Abstract
The ability of vertebrates to ‘remember’ previous infections had once been attributed exclusively to adaptive immunity. We now appreciate that innate lymphocytes also possess memory properties akin to those of adaptive immune cells. In this Review, we draw parallels from T cell biology to explore the key features of immune memory in innate lymphocytes, including quantity, quality, and location. We discuss the signals that trigger clonal or clonal-like expansion in innate lymphocytes, and highlight recent studies that shed light on the complex cellular and molecular crosstalk between metabolism, epigenetics, and transcription responsible for differentiating innate lymphocyte responses towards a memory fate. Additionally, we explore emerging evidence that activated innate lymphocytes relocate and establish themselves in specific peripheral tissues during infection, which may facilitate an accelerated response program akin to those of tissue-resident memory T cells.
The concept of ‘immunological memory’ predates our understanding of the vertebrate immune system. Long before Edward Jenner reported that milkmaids previously exposed to cowpox were protected against smallpox in 17981, the practice of inoculation to ‘immunize’ against an infection can be traced as far back as tenth-century China2. Since then, owing to the many discoveries that span several hundred years, we now appreciate the intricate molecular and cellular events constituting adaptive immunity that are critical for the generation of immunological memory. Nevertheless, in the past decade, the idea of immune memory being an exclusive trait of the adaptive immune system has been challenged by various studies demonstrating the existence of non-B-cell- and non-T-cell-mediated immune memory, thus blurring the line between innate and adaptive immunity.
Regardless of their innate or adaptive origins, immune memory cells must possess several cardinal features that explain their enhanced ability to provide protection compared to naive cells. First, the numerical abundance that results from clonal expansion provides quantitative strength to ensure that memory cells have a higher probability of encountering foreign invaders. Second, molecular adaptations through metabolic, epigenetic, and transcriptional reprogramming should ‘poise’ memory cells for a more rapid or robust effector response upon re-encountering a pathogen. And finally, tissue redistribution of memory cells positions them for an accelerated response against pathogens that breach the barrier surface. Together, these three features of—quantity, quality, and location—are interdependent and exquisitely shaped by early and highly coordinated priming events involving various cellular and molecular signals during the primary encounter with a pathogen.
In this Review, we discuss this newfound appreciation of an innate cell’s ability to ‘remember’ previous insults that has reshaped our understanding of immunological memory, with a special focus on innate lymphocytes (natural killer (NK) cells and innate lymphoid cells (ILCs)). We will expand upon the three cardinal features of immune memory and highlight parallels between innate immune cells and their adaptive T cell siblings at the population level. We will provide an overview of the shared and distinct adaptive features that distinguish innate from adaptive memory, as well as the downstream molecular consequences of these processes that enhance their functionality. Last, we will discuss the emerging evidence that a commonality exists between tissue-resident innate memory cells and tissue-resident memory T cells. A broader understanding of how these responses are orchestrated will provide an opportunity to unleash the full potential of cellular and molecular components of immune memory.
Advantage of numerical abundance
Numerical abundance is one of the key factors in the memory cell’s superior ability to respond to an infection compared with the naive cell. Naive T cells of a given epitope are incredibly sparse and must actively search for their cognate antigen before being primed in secondary lymphoid organs (SLOs) of the host by dendritic cells (DCs)—a mode of surveillance that is relatively slow and inefficient. By contrast, as a result of clonal expansion, memory T cells are much more abundant than naive T cells. When equipped with a greater number of epitope-specific memory T cells, the host enjoys a distinct advantage when re-encountering a pathogen, resulting in a much quicker, stronger, and focused surveillance.
As with T cells, certain innate lymphocytes are capable of clonal expansion, with the earliest and best-studied evidence originating from NK cell responses to mouse cytomegalovirus (MCMV)3-5. In this setting, engagement of the activating receptor Ly49H (encoded by Klra8 and expressed by a subset of NK cells in C57BL/6 mice) with MCMV-derived glycoprotein m157 on infected cells6,7 triggers selective clonal expansion of Ly49H+ NK cells that has been measured to upwards of 10,000-fold compared to NK cells that failed to encounter MCMV-derived ligand3,8 (Fig. 1). Moreover, akin to the subsequent contraction phase observed in their adaptive T and B cell siblings, many of these clonally expanded ‘effector’ Ly49H+ NK cells undergo apoptosis following the acute phase of infection, leaving a pool of long-lived memory cells that can readily respond to reinfection3. Since these early studies, antigen-specific NK cell expansion has also been observed in the response of Rhesus macaques (Macaca mulatta) to simian immunodeficiency virus (SIV)9, and in human responses to CMV (HCMV)10-12 and malaria (Plasmodium falciparum)13,14. During HCMV infection, the expansion of human NKG2C+ NK cells is thought to be analogous to MCMV-specific Ly49H+ NK cells in mice10. These ‘adaptive’ NKG2C+ NK cells are clonal15 and require HCMV UL-40-derived peptide, presented by non-classical HLA-E on HCMV-infected cells, to expand16-19. However, in the context of SIV and malaria, specific ligands and antigens or receptors involved in clonal expansion of NK cells have yet been clearly identified. The antigen-specific responses and clonal expansion described above are in stark contrast to the innate memory recently described in myeloid cells and non-immune cells, in which numerical advantage is not a factor in recall responses20,21.
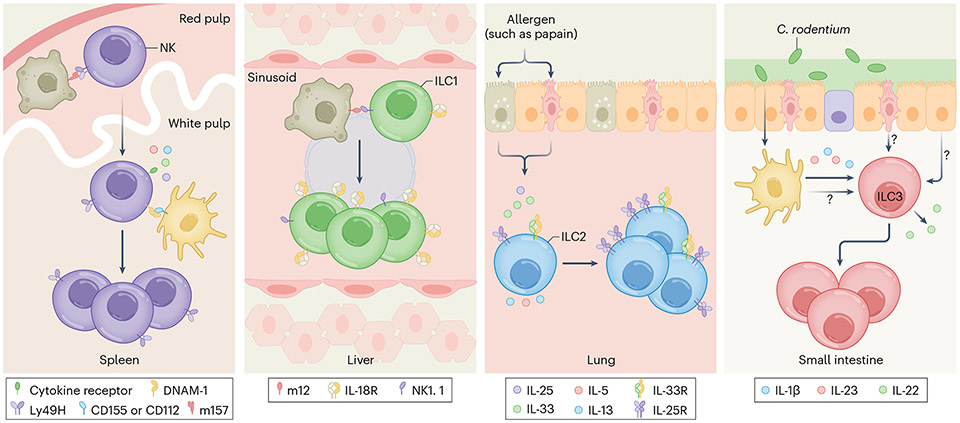
Fig. 1 ∣. Advantage of numerical abundance.
