Abstract
Replication of the Epstein-Barr viral (EBV) genome occurs once per cell cycle during latent infection. Similarly, plasmids containing EBV’s plasmid origin of replication, oriP, are replicated once per cell cycle. Replication from oriP requires EBV nuclear antigen 1 (EBNA-1) in trans; however, its contributions to this replication are unknown. oriP contains 24 EBNA-1 binding sites; 20 are located within the family of repeats, and 4 are found within the dyad symmetry element. The site of initiation of DNA replication within oriP is at or near the dyad symmetry element. We have identified a plasmid that contains the family of repeats but lacks the dyad symmetry element whose replication can be detected for a limited number of cell cycles. The detection of short-term replication of this plasmid requires EBNA-1 and can be inhibited by a dominant-negative inhibitor of EBNA-1. We have identified two regions within this plasmid which can independently contribute to this replication in the absence of the dyad symmetry element of oriP. One region contains native EBV sequences within the BamHI C fragment of the B95-8 genome of EBV; the other contains sequences within the simian virus 40 genome. We have mapped the region contributing to replication within the EBV sequences to a 298-bp fragment, Rep*. Plasmids which contain three copies of Rep* plus the family of repeats support replication more efficiently than those with one copy, consistent with a stochastic model for the initiation of DNA synthesis. Plasmids with three copies of Rep* also support long-term replication in the presence of EBNA-1. These observations together indicate that the latent origin of replication of EBV is more complex than formerly appreciated; it is a multicomponent origin of which the dyad symmetry element is one efficient component. The experimental approach described here could be used to identify eukaryotic sequences which mediate DNA synthesis, albeit inefficiently.
When the human gammaherpesvirus Epstein-Barr virus (EBV) infects primary human B cells, it efficiently induces them to proliferate indefinitely in cell culture, thereby causing immortalization of these cells (53, 54). EBV usually maintains its genome as a plasmid in cells that it infects. To do this, the virus maintains a latent infection in which the viral DNA replicates semiconservatively, once per cell cycle, during S phase (1, 7, 21, 60) as do its host cell’s chromosomes.
During latent infection, replication is initiated from the EBV latent origin of plasmid replication, oriP (17). The virus contributes only one protein, EBV nuclear antigen 1 (EBNA-1) (35, 45, 61), for this feat; the host cell provides all other replication machinery. Plasmids derived from EBV containing oriP replicate semiconservatively each cell cycle in the presence of EBNA-1 as does the viral genome (7, 21, 60). When cells that maintain plasmids containing oriP are grown in the absence of selection, approximately 2 to 6% of those cells will lose their oriP-containing plasmids per cell generation (30, 47, 56, 58). It is not yet clear how oriP plasmids are efficiently maintained in cells. However, the family of repeats (FR) of oriP contributes to the retention of plasmid DNA in EBNA-1-positive cells (32, 40). EBNA-1, the EBV genome, and yeast artificial chromosomes containing the gene encoding EBNA-1 and oriP have in common the ability to associate with metaphase chromosomes in human cells (12, 20, 22, 42, 46, 50). This association may play a critical role in efficiently segregating the viral genome to daughter cells after mitosis.
The latent origin of plasmid replication, oriP, consists of two DNA sequences, the FR and a dyad symmetry element (DS) (47). The FR is composed of 20 copies of a 30-bp repeat, each of which serves as a binding site for a dimer of EBNA-1 (24, 27, 45). The DS contains four partial copies of this repeat sequence which are also bound by EBNA-1 (15, 24, 27, 45). Bidirectional DNA synthesis initiates within or near the DS (17); that is, a replication bubble can be detected in the vicinity of the DS by two-dimensional gel electrophoresis (17). Various derivatives of oriP have been used to characterize the functional elements of this origin. In general, neither the FR nor the DS alone supports replication (35, 47, 57). Multiple copies of the DS do, however, support both short-term and long-term DNA replication in the presence of EBNA-1 (57). Thus, multiple copies of the DS can substitute for the FR, indicating that the DS must have a function distinct from that of the FR.
Derivatives of oriP that contain a minimal FR consisting of seven to nine binding sites for EBNA-1 and a wild-type DS replicate efficiently in both short-term and long-term experiments (8, 57). Also, all four binding sites of EBNA-1 within the DS are not required for replication (8, 23, 58). Derivatives of oriP that contain a wild-type FR and a mutated DS in which two of the four EBNA-1 binding sites are mutated replicate efficiently in both short-term and long-term experiments (23). Derivatives of oriP which lack all of the DS but have selected cellular sequences can also replicate stably in the presence of EBNA-1 (32). These cellular sequences score as having origins of DNA replication by analysis with two-dimensional gels (31). It is not known if these cellular sequences function as origins when in the context of the cell’s chromosomes, or if they have binding sites for EBNA-1.
We have identified a plasmid, FR-BamHI C-Luc, which contains a derivative of oriP that lacks the DS and which replicates in an EBNA-1-dependent manner for a few cell cycles. Wild-type EBNA-1 therefore contributes some function to support the detection of replication of plasmids that are partially defective in DNA synthesis. However, FR-BamHI C-Luc cannot replicate long term in the presence of wild-type EBNA-1. We have characterized FR-BamHI C-Luc’s ability to replicate, using both a functional derivative of EBNA-1, NΔ330-641, and the dominant-negative inhibitor of EBNA-1, NΔ450-641 (29). NΔ330-641 can detectably support short-term replication of FR-BamHI C-Luc, while NΔ450-641 inhibits its EBNA-1-dependent replication in short-term assays. We have mapped within FR-BamHI C-Luc two regions of DNA which independently contribute to its short-term replication. We have further characterized one region which lies within EBV sequences and have designated it Rep*. EBNA-1 can support long-term replication of plasmids which contain three copies of Rep* plus the FR. The assay we have used to identify Rep* may be used to define cellular DNAs which also mediate efficient or even inefficient DNA synthesis.
MATERIALS AND METHODS
Plasmids.
