33 g per ml of supernatant). The precipitated nuclear proteins were collected by centrifugation, dissolved in dialysis buffer (50 mM Tris-HCl [pH 7.9], 1 mM EDTA, 100 mM KCl, 20% glycerol, 3 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride), and dialyzed four times for 2 h each time in 200-fold-excess dialysis buffer. This dialyzed nuclear extract was loaded onto a 2-ml phosphocellulose column preequilibrated with dialysis buffer at 4°C, and the unbound material was recycled three times. The column was washed with 6 ml of dialysis buffer and eluted successively with 2 ml each of the same buffer containing 0.3, 0.5, 0.75, and 1.0 M KCl. Between elutions, the column was washed with 6 ml of the dialysis buffer. The individual fractions were concentrated in a Speed Vac (Savant) to 0.5 ml and dialyzed against 250 ml of dialysis buffer overnight. Each fraction was checked for the presence of PPBP in a gel mobility shift assay with labeled polh B domain oligonucleotide.
In vitro transcription assay.
In vitro transcription was carried out essentially as described earlier (58), with minor modifications. The transcription reaction was carried out in a 50-μl reaction volume with 1.5 μg of the plasmid construct pPolh/CFS containing a C-free cassette (58) as the template and 50 μg of protein prepared as described by Xu et al. from AcNPV-infected Sf21 cells 36 h p.i. The reaction buffer used contained 25 mM Tris (pH 7.9), 60 mM KCl, 0.5 mM EDTA, 1 mM MgCl2, 3 mM DTT, 10% glycerol, 10 mM creatine phosphate, 60 U of RNasin (Promega), 0.6 mM (each) ATP and UTP, 5 μM GTP, and 20 μCi of [α-32P]GTP (∼800 Ci/mmol; DuPont, NEN). The reaction mixture was incubated at 32°C for 30 min. The reaction was stopped by adding 350 μl of stop buffer (50 mM Tris [pH 7.5], 1% sodium dodecyl sulfate, 5 mM EDTA, 25 μg of tRNA per ml) and was extracted twice with phenol and chloroform. RNA was precipitated with 0.15 M sodium acetate and 2 volumes of ethanol and resuspended in 20 μl of loading buffer (80% formamide, 0.01% xylene cyanol, 0.01% bromophenol blue). The transcripts were separated on an 8% polyacrylamide-7 M urea gel, dried, and autoradiographed at −70°C. For a PPBP depletion experiment, the nuclear extract was preincubated for 15 min with unlabeled polh B domain to completely sequester free PPBP before adding it to the reaction mixture.
Construction of wild-type and mutant promoter-reporter gene plasmids.
All DNA manipulations were carried out according to the method of Sambrook et al. (49). For the construction of pAJpol-luc, harboring the wild-type polyhedrin promoter, an EcoRV-BamHI fragment of the transfer vector pVL1393 (30) containing the 92-bp polh promoter was cloned into the HincII-BamHI site of plasmid pAJluc (a derivative of pUC18 carrying the 1,892-bp luc gene ligated at the BamHI site), placing it upstream of the luciferase reporter gene (8). Different oligonucleotides carrying various mutations, as detailed in Table 1, were chemically synthesized and cloned at the HindIII-SalI site of pAJluc to generate the respective mutant promoter plasmids. All promoter mutations and promoter-reporter orientations were confirmed by dideoxy sequencing (50).
TABLE 1.
Sequences and respective plasmid constructs of 65-mer oligonucleotides with promoter mutationsa
| Oligonucleotide | Sequence (−5 to −65) | Construct |
|---|---|---|
| pol | --(−92)----ATCTCGCAAATAAATAAGTATTTTACTGTTTTCGTAACAGTTTTGTAATAAAAAAACCTAT--(−1)-- | pAJpol-luc |
| mutHex | 5′ agctATCTCGCACCCCCCTAAGTATTTTACTGTTTTCGTAACAGTTTTGTCCCCCCAAAACCTAT 3′ | pAJmutHex-luc |
| mutHexOct | 5′ agctATCTCGCACCGCCCGCCTGCGGTTACTGTTTTCGTAACAGTTTTGTCCGCCCAAAACCTAT 3′ | pAJmutHexOct-luc |
| mH3T | 5′ agctATCTCGCAAAGAAATAAGTATTTTACTGTTTTCGTAACAGTTTTGTAATAAAAAAACCTAT 3′ | pAJmH3T-luc |
| mH4A | 5′ agctATCTCGCAAATCAATAAGTATTTTACTGTTTTCGTAACAGTTTTGTAATAAAAAAACCTAT 3′ | pAJmH4A-luc |
| mH5a | 5′ agctATCTCGCAAATACATAAGTATTTTACTGTTTTCGTAACAGTTTTGTAATAAAAAAACCTAT 3′ | pAJmH5A-luc |
| mH6A | 5′ agctATCTCGCAAATAACTAAGTATTTTACTGTTTTCGTAACAGTTTTGTAATAAAAAAACCTAT 3′ | pAJmH6A-luc |
Mutations are underlined. The lower case letters represent a HindIII overhang which was used for cloning into pAJluc. The 5′ end of the complementary oligonucleotide has a SalI overhang. pAJpol-luc has the polyhedrin promoter spanning −1 to −92. The TAAG motif is indicated in boldface. Dashes represent flanking sequences.
In vivo luciferase expression assays.
Lipofectin-mediated transfection of insect cells followed by transient expression of the luciferase reporter gene was carried out as described earlier (12) and quantitated at 60 h p.i.
RESULTS
PPBP binds to the minor groove of DNA.
