Abstract
Honeybees (Apis mellifera) are key pollinators supporting global agriculture and are long-established models for developmental and behavioural research. Recently, they have emerged as models for studying gut microbial communities. Earlier research established that hindguts of adult worker bees harbour a conserved set of host-restricted bacterial species, each showing extensive strain variation. These bacteria can be cultured axenically and introduced to gnotobiotic hosts, and some have basic genetic tools available. In this Review, we explore the most recent research showing how the microbiota establish in the gut and impact bee biology and health. Microbiota members occupy specific niches within the gut where they interact with each other and the host. They engage in cross-feeding and antagonistic interactions, which likely contribute to the stability of the community and prevent pathogen invasion. An intact gut microbiota provides protection against diverse pathogens and parasites and may contribute to processing refractory components of the pollen coat and dietary toxins. Absence or disruption of the microbiota results in altered expression of genes underlying immunity, metabolism, behaviour, and development. In the field, such disruption by agrochemicals may negatively impact bees. These findings demonstrate a key developmental and protective role of the microbiota, with broad implications for bee health.
Introduction
The honeybee, Apis mellifera, has a long history of domestication for honey and wax production, as well as for pollination. In research, honeybees have served as models for developmental plasticity1, cognition2, and social behavior3. More recently, they have emerged as models for gut microbiota studies4,5. The advent of nucleotide sequencing technologies revealed a specific microbial community inhabiting the honeybee gut6–9. Since then, a combination of sequencing and culture-based approaches has been used to characterize the core members of the honeybee gut microbiota, their metabolic capabilities, and their roles in bee health.
The honeybee gut microbiota is relatively simple, dominated by five core bacterial lineages present in all healthy worker bees. These bacteria are acquired orally after emergence from the pupal stage through social interaction and contact with hive compartments4,10, and correspond to clusters within the genera Bifidobacterium, Bombilactobacillus (previously called Lactobacillus Firm-411), Gilliamella, Lactobacillus (previously called Lactobacillus Firm-511) and Snodgrassella4. These core bacteria form a consistent community of about 108 to 109 cells, although their absolute and relative abundances vary with life stage, season and geographic location12–18. Other non-core bacteria are commonly present, and include Bartonella, Commensalibacter and Frischella4. Within each of these genera, between one and five species have been formally characterized19–24, although additional closely related species may remain to be recognized25.
Environmental (for example, Fructobacillus spp.) and pathogenic bacteria (for example, Serratia marcescens, Hafnia alvei and other Enterobacterales) are often present at low abundances in adult bee guts8. Still other bacteria (for example, Apilactobacillus kunkeei and Bombella apis) are associated with larvae, queens and hive compartments26,27. Although dominated by bacteria, some honeybee guts harbour a small proportion of eukaryotes, including fungi28, trypanosomatid parasites (Crithidia and Lotmaria species)29, and microsporidian parasites (Vairimorpha ceranae, previously called Nosema ceranae)30,31. Fungal presence is erratic, and varies among geographic locations, suggesting that fungi are transient in bee guts32.
The five core bacterial lineages appear to have evolved with bees since the origin of the Corbiculata clade, about 80 million years ago33. Most corbiculate bees, including other honeybee (Apis) species native to eastern Asia34, bumblebee species (genus Bombus) worldwide35, and stingless bees (tribe Meliponini) in tropical regions36 retain these core bacteria, although stingless bees have more often gained or lost certain bacteria37–39. Different bee species can also harbour distinct sets of bacteria, such as Frischella perrara and Bartonella apis in Apis spp. and Bombiscardovia and Schmidhempelia in Bombus spp. In general, these bacterial lineages have not been reported outside bees, although some have been detected in non-corbiculate bees, such as carpenter bees40 and euglossine bees39. Thus, the corbiculate bee gut microbiota is dominated by specialized, host-restricted bacterial lineages that have evolved with one another and with hosts for long evolutionary periods.
A stable gut microbiota appears intrinsically associated with honeybee health. It aids in digestion41,42 and detoxification of food components43,44, stimulates the innate immune system45,46, and protects against pathogens47–49. Additionally, the gut microbiota affects key developmental pathways, such as the endocrine signalling pathway which regulates feeding behaviour and weight gain50–52, and olfactory learning and memory acquisition pathways53.
Honeybees have been used as models in efforts to disentangle how gut microorganisms interact with each other, with opportunistic microorganisms, with environmental stressors, and with the host4. Experimental approaches have been enabled by the ability to culture strains of each core species, to perform genetic manipulations on some, and to reintroduce them into microbiota-deprived bees (BOX 1). Earlier research, previously reviewed, has described the honeybee microbiota and metabolic capabilities of the core gut species4,5,47,54. Here, we assess recent research on the roles of the gut microbiota in bee health and disease. We first discuss recent findings on how beneficial bacteria interact with each other and establish themselves in the bee gut. Next, we summarize results from studies showing that the microbiota plays a crucial role in several aspects of bee health. We also examine evidence that environmental stressors can compromise the microbiota, with negative consequences for bees, and preliminary evidence that probiotics might be useful for restoring perturbed gut microbial communities. Finally, we identify knowledge gaps and discuss areas that require further investigation.
Box 1. Approaches for identifying the effects of the gut microbiota on bee biology.
A combination of experimental and genomic approaches have been used to identify the effects of the gut microbiota on honeybees. These approaches include: [b1] the use of microbiota-deprived bees, conventionalized bees, or bees colonized with a single or several isolates, followed by examination of bee phenotypes45,46,48,51,53,61,64,82,88,115. In this context, microbiota-deprived bees refer to newly emerged bees that have been extracted from brood frames at the pupal stage or allowed to emerge on the frame without exposure to hive bees and raised under aseptic conditions in the laboratory. These bees are subjected to minimal exposure to microorganisms, resulting in a reduced presence of microbial colonization and specifically a lack of the usual core lineages10. Complete absence of microorganisms cannot be guaranteed and must be checked for each bee. On the other hand, conventionalized bees are those that have been colonized with the full microbiota obtained from gut homogenates of hive bees, resulting in a typical native and diverse gut microbial community. Effects of these microbiota treatments on gene expression patterns linked to behavioral53,115, developmental50,51, immunity45,46,82,88, and metabolic pathways61,64 have been investigated, as well as the ability of specific bee gut bacteria or the intact microbiota to prevent pathogen proliferation45,46,48,82,88.
Perturbation of the normal microbiota using antibiotics or other stressors, followed by phenotype examination59,123,133,135,138,139,143; for example, honeybees exposed to tetracycline or streptomycin exhibit perturbed gut communities and increased susceptibility to bacterial and fungal infections86,96.
Genetic engineering of bee gut strains to include visual markers or resistance genes57,165,167; for example, the expression of distinct fluorescent proteins in strains of Gilliamella apis and Gilliamella apicola has allowed visualization of spatial niche partitioning within the ileum57.
Heterologous expression of genes from bee gut microbiota to verify gene functions63,143,170; for example, a Bifidobacterium spp. gene that encodes for a glycoside hydrolase family 3 was heterologously expressed in Escherichia coli to identify its role in the metabolism of amygdalin63.
Mark-recapture experiments with hive bees that enable examination of survivorship under natural, field conditions96,125,140,151; for example, honeybees were exposed under laboratory conditions to tetracycline96 or a Roundup formulation151, then returned to their original hives to investigate recovery rates and microbiota resilience.
Genomic sequencing and in vitro experiments to establish symbiont metabolic and antagonism attributes42,43,60,69; for example, the ability of Gilliamella spp. strains to digest pectin and to metabolize diverse sugars was hypothesized from genome sequences and verified experimentally with cultured isolates41,43.
Microbial interactions within the gut
Members of the honeybee microbiota have adopted distinct ecological niches within the hindgut, a nutrient-poor region of the digestive tract consisting of three main sections: ileum, rectum and pylorus (FIG. 1a). Snodgrassella alvi and Gilliamella spp. are more abundant in the ileum, where they form a stable biofilm (FIG. 1c), while Lactobacillus, Bombilactobacillus and Bifidobacterium species are dominant in the rectum4 (FIG. 1d). Frischella perrara, when present, colonizes the pylorus, a region connecting the midgut to the ileum, where it strongly activates the bee immune system55 and induces formation of a melanized scab56 (FIG. 1b). The pylorus, at the junction of the midgut and ileum, offers a distinctive niche, as the site where the Malpighian tubules empty host nitrogenous waste57. While these core bacteria concentrate in specific gut compartments, they can also be found in other compartments in lower abundances9,10.
Fig. 1. Microbial dynamics and spatial organization in the honeybee gut.