The plasmids used in these experiments (critical plasmids are depicted in Fig. 1 to 4) include the vector pCMV-βgal (48) and effector DNA encoding wild-type EBNA-1 (39), NΔ330-641 (29), and NΔ450-641 (40). Reporter DNAs, each containing the aminoglycoside phosphotransferase II gene (9), include oriP-BamHI C-Luc (29); DS-BamHI C-Luc (29); FR-BamHI C-Luc, which was derived from oriP-BamHI C-Luc by an EcoRV-HpaI deletion which removes the DS of oriP; FR-Δ-Luc, which was derived from FR-BamHI C-Luc by removing a SpeI-AvrII fragment which deletes the DS and native EBV sequences up to, but not including, the BamHI C promoter; FR-EBVΔ-Luc, which was derived from FR-BamHI C-Luc by an AvrII-XbaI deletion which removed proximal sequences of the BamHI C promoter; FR-BamHI C-Δ, which was derived from FR-BamHI C-Luc by deleting an XbaI-ClaI fragment which removed the coding region of the luciferase gene; FR-BamHI C-LucΔ, which was derived from FR-BamHI C-Luc by a ClaI-AccI deletion which removed the simian virus 40 (SV40) small tumor antigen (t-antigen) intron and part of the SV40 large tumor antigen (T-antigen) poly(A) addition signal at the 3′ end of the luciferase gene; oriP-Backbone, which was derived from oriP-BamHI C-Luc by a HpaI-HpaI deletion which removed the native EBV sequences downstream of the DS up to and including the BamHI C promoter, the luciferase gene, the SV40 t-antigen intron, and part of the SV40 T-antigen poly(A) addition signal; FR-Backbone, which was constructed by deleting an EcoRV-HpaI fragment from oriP-BamHI C-Luc, which generated a plasmid identical to oriP-Backbone except that the DS was also deleted; FR-λ-Luc, which was derived from oriP-BamHI C-Luc by inserting 2.2 kbp of lambda DNA between SpeI-HindIII, deleting the DS and native EBV sequences up to and including the BamHI C promoter; FR-Δ, which was constructed by deleting an EcoRV-EcoRV fragment from oriP-BamHI C-Luc, which removed the DS, the native EBV sequences up to and including the BamHI C promoter, and the gene encoding luciferase; FR-Δ2, which was generated by deleting an AvrII-AccI fragment from FR-BamHI C-Luc, which removed the proximal sequences of the BamHI C promoter, the luciferase gene, the SV40 t-antigen intron, and the SV40 T-antigen poly(A) addition signal; pΔBal 2 (47); pΔBal 12 (47); 1858, which was derived from FRΔ2 by deleting a DraII-Eco47III fragment; 1859, which was constructed from FRΔ2 by removing a DraIII-Bsu36I fragment; 1860, which was derived from FRΔ2 by removing a DraIII-KpnI fragment; 1861, which was constructed from FRΔ2 by deleting a Bsu36I-KpnI fragment; and 1862, which was derived from FRΔ2 by removing a Bsu36I-Eco47III fragment. Reporter plasmids containing multiple copies of Rep* were generated by amplifying the 298-bp Rep* fragment from FRΔ2 by using two primers: 5′-ACCAGGTCTAGACACTCAGTGTTGGCAAATGTG-3′, which would insert an XbaI site 5′ of Rep*; and 5′-ATGTTACCTAGGCCTAAGGTGTGCAGGCCTAC-3′, which would insert an AvrII site 3′ of Rep*. XbaI-Rep*-AvrII DNA was gel purified and ligated to itself in the presence of XbaI and AvrII to generate dimers of Rep* with both copies of Rep* in the same orientation. A dimer of Rep* was then inserted into 1862 at the SpeI site in the opposite orientation as the copy of Rep* already present in 1862 to generate 1925; 1926 was generated as was 1925 except that the dimer of Rep* was inserted into the SpeI site of 1862 in the same orientation as the copy already present. Control plasmids for the quantitative competitive PCR assay include competitor DNA (29) and oriP-minus (29).
FIG. 1.
Wild-type EBNA-1 and its derivatives. The DNA linking domains (grey boxes) (18, 36), the DNA binding and dimerization domain (hatched box) (3, 6, 28, 38, 49), the internal repeated sequence consisting entirely of glycine and alanine residues (Gly-Gly-Ala), and the nuclear localization sequence (NLS) (2) of EBNA-1 are noted. Regions rich in basic (+) and acidic (−) residues are indicated. The protein products of vectors encoding wild-type EBNA-1 and its derivatives are shown. EBNA-1 contains amino acids (aa) 1 to 641 and is wild-type EBNA-1 of the B95-8 strain of EBV (5). NΔ330-641 contains aa 331 to 641 of EBNA-1 (29), and NΔ450-641 contains the nuclear localization sequence (aa 379 to 386) fused in frame to aa 451 to 641 of EBNA-1 (40).
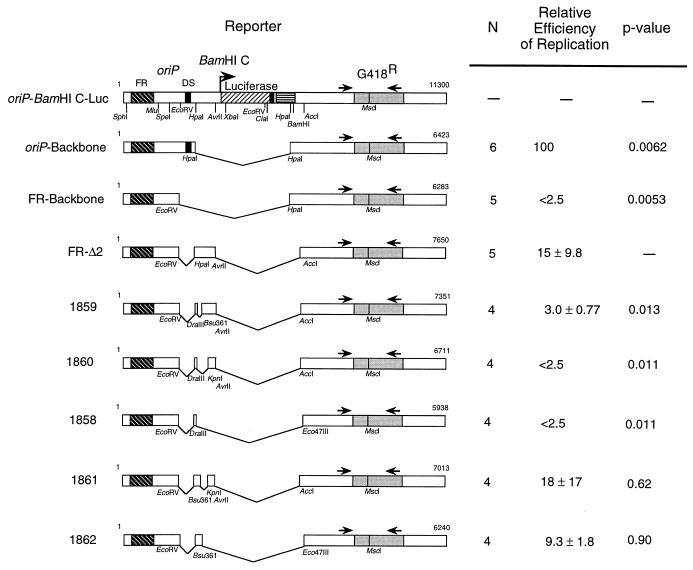
FIG. 4.
A 298-bp region of the EBV genome supports DNA replication in the absence of the DS. Maps of reporter plasmids oriP-BamHI C-Luc, oriP-Backbone, FR-Backbone, FR-Δ2, 1859, 1860, 1858, 1861, and 1862 are shown on the left. N, number of replicates. The efficiency of replication of each reporter by EBNA-1 is expressed as a percentage of DpnI-resistant DNA relative to oriP-Backbone at 96 h, which is set to 100% (100% = 7.9 ± 2.9 copies of DpnI-resistant DNA per transfected cell). Data were analyzed by using the Wilcoxon rank sum test (26) with Mstat (version 1.3, by Norman Drinkwater), and the P value comparing each derivative to FR-Δ2 is noted.
Cell lines.
The cell lines used for the short-term replication assays include 143B, an EBV-negative human osteosarcoma (4), and 143/EBNA-1, which stably expresses EBNA-1 and hygromycin B phosphotransferase (19, 55), conferring resistance to hygromycin (38). The cell lines used for the long-term replication assays include several clones of 143/EBNA-1/oriP-BamHI C-Luc (29), 143/EBNA-1/oriP-Backbone, 143/EBNA-1/FR-Backbone, 143/EBNA-1/1925, and 143/EBNA-1/1926 and one clone of 143/EBNA-1/FR-BamHI C-Luc. The cell lines used in long-term replication assays were generated as described previously (29). All cells were grown in Dulbecco modified Eagle medium (DMEM-HG) containing 10% calf serum, streptomycin sulfate (0.2 mg/ml), and penicillin G potassium (200 U/ml). 143/EBNA-1 cells were also grown in the presence of hygromycin (150 μg/ml). 143/EBNA-1/reporter-based cells were grown in the presence of G418 (600 μg/ml).
Quantitative competitive PCR assay. (i) Introduction of DNA into 143-derived cells.
143B cells or 143/EBNA-1 cells (2 × 107 of each) were resuspended in 1 ml of DMEM-HG–10% calf serum–50 mM HEPES (pH 7.4 to 7.6); 10 μg of each DNA (oriP-BamHI C-Luc or a derivative, oriP-minus, and, for experiments described in Tables 1 and 2, a derivative of EBNA-1 or vector) was electroporated into two samples of 107 cells/0.5 ml. One electroporated sample was plated per 150- by 25-mm plate in 20 ml of DMEM-HG–10% calf serum for 94 to 98 h or for 8, 12, 16, or 20 days. Samples were then harvested and prepared as described previously (29). Briefly, DNA was isolated by the Hirt extraction procedure (25), digested with DpnI to digest any methylated or unreplicated DNA, and digested with BamHI, EcoRV, NruI, AccI, or another appropriate restriction enzyme to linearize the plasmid DNA. Samples were extracted with phenol-chloroform and chloroform, ethanol precipitated, and resuspended in 1× Tris-EDTA at 105 cell equivalents/μl.
TABLE 1.