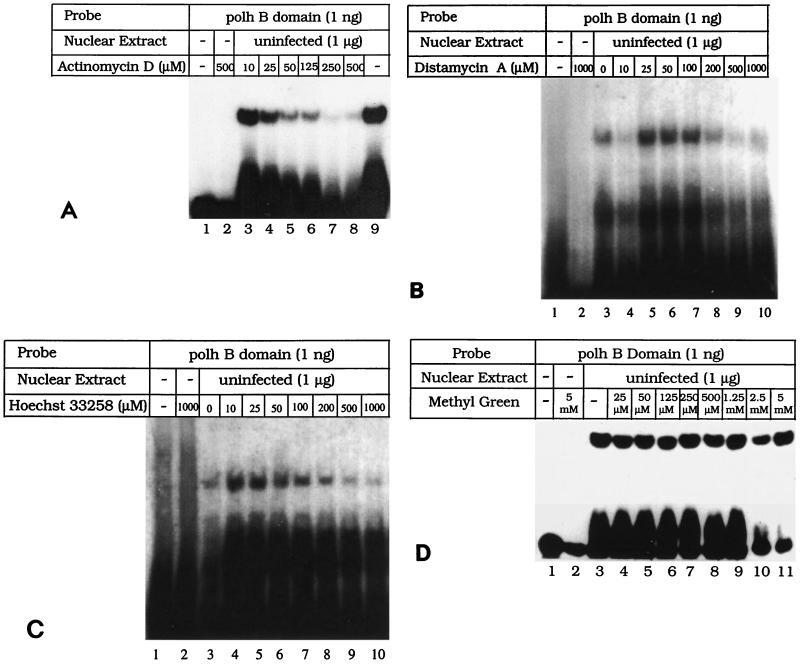
To determine the nature of the interaction between PPBP and the polh promoter sequences, drugs which specifically interact with the major or minor groove of DNA were evaluated for their ability to compete with PPBP for binding. Electrophoretic mobility shift assays (EMSAs) were carried out (2) with the B domain (Fig. 1) of the polh promoter in the presence of increasing concentrations of actinomycin D, a minor groove binding drug (6), and methyl green, a major groove binding drug (21). Formation of the PPBP-DNA complex gradually decreased with increasing concentrations of actinomycin D (Fig. 2A, lanes 3 to 8) compared to that in the control, where no actinomycin D was added (Fig. 2A, lane 9). Actinomycin D alone did not have any effect on the migration of free DNA (Fig. 2A, lane 2). Similarly, increasing concentrations of distamycin A and Hoechst 33258, both of which bind to the minor groove of DNA (5), affected the formation of PPBP-B domain complex (Fig. 2B and C, lanes 4 to 10). Incubation of DNA alone with these drugs did not affect the mobility of DNA (Fig. 2B and C, lanes 2). On the other hand, increasing concentrations of methyl green (Fig. 2D; compare lane 3 with lanes 4 to 11) did not affect the formation of PPBP-DNA complex. Methyl green alone had no effect on migration of the free probe (Fig. 2D, lane 2). These results demonstrate that PPBP, like several other known eukaryotic transcription factors, approaches the promoter region through the minor groove of DNA.
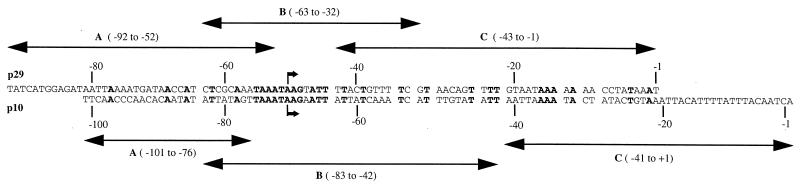
FIG. 1.
Schematic representation of the AcNPV polyhedrin (polh) and p10 promoters. The transcription start points (marked with arrows) are at −50 for the polh (p29) promoter and at −70 for the p10 promoter, and the translation initiation site is at +1. The A, B, and C domains of both promoters are indicated by double-headed arrows. Identical bases of the two promoters are in boldface.
FIG. 2.
PPBP binds through the minor groove of DNA. (A) Formation of polh B domain-PPBP complex is inhibited by actinomycin D. DNA-protein complex obtained with 1 μg of Sf9 cell nuclear extract (lane 9) was incubated with 10 (lane 3), 25 (lane 4), 50 (lane 5), 125 (lane 6), 250 (lane 7), or 500 (lane 8) μM actinomycin D. As controls, labeled polh B domain was incubated either alone (lane 1) or with 500 μM actinomycin D (lane 2). polh B domain spans from −63 to −32 on the p29 promoter, as shown in Fig. 1. (B) Formation of polh B domain-PPBP complex is inhibited by distamycin A. DNA-protein complex obtained with 1 μg of Sf9 cell nuclear extract (lane 3) was incubated with 10 (lane 4), 25 (lane 5), 50 (lane 6), 100 (lane 7), 200 (lane 8), 500 (lane 9), or 1,000 (lane 10) μM distamycin A. As controls, labeled polh B domain was incubated either alone (lane 1) or with 1,000 μM distamycin A (lane 2). (C) Formation of polh B domain-PPBP complex is inhibited by Hoechst 33258 dye. DNA-protein complex obtained with 1 μg of Sf9 cell nuclear extract (lane 3) was incubated with 10 (lane 4), 25 (lane 5), 50 (lane 6), 100 (lane 7), 200 (lane 8), 500 (lane 9), or 1,000 (lane 10) μM Hoechst 33258. As controls, labeled polh B domain was incubated either alone (lane 1) or with 1,000 μM Hoechst 33258 (lane 2). (D) The polh B domain-PPBP complex is unaffected by increasing concentrations of methyl green. Labeled polh B domain was incubated either alone (lane 1), with 5 mM methyl green (lane 2), or with 1 μg of Sf9 cell nuclear extract (lanes 3 to 11). The DNA-protein complex obtained (lane 3) was incubated with 25 (lane 4), 50 (lane 5), 125 (lane 6), 250 (lane 7), 500 (lane 8), 1.25 (lane 9), 2.5 (lane 10), or 5 (lane 11) mM methyl green.
Additional factor(s) possibly interacts and stabilizes PPBP-promoter interaction.
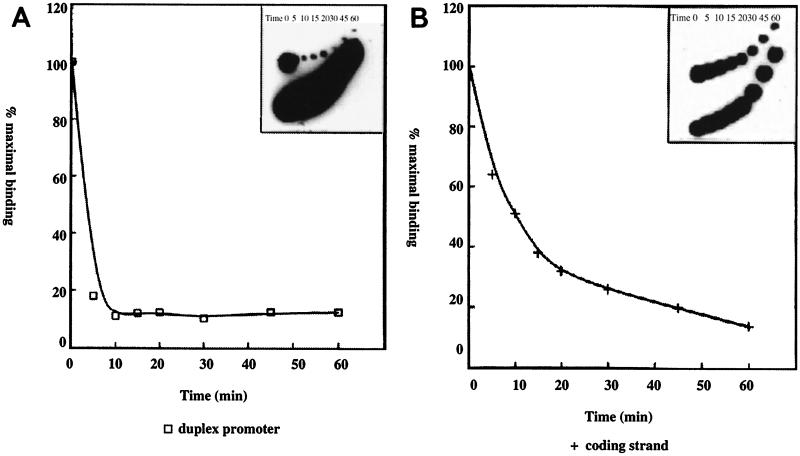
We investigated the role of an additional factor(s) in PPBP-promoter interaction, as previously evident from studies on copurification (2, 14) and analyses of PPBP-promoter complex from transcription-nonpermissive cells (37, 38). Preformed polh-PPBP complex with nuclear extracts from the transcription-nonpermissive Bm5 cell line was challenged with an excess of cold promoter DNA, the reactions were loaded onto a running gel at periods ranging from 0 to 60 min (Fig. 3), and the dissociation of the original complex was plotted as percent maximal binding versus time by using a phosphorimager. It was apparent that the promoter-PPBP complex with Bm5 cell nuclear extract has a half-life of less than 5 min (Fig. 3A) compared to ∼15 min for extract from a permissive cell line, Sf21 (37). Half-life determination of the coding strand-PPBP complex from Bm5 cells gave a value of 15 min (Fig. 3B), which was far less than the 60 min obtained with nuclear extract from the permissive cell line (37). Together these data suggest the importance of an additional factor(s) interacting with PPBP in stabilizing the PPBP-duplex promoter and the PPBP-coding strand complexes in permissive cell lines.