Clonal or clonal-like expansion equips memory cells with a quantitative advantage to quickly respond to secondary infections, in part by increasing the probability of encountering pathogens. For naive splenic Ly49H+ NK cells, signals from engagement of the activating receptor by MCMV-infected cells (gray cells) and co-stimulatory molecules as well as proinflammatory cytokines from myeloid cells (yellow cells) are necessary for clonal expansion (first panel). In the liver, interaction between activating receptor NK1.1 and the MCMV-derived viral ligand m12, expressed by infected cells, initiates clonal-like expansion of ILC1s (second panel). Meanwhile, ILC2s and ILC3s undergo clonal-like proliferation in an antigen-independent manner. Lung ILC2s require the cytokine IL-33, which is released by epithelial cells upon papain challenge (third panel), whereas the expansion of small intestinal ILC3s by C. rodentium are induced by IL-1β, IL-23, and other unidentified signals (fourth panel).
The interaction of Ly49H and m157 itself is required but alone is not sufficient to induce optimal clonal expansion of NK cells. Additional signals from other activating receptors are necessary for the expansion of these virus-specific NK cells, similar to the co-stimulatory signals required for robust expansion of virus-specific T cells22. In mice, interaction of DNAM-1 (encoded by Cd226) on NK cells with its ligands, CD155 and CD112 (expressed on splenic DCs and macrophages during infection), is needed for the optimal clonal expansion program of virus-specific Ly49H+ NK cells (Fig. 1), through downstream signaling kinases Fyn and PKCη (ref. 23). Other receptors, such as NKG2D24 and Ly49D25 in mice, and CD226 in humans, may also support this clonal proliferation program.
In addition to these contact-dependent receptor signals, viral infection triggers a burst of proinflammatory cytokines that are instrumental for the adaptive program of virus-specific NK cells (Fig. 1). The induction of type I interferon (IFN) by plasmacytoid DCs27-29, and likely by stromal30 and myeloid cells31, in response to MCMV is necessary to protect virus-specific NK cells from being killed by neighboring NK cells (a process called fratricide) through the STAT1–STAT2–IRF9 axis, but does not affect the clonal proliferation of Ly49H+ NK cells32. At the same time, interleukin-12 (IL-12), produced by plasmacytoid DCs, classical DCs (cDC1s and cDC2s)27,33, and monocytes34, drives the innate effector program (for example, interferon-γ (IFNγ) secretion) and initiates many aspects of the adaptive features of NK cells by promoting cellular differentiation and proliferation in a STAT4-dependent manner35. Meanwhile, the production of IL-18 by cDC1s36 amplifies IL-12 signaling37, which in turn boosts IFNγ production and induces the expression of CD25 (the high-affinity chain of the IL-2 receptor) to mediate IL-2–STAT5-driven expansion of virus-specific NK cells38. Thus, in many ways, the signals that are required to initiate the clonal expansion program in NK cells parallel those of T cells39. Although we are beginning to understand the contribution and integration of these individual signals40, it remains unclear how they are coordinated in vivo in the context of inflamed or infected tissues.
Along these lines, exactly when and where do antiviral NK cells encounter these signals? Much like naive T cells that are retained in the SLO after antigen engagement, naive NK cells similarly abandon their nomadic lifestyle upon encountering an antigen during viral infection. Early after MCMV infection, Ly49H+ NK cells likely encounter virally infected cells in the red pulp and migrate into the white pulp through the marginal zone41-45, where they then interact with cDC1s36, presumably through an XCL1–XCR1-axis46. Whether these interactions provide some or all of the critical signals that initiate the adaptive program of virus-specific NK cells remains unclear. Interestingly, within the white pulp, neuro–immune crosstalk between Ly49H+ NK cells and adrenergic neurons has recently been suggested to support the optimal expansion of virus-specific NK cells47. Thus, as with T cells, NK cells may need to be primed in SLOs during infection, although the contribution of splenic-specific priming events towards the overall systemic expansion of NK cells and the generation of memory remains to be determined.
This phenomenon poses the question of whether the priming of the adaptive NK cell program is exclusive to SLO sites such as the spleen, or whether priming can occur and/or be initiated in peripheral organs. Indeed, recent studies have suggested that naive T cells can be primed outside of the SLOs, namely in the liver48,49 and the vaginal mucosa50. Moreover, there are examples in which priming of other ILCs in peripheral tissues resulted in clonal or clonal-like expansion (Fig. 1). For instance, liver ILC1s can expand and form memory in an antigen-dependent manner during MCMV infection51. Because ILC1s do not express Ly49H-like NK cells, this process is mediated by the interaction of the activating receptor NK1.1 on liver ILC1s and MCMV-derived protein m12 (refs. 51,52). Notably, both NK cells and ILC1s express NK1.1, and thus it remains unclear to what extent the NK1.1–m12 interaction contributes towards the adaptive features of ILC1s versus NK cells. Other members of the ILC family, namely ILC2s in the lung and ILC3s in the small intestine, have also been recently shown to possess these adaptive features (Fig. 1). Distinct from the antigen-dependency of their group 1 innate lymphoid siblings, ILC2 expansion in the lung is primarily driven by the cytokine IL-33, which is induced by the allergen papain, or Aspergillus53,54. Furthermore, ILC3s can also mount a secondary response following Citrobacter rodentium challenge55. Interestingly, the expansion of ILC3s is not pathogen-specific and requires signals beyond IL-1β and IL-23 as treatment with these cytokines alone in the absence of infection failed to induce memory responses. Thus, the additional signals required to induce ILC3 memory remain an open question. Collectively, these findings suggest that clonal or clonal-like expansion is a conserved trait of all ILCs that provides them with a numerical advantage for heightened protection against secondary infection. Whether in certain settings this expansion may actually represent a maladaptation that drives unwanted inflammation at tissue sites and causes diseases (for example, asthma, inflammatory bowel diseases, or psoriasis) remains to be better understood54,56,57.