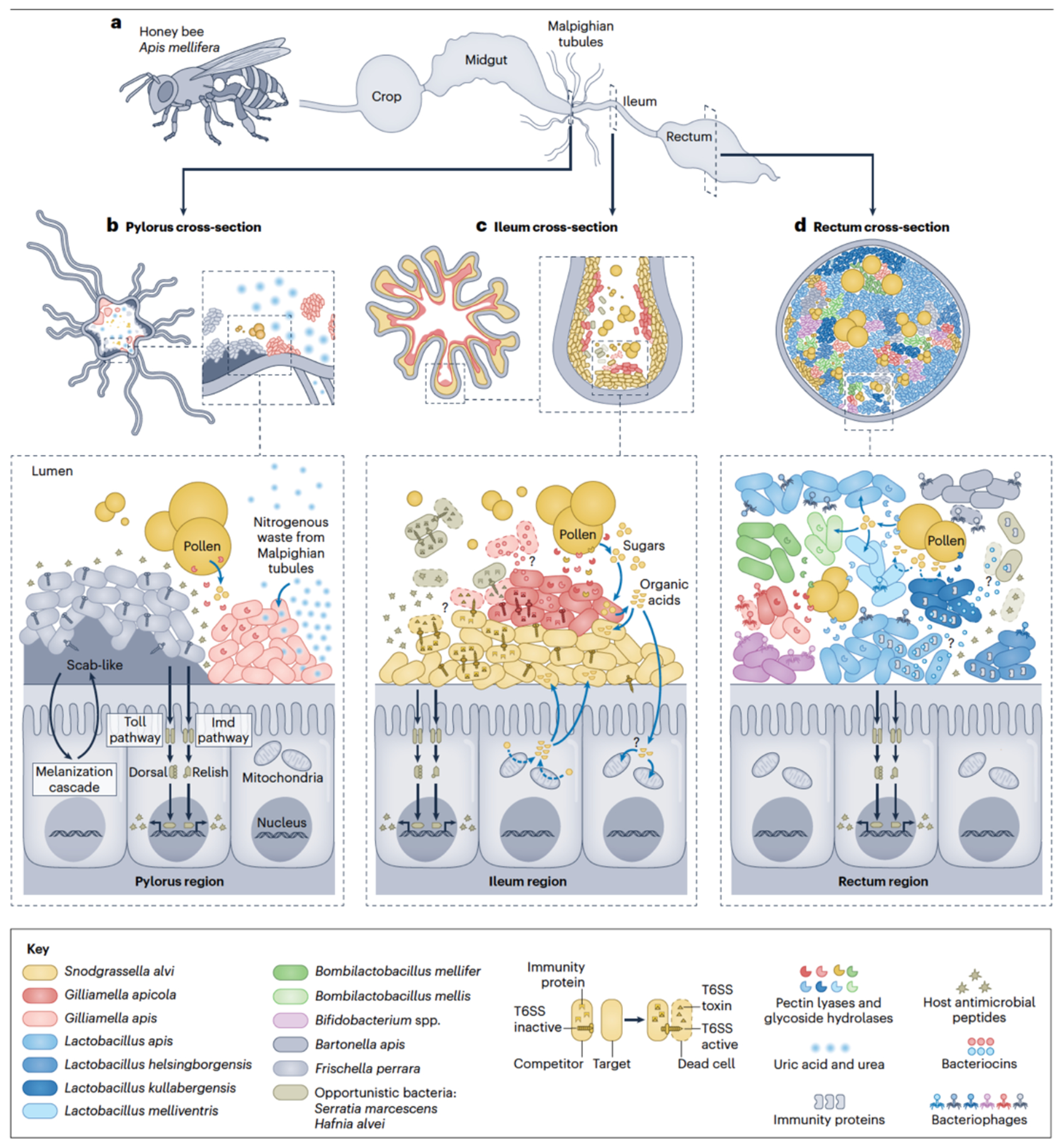
a) Characteristic bacterial communities colonize the distal region of a typical worker honeybee gut (pylorus, ileum, and rectum), based on fluorescence in situ hybridization, localization of fluorescently marked strains, 16S rRNA amplicon sequencing and quantitative PCR studies9,56,57. b) In the pylorus, Frischella perrara activates the host immune system, including humoral immunity (Toll and Imd pathways that lead to the production of antimicrobial peptides) and cellular immunity (melanization cascade that leads to the scab-like phenotype observed in this tissue). Gilliamella apis is involved in the recycling of waste nitrogen and some degradation and fermentation of polysaccharides present in pollen57. c) In the ileum, Snodgrassella alvi and Gilliamella apicola form a stable biofilm165, activate the host immune system45,46, and are potentially involved in cross-feeding with one another and the host64,68. G. apicola produces enzymes for digestion and fermentation of pollen wall components. S. alvi can use host-derived organic acids to independently colonize the bee gut68, but it is unclear whether bees can use bacteria-derived organic acids. d) In the rectum, Lactobacillus spp. and Bifidobacterium spp. are the most abundant bacteria and are involved in digestion and host immune system activation25,42,88. A distinctive bacteriophage community is associated with specific members of the microbiota79–81. Gut microbiota members possess extensive mechanisms for antagonism, such as T6SS and bacteriocins, and these may play a role in community dynamics and pathogen protection. Question marks indicate unconfirmed processes, such as microbial interactions between and within S. alvi and G. apicola strains and absorption of bacteria-derived organic acids by bees. Black arrows indicate activation of host immunity pathways and blue arrows indicate metabolism (degradation, uptake, and/or utilization). Complexities of bee gut morphology are not depicted in this diagram.
Species within the honeybee gut microbiota harbour extensive strain diversity34,58,59. Even within a single bee gut, strains vary in gene repertoires affecting functional attributes, such as the ability to process dietary or host-derived components and to interact with related strains or species and with the host25,41,60. These abilities and interactions are likely to influence the composition of the microbiota, with specific strains adopting specific niches and roles. Potentially, these activities stabilize the microbiota and in turn promote the overall health of bees. Here we summarize findings to date, noting that much remains unknown.
Diversification within the host gut
The genus Gilliamella has diverged into at least two genetically isolated species within the honeybee gut, Gilliamella apis and Gilliamella apicola, and these appear to be associated with distinct gut niches57. Genome analyses and experiments with two representative fluorescently marked strains has shown that G. apis is concentrated in the pylorus region and possesses urease and urea transporters, enabling it to utilize urea derived from bee waste (FIG. 1b), whereas G. apicola colonizes downstream regions of the ileum and cannot use urea57 (FIG. 1c). Whether this metabolic difference is consistent across strains is unclear, but the incidence of Gilliamella-encoded urease genes is greater in the pylorus region57.
Bee-restricted Lactobacillus species are abundant and diverse in honeybee guts25 (FIG. 1d). Investigations of strains representing the closely related species, Lactobacillus apis, Lactobacillus helsingborgensis, Lactobacillus melliventris and Lactobacillus kullabergensis, have shown that these species appear to reduce interspecific competition by partitioning nutrients derived from pollen, including different sugars and plant secondary metabolites61 (FIG. 1d). All four species metabolize simple sugars and acids, but exhibit preferences based on metabolic rates. For example, L. helsingborgensis and L. kullabergensis utilize citrate at higher rates than do L. apis and L. melliventris, and L. helsingborgensis utilizes glucitol at higher rates than the other species61. Regarding plant secondary metabolites, L. apis, L. melliventris and L. kullabergens, but not L. helsingborgensis, contribute to the metabolism of specific glycosylated flavonoids, and L. apis can also metabolize iridoid glycosides61.
Potential cross-feeding interactions
The honeybee microbiota shapes the hindgut environment, for example lowering oxygen and pH levels50. Snodgrassella alvi is an obligate aerobe that forms a biofilm on the stable cuticular lining of the ileum wall (FIG. 1c), depleting oxygen and creating an anaerobic interior lumen where other microbiota members reside. These members, including Gilliamella spp., Lactobacillus spp., and Bifidobacterium spp., produce various carbohydrate-degrading enzymes, such as pectin lyases and glycoside hydrolases41,42 (FIG. 1c and d). These enzymes enable the breakdown of indigestible components from pollen husks (for example, hemicellulose and pectin42,62) and a wide range of nectar-derived metabolites, including disaccharides (for example, cellobiose44), monosaccharides (for example, galactose, mannose, rhamnose, and xylose43,44), and secondary metabolites (for example, cyanogenic glycosides63, flavonoid glycosides61,64 and others65). Microbial digestion of some of these metabolites prevents intoxication43 and promotes parasite protection in different bee species65,66.