FIG. 3.
Half-life of PPBP-polh B domain complex in Bm5 cell line. (A) DNA-protein complex from Bm5 has a shorter half-life. Preformed polh B-PPBP (Bm5) complex was challenged with an excess of cold polh B domain at the times (in minutes) indicated above each lane (inset). The dissociation of the original complex was plotted as percent maximal binding versus time. (B) The polh coding strand-PPBP complex from Bm5 has a shorter half-life. Preformed polh B coding strand-PPBP (Bm5) complex was challenged with an excess of cold coding strand DNA. Reactions were loaded onto a running gel at various time points (in minutes) indicated above each lane (inset). The dissociation of the original complex was plotted as percent maximal binding versus time.
TBP is distinct from PPBP and does not bind the polh promoter.
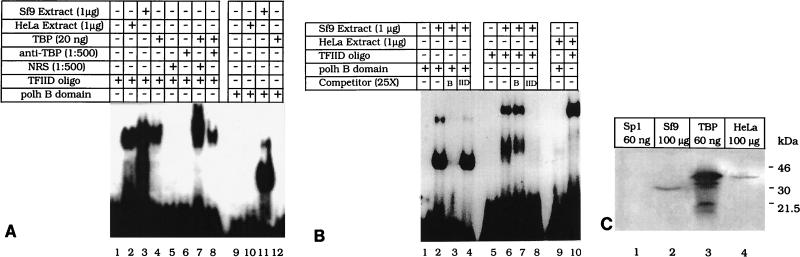
Experiments were designed to determine whether transcription from the polh promoter also directly involves the ubiquitous TBP required for eukaryotic transcription initiation (41, 48). Alternatively, given the apparent similarities between TBP and PPBP (2), we investigated whether the promoter-binding activity of TBP, the crucial step in the nucleation event, has been taken over by PPBP in this system. EMSAs with a consensus TFIID duplex oligonucleotide (5′GCAGAGCATATAAGGTGAGGTAGGA3′) and purified TBP (Promega) were carried out under gel conditions specific for TFIID binding (see Materials and Methods). As expected, HeLa cell extract (1 μg) and purified TBP (20 ng) could bind to the synthetic 25-mer oligonucleotide containing the TFIID cognate sequence (Fig. 4A, lanes 2 and 4) but not to the polh B domain oligonucleotide containing the PPBP cognate sequence motifs (Fig. 4A, lanes 10 and 12). Conversely, Sf9 cell nuclear extract (1 μg) could bind to the B domain to form a PPBP-specific complex (Fig. 4A, lane 11) and also to the TFIID oligonucleotide to form a different complex (Fig. 4A, lane 3). The mobility of the Sf9 cell nuclear extract-TFIID oligonucleotide complex was similar to those of the HeLa cell nuclear extract-TFIID oligonucleotide complex and the purified TBP-TFIID oligonucleotide complex but was distinct from that of the PPBP-polh B domain complex. Figure 4A, lanes 1 and 9, shows the mobilities of the free TFIID oligonucleotide and polh B domain, respectively, in the absence of any extract. The complexes described above were specific in their binding to the respective domains, as seen in Fig. 4B. The PPBP-B domain complex (Fig. 4B, lane 2) could be specifically competed with a 25-fold excess of unlabeled B domain (Fig. 4B, lane 3) but not with a similar excess of the unlabeled TFIID oligonucleotide (Fig. 4B, lane 4). Similarly, the Sf9 cell nuclear extract-TFIID complex (Fig. 4B, lane 6) could be specifically competed with a 25-fold excess of unlabeled TFIID oligonucleotide (Fig. 4B, lane 8) but not with an equal amount of unlabeled polh promoter B domain (Fig. 4B, lane 7). Under the existing binding conditions, HeLa cell nuclear extract could not bind the polh B domain (Fig. 4B, lane 9) whereas it could interact with the consensus TFIID oligonucleotide to give a DNA-protein complex similar in mobility to that obtained with the consensus TFIID oligonucleotide (Fig. 4B; compare lanes 6 and 10). These results clearly demonstrate the presence of a specific TBP-like activity in Sf9 cell nuclear extract (47) distinct from that of PPBP. Western blot analysis (Fig. 4C) with a rabbit antiserum against the C-terminal domain of hTBP (a kind gift from Robert G. Roeder, Rockefeller University) further confirmed the presence of a TBP-like factor in Sf9 cell nuclear extract. Anti-TBP antiserum at a dilution of 1:3,000 gave a specific ∼37-kDa band when crude HeLa cell nuclear extract and commercially available TBP (Promega) were used as positive controls (Fig. 4B, lanes 4 and 3), while the negative control with purified Sp1 (Promega) failed to generate a detectable signal (Fig. 4B, lane 1). A specific ∼30-kDa band was seen with the Sf9 cell nuclear extract (Fig. 4B, lane 2). A similar blot probed with preimmune rabbit serum did not give any signal (data not shown). Supershifting of the consensus TFIID sequence-purified TBP complex with a 1:500 dilution of anti-TBP antibody (Fig. 4A, lane 8) was not observed, which was expected, since these antibodies are against the C-terminal domain of TBP, which is involved in binding to DNA (43, 44). Control lanes with only normal rabbit serum without extract (Fig. 4A, lane 5), only anti-TBP antibody (Fig. 4A, lane 6), or both pure TBP and normal rabbit serum (Fig. 4A, lane 7) as expected showed no complex formation. We also did not observe supershifting of the polhB-PPBP complex with this antiserum (data not shown). These results, while demonstrating the presence of distinct TBP-like activity in Sf9 cells, also strongly argue against TBP having a direct role in polh promoter-driven transcription with respect to its ability to directly contact the polh promoter.