Wild-type EBNA-1 and NΔ330-641 support replication of plasmids oriP-BamHI C-Luc and FR-BamHI C-Luc but not DS-BamHI C-Luc in 143B cells at 96 h postelectroporation
| Effector | Relative efficiency of replicationa
|
||
|---|---|---|---|
| oriP-BamHI C-Luc | FR-BamHI C-Luc | DS-BamHI C-Luc | |
| Vector | <2.1 | <2.1 | <2.1 |
| EBNA-1 | 100 | 43 ± 3.4 | <2.1 |
| NΔ330-641 | 18 ± 0.94 | 2.3 ± 0.19 | <2.1 |
10 μg of each DNA (effector, reporter, and oriP-minus) was introduced into 107 143B cells by electroporation. Low-molecular-weight DNA was isolated after 94 to 98 h, and the relative concentration of DpnI-resistant plasmids was determined by quantitative competitive PCR and was corrected for transfection efficiency of 143B cells as described in Materials and Methods. The relative efficiency of replication by each derivative of EBNA-1 is expressed as a percentage of DpnI-resistant DNA relative to that of oriP-BamHI C-Luc replicated by wild-type EBNA-1, which is set to 100% (100% = 16 ± 14 copies of DpnI-resistant DNA per transfected cell). Data represent an average of two experiments ± standard deviation. The level of DpnI-resistant oriP-minus DNA detected was less than the lowest amount competitor used in all cases: <8.8% when oriP-BamHI C-Luc, oriP-minus, and wild-type EBNA-1 were cotransfected and <2.1% for all other reporter-effector combinations. Data represented as “<” indicate that the level of DpnI-resistant DNA was less than the smallest amount used for the competitor DNA and is therefore reported as less than an average of the smallest amount per transfected cell that could be determined by interpolation from the competitor DNA curve.
TABLE 2.
NΔ450-641 inhibits replication of FR-BamHI C-Luc by wild-type EBNA-1 in 143/EBNA-1 cells
| Effector | Relative efficiency of replicationa
|
|||
|---|---|---|---|---|
| oriP-BamHI C-Luc | oriP-minus | FR-BamHI C-Luc | oriP-minus | |
| Vector | 100 | <2.8 | 24 ± 6.7 | <0.72 |
| NΔ450-641 | 11 ± 7.2 | 1.1 ± 0.14 | 1.3 ± 0.50 | <0.72 |
10 μg of each DNA was introduced into 107 143/EBNA-1 cells by electroporation. Low-molecular-weight DNA was isolated after 94 to 98 h, and the relative concentration of DpnI-resistant plasmids was determined by quantitative competitive PCR and was corrected for transfection efficiency of 143/EBNA-1 cells as described in Materials and Methods. The relative efficiency of replication by each derivative of EBNA-1 is expressed as a percentage of DpnI-resistant DNA relative to that replicated by wild-type EBNA-1, which is set to 100% (100% = 18 ± 6.5 copies of DpnI-resistant DNA per transfected cell). Data represent an average of two (for oriP-BamHI C-Luc) or four (for FR-BamHI C-Luc) experiments ± standard deviation. Data represented as “<” indicate that the level of DpnI-resistant DNA was less than the lowest concentration used for the competitor DNA and is therefore reported as less than an average of the smallest amount per transfected cell that could be determined by interpolation from the competitor DNA curve.
(ii) PCR assay.
The following assay is a modification of the quantitative competitive PCR assay described previously (29). Briefly, five PCRs in a reaction volume of 100 μl were performed per sample, using increasing amounts of linearized competitor DNA per reaction (five of the six following amounts, corresponding to the indicated number of molecules of competitor DNA, per sample: 0.025 pg, approximately 4.9 × 103 molecules; 0.10 pg, 2.0 × 104 molecules; 0.40 pg, 7.9 × 104 molecules; 1.6 pg, 3.2 × 105 molecules, 6.4 pg, 1.3 × 106 molecules, and 26 pg, 5.0 × 106 molecules), 105 cell equivalents of sample DNA, and 2.5 U of Taq polymerase (Boehringer) in final concentrations of 0.2 mM for each deoxynucleoside triphosphate and 0.2 μM for each primer, plus approximately 1 nM for each 32P-end-labeled primer (see below). Each reaction was performed in 500-μl GeneAmp tubes (Perkin-Elmer) and was overlaid with 70 μl of mineral oil. DNA was amplified in a Thermocycler 480 (Perkin-Elmer) under the following conditions: 94°C for 5 min; 22 to 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; 72°C for 10 min; 4°C hold. Then 15 μl of the PCR product from each sample was loaded onto a 1.5% agarose gel, and the gel was electrophoresed in 1× TBE (0.9 M Tris-borate, 1 mM EDTA) overnight at 40 V. The gel was then fixed in 7.5% trichloroacetic acid or in 10% ethanol–10% acetic acid for 30 min and dried, and data were analyzed by using a Molecular Dynamics PhosphorImager.
(iii) End-labeling primers.
Seventy-five picomoles of primer was incubated with 10 U of T4 polynucleotide kinase (New England Biolabs), 1× kinase buffer (New England Biolabs), and 750 μCi of [γ-32P]ATP (125 pmol; Dupont NEN) in a 30-μl total reaction volume for 30 min at 30°C. An additional 10 U of T4 polynucleotide kinase was added, and primers were incubated for another 30 min at 30°C. The primers were then separated from unincorporated label by purification using a QIAquick nucleotide removal kit (Qiagen) according to the manufacturer’s instructions.
The primers (29) used in the PCRs were 5′CGCTTAACAGCGTCAACAGCGTGCC3′, which anneals to the herpes simplex virus thymidine kinase promoter region, and 5′ACGATTCCGAAGCCCAACCTTTCA3′, which anneals to the 3′ nontranscribed region of the gene encoding aminoglycoside phosphotransferase II. Amplification via PCR using these primers generates products of 742 bp for competitor DNA, 964 bp for reporter DNA, and 1,197 bp for oriP-minus DNA.
(iv) Data analysis.
The data from the quantitative competitive PCR assay when radiolabeled primers were used were analyzed as described previously (29) except that end-labeled primers were used to incorporate 32P into the PCR products. These primers allow the simultaneous amplification of the competitor DNA, the reporter DNA, and, if the DpnI digestions had not gone to completion, the oriP-minus DNA. When the number of copies of competitor DNA is equal to the number of copies of reporter DNA in a given PCR, the two templates are amplified with equal efficiency. Thus, the amount of radioactivity incorporated per template molecule (competitor, reporter, or oriP-minus DNA) is the same. For data analysis, a graph of log(molecules of competitor) versus log(PhosphorImager units of competitor/PhosphorImager units of reporter) was generated, and the number of DpnI-resistant molecules per 105 cell equivalents of sample was determined from the inverse log of the intercept. That is, when the counts from the 32P-end-labeled primers incorporated into the amplified competitor DNA are equivalent to the counts incorporated into reporter DNA in the amplified template, the log of 1/1 = 0. Therefore, the inverse log of the intercept equals the number of DpnI-resistant molecules in the sample per 105 cell equivalents, assuming that all cells took up DNA upon transfection. The data presented were corrected for the actual transfection efficiency of each cell line used in this study as described previously (29). The data from the quantitative competitive PCR assays were analyzed by using the Wilcoxon rank sum test (26).
Sequence analysis.
The nucleotide sequence of the parent of FR-BamHI C-Luc, oriP-BamHI C-Luc, was scanned for EBNA-1 binding sites by using four degenerate EBNA-1 consensus binding sites based on the predicted EBNA-1 binding sites within herpesvirus papio (33a, 34), the 20 EBNA-1 binding sites within the FR, the four EBNA-1 binding sites within the DS, and the two EBNA-1 binding sites within the BamHI Q fragment proximal to the F promoter of the B95-8 genome, using a modified (by Ashok Aiyar) version of Signal Scan (44). These four sites are degenerate 1 (5′ GGRHARYMYDYRCYDYCC 3′), degenerate 2 (5′ GGRHARYMYDYRCYD 3′), degenerate 3 (5′ GRHARYMYDYRCYDYCC 3′), and degenerate 4 (5′ GGRHARYVYDYRYYDYC 3′), where R = A or G, Y = C or T, M = A or C, W = A or T, D = A or G or T, H = A or C or T, and V = A or C or G.