Triad of molecular adaptations
Clonal expansion not only serves as a way to produce many memory cells, but also acts as a vehicle for molecular adaptation that endows a select number of long-lived cells to participate in memory responses with heightened functionality, in part owing to the selection of high-affinity and high-avidity cells58,59. Notably, the process of clonal expansion itself is not a prerequisite for molecular adaptation. At the heart of this qualitative difference between naive versus memory cell lies the complex interplay between metabolism, epigenetics, and transcription (Fig. 2). Metabolic reprogramming of antigen-activated lymphocytes provides necessary fuel sources for taxing cellular processes, including epigenetic modifications and transcription, that consequently can modulate additional metabolic pathways by regulating the epigenetic landscape and the expression of metabolic and other important genes, representing both feedback and feed-forward mechanisms.
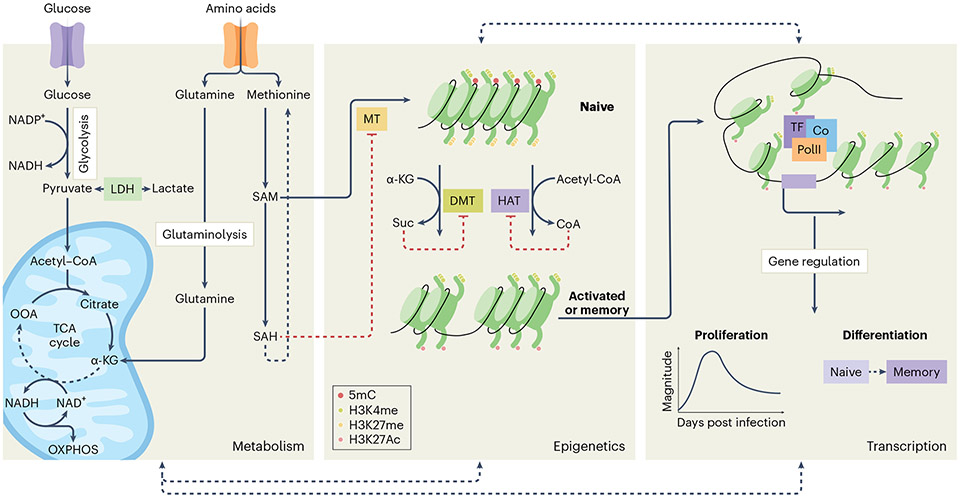
Fig. 2 ∣. Triad of molecular adaptations.
The complex interplay between metabolism, epigenetics, and transcription primes memory cells for qualitative advantages over naive cells. Upon activation, a metabolic shift provides lymphocytes with the necessary fuels for epigenetic modifications that facilitate transcription of genes required for proliferation, differentiation, and effector function. For instance, acetyl-CoA is a substrate for histone acetylation, facilitated by histone acetyltransferases (HAT), whereas SAM is a universal methyl-donor for histone and/or DNA methyltransferases (MT). S-adenosylhomocysteine (SAH), a SAM byproduct, negatively regulates the activity of MTs, whereas α-KG promotes demethylation by histone and/or DNA demethylases (DMT). Altogether, this intimate crosstalk lowers the cell-intrinsic molecular activation threshold in memory cells, in part through phenotypic changes. As a result, memory cells are better equipped to respond more rapidly and robustly to secondary infection than naive cells. LDH, lactate dehydrogenase; OOA, oxaloacetate; suc, succinate; TF, transcription factor.
During activation and clonal expansion, both NK cells and T cells experience substantial metabolic reprogramming to sustain the substrate demand for proliferation as well as differentiation60. For instance, although naive CD8+ T cells primarily rely on oxidative phosphorylation (OXPHOS) for their homeostasis, they shift towards glycolysis and amino acid metabolism (Fig. 2), including glutaminolysis, during activation and differentiation into effector cells63-65, similar to activated NK cells66,67. Deletion of a component of lactate dehydrogenase, the rate-limiting enzyme that mediates the conversion of pyruvate into lactate, is detrimental to both NK cell and CD8+ T cell expansion66, possibly in part owing to the direct regulation of the cell cycle by lactate itself68. However, distinct signals appear to drive the metabolic switch in these cell types, specifically the triggering of the T cell receptor (TCR) and the co-stimulatory molecule CD28 in CD8+ T cells, and the proinflammatory cytokines sensed by NK cells61,66,69. The transcription factor MYC plays a critical role in this metabolic shift by controlling the expression of metabolic enzymes and nutrient transporters, such as GLUT1 and amino acid transporters, to satisfy nutrient demands for cell division and protein synthesis70. This metabolic shift in turn regulates the levels of MYC itself through mTOR signaling and the sensing of endoplasmic reticulum stress67,69,71. Moreover, whereas glutaminolysis acts as an alternative carbon source by breaking down glutamine to fuel the tricarboxylic acid (TCA) cycle in the form α-ketoglutarate (α-KG) in T cells69, this process seems to play a minimal role in maintaining OXPHOS in cytokine-activated NK cells67. Instead, NK cells require the transcription factor hypoxia-inducible factor 1-α for optimal glucose metabolism to support the survival of virus-specific NK cells72 and the effector functions of tumor-infiltrating NK cells73.
In contrast to activated and effector cells, several studies have reported that memory CD8+ T cells are more dependent on fatty-acid oxidation74 rather than glycolysis—a feature shared with memory ILC3s55. Indeed, deletion of a glycerol transporter or enzymes involved in triglyceride synthesis and storage results in impaired survival of memory CD8+ T cells75. However, genetic deletion of the fatty-acid transporter CPT1A, the rate-limiting enzyme of fatty-acid oxidation, in CD8+ T cells had no effect on memory cells. This is in contrast to studies that have used the pharmacological inhibitor of CPT1A, etomoxir; thus, highlighting CPT1A-independent off-target effects by etomoxir that may explain this discrepancy76. Whereas other metabolic traits, such as mitochondrial biomass and spare respiratory capacity, are also increased in memory CD8+ T cells compared with their naive counterparts77, memory Ly49H+ NK cells exhibit a lower number of mitochondria and higher mitochondrial membrane potential compared to naive NK cells78. Similarly, human adaptive NK cells also exhibit higher mitochondrial membrane potential and OXPHOS that is seemingly mediated by the chromatin-modifying enzyme ARID5B79. It is very likely that other memory ILCs experience distinct types of metabolic remodeling, because even myeloid populations that exhibit ‘memory-like’ responses have been shown to undergo metabolic rewiring80-82.