FIG. 4.
PPBP is different from TBP. (A) Labeled TFIID consensus oligonucleotide (lanes 1 to 8) and polh B domain (lanes 9 to 12) were used in gel mobility shift assays either alone (lanes 1 and 9), with 1 μg of HeLa cell nuclear extract (lanes 2 and 10), with 20 ng of purified TBP (Promega) (lanes 4, 7, 8, and 12), or with 1 μg of Sf9 cell nuclear extract (lanes 3 and 11). For the supershift assay TFIID oligonucleotide was incubated either with a 1:500 dilution of normal rabbit serum (NRS) (lane 5) or a 1:500 dilution of anti-TBP serum (lane 6), with normal rabbit serum and 20 ng of purified TBP (lane 7), or with anti-TBP serum and 20 ng of purified TBP (lane 8). (B) Gel retardation assays were carried out with either labeled polh B domain (lanes 1 to 4 and 9) or TFIID consensus oligonucleotide (lanes 5 to 8 and 10). The labeled polh B domain was incubated either alone (lane 1), with 1 μg of Sf9 cell nuclear extract (lanes 2 to 4), or with 1 μg of HeLa cell nuclear extract (lane 9). The DNA-protein complex obtained (lane 2) was competed with 25 ng of unlabeled polh B domain (lane 3) or with 25 ng of unlabeled TFIID domain (lane 4). The labeled TFIID oligonucleotide was incubated either alone (lane 5), with 1 μg of Sf9 cell nuclear extract (lanes 6 to 8), or with 1 μg of HeLa cell nuclear extract (lane 10). The TFIID-Sf9 complex obtained (lane 6) was competed with 25 ng of unlabeled polh B domain (lane 7) or with 25 ng of unlabeled TFIID domain (lane 8). (C) Western blot analysis. Lane 1, 60 ng of purified human Sp1 protein as a negative control; lane 2, 100 μg of Sf9 cell nuclear extract; lanes 3 and 4, 60 ng of purified TBP and 100 μg of HeLa cell nuclear extract, respectively, as positive controls. The blot was probed with a 1:3,000 dilution of anti-hTBP antibody. Protein molecular mass markers are shown on the right.
PPBP is present in phosphocellulose fractions that support in vitro transcription.
The presence of PPBP activity was investigated in the 0.3 and 0.5 M phosphocellulose fractions used in the in vitro transcription system reported for AcNPV late and very late genes (58). By using EMSAs, PPBP was detected in 0.3 and 0.5 M KCl fractions (Fig. 5, lanes 3 and 4), which were shown earlier to support in vitro transcription, but not in the 0.75 and 1.0 M KCl eluates (Fig. 5, lanes 5 and 6), which did not support in vitro transcription of a C-free cassette driven by the polh promoter (58). The physical presence of PPBP in both of the transcription-permissive fractions and its absence from the phosphocellulose fractions which failed to support in vitro transcription from AcNPV late promoters are further pointers to the possible involvement of PPBP in transcription from the baculovirus late and very late gene promoters.
FIG. 5.
In vitro transcription-permissive fractions contain PPBP. Labeled polh B domain was incubated either alone (lane 1); with 1 μg of crude Sf9 cell nuclear extract (lane 2); or with 0.3 (lane 3), 0.5 (lane 4), 0.75 (lane 5), or 1.0 (lane 6) M KCl phosphocellulose fractions.
PPBP is directly involved in in vitro transcription from the polh promoter.
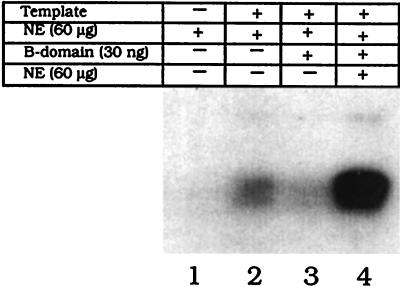
To demonstrate that PPBP is involved in transcription from the polh promoter, an in vitro transcription assay with Sf9 cell nuclear extracts prepared 36 h p.i. was carried out. A specific transcript corresponding to the C-free cassette (a kind gift from Linda A. Guarino, Texas A&M University) was produced from an AcNPV-infected extract (Fig. 6, lane 2). The same transcript was not generated when transcription was carried out in the absence of template (Fig. 6, lane 1). Transcription was significantly reduced (Fig. 6, lane 3) when PPBP was specifically sequestered out, in the presence of an excess of unlabeled oligonucleotide containing PPBP-binding motifs, and was thus not available for binding to the polh promoter driving transcription of the C-free cassette. Transcription was restored when this reaction mixture was replenished with infected nuclear extract containing PPBP (Fig. 6, lane 4). An enhanced regaining of transcriptional activity above normal, upon replenishment, is probably due to an increase in the concentration of other factors in the extract that promote reinitiation. The amount of unlabeled B domain required to completely deplete PPBP from the crude extract was determined in a parallel EMSA (data not shown). These results unequivocally demonstrate that PPBP is not only present in in vitro transcription-permissive extracts but is recruited during the basal transcription assembly process.
FIG. 6.
PPBP is required for in vitro transcription from the polyhedrin promoter. pPolh/CFS template (1.5 μg) harboring the C-free cassette (58) was transcribed in the presence of 60 μg of AcNPV-infected Sf21 cell nuclear extract (NE) (lane 2) collected 36 h p.i. Lane 3 shows the transcription reaction carried out after sequestering PPBP with 30 ng of polh B domain. Lane 4 shows the transcription reaction after first specifically mopping out PPBP from the reaction mixture with 30 ng of polh B domain and then replenishing it with 60 μg of fresh nuclear extract. Lane 1 is the reaction carried out in the absence of template DNA.
PPBP-polh promoter interaction is required for transcription in vivo.