The 1,773-bp fragment between the HpaI and AvrII sites of oriP-BamHI C-Luc and the 1,161-bp region between the ClaI and HpaI sites of oriP-BamHI C-Luc that partially substituted for the DS in FR-BamHI C-Luc were also scanned for potential EBNA-1 dimer binding sites, using all synthetic EBNA-1 half sites identified by Ambinder et al. which bound EBNA-1 with a 10% or greater efficiency relative to a consensus site from the FR in gel shift assays in vitro (3), using the FINDPATTERNS program within the Genetics Computer Group package (14).
DNA blotting.
DNA blotting was performed as previously described (29, 30, 51).
Gel shift assays.
Electrophoretic mobility shift assays were performed as previously described (36, 37).
RESULTS
Transient replication of plasmids containing derivatives of oriP which lack either the DS or the FR.
During our analysis of functional derivatives of EBNA-1 (29), we performed experiments to determine whether wild-type EBNA-1 or its functional derivative, NΔ330-641 (Fig. 1), could support replication of a plasmid that contained a derivative of oriP that had the DS but lacked the FR, DS-BamHI C-Luc. In these experiments, NΔ330-641, like wild-type EBNA-1, did not detectably replicate DS-BamHI C-Luc (Table 1). We also tested whether wild-type EBNA-1 or NΔ330-641 could replicate a plasmid that contained the FR but lacked the DS, FR-BamHI C-Luc. Surprisingly, wild-type EBNA-1 supported the replication of FR-BamHI C-Luc 43% as well as it supported replication of oriP-BamHI C-Luc in 143B cells at 96 h postelectroporation. This result was unexpected based on studies which demonstrated that plasmids containing the FR alone could not support replication in the presence of EBNA-1 (35, 47). In the absence of EBNA-1, neither oriP-BamHI C-Luc nor FR-BamHI C-Luc replicated detectably at 96 h postelectroporation (Table 1). These results indicate that the synthesis of DpnI-resistant DNA was dependent on EBNA-1 and therefore probably resulted from bona fide replication of that DNA, not from repair of it. In these experiments, a mutant of EBNA-1, NΔ330-641, supported the replication of FR-BamHI C-Luc to 13% of the level that it supported replication from wild-type oriP at 96 h postelectroporation in 143B cells. These results indicate that wild-type EBNA-1 and its derivative, NΔ330-641, through their association with the FR of oriP, support replication of a plasmid lacking the site where the initiation of DNA replication normally occurs.
Analysis of the effects of a dominant-negative inhibitor of EBNA-1 on replication of plasmids containing a derivative of oriP.
To characterize further this unexpected EBNA-1-dependent replication of FR-BamHI C-Luc, we tested whether a derivative of EBNA-1 that can efficiently inhibit replication by wild-type EBNA-1 of plasmids which contains oriP could also inhibit the EBNA-1-dependent replication of FR-BamHI C-Luc. To this end, we cotransfected the dominant-negative inhibitor, NΔ450-641 (Fig. 1), or vector DNA, FR-BamHI C-Luc or oriP-BamHI C-Luc DNA and oriP-minus DNA into 143/EBNA-1 cells and measured replication at 96 h postelectroporation by quantitative competitive PCR analysis (Table 2). FR-BamHI C-Luc replicated in cells that constitutively express wild-type EBNA-1 with approximately 24% of the efficiency of oriP-BamHI C-Luc. NΔ450-641 inhibited replication of FR-BamHI C-Luc supported by wild-type EBNA-1 by 95% at 96 h postelectroporation in 143/EBNA-1 cells. These results confirmed that replication of FR-BamHI C-Luc which lacks the dyad symmetry element required EBNA-1 and indicated that the dominant-negative inhibitor, NΔ450-641, likely functions through disrupting a critical role of EBNA-1 that occurs through the FR.
Limited replication of plasmids with a mutant oriP in the presence of wild-type EBNA-1.
To determine whether FR-BamHI C-Luc could replicate and be maintained as a plasmid over a period of several months in the presence of wild-type EBNA-1, we attempted to generate G418-resistant clonal 143/EBNA-1 cell lines that stably maintained FR-BamHI C-Luc as a plasmid. Ten micrograms of FR-BamHI C-Luc DNA was electroporated into 143/EBNA-1 cells, and cells were subjected to selection in G418 as described previously (29). After 2 weeks of selection, G418-resistant colonies of 143/EBNA-1 cells transfected with FR-BamHI C-Luc arose with an efficiency similar to that of those transfected with oriP-BamHI C-Luc (6.3 and 8.3%, respectively [data not shown]). However, only 1 of 56 drug-resistant colonies carrying FR-BamHI C-Luc could be expanded. Eleven of twelve drug-resistant colonies carrying oriP-BamHI C-Luc picked after 2 weeks of selection were expanded successfully (29). Plasmid copies of FR-BamHI C-Luc were not detected by Southern analysis of Hirt extracts of 143/EBNA-1/FR-BamHI C-Luc cells derived from the single G418-resistant colony (data not shown), indicating that integration had likely occurred. These results indicate that although FR-BamHI C-Luc can be replicated in the presence of wild-type EBNA-1 for short periods, it has an inefficient origin of replication and/or cannot be maintained as a plasmid in proliferating host cells over longer times.
Transient drug resistance similar to that of cells containing FR-BamHI C-Luc has been observed with other plasmids that contain the FR and a drug resistance marker (47). Previously, this transient drug resistance had been attributed to an increased transcription of the gene (and, therefore, protein expression) conferring drug resistance either because the FR acts as an enhancer in the presence of EBNA-1 or because the plasmid itself is retained in the cells for a prolonged period in the presence of EBNA-1 (40, 47). While these two possible activities of EBNA-1 and the FR may contribute to prolonged drug resistance, the ability of some plasmids that contain the FR to replicate inefficiently, documented here, could also contribute to transient drug resistance.
Not all plasmids that contain the FR replicate in the presence of EBNA-1.
During the initial studies designed to map the latent origin of replication of EBV (47), we analyzed two plasmids, pΔBal 2 and pΔBal 12, that contain the FR but lack the DS of oriP (Table 3). In these studies, short-term replication of these plasmids was not detected as measured by DNA blotting (47). pΔBal 2 and pΔBal 12 contain EBV sequences including the FR in a pKan2 (58) background. These two plasmids were generated from the same parental plasmid used to generate oriP-BamHI C-Luc and, subsequently, FR-BamHI C-Luc. Therefore, we analyzed pΔBal 2 and pΔBal 12 in short-term replication assays with 143/EBNA-1 cells by quantitative competitive PCR (Table 3). pΔBal 2 and pΔBal 12 replicated less than 1% as efficiently as oriP-BamHI C-Luc and less than 6 and 4% as efficiently, respectively, as FR-BamHI C-Luc at 96 h postelectroporation in 143/EBNA-1 cells. Similarly, FR-Backbone replicated approximately 4% as efficiently as FR-BamHI C-Luc (P = 0.0015) or less than 1% as efficiently as oriP-BamHI C-Luc (Table 3). These studies indicate that the FR in the context of pΔBal 2, pΔBal 12, and FR-Backbone supports short-term replication significantly less well than in the context of FR-BamHI C-Luc. Therefore, these findings indicated that sequences that support replication in the absence of the dyad symmetry element in cis, in the presence of EBNA-1 in trans, were likely to exist in the 4,877 bp of DNA present in FR-BamHI C-Luc and lacking in pΔBal 2, pΔBal 12, or FR-Backbone.