The metabolic rewiring of CD8+ T cells and NK cells during viral infection not only fulfills their energy and nutrient demands, but also mediates epigenetic reprogramming that facilitates optimal function and differentiation. Metabolic intermediates can act as substrates for epigenetic modifications, as well as regulators of epigenetic enzyme activity (Fig. 2). For instance, acetyl-CoA, a metabolite derived from carbohydrates, fatty acids, and amino acid catabolism, acts as a substrate for histone acetylation by the histone acetyltransferases (HATs), including p300 and CBP, in addition to its role as a fuel source for the TCA cycle83. Similarly, the catabolism of the amino acid methionine results in the production of S-adenosylmethionine (SAM), a universal methyl-donor for nucleotide and histone methylation84, whereas the byproduct of glutaminolysis, α-KG, regulates enzyme activities that promote demethylation85. As such, disruption of some of these key metabolic pathways can immensely impact the epigenetic landscape and functions of a variety of cell types. Moreover, there are many examples of histone modifications mediated by metabolic intermediates that have recently been discussed elsewhere86.
Naive CD8+ T cells possess epigenetic features of ‘quiescence’ that are distinct from those of effector and memory cells, which may explain the distinct levels of responsiveness between these differentiation states. Histone profiling of differentiating CD8+ T cells suggests that many regulatory regions associated with loci of key factors critical for effector and memory fates, such as the transcription factors Tbx21 (which encodes T-bet), Id2, and Id3, among others, are kept in a ‘bivalent’ state in naive CD8+ T cells in which they are marked by the specific histone marks trimethylated histone H3 K4 (H3K4me3) and H3K27me3, but not acetylated H3 K27 (H3K27ac), which are indicative of a poised but inactive transcriptional state87,88. Upon activation, naive CD8+ T cells experience substantial epigenetic reprogramming that coincides with changes in metabolic programs89. For example, blocking pyruvate transport into the mitochondria for acetyl-CoA conversion impairs effector T cell differentiation while paradoxically promoting memory generation by altering metabolic pathways that affect the availability of acetyl-CoA for histone acetylation90. Disruption of other metabolic enzymes, including lactate dehydrogenase, may similarly affect T cell fate, in part through epigenetic mechanisms65,91. Meanwhile, deletion of a component of p300/CBP results in the loss of the naive cell’s ability to expand and differentiate into effector and memory cells92, whereas inhibition of EZH2 (refs. 93,94) or SUV39H1 (ref. 95), which methylate H3K27 and H3K9, respectively, selectively impairs effector, but not memory, differentiation. Together, these studies underscore the importance of active epigenetic regulation by metabolic pathways and epigenetic enzymes in promoting optimal cytotoxic lymphocyte responses to infection.
Much like CD8+ T cells, NK cells also undergo significant epigenetic remodeling in response to viral infection96. These changes are primarily induced by proinflammatory cytokine signals early after infection that result in a distinct yet coordinated epigenetic reprogramming of NK cells (Fig. 2). During MCMV infection, type I IFN signaling through its downstream transcription factor STAT1 triggers chromatin modifications at gene promoters, in part through deposition of H3K4me3 (ref. 40). The specific mechanisms by which STAT1 mediates this promoter-centric chromatin remodeling are not entirely clear but may be due to preferential binding of STAT1 to promoter regions, which affects the ability of the MLL complex to methylate H3K4 in these regions96. By contrast, the IL-12–STAT4 axis predominantly remodels enhancers by acting as a pioneering factor to prime de novo enhancers through induction of chromatin accessibility and recruitment of p300/CBP for H3K27 acetylation, as deletion of STAT4 results in loss of enhancer accessibility as well as p300 binding40,96,97. Moreover, IL-12–STAT4-mediated chromatin remodeling may be enhanced by the synergistic cooperation between IL-12 and the IL-18–NF-κB and IL-2–IL-15–STAT5 axes40, signals known to be critical in driving both innate and adaptive features of NK cells. However, little is known about how antigen receptor signaling, along with co-stimulatory receptor triggering, impacts chromatin remodeling—all of which could potentially be driven by the crosstalk of NFAT, NF-κB, and/or AP-1 factors.
Whereas the early cytokine-driven histone remodeling of NK cells is becoming better understood, much less is known about the regulation of epigenetic reprogramming during their differentiation in vivo throughout the course of an infection. Naive NK cells possess significant epigenetic and transcriptomic features (likely acquired during their development and education)98 that bear striking resemblance to memory CD8+ T cells, as assessed by chromatin accessibility96. Furthermore, memory Ly49H+ NK cells and adaptive NKG2C+ NK cells are enriched in AP-1 footprints compared with their naive counterparts15,96, a feature that may be imprinted early following antigen-receptor engagement. Importantly, this AP-1 footprint is a common epigenetic signature that is shared with not only memory CD8+ T cells, but also other types of memory cells99. AP-1 itself is a critical factor for cell-type-specific enhancer selection through its interaction with other transcription factors and the chromatin-remodeling SWI/SNF complex100, and has recently been implicated in shaping the three-dimensional (3D) enhancer landscape101. Notably, in CD4+ T cells, AP-1-mediated 3D chromatin remodeling has been reported to be dependent on ETS-mediated recruitment of CTCF102,103. Our own analysis of the 3D chromatin structure of differentiating MCMV-specific CD8+ T cells and Ly49H+ NK cells also suggests that AP-1 factors are involved in mediating dynamic changes in the genome architecture during the transition from naive to memory cells in these two cell types104. Thus, the rapid responsiveness of memory cells across different cell types may be explained by the shared epigenetic identity imparted through the cooperation between transcription factors and chromatin organizers.
The intricate relationship between transcription factors and epigenetic remodeling facilitates the expression and/or binding of other transcription factors that regulate cellular differentiation, function, and phenotype (Fig. 2). Both NK cells and CD8+ T cells share many transcriptional programs, including those mediated by the T-box family of transcription factors, T-bet and EOMES, which are indispensable for the development of NK cells and the maintenance of memory CD8+ T cells105-107. ILC1 development is similarly dependent on T-bet, but not EOMES108. Other transcription factors that control effector and memory fates of CD8+ T cells, such as BACH2109, ZEB2110, BLIMP-1111, ID2112, and TCF7113, also regulate NK cell maturation at steady state109,114-119, further highlighting a shared transcriptional program between these innate and adaptive cytotoxic lymphocytes. Interestingly, a master transcription factor of T cell identity, BCL11B, has been shown to be similarly critical for adaptive NK cell differentiation in humans and mice120.