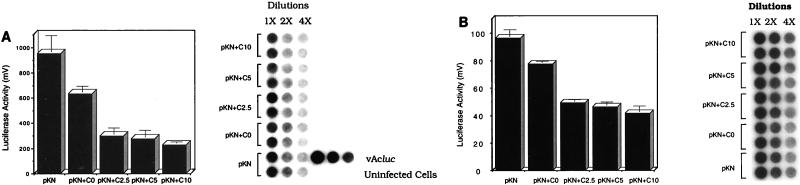
To provide evidence for the functional significance of PPBP vis-a-vis transcription from the polyhedrin promoter, in vivo competition of PPBP binding in transient expression assays was performed (Fig. 7A). The construct pKNluc, which carries the luc gene under the polh promoter together with the promoter-flanking sequences encompassing the EcoRI I fragment of AcNPV, was used as the reporter plasmid. Interaction of PPBP with the polh sequence in pKNluc was competed for by cotransfection of different amounts of the construct pAJpol, which carries only the 92-bp polh promoter. The total transfected plasmid was normalized to 20 μg with pUC18, with 10 μg of pKNluc used in all cotransfections. There was about 50% reduction in luciferase activity assayed 60 h p.i. when 2.5 μg of the competitor plasmid was added. A minor reduction beyond this decline in luciferase activity was observed even when 5 and 10 μg of the competing plasmid were used. Interestingly, there was also a minor but consistent reduction in luciferase expression when only pUC18 was used as a competitor. Analysis of the pUC18 sequence revealed the presence of two AATAAA motifs in the vector backbone that are identical to the sequences binding PPBP within the polh promoter. These sequences may therefore act as weak competitors of PPBP binding by pUC18 in vivo, accounting for the reduction in luc expression. Sequestering of PPBP from the polh promoter, therefore, causes a reduction in reporter gene expression, suggesting a function for this host protein in polh promoter-driven transcription.
FIG. 7.
PPBP-polh promoter interaction is required for transcription in vivo. (A) Luciferase activity of the reporter construct pKNluc (pKN) is decreased in the presence of increasing amounts of the competitor plasmid pAJpol (C). The total amount of transfected plasmid DNA was normalized to 20 μg with pUC18. The bars are labeled as follows: pKN, 10 μg of pKNluc in the absence of both competitor and pUC18; pKN+CO, 10 μg of pKNluc cotransfected with 10 μg of pUC18 in the absence of competitor; pKN+C2.5, 10 μg of pKNluc cotransfected with 2.5 μg of competitor; pKN+C5, 10 μg of pKNluc cotransfected with 5 μg of competitor; pKN+C10, 10 μg of pKNluc cotransfected with 10 μg of competitor. Southern hybridization of a DNA dot blot of transfected cells is shown in the right-hand panel. Replicates of three dilutions of cells from each transfection set were blotted and probed with the radiolabeled luc gene. Uninfected cells and vAcluc (a recombinant virus carrying the luc gene in place of the polyhedrin gene)-infected cells are shown as negative and positive controls, respectively. (B) Luciferase activity of the reporter construct pKNluc (pKN) is decreased in the presence of increasing amounts of the competitor plasmid pAJpol (C) and in the presence of 1 μg of α-amanitin/ml added 8 h posttransfection. The bars are labeled as in panel A. The right-hand panel shows a dot blot of cells from each transfection set with the radiolabeled luc fragment as a probe. Error bars indicate standard deviations.
In a parallel experiment (Fig. 7B), in vivo competition of PPBP binding was carried out in transient expression assays in the presence of α-amanitin. Late and very late gene transcription in AcNPV is dependent upon an α-amanitin-insensitive virus-encoded or virus-modified RNA polymerase. Sf9 cells were first infected with AcNPV and 36 h later were cotransfected with the reporter and competitor plasmids as well as pUC18. Medium containing α-amanitin (1 μg/ml) was added to the wells 8 h posttransfection, and the cells were assayed for luciferase activity 24 h after the addition of α-amanitin. As expected for very late AcNPV gene transcription, there was no difference in the results obtained in the presence or absence of α-amanitin.
Mutations in the PPBP cognate sequence motifs within the polh promoter abolish expression from the polyhedrin promoter in vivo.
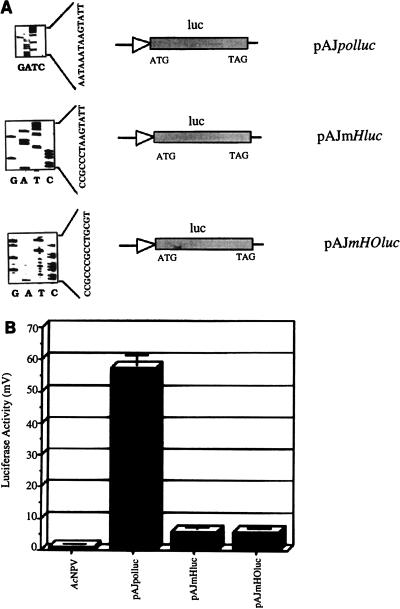
To demonstrate that binding of PPBP to its cognate sequences, essentially the octa- and hexamotifs within the initiator promoter, is critical for transcription in vivo, three sets of 65-mer complementary oligonucleotides with appropriate restriction sites spanning the polyhedrin promoter (−5 to −65) were designed (Table 1). These oligonucleotides represented either the wild-type polyhedrin promoter or mutated PPBP cognate motifs where the hexamotif (CCGCCC in place of AATAAA) and/or the octamotif (GCCTGCGG in place of TAAGTATT) was altered. In vitro binding analyses of these 65-mer oligonucleotides on a gel mobility shift assay failed to generate PPBP-promoter complex for the mutant derivatives of the promoter (data not shown), as reporter earlier (2). These oligonucleotides were then cloned into pAJluc, a pUC18-based plasmid carrying a promoterless luc gene, so as to drive the transcription of the luc reporter gene. The identities of these promoter constructs were confirmed by dideoxy sequencing (Fig. 8A).
FIG. 8.
Mutations within the hexa- and octamotifs recognized by PPBP abolish polyhedrin promoter activity. (A) Constructs carrying the unmutated and mutated polyhedrin promoters are depicted. The sequencing gels showing the mutations are on the left. (B) Transient luciferase expression measured 60 h p.i. in a luminometer. Equal amounts of plasmid DNA constructs were transfected into the insect cells.
Transient expression assays of the luciferase reporter gene were carried out with the recombinant promoter fusion constructs. Mutation of the PPBP-binding motifs resulted in near-zero (Fig. 8B) luciferase expression compared to that of the unmutated wild-type polyhedrin promoter construct. Luciferase expression above the cutoff limit was undetectable for both the hexamotif-octamotif mutant construct (comprising the transcription start point) and the hexamotif (alone) mutant construct. This knock-out data directly demonstrates that in a situation where binding of PPBP is eliminated due to the absence of cognate sequence motifs, in vivo expression of a reporter gene from the polh promoter is also abolished.