TABLE 3.
cis-acting sequences within a 4,877-bp fragment of FR-BamHI C-Luc substitute for the DS in short-term replication assays
| Reporter | Relative efficiency of replicationa
|
nb | |
|---|---|---|---|
| Reporter | oriP-minus | ||
| oriP-BamHI C-Luc | 100 | <2.0 | 7 |
| FR-BamHI C-Luc | 13 ± 3.1 | <0.50 | 8 |
| oriP-Backbone | 130 ± 115 | <0.50 | 5 |
| FR-Backbone | 0.54 ± 0.058 | <0.50 | 6 |
| pΔBal 2 | 0.81 ± 0.27 | <0.50 | 2 |
| pΔBal 12 | <0.50 | <0.50 | 3 |
| DS-BamHI C-Luc | <0.50 | <0.50 | 5 |
10 μg of each DNA (reporter and oriP-minus DNA) was introduced into 107 143/EBNA-1 cells by electroporation. Low-molecular-weight DNA was isolated after 94 to 98 h, and the relative concentration of DpnI-resistant plasmids was determined by quantitative competitive PCR and was corrected for the transfection efficiency of 143B/EBNA-1 cells as described in Materials and Methods. The relative efficiency of replication is expressed as a percentage relative to oriP-BamHI C-Luc, which is set to 100% (100% = 26 ± 20 copies of DpnI-resistant DNA per transfected cell). Data represents an average ± standard deviation. Data represented as “<” indicate that the level of DpnI-resistant DNA was less than the smallest amount used for the competitor DNA and is therefore reported as less than an average of the smallest amount per transfected cell that could be determined by interpolation from the competitor DNA curve.
n, number of times the reporter plasmid was tested during the mapping analysis.
In these experiments, DS-BamHI C-Luc, which lacks the FR but maintains the DS of oriP (in the context of oriP-BamHI C-Luc), does not detectably replicate (less than 0.5 to 2% of the efficiency of oriP-BamHI C-Luc and less than 4 to 5% of the efficiency of FR-BamHI C-Luc) (Tables 1 and 3) at 96 h postelectroporation. In other words, although the 4,877-bp fragment contributes to replication of an FR-positive, DS-negative plasmid, FR-BamHI C-Luc, it does not contribute to detectable replication of a DS-positive, FR-negative plasmid, DS-BamHI C-Luc. These results indicate that the FR contributes a function(s) that is not provided by the DS in the context of these heterologous sequences.
In addition, in this experiment, oriP-Backbone was replicated as efficiently as oriP-BamHI C-Luc at 96 h postelectroporation (P = 0.81). This finding indicates that those sequences which contribute to the short-term replication of FR-BamHI C-Luc do not detectably affect the overall efficiency of short-term replication of a plasmid containing oriP. In other words, short-term replication from wild-type oriP is not enhanced by the presence of this 4,877-bp fragment. It is not known whether these sequences affect the efficiency of replication during many cell cycles.
Identification of DNA sequences that partially substitute for the DS.
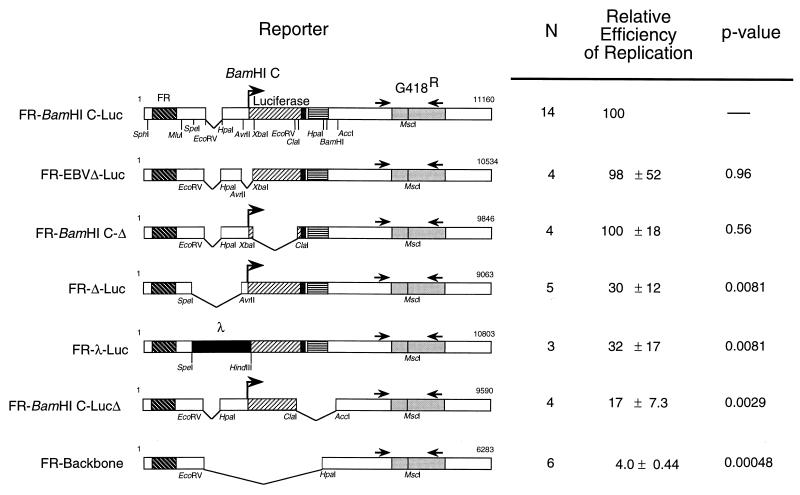
Five derivatives of FR-BamHI C-Luc were used to map the sequences within FR-BamHI C-Luc that contribute to its short-term replication (Fig. 2). Deletions were made within the 4,877 bp of FR-BamHI C-Luc that are not present in FR-Backbone, which served as the negative control in these experiments. Two sets of sequences, those surrounding the BamHI C promoter and those within the luciferase open reading frame, when deleted had no effect on short-term replication (FR-EBVΔ-Luc [P = 0.96] and FR-BamHI C-Δ [P = 0.56], respectively [Fig. 2]). Two sets of sequences, the EBV sequences adjacent to the position of the DS and sequences surrounding and including the SV40 t-antigen intron, when deleted independently led to significant reductions in replication (FR-Δ-Luc [P = 0.0081] and FR-BamHI C-LucΔ [P = 0.0029], respectively [Fig. 2]). In one case, this reduction was shown to result from the absence of the deleted sequences and not the contextual effects of the deletion because substitution of phage lambda sequences for the deleted EBV sequences failed to restore replication (compare FR-Δ-Luc and FR-λ-Luc [P = 0.0081] in Fig. 2). These analyses indicate that only specific sequences can contribute to the short-term replication of plasmids containing the FR in the presence of EBNA-1.
FIG. 2.
Sequences within nt 9132 to 10905 of the EBV genome and within the t-antigen intron and the T-antigen poly(A) addition signal of SV40 contribute to short-term replication of FR-BamHI C-Luc by EBNA-1. The amount of DpnI-resistant DNA of each derivative of oriP-BamHI C-Luc detected at 96 h postelectroporation was compared to that of FR-BamHI C-Luc, using quantitative competitive PCR as described in Materials and Methods. Maps of reporter plasmids tested are shown on the left. N, number of replicates (a subset of the data from Table 3 and Fig. 3 is included). The efficiency of replication of each reporter by EBNA-1 is expressed as a percentage of DpnI-resistant DNA relative to FR-BamHI C-Luc, which is set to 100% (100% = 3.4 ± 1.2 copies of DpnI-resistant DNA per transfected cell). In these experiments, FR-BamHI C-Luc replicates 14% as efficiently as does oriP-BamHI C-Luc. Data were analyzed by using the Wilcoxon rank sum test (26) with Mstat (version 1.3, by Norman Drinkwater) and the P value comparing each derivative to FR-BamHI C-Luc is noted.
Restoration of the function of the DS by addition of specific DNA sequences.
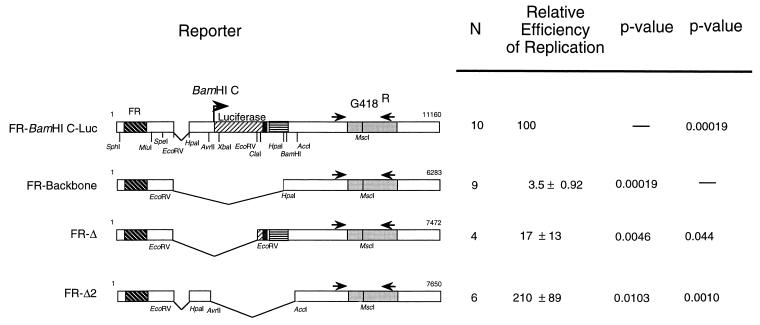
The DNA sequences defined by deletional analyses which contribute to short-term replication of FR-BamHI C-Luc were introduced into the negative control, FR-Backbone, to test their ability to restore replication of this plasmid. Each of the two sequences independently supported replication of the plasmids in the presence of EBNA-1 at 96 h postelectroporation (P = 0.044 and 0.0010 for FR-Δ and FR-Δ2, respectively [Fig. 3]). In fact, FR-Δ2 supported replication more efficiently than did FR-BamHI C-Luc (P = 0.010), indicating that the context of cis-acting elements can affect their ability to facilitate the initiation of DNA replication. These experiments indicate that this assay for short-term replication supported by EBNA-1 can be used to identify DNA sequences that functionally substitute for oriP’s DS, presumably to permit initiation of DNA synthesis.