During infection, the transcriptional circuitries that govern virus-specific NK cells and CD8+ T cells also exhibit a high degree of overlap, as highlighted by the shared requirement of STAT transcription factors for the optimal responses of these sibling lymphocytes, and may involve complex interplay between additional lineage-defining and signal-driven transcription factors97,121. Antigen receptor triggering in T cells induces an AP-1-dependent transcriptional program through the cooperation of AP-1 with interferon regulatory factor 4 (IRF4), which may be regulated by the affinity of antigen–receptor interactions122,123, and this cooperative interaction has a critical role in controlling the transcription-factor network that is essential for T cell subset differentiation122-126. Importantly, AP-1 family members (such as BATF) also act as pioneering factors that shape the chromatin landscape of activated T cells, although this activity is independent of IRF4103. It is unclear whether an affinity- or avidity-dependent transcriptional program exists in NK cells18,59. In contrast to T cells, in which IRF4 expression is triggered by antigen receptor signaling, the expression of IRF4 in MCMV-specific NK cells relies on the synergistic cooperation between activating receptor and proinflammatory cytokine signals. Our recent study provides evidence that the transcriptional program orchestrated by IRF4 is critical for the survival and the differentiation of virus-specific NK cells in part by regulating nutrient uptake essential for the adaptive NK cell response127.
Notably, NK cells at steady state do express IRF8, which shares significant homology with IRF4 and can partner with AP-1 factors128. During infection, IRF8 can be further induced in NK cells by IL-12 and IL-18, and IRF8-deficient Ly49H+ NK cells failed to expand owing to their inability to upregulate ZBTB32, a cell cycle regulator that promotes proliferation by suppressing BLIMP-1 in NK cells129,130. Whether IRF8-mediated induction of ZBTB32 is dependent on AP-1 factors is yet to be determined. In addition to its regulation of the IRF8–ZBTB32 arm, the IL-12–STAT4 program is also required for the induction of RUNX family members, namely RUNX1 and RUNX3, which along with CBFβ support the proliferation program of MCMV-specific Ly49H+ NK cells for optimal generation of memory131. Although the mechanism by which RUNX factors orchestrate this process in NK cells is unclear, RUNX3 in CD8+ T cells appears to function through both transcriptional and epigenetic mechanisms132-134. Furthermore, although less is known about the transcriptional regulation for the maintenance of memory NK cells, both memory NK cells and CD8+ T cells depend on the cytokine IL-15 and its downstream transcription factor STAT5 for their survival135. Altogether, the intimate crosstalk between metabolism, epigenetics, and transcription orchestrates the cell-intrinsic qualitative differences that render memory cells superior to naive cells in the host defense against pathogens.
Convenience of tissue redistribution
A major immunosurveillance mechanism of NK cells and CD8+ T cells relies on the identification and elimination of infected or transformed target cells. As such, the continuous patrolling of these cytotoxic lymphocytes provides an effective, yet relatively delayed, mode of surveillance. As previously discussed, naive CD8+ T cells must be primed in SLOs such as lymph nodes, undergo clonal expansion, and then be actively recruited to infected or inflamed peripheral tissues—a lengthy and taxing process that may take several days to complete. Although the speed of the response is faster for NK cells because of their ability to execute innate effector functions without pre-sensitization, they still need to actively migrate from the blood into the affected tissues before eliminating infected cells.
In recent years, many studies have highlighted the ability of memory T cells to adapt and reside in tissues for extended periods of time. This ability is a convenient mechanism for the host to provide immediate protection at sites of pathogen entry, as these tissue-resident memory T (TRM) cells can quickly respond to local infection without having to migrate136. Thus, the capacity of TRM cells to persist outside of lymphoid organs is analogous to the ability of ILC1s, ILC2s, and ILC3s (helper ILCs) to preferentially inhabit peripheral tissues. Importantly, although both TRM and helper ILCs are tissue-resident lymphocytes that provide local immune protection, helper ILCs arise early during organism development, seed the tissues, and maintain their identity through self-renewal and proliferation137, whereas TRM cells originate from activated T cells primed in the SLO and have subsequently traveled through circulation into tissues136. However, there are studies that have described the mobilization of ILC2s from other tissues that subsequently acquire residency in the lung upon infection138,139 (Fig. 3). Nevertheless, despite their distinct ontogeny and mechanisms of tissue recruitment and seeding, TRM cells and helper ILCs share many features, including their dependency on the transcription factors Hobit33,140 and T-bet108,141, among others.
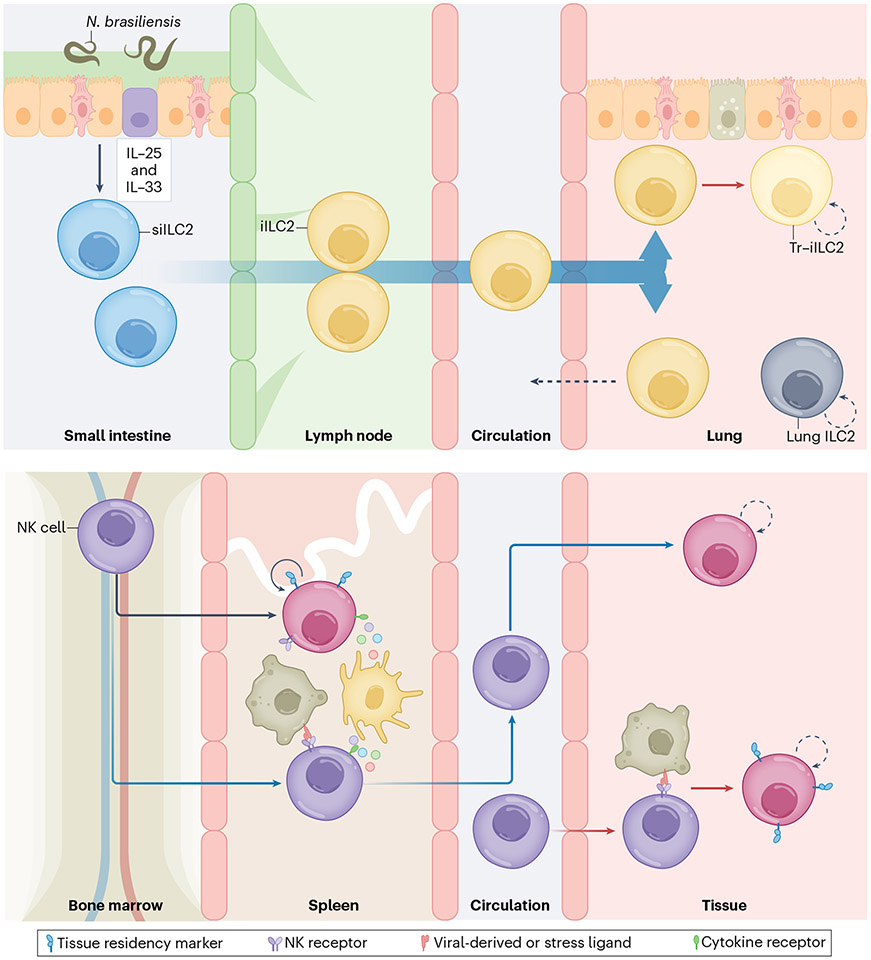
Fig. 3 ∣. Convenience of tissue redistribution.