As byproducts from bacterial metabolism within the bee gut, short-chain fatty acids (organic acids) are produced, including acetate, butyrate, formate, lactate, pyruvate and succinate44,64. Some of these are used as energy sources by S. alvi50, which cannot use carbohydrates directly, and potentially by the host (FIG. 1c). Such cross-feeding interactions may play a role in shaping bee gut microbial communities64. At least some cross-feeding can occur between Gilliamella and Snodgrassella strains, which coexist in the ileum biofilm7 and possess complementary metabolic capabilities67. Carbohydrate metabolism by G. apicola leads to accumulation of pyruvate, and this is utilized, at least in part, by S. alvi, based on metabolomic analyses of cultures64. In vitro assays show that S. alvi growth is mildly enhanced when provided with supernatant from G. apicola cultures64. However, cross-feeding is not required as S. alvi can use host-derived organic acids, such as citrate, glycerate and 3-hydroxy-3-methylglutarate as carbon sources within the bee gut68 (FIG. 1c), and monoinoculation with S. alvi leads to robust colonization33,45,48,64,67.
Metabolomics on sections of the honeybee gut reveal higher levels of aromatic amino acids in the ileum and rectum, but not in the midgut, of conventionalized bees compared to microbiota-deprived bees50. S. alvi, for example, cannot colonize the ileum without intact amino acid biosynthetic pathways69. Bombilactobacillus spp. and Lactobacillus spp. are auxotrophic for several amino acids42 and may take advantage of amino acids produced by G. apis and S. alvi. However, these are not essential, as single strains of Bombilactobacillus spp. and Lactobacillus spp. can colonize the bee gut in the absence of other community members48,64,70.
Antagonistic interactions within the gut
The ability to colonize niches in the bee gut depends on host-defined factors, but also appears to reflect the outcomes of antagonistic interactions among strains and species. These interactions can be contact-dependent or contact-independent. Genes underlying antagonism are some of the most dynamic in the genomes, showing rapid evolution and gain and loss across strains60,62,67,71.
Type 6 secretion systems (T6SSs) confer survival advantages to bacteria in microbial communities by delivering toxins that kill competing bacteria in a contact-dependent way (interference competition) and by improving acquisition of essential micronutrients (exploitation competition)72. Based on genome sequences, many Gram-negative gut symbionts of A. mellifera (for example, S. alvi, Gilliamella spp., and F. perrara), as well as of Apis cerana (for example, Apibacter spp.) and Bombus spp. (for example, Schmidhempelia spp.), carry genes encoding T6SSs, associated Rhs toxins, and their respective immunity genes60,71 (FIG. 1b and c). In some S. alvi strains, two independently acquired T6SSs are present and appear to differ in function, as only one is associated with presence of Rhs toxins60. In a global mutagenesis study of S. alvi, immunity gene mutants failed to colonize guts, verifying inter-strain toxicity of the Rhs toxins, whereas mutants in genes of the T6SS itself were favoured, indicating that the production of the structure is costly69.
In F. perrara, the T6SS machinery may interact with both the host and other bacteria in the bee gut73. The expression of T6SS genes, as well as pilus, colibactin, and aryl polyene (APE) biosynthesis genes, is regulated by a DNA-binding protein, the integration host factor (IHF). The deletion of ihf impairs the ability of F. perrara to colonize the pylorus and form the scab phenotype, suggesting some direct host interaction73. Deletion of IHF-regulated genes leads to impaired gut colonization, and/or abolishes scab development73. In the presence of a defined community, F. perrara mutants lacking T6SS-2 or APE biosynthesis show reduced colonization, suggesting their advantage in interactions with other bee gut symbionts73.
Some bee bacterial pathogens, such as Serratia marcescens, encode T6SSs that can antagonize closely related S. marcescens strains and Escherichia coli (FIG. 1c). In vitro experiments examining the effects of T6SSs from bee-associated S. marcescens on Gilliamella spp. and S. alvi revealed only weak impact on specific Gilliamella spp. strains48. However, S. marcescens T6SSs potentially target Gram-positive bacteria74, which are abundant in the bee gut.
The roles of T6SSs in the bee gut are not fully defined. Their erratic presence and rapid evolution across strains within species are consistent with roles in ongoing antagonistic coevolution among competing community members. Potentially, this microbial warfare contributes to the stability of the community, which may in turn provide protection of hosts against invasive pathogens75.
Bacteriocins are small peptides that exhibit contact-independent antimicrobial properties, resulting in antagonism between bacterial strains or species and thus influencing the composition of gut microbial communities76. Although little is known about the roles of bacteriocins in bee gut microbial communities (FIG. 1c and d), bee-associated Lactobacillus spp. and Apilactobacillus kunkeei. strains possess genes encoding bacteriocins and respective immunity genes62,77. Different bacteriocins are found in Lactobacillus spp. strains associated with bumblebees (for example, lactococcin 972 homologues) versus honeybees (for example, helveticin J homologues)62 (FIG. 1d).
Bacteriophages can mediate antagonistic and beneficial interactions within bacterial communities, including gut communities78. The honeybee gut community includes phages that have coevolved with the core bacterial lineages79–81. At least some of these phages are likely highly specific: for example, matching CRISPR spacers are found across Gilliamella spp. genomes within recombining species clusters but not across distinct clusters, such that G. apis versus G. apicola do not share spacers57. The most abundant phages target major core members of the bee gut microbiota, such as Bifidobacterium, Gilliamella and Lactobacillus species, but also non-core members such as Bartonella species79–81 (FIG. 1d). These include both temperate and lytic phages representing undescribed families or genera within Siphoviridae, Myoviridae, and Podoviridae, as well as some Microviridae, Inoviridae and Caudovirales. The roles of phages within the bee gut community remain to be elucidated.
Functions in bee biology and health
Pathogen protection and immune system
Several experimental studies have demonstrated that the gut microbiota can protect honeybees against pathogens47, including opportunistic bacterial45,46,48,82 and fungal pathogens83–86, and potentially against RNA viruses87 (FIG. 2). These studies have used gnotobiotic honeybees with defined communities or bees with native microbiota disrupted by antibiotics or other agents to investigate effects of the overall community or of specific core members on susceptibility to subsequent pathogen challenge. Often the mechanisms of protection remain unidentified, but studies suggest that enhanced resistance can result from stimulation of host immune pathways45,46,82,88, competition for space and/or nutrients48, physical barrier protecting the gut wall from pathogen invasion48, or production of antimicrobial metabolites83.
Fig. 2. The roles of the honeybee gut microbiota in pathogen protection.
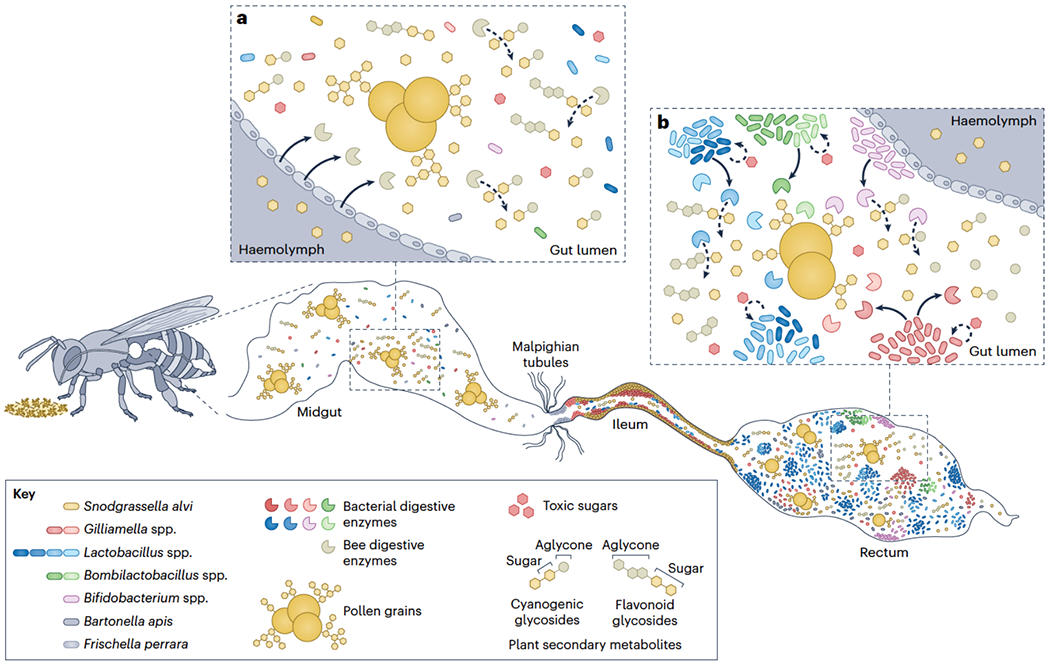
Members of the microbiota protect honeybees from prokaryotic and eukaryotic pathogens. Protection may arise from activation of bee innate immune pathway, as in the case of protection by Snodgrassella alvi against Serratia marcescens46 and potentially Escherichia coli45, and Lactobacillus spp. against Hafnia alvei88 and potentially trypanosomatids49. Protection can occur also from production of antimicrobial molecules (for example, Bombella apis protection against Aspergillus flavus83; Apilactobacillus kunkeei protection against Paenibacillus larvae and Melissococcus plutonius77,100), or from formation of a stable biofilm that forms a physical barrier on the gut wall48. An intact microbiota provides greater protection48. The top left shows a worker honeybee with a simplified image of the gut. The top right shows a piece of frame comb from a hive, in which cells have different contents, including larvae (brown), pollen (yellow), and nectar (orange). Solid arrows indicate activation of specific immunity pathways, solid lines indicate inhibition of specific pathogens, and dashed lines indicate potential inhibition of specific pathogens. Complexities of bee gut morphology are not depicted in this diagram.