Mutations of individual bases within the AATAAA motif abolish reporter gene expression from the polyhedrin promoter in vivo.
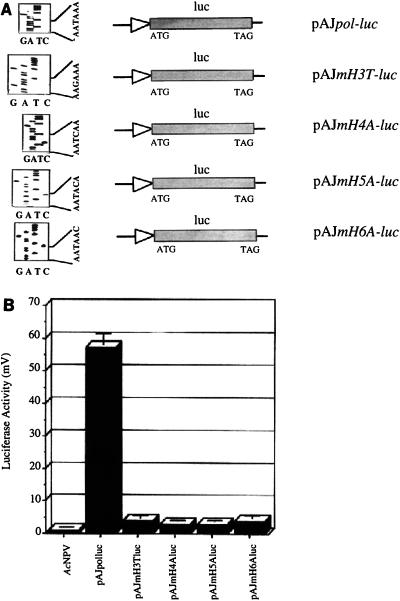
Having demonstrated the importance of the hexamotif alone in in vivo transcription, we investigated mutation of this sequence and its corresponding effect on transcription. Sequence alignment of the polyhedrin promoter and another very late promoter, p10, revealed that the hexamotifs in the two promoters share four bases (TAAA), at positions −52 to −55 and −72 to −75 in the polyhedrin and p10 promoters (16), respectively. Since both promoters can bind PPBP and the PPBP-p10 duplex promoter interaction also involves the hexamotif (16), it is conceivable that these last four bases alone may be involved. Four sets of complementary oligonucleotides spanning the polyhedrin promoter from −5 to −65, with appropriate restriction sites to facilitate cloning, were synthesized. Within these oligonucleotides the hexamotif AATAAA was mutated to either AAGAAA, AATCAA, AATACA, or AATAAC (mutations are underlined) to give rise to mH3T, mH4A, mH5A, and mH6A constructs, respectively. From in vitro binding analyses of these 65-mer oligonucleotides it was apparent that none of the mutations give rise to a DNA-protein complex similar in mobility to the PPBP complex but instead give complexes differing in intensity and/or mobility (data not shown), although the same amounts of nuclear extract and equal amounts of labeled oligonucleotides were used. These oligonucleotides were then cloned into the promoterless luciferase vector, pAJluc, to drive the expression of the luc reporter gene. Mutations were confirmed by dideoxy sequencing (Fig. 9A). These mutated plasmid reporter constructs were used as before for transient expression assays, and luc expression levels were compared with respect to the original unmutated construct, pAJpol-luc. A drastic reduction in luciferase expression was observed with all of the mutations (Fig. 9B). These results demonstrate the importance of the individual nucleotides within the hexanucleotide motif in terms of both complex formation in vitro and expression of the reporter gene in vivo.
FIG. 9.
Individual bases within the hexamotif regulate polyhedrin promoter activity. (A) Plasmid constructs with point mutations within the polyhedrin promoter are shown. The sequencing gels showing the mutations are shown on the left. (B) Transient luciferase expression measured 60 h p.i. in a luminometer. Equal amounts of plasmid DNA constructs were transfected into the insect cells.
DISCUSSION
Many cellular genes do not contain a canonical TATA box-like sequence and are far simpler in their organization. Several initiator-containing promoters (51), which probably represent the simplest promoters known so far and which are sufficient for accurate basal transcription both in vitro and in vivo, have now been identified (3, 60). A core tetranucleotide motif, CAGT, has been located at the RNA start site of the transregulator gene ie-1 of AcNPV (45), which is involved in its transcription and also resembles the proposed consensus for arthropod transcriptional initiator elements (4). Functional analyses of 80 random and mutant initiator elements (18) identified a loose consensus sequence (Py Py A+1 N T/A Py Py) for such initiator promoter elements. The 18-bp sequence surrounding the baculovirus polyhedrin gene transcription initiation point, comprised of the hexanucleotide (AATAAA) and octanucleotide (TAAGTATT) motifs, represents an initiator-like sequence and also has considerable homology with the initiator consensus sequence.
We reported earlier the identification of a host factor, PPBP, from Sf9 insect cells which displayed unusual characteristics with respect to affinity, specificity, and cognate sequence requirements (2). PPBP binding was abolished upon dephosphorylation, and it showed sequence-specific ssDNA-binding activity restricted to the coding strand. The half-life of the PPBP-coding strand interaction was higher than that of the PPBP-duplex promoter interaction (37, 38). The enhanced stability of PPBP-polh interaction induced by an additional factor(s) in a transcription-permissive cell line, as opposed to that in the nonpermissive Bm5 cell line reported in this study, is similar to the role played by a number of transcription activators in stabilizing TFIID-promoter complexes (22, 54). Furthermore, affinity purification of PPBP yielded a comigrating protein of ∼30 kDa on a silver-stained gel (2, 14). It is possible that PPBP is an initiator-binding protein which, like TBP (25) and other transcription factors, such as human cytomegalovirus IE2 protein (24), HMG-1Y protein (52), etc., contacts DNA through the minor groove. PPBP therefore represents a rare example of an initiator-binding protein with such a high salt tolerance and dual binding activities (2, 37).
Our results showing that 0.3 and 0.5 M KCl fractions supporting late and very late gene transcription in vitro (58) also have PPBP activity point to the central role of PPBP in polh transcription. In vitro transcription of the C-free RNA reporter cassette was affected when PPBP was sequestered and unavailable for binding to the polh promoter. However, transcription was immediately restored upon the addition of extract containing PPBP, thus categorically demonstrating that the host factor PPBP is not merely involved in polh transcription but is absolutely necessary. In in vivo binding site knock-out experiments, where PPBP binding was abrogated, luciferase reporter gene expression driven from the polh promoter was not detected. The in vivo mopping (13) of PPBP by plasmids carrying PPBP-binding sites and the consequent downregulation of polh promoter-driven luc expression further document the necessity of PPBP-polh promoter interaction in transcription from this promoter. However, it is interesting to note that in the absence of either the initiator sequence or PPBP, transcription from the polh promoter still proceeds, albeit at a reduced level.