The ability of memory cells to reside in tissues facilitates an accelerated immune response at the site of pathogen entry without having to migrate. Top, small-intestine ILC2s (siILC2s) have been shown to differentiate and migrate into the lung in response to Nippostrongylus brasiliensis in a sphingosine-1-phosphate-dependent manner (blue gradient). A small fraction of these iILC2s then stays in the lung (Tr-iILC2s) and co-exists with lung ILC2s, whereas other iILC2s migrate back into the small intestine. Bottom, in addition to tissue-resident helper ILCs, splenic-tissue-resident NK cells that contribute to the MCMV-specific response have been described (black arrow). Furthermore, although other tissue-resident NK cells can be generated in response to MCMV and other infections, it is unclear whether they are primed elsewhere, such as SLOs (blue arrow), or in the tissues in which they ultimately reside (red arrow).
The ability to establish tissue residency is not just a characteristic of helper ILCs and TRM cells, but is also a feature of circulating NK cells. A couple of recent studies have suggested that MCMV infection drives clonal expansion of spleen-resident NK cells and promotes the recruitment of NK cells that reside long-term in the salivary gland and the lungs46,142. Notably, CMV infection in both mice and humans results in virus latency, and CMV hides within organs including the salivary gland, where it can be reactivated and shed143,144. Thus, it is unclear whether the initial long-term retention of NK cells in these tissues is driven by the need to continuously control the virus. Although one study has proposed that these tissue-resident NK cells aid the rapid activation of CD8+ T cells by facilitating T–DC interactions during viral infection, another suggests that these NK cells remain in the tissue after viral clearance to limit pathology mediated by CD4+ T cells46,142. Moreover, the recruitment and residence of long-lived NK cells in tissue have not only been observed in systemic MCMV infection, but also during acute local skin infection with Vaccinia virus or Staphylococcus aureus, with tissue-resident NK cells exhibiting a distinct transcriptional profile from that of circulating NK cells, and mediating an accelerated effector response upon reinfection (unpublished observations, Torcellan and Gasteiger) (Fig. 3).
Many exciting questions remain to be resolved regarding memory in innate lymphocytes from peripheral tissues. For instance, does the priming of tissue-resident NK cells first occur in situ or elsewhere? How are tissue-resident memory innate lymphocytes first recruited into the tissues, and what are the signals required for the recruitment or residency program? Are there specific roles for tissue-resident NK cells and other memory ILCs during tissue homeostasis and/or local infection? What are the metabolic, epigenetic, and transcriptional programs that drive tissue residency of NK cells and other memory ILCs, and are these programs conserved or distinct across different tissues? Finally, what transcription factors maintain the identity of tissue-resident memory ILCs, and are these factors and programs shared with TRM cells? Importantly, tissue-resident signatures have also been observed in NK cells isolated from various human tissues and have been associated with positive clinical outcomes, suggesting that there is a conserved tissue-residency program within ILCs across species that may contribute to individual fitness145,146.
Concluding remarks
We now appreciate that innate immune cells possess characteristics that were previously thought to be exclusive to the adaptive immune system. Exciting efforts to harness the memory properties of innate lymphocytes have shown promising results in the clinic, as in the case of bone marrow transplantation or in treatments for various malignancies147-151. Although current endeavors using the memory properties of ILCs have been somewhat limited to cytokine-induced memory-like NK cells, other NK-cell-based therapeutics, including chimeric antigen receptor NK cells and NK cell engagers, may benefit from exploiting the adaptive features of NK cells152,153.
Beyond utilizing NK cells in the clinic, harnessing helper ILCs could improve strategies for anti-tumor immunity as well. However, the role of helper ILCs in malignancy is still unclear, with emerging studies only beginning to unravel their complex interactions with the tumor microenvironment154. Whether the memory features of helper ILCs can be harnessed to improve anti-tumor immunity in a context-dependent manner requires further investigation but holds potential for future therapeutics. As we continue to explore the cellular and molecular mechanisms that govern these adaptive features and transition from population-level approaches to the single-cell level, we hope to unravel new complexities and gain a more nuanced understanding of the intricate workings of the innate and adaptive immune system, with the ultimate goal of developing new therapies or immunization strategies to combat infectious diseases and other pathologies.