The gut microbiota has a major impact on bee immunity. Colonization by the whole gut community or by single community members upregulates the expression of host immunity genes, such as those encoding antimicrobial peptides (AMPs)45,46,55,82,88 and the melanization cascade55. The honeybee innate immune system provides protection against opportunistic bacteria, fungi, and parasites89 and is broadly categorized into humoral and cellular immunity. Both are initiated by pattern recognition receptors that recognize molecules such as peptidoglycan and lipopolysaccharides from the bacterial outer membrane. Humoral immunity involves the production of AMPs, such as abaecin, apidaecin, defensin, and hymenoptaecin, that circulate in different body regions. Cellular immunity involves phagocytosis, nodulation, and encapsulation, often accompanied by melanization. The latter involves the activation of phenoloxidase, which results in the formation of melanin able to encapsulate and kill invading microorganisms89,90. The details of how the bee gut microbiota influences the immune system are still unclear.
Protection against bacterial pathogens
Compared to microbiota-deprived bees, bees colonized with a conventional microbiota, with single native gut bacterial strains, or with defined communities of several native bacterial strains show improved survivorship following exposure to opportunistic bacterial pathogens, including S. marcescens46,48, Hafnia alvei82,88, and potentially E. coli45 (FIG. 2).
S. marcescens and H. alvei are broad-range pathogens that cause sepsis in animals, including humans. Although not widely recognized as bee pathogens, they can kill workers following oral ingestion or wounding82,88,91–93. In contrast to larval pathogens, adult pathogens are not conspicuous in hives because sick workers abandon the hive to avoid spreading disease94,95. However, loss of adult workers can cause colonies to collapse. Oral exposure to S. marcescens causes high rates of mortality in microbiota-deprived bees and in microbiota-perturbed bees but not in bees with a conventional microbiota or with a defined community of core gut bacteria48,96. Partial protection is observed in bees monocolonized with single core bacterial strains48. A conventional microbiota also limits proliferation of H. alvei, potentially by stimulating the bee immune system82,88. Specific strains of L. apis induce expression of genes regulating the Toll pathway, causing increased production of host antimicrobial peptides, such as apidaecin, which strongly inhibits H. alvei in vitro88.
Paenibacillus larvae and Melissococcus plutonius are the causal agents of foulbrood diseases97,98, and are larval pathogens that can spread between hives in the guts of asymptomatic adult bees. Some non-core microbiota members found in larvae and at low abundances in the adult gut, such as Apilactobacillus kunkeei, may contribute protection against P. larvae and/or M. plutonius77,99,100 (FIG. 2).
Protection against eukaryotic pathogens
The first experimental demonstration of a beneficial effect of bee gut microbiota was microbiota-dependent protection against the trypanosomatid Crithidia bombi in Bombus terrestris101,102. Similar results were later found for Bombus impatiens103. C. bombi has been shown to infect bumble bees by using its flagellum to attach to the ileum wall104, raising the possibility that the biofilm formed in the ileum by S. alvi and Gilliamella spp. may serve as a protective physical barrier. In vitro studies with bumblebee- and honeybee-associated Lactobacillus species suggest the production of metabolites that inhibit Crithidia spp.49,105.
Bombella apis (previously called Parasaccharibacter apium), a bacterial symbiont associated with honeybee larvae, inhibits two fungal pathogens in vitro, Beauveria bassiana and Aspergillus flavus, and protects larvae against A. flavus83, but not against the bacterial pathogen M. plutonius106. Fungal protection is probably achieved through the production of specific antifungal metabolites (FIG. 2).
Whether the gut microbiota protects against members of the microsporidian Vairimorpha genus (formerly Nosema), the most common eukaryotic parasite of honeybees, remains unclear. In contrast to trypanosomatids, which infect hosts through the hindgut, Vairimorpha spp. invade the host through the wall of the midgut, potentially limiting the protection by the hindgut community. Monocolonization of honeybees with S. alvi strains has shown some reduction in V. ceranae spore loads84 and increased bee survival85. Disruption of the microbiota using antibiotics has been shown to increase V. ceranae spore loads86; conversely, V. ceranae infection itself can lead to microbial dysbiosis107.
Protection against viruses
RNA viruses are common and harmful pathogens of honeybees. There is limited experimental evidence suggesting that the core gut microbiota plays a role in viral tolerance. Studies have shown that microbiota-deprived bees had lower survival rates compared to conventionalized bees when exposed to Deformed Wing Virus87. However, viral titres were not affected in these bees87. Other studies have found correlations between viral infection and the composition or size of the gut microbiota108,109. Further investigation is needed to confirm, and if so, elucidate the mechanisms and dynamics underlying microbiome-mediated protection against viruses, whether the protection primarily arises from the microbiome’s role in enhancing bee health and immune system function, or from direct mechanisms within the microbiome that contribute to viral tolerance or resistance.
Role in development and behavior
Adult worker honeybees undergo distinct developmental changes after emergence from the pupal stage, and these are accompanied by weight gain and behavioural shifts, which are in turn linked to changes in expression of key developmental genes including those affecting juvenile hormone titres, insulin signalling, and vitellogenin1,3. Recent experimental studies suggest that the gut microbiota can influence these aspects of bee biology (FIG. 3).
Fig. 3. The roles of the honeybee gut microbiota in development and behaviour.
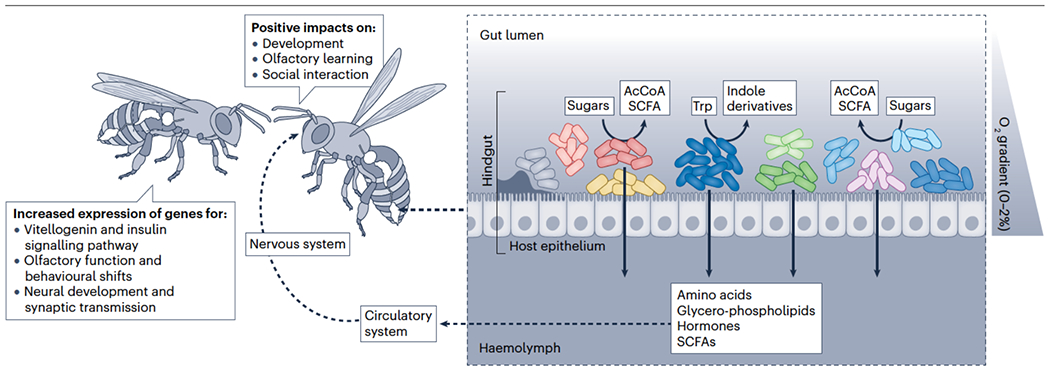
An intact microbiota is associated with increased expression of genes for vitellogenin and insulin signalling pathway, olfactory functions and behavioural shifts, neural development and synaptic transmission, and increased abundance of amino acids, glycerophospholipids, hormones and short-chain fatty acids in the gut, haemolymph and/or brain tissues50,53,64,113,115. These metabolites are associated with gut physiology, oxygen concentrations, pH, redox potential, and with regulation of developmental and behavioural genes, olfactory learning, and social interactions50,53,113,115. AcCoA, acetyl coenzyme A; SCFA, short-chain fatty acid. Curved solid arrows indicate microbial metabolism, straight solid arrows indicate host uptake of microbial-derived byproducts, and dashed arrows indicate movement of microbial-derived metabolites within the honeybee body. Colours of the bacterial taxa correspond to those in Figure 1.