FIG. 3.
Sequences within nt 9132 to 10905 of the EBV genome and within the SV40 t-antigen intron and the SV40 T-antigen poly(A) addition signal can rescue short-term replication of FR-Backbone by EBNA-1. Maps of reporter plasmids FR-BamHI C-Luc, FR- Backbone, FR-Δ, and FR-Δ2 are shown on the left. N, number of replicates (a subset of the data from Table 3 and Fig. 2 is included). The efficiency of replication of each reporter by EBNA-1 is expressed as a percentage of DpnI-resistant DNA relative to FR-BamHI C-Luc, which is set to 100% (100% = 4.5 ± 0.1.8 copies of DpnI-resistant DNA per transfected cell). In these experiments, FR-BamHI C-Luc replicates 13% as efficiently as does oriP-BamHI C-Luc. Data were analyzed by using the Wilcoxon rank sum test (26) with Mstat (version 1.3, by Norman Drinkwater), and the P value comparing each derivative to FR-BamHI C-Luc or FR-Backbone is noted.
A 298-bp region of the EBV genome, Rep*, supports short-term replication in the absence of the DS.
Five derivatives of FR-Δ2 containing deletions within the 1,773-bp fragment of FR-Δ2 were generated to identify the sequences that contribute to its short-term replication (Fig. 4). Three plasmids, 1858, 1859, and 1860, were defective for short-term replication in the presence of EBNA-1 relative to FR-Δ2 (P = 0.011, 0.013, and 0.011, respectively). Each of these plasmids lacked a 298-bp region of DNA, designated Rep*, between a DraIII and a Bsu36I site. In contrast, two plasmids which contained Rep*, 1861 and 1862, replicated with an efficiency similar to that of FR-Δ2 (P = 0.62, and 0.90, respectively). These results indicate that sequences between nucleotides (nt) 9370 and 9668 of the EBV genome plus the FR contribute to replication of a plasmid that lacks the DS in the presence of EBNA-1.
Multiple copies of Rep* plus the FR enhance replication in the presence of EBNA-1.
Plasmids containing Rep* support short-term replication less efficiently than do plasmids containing the DS, an efficient site of initiation of DNA replication. We hypothesized that placing multiple low-efficiency sequences on a single plasmid would increase the probability of that plasmid being replicated during a given cell cycle. To test this model, we generated two plasmids, 1925 and 1926, which contain three copies of Rep*, with two copies in either the opposite or the same orientation, respectively, as the first copy (see Materials and Methods). We compared the levels of replicated 1925 and 1926 DNA to that of 1862, which contains a single copy of Rep*, at 96 h postelectroporation (Table 4). Both plasmids containing three copies of Rep* were replicated more efficiently than was a plasmid that contained a single copy of Rep* (P = 0.021). These results indicate that multiple copies of a sequence that inefficiently supports replication can increase the efficiency of replication in a given cell cycle.
TABLE 4.
Multiple copies of Rep* enhance short-term replication of plasmids in 143/EBNA-1 cells
| Reporter | Copies/cella | P valueb |
|---|---|---|
| oriP-Backbone | 12 ± 7.3 | 0.021 |
| FR-Backbone | <0.2c | 0.014 |
| 1862 | 1.4 ± 0.37 | |
| 1925 | 8.3 ± 6.9 | 0.021 |
| 1926 | 4.2 ± 1.1 | 0.021 |
10 μg each of reporter DNA and oriP-minus was introduced into 107 143/EBNA-1 cells by electroporation. Low-molecular-weight DNA was isolated after 94 to 98 h, and the relative concentration of DpnI-resistant plasmids was determined by quantitative competitive PCR and was corrected for transfection efficiency of 143/EBNA-1 cells as described in Materials and Methods. Data represent an average of four experiments ± standard deviation.
Data were analyzed by using the Wilcoxon rank sum test (26) with Mstat (version 1.3, by Norman Drinkwater), and the P value comparing each derivative to 1862 is noted.
The level of DpnI-resistant DNA was less than the lowest concentration used for the competitor DNA and is therefore reported as less than an average of the smallest amount per transfected cell that could be determined by interpolation from the competitor DNA curve.
Based on the promising results of the short-term replication assay, we asked whether multiple copies of Rep* plus the FR could now support long-term replication in the presence of EBNA-1 in two separate experiments. (As described above, FR-BamHI C-Luc, which contained only one copy of Rep*, was defective for long-term replication.) In the first experiment (Table 5), oriP-Backbone, FR-Backbone, 1925, or 1926 plus oriP-minus were introduced into cells by electroporation. The cells were then grown in the absence of selection, and low-molecular-weight DNA was isolated 8, 12, 16, and 20 days after introduction of the reporter DNA into cells. In the absence of selection, oriP-Backbone as well as 1925 and 1926 was lost from cells over time. This loss is reflected in a decrease in the average number of copies of reporter DNA per transfected cell during the course of the experiment (Table 5). However, the absolute rate of loss of plasmids which contained three copies of Rep* was greater than that of plasmids which contained the DS (Table 5). 1925 and 1926 were lost from cells 1.9 and 1.8 times as rapidly, respectively, as was oriP-Backbone. In spite of the increased rate of loss, replicated 1925 and 1926 DNA could still be detected 20 days postelectroporation. In this experiment, although the copies of Rep* are oriented differently in 1925 and 1926, the two plasmids behave similarly. In fact, during the time course of this experiment, the absolute amount of replicated 1925 and 1926 would have increased more than 2,500-fold, were all progeny cells to have been saved and grown. In contrast, FR-Backbone was not detected over the course of the assay (<0.20 copy per transfected cell at each time point [data not shown]) and was defective for replication as early as 96 h in other experiments (Tables 3 and 4; Fig. 2 to 4). In a second experiment to test long-term replication, we generated G418-resistant, clonal 143/EBNA-1 cell lines that stably maintained either oriP-Backbone, FR-Backbone, 1925, or 1926. Ten micrograms of each reporter DNA was electroporated into 143/EBNA-1 cells, and cells were subjected to selection in G418 as described previously (29). After 2 weeks of selection, G418-resistant colonies of 143/EBNA-1 cells transfected with 1925, 1926, and oriP-Backbone arose 17 to 97 times more efficiently than did G418-resistant colonies of 143/EBNA-1 cells transfected with FR-Backbone. Cell lines containing 1925 or 1926 were difficult to grow and took longer to expand than did cell lines containing oriP-Backbone or FR-Backbone. Once cell lines containing each reporter DNA were generated, low-molecular-weight DNA was isolated from a subset of the lines and the number of copies of plasmid DNA per cell was measured by quantitative competitive PCR. The low-molecular-weight DNA did not contain detectable levels of FR-Backbone (<0.05 copy per cell) in 143/EBNA-1/FR-Backbone cell lines (n = 2), which is consistent with integration of the plasmid. Approximately one to nine copies of oriP-Backbone per cell were detected in 143/EBNA-1/oriP-Backbone cell lines (n = 6), and approximately one to five copies of 1925 or 1926 per cell were detected in 143/EBNA-1/1925 or 143/EBNA-1/1926 cell lines (n = 1 and 4, respectively). The low-molecular-weight DNAs from these cell lines were also analyzed on DNA blots probed with FR-Backbone to determine whether the reporter plasmids had rearranged. Cell lines containing oriP-Backbone, 1925, or 1926 each contained plasmid DNA of unit length when digested with a restriction enzyme which recognized a unique site on that plasmid (data not shown). FR-Backbone was not detected in the low-molecular-weight DNA isolated from the 143/EBNA-1/FR-Backbone cell lines, consistent with integration of that plasmid (data not shown). Together, these results indicate that multiple copies of Rep* plus the FR can support replication for 25 or more generations in the presence of EBNA-1.