Acknowledgements
The authors apologize to all those whose significant and valuable contributions could not be cited or discussed in this manuscript owing to space limitations. We thank members of the Sun lab for helpful feedback on the manuscript. J.C.S. was supported by the Ludwig Center for Cancer Immunotherapy, the American Cancer Society, the Burroughs Wellcome Fund, and the NIH (AI100874, AI130043, AI155558, and P30CA008748).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Riedel S. Edward Jenner and the history of smallpox and vaccination. Proc. (Bayl. Univ. Med Cent.) 18, 21–25 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boylston A. The origins of inoculation. J. R. Soc. Med 105, 309–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun JC, Beilke JN & Lanier LL Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dokun AO et al. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol 2, 951–956 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Daniels KA et al. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med 194, 29–44 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown MG et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292, 934–937 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Arase H, Mocarski ES, Campbell AE, Hill AB & Lanier LL Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Grassmann S. et al. Distinct surface expression of activating receptor Ly49H drives differential expansion of NK cell clones upon murine cytomegalovirus infection. Immunity 50, 1391–1400 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Reeves RK et al. Antigen-specific NK cell memory in rhesus macaques. Nat. Immunol 16, 927–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlums H. et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Verges S. et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl Acad. Sci. USA 108, 14725–14732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J. et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ty M. et al. Malaria-driven expansion of adaptive-like functional CD56-negative NK cells correlates with clinical immunity to malaria. Sci. Transl. Med 15, eadd9012 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart GT et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J. Exp. Med 216, 1280–1290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruckert T, Lareau CA, Mashreghi MF, Ludwig LS & Romagnani C Clonal expansion and epigenetic inheritance of long-lasting NK cell memory. Nat. Immunol 23, 1551–1563 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasec P. et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Heatley SL et al. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. J. Biol. Chem 288, 8679–8690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammer Q. et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol 19, 453–463 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Ulbrecht M. et al. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol 164, 5019–5022 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Netea MG et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol 20, 375–388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naik S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L & Flies DB Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nabekura T. et al. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity 40, 225–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nabekura T. et al. Cutting edge: NKG2D signaling enhances NK cell responses but alone is insufficient to drive expansion during mouse cytomegalovirus infection. J. Immunol 199, 1567–1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabekura T & Lanier LL Activating receptors for self-MHC class I enhance effector functions and memory differentiation of NK cells during mouse cytomegalovirus infection. Immunity 45, 74–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu LL et al. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. 15, 1088–1099 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krug A. et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21, 107–119 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Abbas A. et al. The activation trajectory of plasmacytoid dendritic cells in vivo during a viral infection. Nat. Immunol 21, 983–997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valente M. et al. Novel mouse models based on intersectional genetics to identify and characterize plasmacytoid dendritic cells. Nat. Immunol 24, 714–728 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lio CW et al. cGAS–STING signaling regulates initial innate control of cytomegalovirus infection. J. Virol 90, 7789–7797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paijo J. et al. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog. 12, e1005546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madera S. et al. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med 213, 225–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weizman OE et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolle A. et al. IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J. Clin. Invest 124, 5305–5316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun JC et al. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med 209, 947–954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrews DM, Scalzo AA, Yokoyama WM, Smyth MJ & Degli-Esposti MA Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol 4, 175–181 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Chaix J. et al. Cutting edge: priming of NK cells by IL-18. J. Immunol 181, 1627–1631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedemann GM et al. Divergent role for STAT5 in the adaptive responses of natural killer cells. Cell Rep. 33, 108498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaech SM & Cui W Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiedemann GM et al. Deconvoluting global cytokine signaling networks in natural killer cells. Nat. Immunol 22, 627–638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekiaris V. et al. Ly49H+ NK cells migrate to and protect splenic white pulp stroma from murine cytomegalovirus infection. J. Immunol 180, 6768–6776 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Dokun AO, Chu DT, Yang L, Bendelac AS & Yokoyama WM Analysis of in situ NK cell responses during viral infection. J. Immunol 167, 5286–5293 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Andrews DM et al. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J. Immunol 166, 1796–1802 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Salazar-Mather TP, Ishikawa R & Biron CA NK cell trafficking and cytokine expression in splenic compartments after IFN induction and viral infection. J. Immunol 157, 3054–3064 (1996). [PubMed] [Google Scholar]
- 45.Hsu KM, Pratt JR, Akers WJ, Achilefu SI & Yokoyama WM Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol 90, 33–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flommersfeld S. et al. Fate mapping of single NK cells identifies a type 1 innate lymphoid-like lineage that bridges innate and adaptive recognition of viral infection. Immunity 54, 2288–2304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz-Salazar C. et al. Cell-intrinsic adrenergic signaling controls the adaptive NK cell response to viral infection. J. Exp. Med 217, e20190549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benechet AP et al. Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming. Nature 574, 200–205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kern M. et al. Virally infected mouse liver endothelial cells trigger CD8+ T-cell immunity. Gastroenterology 138, 336–346 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Wang Y. et al. Vaginal type-II mucosa is an inductive site for primary CD8+ T-cell mucosal immunity. Nat. Commun 6, 6100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weizman OE et al. Mouse cytomegalovirus-experienced ILC1s acquire a memory response dependent on the viral glycoprotein m12. Nat. Immunol 20, 1004–1011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguilar OA et al. A viral immunoevasin controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell 169, 58–71 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Gonzalez I. et al. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity 45, 198–208 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Verma M. et al. The molecular and epigenetic mechanisms of innate lymphoid cell (ILC) memory and its relevance for asthma. J. Exp. Med 218, e20201354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serafini N. et al. Trained ILC3 responses promote intestinal defense. Science 375, 859–863 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naik S & Fuchs E Inflammatory memory and tissue adaptation in sickness and in health. Nature 607, 249–255 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halper-Stromberg A & Jabri B Maladaptive consequences of inflammatory events shape individual immune identity. Nat. Immunol 23, 1675–1686 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Busch DH & Pamer EG T cell affinity maturation by selective expansion during infection. J. Exp. Med 189, 701–710 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams NM et al. Cytomegalovirus infection drives avidity selection of natural killer cells. Immunity 50, 1381–1390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chapman NM, Boothby MR & Chi H Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol 20, 55–70 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Frauwirth KA et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Ma EH et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Xu K. et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phan AT et al. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45, 1024–1037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheppard S. et al. Lactate dehydrogenase A-dependent aerobic glycolysis promotes natural killer cell anti-viral and anti-tumor function. Cell Rep. 35, 109210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loftus RM et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat. Commun 9, 2341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu W. et al. Lactate regulates cell cycle by remodeling the anaphase promoting complex. Nature 616, 790–797 (2023). [DOI] [PubMed] [Google Scholar]