In some studies comparing microbiota-deprived bees to conventionalized bees, the former exhibited reduced weight gain during early adulthood50,52 and abnormal guts characterized by elevated oxygen and pH levels. These gut changes are expected in the absence of oxygen depletion by S. alvi and short chain fatty acid production by Gilliamella spp. and other fermenters50. Microbiota-deprived bees also had suppressed expression of developmental genes, including vitellogenin and genes involved in the insulin pathway in head, abdomen, or whole bee body samples50,51, and changes in levels of other hormones, such as prostaglandins and juvenile hormone III derivatives in gut samples64.
While these effects are usually attributed to the complete microbiota, in some instances, they have been attributed to specific symbionts. For instance, monocolonization by Bifidobacterium asteroides elevates the gut concentration of juvenile hormone III derivatives64. Juvenile hormone III is a key regulator of insect growth, development, and reproduction. In honeybees, it governs the transition from nurse bees to forager bees1,110, a process influenced by nutrition111 and potentially impacted by gut bacterial metabolism. Juvenile hormone III derivatives can affect insect gut functioning112, but their roles in the bee gut are not known.
The gut microbiota also seems to influence bee behaviour. Proboscis extension response assays, which measure feeding reactions to gustatory or olfactory stimuli, have been used to study the roles of the gut microbiota on sucrose sensitivity, olfactory learning and memory abilities of honeybees. The full native gut microbiota, with its high strain diversity, appears to play a role in normal taste-related behaviour in honeybees. Conventionalized bees are more sensitive to lower doses of sucrose compared to microbiota-deprived bees50,113. However, this effect was not observed for a defined community of specific native strains70. On the other hand, honeybees colonized with a conventional or defined microbiota of native bacterial strains exhibit higher learning rates than microbiota-deprived or antibiotic-treated bees53,70.
In bumblebees, the gut microbiota seems to drive individual memory variation, with one study showing a positive correlation between the abundance of Lactobacillus apis and memory retention114. Bumblebees supplemented with a strain of L. apis displayed improved long-term memory retention, based on a visual discrimination foraging test114. This was accompanied by increased levels of glycerophospholipids in the haemolymph, which is associated with enhanced long-term memory114.
Some experimental evidence suggests potential routes by which the bee gut microbiota could affect brain function and thus behaviour. A direct connection between the bee gut and the nervous system could be mediated by the haemolymph metabolome, which is shaped in part by the gut microbiota50,53,115,116 (FIG. 3). Metabolomic analyses demonstrate distinct profiles in the gut50,64, the haemolymph and brain tissues53,115 of microbiota-colonized bees compared to microbiota-deprived bees. These profiles show consistent increased levels of amino acids and intermediates of amino acid metabolism. For example, specific members of the microbiota may impact the metabolism of tryptophan, an essential amino acid for honeybees, in the gut and haemolymph samples53. When supplemented with tryptophan, a specific strain of L. apis promoted memory in honeybees, possibly by transforming tryptophan to indole derivatives that activate the host aryl hydrocarbon receptor53. Additionally, the gut microbiota appears to influence carbohydrate and glycerophospholipid metabolisms in the haemolymph113.
Bee gut bacteria have been reported to alter brain neurotransmitter levels directly. The levels of biogenic amines with inhibitory effects on sensory sensitivity, such as dopamine and serotonin, are downregulated in bees monocolonized with Bombilactobacillus, Gilliamella, and Lactobacillus species113.
Brain transcriptomes of conventionalized bees and bees monocolonized with Bifidobacterium, Bombilactobacillus and Lactobacillus species exhibit elevated expression of genes related to olfactory functions (for example, odorant binding proteins and receptors) and/or genes affecting caste determination and age polyethism, which is the phenomenon whereby an animal shows different behaviour at different ages (for example, genes underlying the major royal jelly protein)53,113,115. Moreover, differentially spliced genes in the brains of bees monocolonized with Bombilactobacillus and Lactobacillus species are enriched for neural development and synaptic transmission pathways113.
The honeybee gut microbiota also appears to modulate the colony social network, influencing interactions between nestmates through changes in chromatin accessibility and amino acid biosynthesis115. Bees colonized with a conventional microbiota have increased head-to-head interactions among nestmates and exhibit greater specialization and stronger social ties with specific subsets of nestmates compared to microbiota-deprived bees115. These effects may be linked to higher levels of specific brain metabolites, such as serine and ornithine, which are known to be involved in synaptic transmission117 and correlated with the numbers of nestmate interactions115. These findings highlight a potential role of the gut microbiota in promoting and organizing social interactions within the honeybee colony115.
It is important to note that studies on microbiota effects on bee development and behaviour are challenged by the fact that larval development occurs under varying hive conditions, which have been shown to affect adult phenotypes118. Moreover, we note that only some of these results have been replicated, so their generality among bee genotypes, microbiota strains, and environmental conditions, is not yet certain.
Role in nutrition and detoxification
The microbiota primarily colonizes the bee hindgut (ileum and rectum) (FIG. 4). Readily accessible nutrients, such as sugars in nectar and amino acids in pollen germ cells, are processed and absorbed in the midgut, leaving primarily refractory components of the pollen coat to enter the ileum (FIG. 4a), along with the nitrogenous waste products of the Malpighian tubules (FIG. 1b). Genomic analyses have demonstrated that specific members of the bee gut microbiota have extensive capabilities for digestion of polysaccharides and for transport and metabolism of the released sugars. Genes for pectin lyases, glycoside hydrolases and sugar transport and utilization are found in bee-restricted Bifidobacterium spp., Bombilactobacillus spp., Gilliamella spp., and Lactobacillus spp., and presence of these genes varies among strains41,42,62 (FIG. 4b). Specific strains of these four bacterial genera can uptake and metabolize mannose, arabinose, xylose and rhamnose, sugars known to be toxic for bees if accumulated in the gut43,44. Genes underlying these capabilities were probably acquired from members of the phylum Bacillota (formerly called Firmicutes) through horizontal gene transfer43.
Fig. 4. The roles of the honeybee gut microbiota in digestion and detoxification.
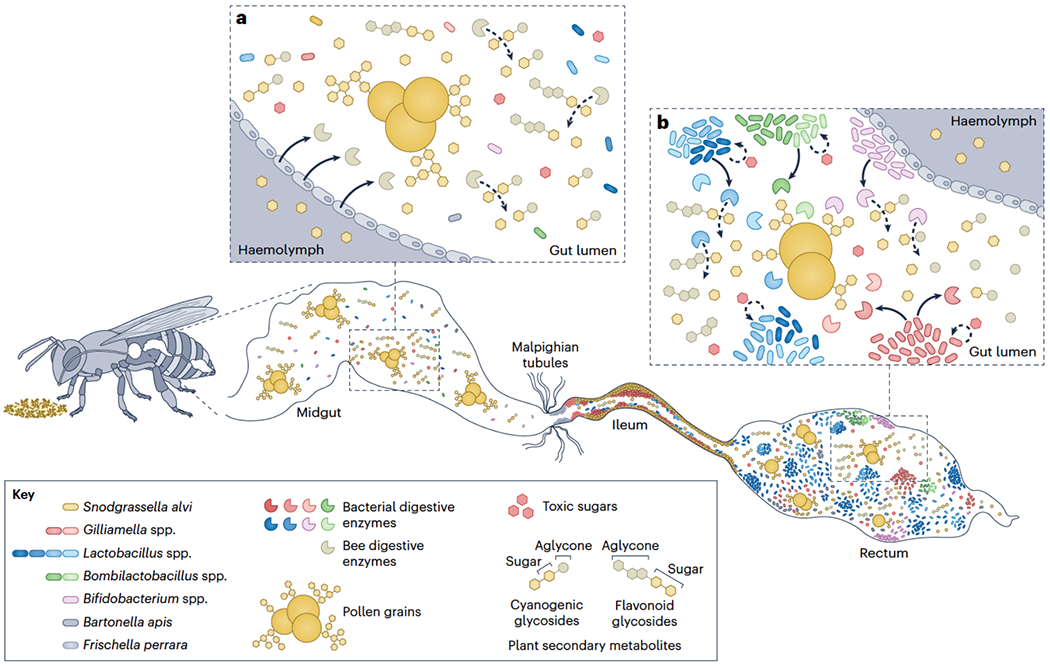
a) Easy-to-digest components from nectar (for example, some polysaccharides and simple sugars) and pollen (for example, amino acids, lipids, vitamins) are absorbed or metabolized by bee enzymes in the midgut. Metabolism of polysaccharides from the pollen coat releases several simple sugars that may be metabolized (for example, fructose and glucose) or not (for example, arabinose, galactose, mannose, and rhamnose) by bee enzymes. b) Hard-to-digest components, including refractory polysaccharides41, toxic sugars43,44 and plant secondary metabolites like flavonoid and cyanogenic glycosides61,63,65, are primarily metabolized by specific strains of major members of the native microbiota (for example, Gilliamella spp., Bifidobacterium spp., Bombilactobacillus spp., and Lactobacillus spp.) in the ileum and rectum, through the production of pectin lyases and glycoside hydrolases42,64. Solid arrows indicate production and release of digestive enzymes, and dashed arrows indicate microbial metabolism of toxic sugars and plant secondary metabolites.