TABLE 5.
1925 and 1926 are lost more rapidly from 143/EBNA-1 cells than is oriP-Backbone over a 12-day time course
| Days postelectroporation | % Relative to oriP-Backbone on day 8a
|
||
|---|---|---|---|
| oriP-Backbone | 1925 | 1926 | |
| 8 | 100 ± 31 | 100 ± 59 | 100 ± 44 |
| 12 | 63 ± 40 | 55 ± 40 | 40 ± 26 |
| 16 | 62 ± 28 | 18 ± 7.7 | 17 ± 8.0 |
| 20 | 48 ± 17 | 11 ± 1.9 | 14 ± 3.6 |
10 μg of each DNA (oriP-Backbone, 1925, or 1926) and oriP-minus were introduced into 107 143/EBNA-1 cells by electroporation. Low-molecular-weight DNA was isolated at 8, 12, 16, and 20 days postelectroporation, and the number of DpnI-resistant plasmids per transfected cell was determined by quantitative competitive PCR and was corrected for the transfection efficiency of 143/EBNA-1 cells as described in Materials and Methods. The efficiency of replication of each reporter is expressed as a percentage of the amount of DpnI-resistant DNA per transfected cell relative to that present at day 8 for each reporter construct. For oriP-Backbone, 1925, and 1926, 100% = 12, 2.2, and 2.5 copies of DpnI-resistant DNA per transfected cell, respectively. The level of DpnI-resistant oriP-minus DNA was less than the smallest amount used for the competitor DNA for all samples (less than 0.20 copies per transfected cell). Data represent an average of three experiments for each time point ± standard deviation.
Analysis of the two regions of FR-BamHI C-Luc that positively contribute to short-term replication for EBNA-1 binding sites.
To determine whether any potential EBNA-1 binding sites were present in the sequences deleted from FR-Δ-Luc and FR-BamHI C-LucΔ, sequence analysis was performed. The 1,161- and 1,773-bp fragments which contribute positively to replication of FR-BamHI C-Luc were scanned for EBNA-1 binding sites by using all synthetic half sites identified by Ambinder et al. which were efficiently bound by EBNA-1 in vitro (3), using FINDPATTERNS within the Genetics Computer Group package (14). Also, oriP-BamHI C-Luc was scanned for EBNA-1 consensus binding sites with a modified version of Signal Scan (44). Sequence analyses did not identify possible EBNA-1 binding sites in either fragment. Together, these sequence analyses indicate that the cis-acting sequences present in the 1,161- and 1,773-bp fragments that positively contribute to replication of FR-BamHI C-Luc are unlikely to be bound by EBNA-1 directly. Sequence analysis did identify potential stem-loop structures within Rep*; whether these potential secondary structures contribute to the function of Rep* is not known.
To determine whether any EBNA-1 binding sites not recognized by sequence analysis were present within Rep* (which lies within the 1,773-bp fragment described above), electrophoretic mobility shift assays were performed as previously described (36, 37). The ability of 6.25, 25, 100, or 400 fmol of a derivative of EBNA-1 containing the DNA binding and dimerization domain, NΔ399 (36), to bind to approximately 20 fmol of a probe consisting of the 298-bp Rep*, a 322-bp probe that lacked EBNA-1 binding sites, a 353-bp probe with one EBNA-1 binding site, or a 400-bp probe with two EBNA-1 binding sites was tested. NΔ399 did not shift either the zero-binding-site probe or Rep*, whereas NΔ399 bound both the one- and two-EBNA-1-binding-site probes (in the presence of 100 fmol of NΔ399, 0.39, 0.10, 13, and 23% of the probe shifted, respectively [data not shown]). Together, these results indicate that the identified sequences which support replication in the absence of the DS are not bound by EBNA-1.
DISCUSSION
The replication of oriP plasmids supported by EBNA-1 requires only one viral cis-acting sequence and one viral trans-acting factor but is still complex. oriP has two required elements (35, 47). The DS of oriP is at or near a bidirectional origin of DNA synthesis (17), while the FR of oriP appears to be required for the maintenance of the plasmid (32) such that it can be replicated in sequential S phases. EBNA-1 binds both of these elements (45) and is required for detection of replication of oriP-containing plasmids at 96 h postelectroporation (29, 47, 59) (Tables 1 to 3). Although EBNA-1 is thus an origin binding protein, it does not have intrinsic DNA-dependent ATPase or helicase activities (16, 39) found in other origin binding, viral replication proteins such as the T antigen of SV40 (52), E1 of bovine papillomavirus (33), or UL9 of herpes simplex virus (10, 11). One possible contribution of EBNA-1 to the replication of oriP replicons is to retain the replicated DNA in cells (40) and, we hypothesize, to mediate its segregation to daughter cells. We have used this feature of EBNA-1 to develop a sensitive assay to define DNA sequences that support replication independent of the DS, albeit less efficiently. These plasmids contain DNA sequences which permit them to replicate less efficiently than plasmids that contain the wild-type DS and are still dependent on EBNA-1 for their replication (Tables 1 to 4). Two sequences which contribute to the plasmid’s replication have been mapped within the DS-minus plasmid FR-BamHI C-Luc (Fig. 2 and 3). One of these sequences was further characterized and found to contain a 298-bp fragment, Rep*, which supports replication in the absence of the DS (Fig. 4).
It is important to note that the plasmid used by Gahn and Schildkraut to map the site of initiation of DNA replication within oriP to at or near the DS also contained approximately half of the sequences present within Rep* (17). Their mapping studies would not have distinguished efficient initiation events at the DS from those possibly occurring infrequently within that portion of Rep*. Thus, all models considered for the role of Rep* in replication may be applicable to replication of EBV.
The mechanism by which Rep* substitutes for the DS is not yet clear. Rep* does not contain EBNA-1 binding sites or the nonamer located within the DS that is protected in a cell cycle-dependent manner as measured by in vivo footprinting (41). However, this region is likely to contain binding sites recognized by cellular factors involved in chromosomal DNA synthesis. Because these sequences lack binding sites for EBNA-1, we propose that EBNA-1 supports the inefficient replication of the mutant oriPs that contain them, not by affecting initiation of DNA synthesis but by affecting plasmid maintenance in proliferating cells over the 96 h of the assay. In this model, the decreased efficiency of replication associated with Rep* results from its supporting initiation of DNA synthesis less efficiently than does the DS of oriP. At 96 h postelectroporation, plasmid 1862, with one copy of Rep*, has approximately 9.3% of the replicated molecules that oriP-Backbone, with one copy of the DS, does (Fig. 4). If three cell doublings occurred during these 96 h, Rep* would have supported each round of DNA synthesis about 45% as efficiently as did the DS, assuming that these plasmids are maintained comparably. A plasmid with three copies of Rep* such as 1925 or 1926 should support initiation of DNA synthesis at approximately 83% of the level of the dyad if the contribution of one copy of Rep* excludes the other copies from contributing during a given S phase as has been found for the DS (30, 43, 56, 57). This model predicts that over 12 generations about 89% (0.8312 = 0.11) of 1925 and 1926 will be lost as a result of failure to initiate DNA synthesis. The measurements in Table 5 are consistent with this prediction and therefore support the model that EBNA-1 affects primarily or, perhaps, exclusively the maintenance of oriP plasmids.