In a simplistic model implicating PPBP in polyhedrin transcription, this host factor, after scanning the promoter, binds to the initiator element. It then recruits other factors, perhaps via the TBP and/or virus-specific factors (LEFs?), and then switches over to a ssDNA-binding regime, keeping the coding strand in place for repeated rounds of transcription. The observed half-lives of 15 and 60 min of PPBP duplex promoter and ssDNA-binding activity, respectively, support this scenario. The observation of a putative helicase activity and DNA-dependent ATPase activity in partially purified PPBP (data not shown) makes possible the unwinding of DNA required between the ds- and ssDNA-binding events without PPBP moving away from the DNA. The promoter-binding role of TBP, although present in these cells, has apparently been taken over by PPBP, which makes direct contact with the promoter. In the absence of the initiator motif and consequently of the initiator-binding protein PPBP, an Sp-like factor(s) which is also present in these cells (17) may help in recruiting TBP functionally at the start site, thus allowing basal levels of transcription. This provides a possible explanation for the low level of transcription noted during in vitro and in vivo transcription mopping assays (Fig. 6 and 7). These results fit well with a proposed model (31) for transcription initiation from initiator-containing promoters through an initiator-binding protein, thereby making PPBP an important component of the transcription initiation complex.
Subtractive hybridization or marker rescue experiments have identified a number of lef and vlf gene products encoded by the viral genome (26, 34, 55). These genes may be directly or indirectly involved in polyhedrin promoter activation and must function in conjunction with cellular factors. Direct interaction and involvement in transcription from late and very late promoters so far has not been demonstrated for any of these factors. A 38-kDa host factor (13) which is required for the enhancer function of the recently reported hr1 enhancer element (12) is the other example of a host factor modulating polh gene expression. A Trichoplusia ni host cell-specific factor, hcf1, has been reported to be involved in differential gene expression from late gene promoters in two different cell lines (28). PPBP, therefore, constitutes the only host factor that binds to transcriptionally important motifs within the baculovirus very late polyhedrin and p10 gene promoters (16) and is the likely candidate to be involved in cross-talk with other accessory transcription factors, including the lef gene products. The identity of these factors and the nature of their cross-talk with PPBP remain important questions.
The fact that the virus recruits a host factor for transcribing one of the most important viral genes then raises the fundamental question of the function of PPBP per se within the insect cell. The fact that the TAAG motif of the polh promoter has possibly originated from the host genome (9) explains why this motif is recognized by host PPBP. The PPBP-binding motif AATAAA within the polh initiator promoter, though identical to the polyadenylation signal (56), is clearly not such a signal in the context of the polyhedrin-flanking open reading frames (1) in the viral genome. AATAAA motifs are also spread throughout the AcNPV sequence (57) but do not act as transcription start sites unless they are in the neighborhood of the transcription start point, as in the case of the polh promoter. The eukaryotic cleavage-polyadenylation consists of four polypeptides, one of which is ∼30 kDa in molecular mass, which previously escaped detection (19). It is tempting to suggest that PPBP may belong to the polyadenylation specificity factor (CPSF) complex. Direct linkage between mRNA processing and transcription via the CTD of RNA polymerase II has been established (33, 53), including the recruitment by TFIID of one of the factors of the CPSF complex to the preinitiation complex (7). While we are in the process of documenting the natural role of the host factor PPBP, our results provide a novel insight into host-parasite interaction (15) at the transcription level during viral pathogenesis (35, 42).
ACKNOWLEDGMENTS
S.G. and A.J. contributed equally to this work.
We thank Sandip K. Basu, National Institute of Immunology, New Delhi, for critically reviewing this manuscript. We thank Narendra K. Tuteja, ICGEB, New Delhi, India, for the HeLa cell nuclear extracts used in this study.
A.J. and S.H. were recipients of a research fellowship from the Council for Scientific and Industrial Research, Government of India. This work was supported by a grant (SP/SO/D-45/95) to S.E.H. from the Department of Science and Technology, Government of India.
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Burma S, Mukherjee B, Jain A, Habib S, Hasnain S E. An unusual 30-kDa protein binding to the polyhedrin gene promoter of Autographa californica nuclear polyhedrosis virus. J Biol Chem. 1994;269:2750–2757. [PubMed] [Google Scholar]
- 3.Chen H, Vinnakota R, Flint S J. Intragenic activating and repressing elements control transcription from the adenovirus IVa2 initiator. Mol Cell Biol. 1994;14:676–685. doi: 10.1128/mcb.14.1.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- 5.Chiang S Y, Welch J, Rauscher III F J, Beerman T A. Effects of minor groove binding drugs on the interaction of TATA box binding protein and TFIIA on DNA. Biochemistry. 1994;33:7033–7040. doi: 10.1021/bi00189a003. [DOI] [PubMed] [Google Scholar]
- 6.Copenhaver G P, Putnam C D, Denton M L, Pikaard C S. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994;22:2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 8.deWet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen P D, Rice W C, Miller D W, Miller L K. Bidirectional transcription from a solo long terminal repeat of the retrotransposon TED: symmetrical RNA start sites. Mol Cell Biol. 1986;6:1599–1607. doi: 10.1128/mcb.6.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs L Y, Woods M S, Weaver R F. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;48:641–646. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grula M A, Buller P L, Weaver R F. α-Amanitin-resistant viral RNA synthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvae and Spodoptera frugiperda cells. J Virol. 1981;38:916–921. doi: 10.1128/jvi.38.3.916-921.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habib S, Pandey S, Chatterji U, Burma S, Ahmad R, Jain A, Hasnain S E. The bifunctional AcMNPV homologous region sequence (hr1) enhancer and ori functions have different sequence requirements. DNA Cell Biol. 1996;15:737–747. doi: 10.1089/dna.1996.15.737. [DOI] [PubMed] [Google Scholar]
- 13.Habib S, Hasnain S E. A 38 kDa host factor interacts with functionally important motifs within the AcMNPV homologous region (hr1) DNA sequence. J Biol Chem. 1996;271:28250–28258. doi: 10.1074/jbc.271.45.28250. [DOI] [PubMed] [Google Scholar]
- 14.Hasnain S E, Habib S, Jain A, Burma S, Mukherjee B. A host factor with single-stranded DNA binding activity is involved in transcription from the baculovirus polyhedrin promoter. Methods Enzymol. 1996;274:20–32. doi: 10.1016/s0076-6879(96)74005-3. [DOI] [PubMed] [Google Scholar]
- 15.Hasnain S E, Jain A, Habib S, Ghosh S, Chatterji U, Ramachandran A, Das P, Venkaiah B, Pandey S, Liang B, Ranjan A, Natarajan K, Azim C A. Involvement of host factors in transcription from baculovirus very late promoters—a review. Gene. 1997;190:113–118. doi: 10.1016/s0378-1119(96)00827-x. [DOI] [PubMed] [Google Scholar]
- 16.Jain A, Hasnain S E. A 30-kDa host protein binds to two very-late baculovirus promoters. Eur J Biochem. 1996;239:384–390. doi: 10.1111/j.1432-1033.1996.0384u.x. [DOI] [PubMed] [Google Scholar]
- 17.Jain, A., A. Ramachandran, S. Ghosh, U. Chatterji, K. Natarajan, C. A. Azim, S. Burma, and S. E. Hasnain. Sp1-like transcription factors present in insect cells support basal levels of gene expression from the baculovirus polyhedrin initiator promoter. Submitted for publication.