The ability of the gut microbiota to metabolize plant polysaccharides and other dietary components has potential consequences for both bee nutrition and for detoxification. At least some of the released short chain fatty acids from bacterial metabolism are taken up by hosts, and these, especially butyrate, dominate in the bee haemolymph50. However, the extent to which bacterial digestion of pollen coats contributes to bee nutrition is currently unknown.
Protein is often limited in bee diets119, and the microbiota has potential for contributing to the bee nitrogen budget through the recycling of nitrogenous waste that enters from the Malpighian tubules at the midgut-ileum junction. S. alvi and G. apis have genes for urea utilization57, and several bee gut bacteria have complete pathways for amino acid biosynthesis42. Although uptake of amino acids in the hindgut is not documented in bees, absorption could occur through unknown mechanisms such as backflow into the midgut extraperitrophic space. In comparisons between microbiota-deprived bees and conventionalized bees, the latter usually exhibit increases in amino acids and/or amino acid derivatives in both the hindgut50,64 and the haemolymph50,53,115.
Bee gut bacteria have been shown to play a role in metabolizing recalcitrant plant secondary metabolites, including flavonoid glycosides64, cyanogenic glycosides63, and others65, primarily by deglycosylation of these metabolites (FIG. 4b). The consequences of the release of aglycones are understudied, but some studies point to activation or deactivation of these bioactive products. For example, the full metabolism of amygdalin, a cyanogenic glycoside found in almond-pollinated trees, is only possible in the presence of the microbiota63. While bee enzymes can metabolize amygdalin into an intermediate, prunasin, this intermediate accumulates in the bee gut only if the microbiota is absent. Microbial metabolism of prunasin leads to the release of hydrogen cyanide, a toxic chemical for aerobic organisms. Whether this full metabolism is toxic for bees deserves investigation, but it seems that naturally occurring amygdalin concentrations in nectar and pollen do not affect the microbiota and may even prevent parasite proliferation under hive conditions66.
Other nectar secondary metabolites, such as tiliaside from linden trees and unedone from strawberries, are known to be metabolized (glycosylated or deglycosylated) by bumblebees and their gut microbiota65. This metabolism leads to activation or inactivation of activity against C. bombi. For example, deglycosylation of tiliaside by host or microbial enzymes is required for activity during gut passage65. The aglycone unedone, on the other hand, has both in vitro and in vivo antiparasitic activity. Bee enzymes glycosylate and therefore inactive unedone in the midgut, and microbial enzymes deglycosylate and reactivate it in the hindgut65.
Potentially, the microbiota interacts with hosts to promote or limit processing of dietary components and other chemicals. For example, microbiota-deprived and antibiotic-treated bees show reduced expression of cytochrome P450 genes in the midgut and increased accumulation of pesticides in their bodies120. Overall, there is still limited evidence on how the bee gut microbiota play a role in detoxifying xenobiotics.
Impact of agricultural practices
Honeybees are often exposed to agrochemicals used in beekeeping (for example, antibiotics and acaricides) or in agriculture (for example, pesticides). Sometimes these chemicals, particularly insecticides, directly harm bees, while others may have sublethal impacts, including impacts mediated by disruption of the microbiota.
Beekeeping practices
Early studies on the effects of agrochemicals on the bee gut microbiota concerned antibiotic exposure121. Antibiotics are used in beekeeping for the prevention or treatment of larval infections, such as those causing foulbrood diseases. However, due to their broad spectrum of action, antibiotics can also impact the adult or larval gut microbiota. For instance, tetracycline, widely used in beekeeping in some countries since the 1950’s, has been shown to decrease the abundance of core gut bacteria, including S. alvi and species of Bifidobacterium, Lactobacillus and Bombilactobacillus (FIG. 5), and increase mortality rates within the hive environment and susceptibility to S. marcescens96. Other studies have corroborated the impacts of tetracycline on the adult bee microbiota122,123. These impacts can occur despite high levels of tetracycline resistance in some core bee gut bacteria121.
Fig. 5. Beekeeping and agricultural practices affecting honeybee gut communities.
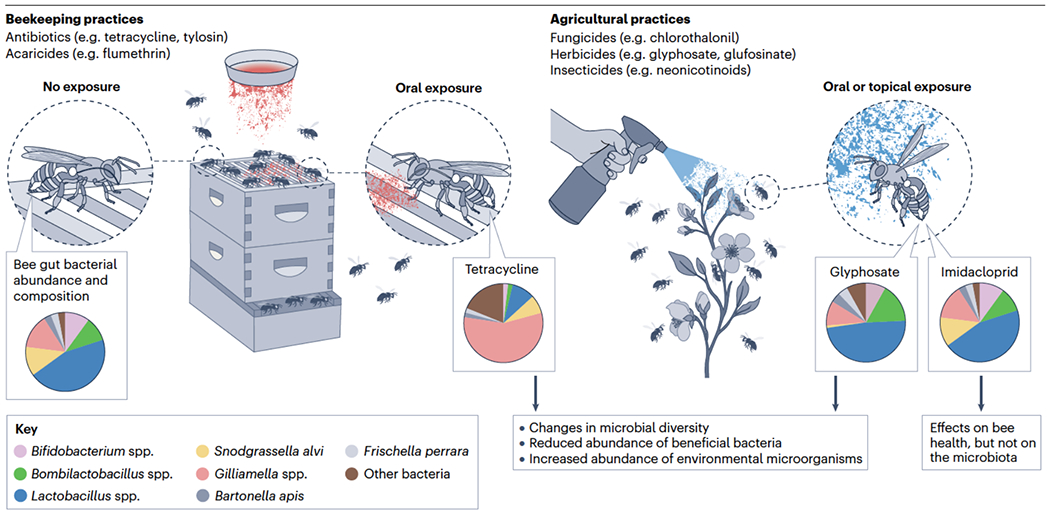
In beekeeping, the overuse of antibiotics and acaricides for the treatment of larval infections and mite infestation, respectively, can negatively impact the abundance and composition of beneficial bacteria in the adult worker bee microbiota, with consequences for bee health, such as increased susceptibility to infections and higher mortality rates125,130. Similarly, the indiscriminate use of fungicides, herbicides, and insecticides in agriculture, can negatively impact the adult worker bee microbiota, but effects are highly variable depending on the compounds involved and exposure level138,140,151. From left to right, pie charts illustrate the relative abundance of bee gut bacteria under normal conditions4, and under exposure to tetracycline96, glyphosate151 and imidacloprid140.
Tylosin is another antibiotic commonly used in beekeeping that has detrimental effects on the bee gut microbiota in both laboratory124 and hive conditions123,125,126, and increases susceptibility to S. marcescens in the laboratory125. Additionally, mixtures of penicillin-streptomycin lead to increased susceptibility to V. ceranae and downregulate the expression of host antimicrobial peptides, such as abaecin, defensin-1, and hymenoptaecin, under laboratory conditions86.
Not surprisingly, antibiotic treatment reduces bacterial loads in the larval gut, and impacts nutrient metabolism, body weight gain, development, and immune competence of larvae127. The expression of host antimicrobial peptides abaecin, apidaecin, defensin-1 and hymenoptaecin, for example, are reduced at specific stages of larval development upon exposure to penicillin-streptomycin127.
Antibiotics potentially have direct negative effects on bees126,128, which can be difficult to distinguish from those arising due to impacts on the microbiota. However, in a control experiment on microbiota-deprived bees in the lab, tetracycline had no negative impact on bee survival96. Also, the increased susceptibility to S. marcescens caused by antibiotic exposure echoes that of microbiota-deprived bees48, consistent with a role of microbiota perturbation.