Approximately 94 to 98% of EBNA-1-positive cells containing wild-type oriP plasmids will retain that DNA after each cell cycle in the absence of selection (30, 47, 56, 59). Thus, for those cells maintaining a single copy of an oriP-containing plasmid, the DS will minimally support the initiation of DNA synthesis during 94 to 98% of S phases. Our identification of multiple independent sequences which inefficiently support short-term replication as well as the demonstration that multiple copies of Rep* can partially substitute for the DS supports a model for stochastic initiation events during DNA synthesis in mammals. While initiation of DNA replication from the DS occurs efficiently each S phase, we propose that the sequences identified in this study support initiation of DNA synthesis on the plasmid with a reduced probability relative to that of the DS. This reduced efficiency of firing during any one S phase leads to an overall reduction in the level of replicated DNA detected relative to that of wild-type oriP-containing plasmids after the three to four S phases which occur during 96 h of the assay. This model is consistent with the multiple sites for initiation of DNA synthesis observed in the studied mammalian origins of DNA replication acting combinatorially to produce an efficient origin of replication (13). Initiation of DNA replication at each site is inefficient, but collectively the sites mediate efficient initiation and thus define an origin. The proximity of Rep* relative to the DS is consistent with the initiation of latent replication of EBV being stochastic and dictated by a number of elements, each of which is recognized with less than 100% efficiency during any given cell cycle. Our model also underscores the remarkable compactness and efficiency of the 140-bp DS of oriP.
ACKNOWLEDGMENTS
We thank Gretchen Anderson, Toni Gahn, Todd Hopkins, Mary Kay Koeller, and Rebecca Wisniewski for help making reagents used in this study. We also thank Ashok Aiyar for assistance with sequence analysis and Dave Mackey and Paul Lambert for suggestions to improve the manuscript.
This research was supported by Public Health Service grants CA-22443, CA-07175, and T32-CA-09135.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M, Chang Y-N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacchetti S, Graham F L. Transfer of the gene for thymidine kinase to thymidine kinase-deficient human cells by purified herpes simplex viral DNA. Proc Natl Acad Sci USA. 1977;74:1590–1594. doi: 10.1073/pnas.74.4.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Séguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev A, Barwell J A, Pfuetzner R A, Bochkareva E, Frappier L, Edwards A M. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 7.Carroll S M, Trotter J, Wahl G M. Replication timing control can be maintained in extrachromosomally amplified genes. Mol Cell Biol. 1991;11:4779–4785. doi: 10.1128/mcb.11.9.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chittenden T, Lupton S, Levine A J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989;63:3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colbère-Garapin F, Horodniceanu F, Kourilsky P, Garapin A-C. A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol. 1981;150:1–14. doi: 10.1016/0022-2836(81)90321-1. [DOI] [PubMed] [Google Scholar]
- 10.Crute J J, Mocarski E S, Lehman I R. A DNA helicase induced by herpes simplex virus type 1. Nucleic Acids Res. 1988;16:6585–6596. doi: 10.1093/nar/16.14.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crute J J, Tsurumi T, Zhu L A, Weller S K, Olivo P D, Challberg M D, Mocarski E S, Lehman I R. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delecluse H-J, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm G W. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J Virol. 1993;67:1292–1299. doi: 10.1128/jvi.67.3.1292-1299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePamphilis M L. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frappier L, O’Donnell M. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J Virol. 1992;66:1786–1790. doi: 10.1128/jvi.66.3.1786-1790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frappier L, O’Donnell M. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991;266:7819–7826. [PubMed] [Google Scholar]
- 17.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith K, Bendell L, Frappier L. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1993;67:3418–3426. doi: 10.1128/jvi.67.6.3418-3426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 20.Grogan E A, Summers W P, Dowling S, Shedd D, Gradoville L, Miller G. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampar B, Tanaka A, Nonoyama M, Derge J G. Replication of the resident repressed Epstein-Barr virus genome during the early S phase (S-1 period) of nonproducer Raji cells. Proc Natl Acad Sci USA. 1974;71:631–633. doi: 10.1073/pnas.71.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris A, Young B D, Griffin B E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt’s lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hearing J, Mulhaupt Y, Harper S. Interaction of Epstein-Barr virus nuclear antigen 1 with the viral latent origin of replication. J Virol. 1992;66:694–705. doi: 10.1128/jvi.66.2.694-705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 26.Hollander M, Wolfe D A. Nonparametric statistical methods. In: Bradley R A, Hunter J S, Kendall D G, Watson G S, editors. Wiley series in probability and mathematical statistics. New York, N.Y: John Wiley & Sons, Inc.; 1973. pp. 67–82. [Google Scholar]
- 27.Hsieh D-J, Camiolo S M, Yates J L. Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J. 1993;12:4933–4944. doi: 10.1002/j.1460-2075.1993.tb06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue N, Harada S, Honma T, Kitamura T, Yanagi K. The domain of Epstein-Barr virus nuclear antigen 1 essential for binding to oriP region has a sequence fitted for the hypothetical basic-helix-loop-helix structure. Virology. 1991;182:84–93. doi: 10.1016/0042-6822(91)90651-q. [DOI] [PubMed] [Google Scholar]
- 29.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirchmaier A L, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krysan P J, Calos M P. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol Cell Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lentz M R, Pak D, Mohr I, Botchan M. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation sites. J Virol. 1993;67:1414–1423. doi: 10.1128/jvi.67.3.1414-1423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Loeb, D. Personal communication.
- 34.Loeb D D, Sung N S, Pesano R L, Sexton C J, Hutchinson III C, Pagano J S. Plasmid origin of replication of herpesvirus papio: DNA sequence and enhancer function. J Virol. 1990;64:2876–2883. doi: 10.1128/jvi.64.6.2876-2883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackey D, Middleton T, Sugden B. Multiple regions within EBNA1 can link DNAs. J Virol. 1995;69:6199–6208. doi: 10.1128/jvi.69.10.6199-6208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackey D, Sugden B. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J Biol Chem. 1997;272:29873–29879. doi: 10.1074/jbc.272.47.29873. [DOI] [PubMed] [Google Scholar]
- 38.Middleton T, Sugden B. A chimera of EBNA1 and the estrogen receptor activates transcription but not replication. J Virol. 1992;66:1795–1798. doi: 10.1128/jvi.66.3.1795-1798.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niller H H, Glaser G, Knüchel R, Wolf H. Nucleoprotein complexes and DNA 5′-ends at oriP of Epstein-Barr virus. J Biol Chem. 1995;270:12864–12868. doi: 10.1074/jbc.270.21.12864. [DOI] [PubMed] [Google Scholar]
- 42.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 43.Platt T H K, Tcherepanova I Y, Schildkraut C L. Effect of number and position of EBNA-1 binding sites in Epstein-Barr virus oriP on the sites of initiation, barrier formation, and termination of replication. J Virol. 1993;67:1739–1745. doi: 10.1128/jvi.67.3.1739-1745.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prestridge D S. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS. 1991;7:203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- 45.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 46.Reedman B, Klein G. Cellular localization of an Epstein-Barr (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int J Cancer. 1973;11:499–520. doi: 10.1002/ijc.2910110302. [DOI] [PubMed] [Google Scholar]
- 47.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schleiss M R, Degnin C R, Geballe A P. Translational control of human cytomegalovirus gp48 expression. J Virol. 1991;65:6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah W A, Ambinder R F, Hayward G S, Hayward S D. Binding of EBNA-1 to DNA creates a protease-resistant domain that encompasses the DNA recognition and dimerization functions. J Virol. 1992;66:3355–3362. doi: 10.1128/jvi.66.6.3355-3362.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 52.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugden B. Latent infection of B lymphocytes by Epstein-Barr virus. Semin Virol. 1994;5:197–205. [Google Scholar]
- 54.Sugden B, Mark W. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J Virol. 1977;23:503–508. doi: 10.1128/jvi.23.3.503-508.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphocytes transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugden B, Warren N. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol Biol Med. 1988;5:85–94. [PubMed] [Google Scholar]
- 57.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer activation functions of Epstein-Barr Virus nuclear antigen 1. In: Kelly T, Stillman B, editors. Eukaryotic DNA replication. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 197–205. [Google Scholar]
- 60.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]