- 69.Wang R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Littwitz-Salomon E. et al. Metabolic requirements of NK cells during the acute response against retroviral infection. Nat. Commun 12, 5376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong H. et al. The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol 20, 865–878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Victorino F. et al. HIF1α is required for NK cell metabolic adaptation during virus infection. eLife 10, e68484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ni J. et al. Single-cell RNA sequencing of tumor-infiltrating NK cells reveals that inhibition of transcription factor HIF-1α unleashes NK cell activity. Immunity 52, 1075–1087 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Pearce EL et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui G. et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell 161, 750–761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raud B. et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 28, 504–515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van der Windt GJ et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Sullivan TE, Johnson LR, Kang HH & Sun JC BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43, 331–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cichocki F. et al. ARID5B regulates metabolic programming in human adaptive NK cells. J. Exp. Med 215, 2379–2395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arts RJ et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bekkering S. et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell 172, 135–146 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Cheng SC et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wellen KE et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mentch SJ et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carey BW, Finley LW, Cross JR, Allis CD & Thompson CB Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dai Z, Ramesh V & Locasale JW The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet 21, 737–753 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu B. et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Russ BE et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8+ T cell differentiation. Immunity 41, 853–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soriano-Baguet L & Brenner D Metabolism and epigenetics at the heart of T cell function. Trends Immunol. 44, 231–244 (2023). [DOI] [PubMed] [Google Scholar]
- 90.Wenes M. et al. The mitochondrial pyruvate carrier regulates memory T cell differentiation and antitumor function. Cell Metab. 34, 731–746 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tyrakis PA et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Piccirillo AR, Cattley RT, D’Cruz LM & Hawse WF Histone acetyltransferase CBP is critical for conventional effector and memory T-cell differentiation in mice. J. Biol. Chem 294, 2397–2406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kakaradov B. et al. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gray SM, Amezquita RA, Guan T, Kleinstein SH & Kaech SM Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46, 596–608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pace L. et al. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359, 177–186 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Lau CM et al. Epigenetic control of innate and adaptive immune memory. Nat. Immunol 19, 963–972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sciume G. et al. Rapid enhancer remodeling and transcription factor repurposing enable high magnitude gene induction upon acute activation of NK cells. Immunity 53, 745–758 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mujal AM, Delconte RB & Sun JC Natural killer cells: from innate to adaptive features. Annu. Rev. Immunol 39, 417–447 (2021). [DOI] [PubMed] [Google Scholar]
- 99.Larsen SB et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell 28, 1758–1774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vierbuchen T. et al. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell 68, 1067–1082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wolf BK et al. Cooperation of chromatin remodeling SWI/SNF complex and pioneer factor AP-1 shapes 3D enhancer landscapes. Nat. Struct. Mol. Biol 30, 10–21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chandra A. et al. Quantitative control of Ets1 dosage by a multi-enhancer hub promotes TH1 cell differentiation and protects from allergic inflammation. Immunity 56, 1451–1467 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pham D. et al. Batf pioneers the reorganization of chromatin in developing effector T cells via Ets1-dependent recruitment of Ctcf. Cell Rep. 29, 1203–1220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Santosa EK, Lau CM, Sahin M, Leslie CS & Sun JC 3D Chromatin dynamics during innate and adaptive immune memory acquisition. Preprint at bioRxiv 10.1101/2023.01.16.524322 (2023). [DOI] [Google Scholar]
- 105.Gordon SM et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Intlekofer AM et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Joshi NS et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klose CSN et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- 109.Roychoudhuri R. et al. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol 17, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Omilusik KD et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med 212, 2027–2039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xin A. et al. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol 17, 422–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang CY et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol 12, 1221–1229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou X. et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33, 229–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van Helden MJ et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med 212, 2015–2025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kallies A. et al. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood 117, 1869–1879 (2011). [DOI] [PubMed] [Google Scholar]
- 116.Jeevan-Raj B. et al. The transcription factor Tcf1 contributes to normal NK cell development and function by limiting the expression of granzymes. Cell Rep. 20, 613–626 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Delconte RB et al. The helix-loop-helix protein ID2 governs NK cell fate by tuning their sensitivity to interleukin-15. Immunity 44, 103–115 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Imianowski CJ et al. BACH2 restricts NK cell maturation and function, limiting immunity to cancer metastasis. J. Exp. Med 219, e20211476 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li S. et al. The transcription factor Bach2 negatively regulates murine natural killer cell maturation and function. eLife 11, e77294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Holmes TD et al. The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation. Sci. Immunol 6, eabc9801 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kallal LE & Biron CA Changing partners at the dance: variations in STAT concentrations for shaping cytokine function and immune responses to viral infections. JAKSTAT 2, e23504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li P. et al. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Man K. et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Schraml BU et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 460, 405–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kurachi M. et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol 15, 373–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yao S. et al. Interferon regulatory factor 4 sustains CD8+ T cell expansion and effector differentiation. Immunity 39, 833–845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Santosa EK et al. Control of nutrient uptake by IRF4 orchestrates innate immune memory. Nat. Immunol 10.1038/s41590-023-01620-z (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Glasmacher E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1–IRF complexes. Science 338, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Adams NM et al. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity 48, 1172–1182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Beaulieu AM, Zawislak CL, Nakayama T & Sun JC The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat. Immunol 15, 546–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rapp M. et al. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci. Immunol 2, eaan3796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cruz-Guilloty F. et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med 206, 51–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang D. et al. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Milner JJ et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun JC & Lanier LL NK cell development, homeostasis and function: parallels with CD8+ T cells. Nat. Rev. Immunol 11, 645–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Masopust D & Soerens AG Tissue-resident T cells and other resident leukocytes. Annu Rev. Immunol 37, 521–546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vivier E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 138.Huang Y. et al. IL-25-responsive, lineage-negative KLRG1hi cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol 16, 161–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Huang Y. et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mackay LK et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Mackay LK et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015). [DOI] [PubMed] [Google Scholar]
- 142.Schuster IS et al. Infection induces tissue-resident memory NK cells that safeguard tissue health. Immunity 56, 531–546 e536 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Krmpotic A, Bubic I, Polic B, Lucin P & Jonjic S Pathogenesis of murine cytomegalovirus infection. Microbes Infect. 5, 1263–1277 (2003). [DOI] [PubMed] [Google Scholar]
- 144.Crough T & Khanna R Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol Rev 22, 76–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kirchhammer N. et al. NK cells with tissue-resident traits shape response to immunotherapy by inducing adaptive antitumor immunity. Sci. Transl. Med 14, eabm9043 (2022). [DOI] [PubMed] [Google Scholar]
- 146.Brownlie D. et al. Expansions of adaptive-like NK cells with a tissue-resident phenotype in human lung and blood. Proc. Natl Acad. Sci. USA 118, e2016580118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Berrien-Elliott MM et al. Hematopoietic cell transplantation donor-derived memory-like NK cells functionally persist after transfer into patients with leukemia. Sci. Transl. Med 14, eabm1375 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Marin ND et al. Memory-like differentiation enhances NK cell responses to melanoma. Clin. Cancer Res 27, 4859–4869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Romee R. et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med 8, 357ra123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cichocki F. et al. Adaptive NK cell reconstitution is associated with better clinical outcomes. JCI Insight 4, e125553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boudreau JE et al. KIR3DL1/HLA-B subtypes govern acute myelogenous leukemia relapse after hematopoietic cell transplantation. J. Clin. Oncol 35, 2268–2278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu E. et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med 382, 545–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gauthier L. et al. Control of acute myeloid leukemia by a trifunctional NKp46-CD16a-NK cell engager targeting CD123. Nat. Biotechnol 10.1038/s41587-022-01626-2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jacquelot N, Seillet C, Vivier E & Belz GT Innate lymphoid cells and cancer. Nat. Immunol 23, 371–379 (2022). [DOI] [PubMed] [Google Scholar]