Acaricides, such as flumethrin, are commonly used in beekeeping for the treatment and/or prevention of infestation by mites, primarily Varroa destructor. V. destructor attaches to the bee exoskeleton and feeds on fat bodies and haemolymph, thereby spreading viruses, such as Deformed Wing Virus, which also impact bee health129. Flumethrin exposure leads to overexpression of immune- and detoxification-related genes, and decreases microbial abundance and diversity in the larval gut130, but it seems to have limited effects on the adult gut microbiota composition131. Other acaricides used to control V. destructor, such as coumaphos and tau-fluvalinate, can affect microbial diversity associated with adult honeybees132,133. Potential safer alternatives for combating V. destructor include the use of menthol, thymol, and oxalic acid134, found naturally in honey and plants. Oxalic acid, however, has been shown to inhibit growth of specific Lactobacillus species in vitro134, and impact microbial composition, including reduction in strain richness, in adult honeybees135.
Agrochemicals encountered by foragers
In addition to exposure to chemicals used in beekeeping, foragers can be exposed to agrochemicals, such as insecticides, herbicides, and fungicides used on crops (FIG. 5). Foragers deliver these back to hives, where they can accumulate in food stores, thus exposing larvae and young bees.
Insecticides affect bees primarily through direct toxicity, but sublethal effects including gut microbial perturbations have also been detected. Neonicotinoids are widely used broad-spectrum neurotoxic insecticides136 that are less toxic to mammals than are long-standing insecticides such as carbamates, organophosphates, and pyrethroids137. Neonicotinoids, such as acetamiprid, sulfoxaflor and thiacloprid, can affect microbial diversity in the bee gut, though these effects may reflect other impacts on bee physiology, as exposure reduces survivorship and appetite135,138,139. Other experimental studies on both honeybees and bumblebees found no impacts of imidacloprid on the gut microbiota and little or no ability of the microbiota to metabolize imidacloprid140,141 (FIG. 5).
Some herbicides have antimicrobial properties and can indirectly affect bees through effects on the microbiota. Glyphosate, the most used herbicide globally, inhibits an enzyme in the shikimate pathway (5-enolpyruvylshikimate-3-phosphate synthase) that is required for the production of essential amino acids in plants and most microorganisms. Experimental studies have shown that some core bee gut bacteria are susceptible to glyphosate. The gut microbiota species most consistently impacted by glyphosate exposure is S. alvi, with this effect being dose-dependent124,142–145 (FIG. 5). Impacts on gut microbiota composition are observed when honeybees are exposed to glyphosate concentrations documented in nectar and pollen of recently exposed plants146, and can also occur in bumblebees, in which effects appear milder and less persistent147–149.
Glyphosate exposure associated with microbiota disruption can impact the expression of host antimicrobial peptides, including apidaecin, defensin and hymenoptaecin150 and lead to increased susceptibility to S. marcescens143,151, but not to V. ceranae142,144. Exposure also promotes Deformed Wing Virus replication and decreases vitellogenin expression144. Additionally, glyphosate exposure impacts bee physiology, including antioxidant and detoxification systems, learning and memory, and behaviour, which has been extensively reviewed152. The extent to which these effects are direct or mediated by the gut microbiota is unknown.
Fungicides also can affect the honeybee gut microbiota. Chlorothalonil, a non-systemic organochlorine fungicide and one of the most used fungicides in agriculture, can perturb gut bacterial communities of adult bees132 and increase susceptibility to V. ceranae infection153. Chronic exposure to field-realistic concentrations of azoxystrobin, a broad-spectrum fungicide commonly used in agriculture, impacts both fungal and bacterial communities in the honeybee gut, and can result in an increase in the relative abundance of Serratia154.
Antibiotics are not only used in beekeeping but also in agriculture to control bacterial pathogens in plant crops, and bees can be exposed to antibiotics during foraging activities. A study comparing antibiotic resistance genes in gut bacteria of honeybees from the United States and Norway revealed a high incidence of streptomycin resistance in the U.S. samples, where streptomycin is sprayed on fruit trees to protect against fire blight (Erwinia amylovora) but not in samples from Norway, where streptomycin is not used155.
Nutritional impacts on microbiota
Diet is widely documented to affect the composition of animal gut microbiota. Honeybees experience extensive variation in diet due to varying availability of flowering plant species or to artificial dietary supplementation by beekeepers. Several studies show that these dietary variables can affect the honeybee gut microbiota and increase susceptibility to pathogens. For example, sucrose supplementation appears to lower abundance of core gut species in relation to potentially pathogenic Serrafia spp. in bee guts156. Bees fed nutritionally poor-quality pollen exhibit lower abundances of Bombilactobacillus, Lactobacillus and Bifidobacterium species, and higher abundance of the non-core species Bartonella apis, than do bees fed polyfloral pollen157. Feeding on poor-quality pollen also results in lower expression of vitellogenin and immunity genes, and increased proliferation of Vairimorpha157.
Beekeepers often provide hives with protein supplements containing products, such as soy protein or casein, absent from the natural bee diet. In a recent study, young adult bees with conventional microbiota were given either dietary supplements or pollen for 14 days, then sampled to examine microbiota size and composition as well as expression of genes involved in development and immunity158. In bees given the artificial diets, gut communities were larger in absolute numbers of bacteria, but showed lower diversity of sequence variants, lower evenness, and higher incidence of bacteria atypical for bee guts, such as Streptococcus spp. and Staphylococcus spp. The artificial diet also resulted in lower expression of juvenile hormone esterase and vitellogenin and in higher susceptibility to the pathogen S. marcescens. These results were largely consistent across hives at two locations.
Potential for probiotics in bees
The use of probiotics aimed at treating or preventing microbial infections in hives is common in beekeeping. Recent reviews have summarized the studies in the bee probiotics field51,159. Most commercially available bee probiotics consist of non-native microorganisms, including bacteria and fungi from the food industry, which are marketed as promoting bee health, though they do not stably colonize bees51,160. An alternative approach involves probiotics consisting of native microorganisms that colonize and persist in the bee gut51. Orally delivered gut homogenates are one way to transfer bacteria from healthy worker bees to bees lacking microbiota or with perturbed microbiota. Gut homogenate treatments lead to stable colonization in young bees under laboratory conditions, but potentially introduce pathogens from donor bees. Defined communities of isolates of native core bacteria are another approach48,70,82,125. Such defined communities can counteract perturbations caused by agrochemicals and other environmental stressors and prevent the proliferation of opportunistic pathogens that often follow perturbation48,82,125. However, these studies have been primarily conducted in laboratory settings, and further hive-level studies are necessary to evaluate the efficacy of probiotics for beekeeping.
Probiotic approaches may be effective ways to prevent or treat P. larvae in hives. In a study of two control hives and two hives treated with a bacterial consortium consisting of A. kunkeei, Lactiplantibacillus plantarum and Lacticaseibacillus rhamnosus, the treated hives seemed to have lower levels of P. larvae and less immune dysregulation161. However, treating P. larvae-infected hives with a probiotic mixture of hive- and gut-associated Lactobacillus spp. and Bifidobacterium spp. strains, with or without antibiotic treatment, did not improve colony fitness162,163. These varying outcomes among studies may reflect differences in experimental design, execution, or the condition of the study hives. Delivery methods may also influence the impact of probiotics on hive fitness164.
Engineered bee gut strains offer an alternative strategy to improve bee health165,166. Snodgrassella alvi was engineered to express double-stranded RNA targeting Deformed Wing Virus and V. destructor through the bee or mite RNA interference pathways167. In bees colonized with the engineered S. alvi strain in the laboratory, viral proliferation was suppressed, and mites suffered elevated mortality167. Similarly, S. alvi was engineered to express double-stranded RNA targeting the microsporidian parasite V. ceranae, with different essential genes selected in two independent studies84,85. In both studies, bees monocolonized with the engineered S. alvi strain in the laboratory had reduced V. ceranae. spore loads.
Although some results are promising, the potential for using probiotics in honeybees is still unclear, particularly under field conditions.
Summary and future directions
Studies to date support a substantial influence of the honeybee gut microbiota on host digestion, detoxification, behaviour, pathogen protection, and immune system. Bees deprived of their normal microbiota, and bees in which the microbiota is disrupted by chemicals, show a range of health deficits including changes in feeding behaviour, greater susceptibility to pathogens, and higher mortality in the hive itself. Experimental colonization of gnotobiotic hosts with single or multiple microbiota members can restore at least some benefits of the full bee microbiota.