- 18.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenny A, Hauri H-P, Keller W. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol. 1994;14:8183–8190. doi: 10.1128/mcb.14.12.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidd M, Emery V C. The use of baculoviruses as expression vectors. Appl Biochem Biotechnol. 1993;42:137–159. doi: 10.1007/BF02788049. [DOI] [PubMed] [Google Scholar]
- 21.Kim S K, Norden B. Methyl green: a DNA major-groove binding drug. FEBS Lett. 1993;315:61–64. doi: 10.1016/0014-5793(93)81133-k. [DOI] [PubMed] [Google Scholar]
- 22.Kingston R E, Green M E. Modeling eukaryotic transcriptional activation. Curr Biol. 1994;4:325–332. doi: 10.1016/s0960-9822(00)00071-3. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lang D, Stamminger T. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 1994;22:3331–3338. doi: 10.1093/nar/22.16.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D K, Horikoshi M, Roeder R G. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- 26.Lu A, Miller L K. Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis virus. J Virol. 1994;68:6710–6718. doi: 10.1128/jvi.68.10.6710-6718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu A, Miller L K. Differential requirements for baculovirus late expression factor genes in two cell lines. J Virol. 1995;69:6265–6272. doi: 10.1128/jvi.69.10.6265-6272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckow V A. Cloning and expression of heterologous genes in insect cells with baculovirus vectors. In: Prokop A, Bajpai R K, Ho C, editors. Recombinant DNA technology and applications. New York, N.Y: McGraw-Hill; 1991. pp. 97–152. [Google Scholar]
- 30.Luckow V A, Summers M D. High level expression of nonfused foreign genes with Autographa californica nuclear polyhedrosis virus expression vector. Virology. 1989;170:31–39. doi: 10.1016/0042-6822(89)90348-6. [DOI] [PubMed] [Google Scholar]
- 31.Martinez E, Chiang C M, Ge H, Roeder R G. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuura Y, Possee R D, Overton H A, Bishop D H L. Baculovirus expression vectors: the requirements for high level expression of proteins including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 33.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 34.McLachlin J R, Miller L K. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol. 1994;68:7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda G, Schuppli D, Barrerra I, Hausherr C, Sogo J M, Weber H. Recognition of bacteriophage Qbeta plus strand as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J Mol Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 36.Morris T D, Miller L K. Mutational analysis of a baculovirus major late promoter. Gene. 1994;140:147–153. doi: 10.1016/0378-1119(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee B, Burma S, Hasnain S E. The 30-kDa protein binding to the “initiator” of the baculovirus polyhedrin promoter also binds specifically to the coding strand. J Biol Chem. 1995;270:4405–4411. doi: 10.1074/jbc.270.9.4405. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee B, Burma S, Talwar G P, Hasnain S E. Transcriptional regulation of cell line-dependent, baculovirus-mediated expression of foreign genes. DNA Cell Biol. 1995;14:7–14. doi: 10.1089/dna.1995.14.7. [DOI] [PubMed] [Google Scholar]
- 39.Ooi B G, Rankin C, Miller L K. Downstream sequences augment transcription from the essential initiation site of baculovirus polyhedrin gene. J Mol Biol. 1989;210:721–736. doi: 10.1016/0022-2836(89)90105-8. [DOI] [PubMed] [Google Scholar]
- 40.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman and Co.; 1992. [Google Scholar]
- 41.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 42.Pedulla M L, Lee M H, Lever D C, Hatfull G F. A novel host factor for integration of mycobacteriophage L5. Proc Natl Acad Sci USA. 1996;93:15411–15416. doi: 10.1073/pnas.93.26.15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson M G, Tanese N, Pugh B G, Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990;248:1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- 44.Possee R D, Howard S C. Analysis of the polyhedrin gene promoter of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 1987;5:10233–10248. doi: 10.1093/nar/15.24.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pullen S S, Friesen P D. The CAGT motif functions as an initiator element during early transcription of the baculovirus transregulator ie-1. J Virol. 1995;69:3575–3583. doi: 10.1128/jvi.69.6.3575-3583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rankin C, Ooi B G, Miller L K. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene. 1988;70:39–50. doi: 10.1016/0378-1119(88)90102-3. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen C, Rohrmann G F. Characterization of the Spodoptera frugiperda TATA-binding protein: nucleotide sequence and response to baculovirus infection. Insect Biochem Mol Biol. 1994;24:699–708. doi: 10.1016/0965-1748(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 48.Roeder R G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 52.Solomon M J, Strauss F, Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A·T base pairs in duplex DNA. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinmetz E. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 54.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 55.Todd J W, Passarelli A L, Miller L K. Eighteen baculovirus genes, including lef-11, p35, 39K, and p47, support late gene expression. J Virol. 1995;69:968–974. doi: 10.1128/jvi.69.2.968-974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 57.Westwood J A, Jones I M, Bishop D H L. Analyses of alternative poly(A) signals for use in baculovirus expression vectors. Virology. 1993;195:90–99. doi: 10.1006/viro.1993.1349. [DOI] [PubMed] [Google Scholar]
- 58.Xu B, Yoo S, Guarino L A. Differential transcription of baculovirus late and very late promoters: fractionation of nuclear extracts by phosphocellulose chromatography. J Virol. 1995;69:2912–2917. doi: 10.1128/jvi.69.5.2912-2917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang L, Stetler D A, Weaver R F. Structural comparison of the Autographa californica nuclear polyhedrosis virus induced RNA polymerase and the three nuclear RNA polymerases from the host, Spodoptera frugiperda. Virus Res. 1991;20:251–264. doi: 10.1016/0168-1702(91)90079-b. [DOI] [PubMed] [Google Scholar]
- 60.Yoo W, Martin M E, Folk W R. PEA1 and PEA3 enhancer elements are primary components of the polyomavirus late transcription initiator element. J Virol. 1991;65:5391–5400. doi: 10.1128/jvi.65.10.5391-5400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]