Although considerable evidence points to benefits of gut symbionts for bees, the molecular mechanisms behind these effects are largely unknown. For example, specific members of the gut microbiota have been shown to prevent pathogen proliferation and to protect hosts from pathogen-induced mortality, but it is unknown whether protection results from host immune responses and/or direct interactions between microorganisms. Final effectors of immunity pathways (for example, antimicrobial peptides) are upregulated in specific bee body tissues, but the identities of the microbial effectors that trigger these pathways are unknown. Biofilm formation, as observed for S. alvi in the ileum, appears to be a critical component of successful colonization69, but the triggers for biofilm formation and whether and how biofilm blocks pathogens have not been determined. Genomic analyses have shown the potential abilities of the core bacteria to interact with each other by contact-dependent (for example, T6SS) or contact-independent (for example, bacteriocins) ways. Future studies should focus on elucidating these molecular mechanisms.
Studies of the roles of the bee microbiota in toxin metabolism are still incipient. Although some studies have investigated how xenobiotics, including agrochemicals and specific plant secondary metabolites, are metabolized, the consequences of such metabolism for bee health are largely unknown. The impacts of agrochemicals on gut microbial communities may stem from bee mechanisms (for example, cytochrome P450s) or from metabolic capabilities (for example, hydrolases) of specific gut symbionts.
Another recent research direction involves the gut-brain axis. Honeybees have long been used as models for studying behaviour, ranging from cognition to social interactions, and behavioural assays are well developed. Recent studies have taken advantage of these behavioural assays and gnotobiotic bees to explore the roles of the microbiota in taste, olfactory learning, and colony social network, and in shaping transcriptomic and metabolomic profiles in different compartments of the bee body. Results suggest that members of the native microbiota act together to shape bee behaviour. Linking effects of the microbiota on behaviours to changes in gene expression and metabolites in the haemolymph and brain tissues115, is a promising next step to fill the causation gap in this emerging field.
For both fundamental and applied research goals, one challenge is the variability among the bees themselves. While an advantage of studying honeybees is their global distribution and the opportunity to study them under natural hive conditions, these same factors introduce complications. A. mellifera varies genetically, with different breeds or subspecies in different regions. It also varies according to environmental conditions, such as nectar and pollen sources and quantities, season, climate and weather, and exposure to environmental toxins and pathogens. Hives from the same apiary often differ in genetics and physiological condition. For example, nutrient scarcity during larval development, which occurs when floral resources are limited, can have major consequences for the metabolism, behaviour and development of the resulting adult workers118,168. Researchers use honeybee colonies typical for their geographic area and perform experiments during different seasons. In the future, it will be important to replicate results for bees from different genetic and environmental backgrounds to understand how these variables affect the roles of the gut microbiota.
Honeybees are exposed to environmental stressors encountered in hives and their surroundings. These stressors, including anthropogenic chemicals and long-distance transport, often impact the gut microbiota. However, most studies to date have limitations. Usually, they examine only relative abundances of gut community members, whereas measures of absolute abundances, using quantitative PCR or other approaches, are needed for robust interpretations. Moreover, agrochemicals are usually deployed along with co-formulants, but these are rarely investigated though they sometimes exert stronger impacts than the active ingredient. For example, pure glyphosate does not lead to increased susceptibility to Vairimorpha, but a glyphosate-based formulation does169. Future studies aimed at evaluating impacts should consider both active chemicals and co-formulants. Another question rarely examined in these studies is community resilience, that is, how long detrimental impacts persist after perturbation. Moreover, identifying impacts on gut microbiota is not meaningful without examining whether these extend to effects on bee health.
Most research on the bee microbiota has focused on the honeybee, A. mellifera, with a more limited number of studies on the commercially available bumblebees, B. impatiens and B. terrestris. Little is known about factors affecting the microbiota of other bee species, many of which are declining in numbers. More research on microbiomes of a diversity of bee species will undoubtedly lead to new discoveries and potentially contribute to the conservation of wild pollinators.
Acknowledgements
This work was supported by the US National Institutes of Health (award R35GM131738) and the USDA National Institute of Food and Agriculture (award 2018-67013-27540) to N.A.M.
Footnotes
Competing interests
N.A.M. is an author on a patent application (US20220152128A1) for using native bee gut bacteria as bee probiotics. E.V.S.M. declares no competing interests.
References
- 1.Robinson GE, Page RE Jr, Strambi C & Strambi A Hormonal and genetic control of behavioral integration in honey bee colonies. Science 246, 109–112 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Menzel R. The honeybee as a model for understanding the basis of cognition. Nat. Rev. Neurosci 13, 758–768 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Zayed A & Robinson GE Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu. Rev. Genet 46, 591–615 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Kwong WK & Moran NA Gut microbial communities of social bees. Nat. Rev. Microbiol 14, 374–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero S, Nastasa A, Chapman A, Kwong WK & Foster LJ The honey bee gut microbiota: strategies for study and characterization. Insect Mol. Biol 28, 455–472 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Cox-Foster DL et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Martinson VG et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol 20, 619–628 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Moran NA, Hansen AK, Powell JE & Sabree ZL Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7, e36393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinson VG, Moy J & Moran NA Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol 78, 2830–2840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powell JE, Martinson VG, Urban-Mead K & Moran NA Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol 80, 7378–7387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J. et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol 70, 2782–2858 (2020). [DOI] [PubMed] [Google Scholar]
- 12.D’Alvise P. et al. The impact of winter feed type on intestinal microbiota and parasites in honey bees. Apidologie 49, 252–264 (2018). [Google Scholar]
- 13.Corby-Harris V, Maes P & Anderson KE The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS One 9, e95056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothman JA, Carroll MJ, Meikle WG, Anderson KE & McFrederick QS Longitudinal effects of supplemental forage on the honey bee (Apis mellifera) microbiota and inter- and intra-colony variability. Microb. Ecol 76, 814–824 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Kapheim KM et al. Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera). PLoS One 10, e0123911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J. et al. Honey bee genetics shape the strain-level structure of gut microbiota in social transmission. Microbiome 9, 225 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kešnerová L. et al. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 14, 801–814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones JC et al. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol 8, 441–451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olofsson TC, Alsterfjord M, Nilson B, Butler È & Vásquez A Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol 64, 3109–3119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel P, Kwong WK & Moran NA Frischella perrara gen. nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. Int. J. Syst. Evol. Microbiol 63, 3646–3651 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Kešnerová L, Moritz R & Engel P Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol 66, 414–421 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ludvigsen J, Porcellato D, Amdam GV & Rudi K Addressing the diversity of the honeybee gut symbiont Gilliamella: description of Gilliamella apis sp. nov., isolated from the gut of honeybees (Apis mellifera). Int. J. Syst. Evol. Microbiol 68, 1762–1770 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Milani C. et al. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol 80, 6290–6302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong WK & Moran NA Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol 63, 2008–2018 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Ellegaard KM & Engel P Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun 10, 446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Shotgun metagenomics has allowed the assessment of the honeybee gut microbiota diversity, showing that previously described phylotypes contain sequence-discrete populations or species, which tend to co-exist in individual bees and show age-specific abundance profiles.
- 26.Parish AJ, Rice DW, Tanquary VM, Tennessen JM & Newton ILG Honey bee symbiont buffers larvae against nutritional stress and supplements lysine. ISME J. 16, 2160–2168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell JE, Eiri D, Moran NA & Rangel J Modulation of the honey bee queen microbiota: Effects of early social contact. PLoS One 13, e0200527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callegari M. et al. Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. npj Biofilms Microbiomes 7, 42 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gómez-Moracho T. et al. Experimental evidence of harmful effects of Crithidia mellificae and Lotmaria passim on honey bees. Int. J. Parasitol 50, 1117–1124 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Tokarev YS et al. A formal redefinition of the genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and reassignment of species based on molecular phylogenetics. J. Invertebr. Pathol 169, 107279 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Grupe AC 2nd & Quandt CA A growing pandemic: A review of Nosema parasites in globally distributed domesticated and native bees. PLoS Pathog. 16, e1008580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decker LE et al. Higher variability in fungi compared to bacteria in the foraging honey bee gut. Microb. Ecol 85, 330–334 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Kwong WK et al. Dynamic microbiome evolution in social bees. Sci Adv 3, e1600513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; The distribution and phylogenies of the core bacterial lineages in the microbiota of social bees suggests that these lineages have coevolved with bee hosts since the origin of the Corbiculata clade, about 80 million years ago.
- 34.Ellegaard KM, Suenami S, Miyazaki R & Engel P Vast differences in strain-level diversity in the gut microbiota of two closely related honey bee species. Curr. Biol 30, 2520–2531.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammer TJ, Le E, Martin AN & Moran NA The gut microbiota of bumblebees. Insectes Soc. 68, 287–301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarton-Lohéac G et al. Deep divergence and genomic diversification of gut symbionts of neotropical stingless bees. MBio e0353822 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerqueira AES et al. Extinction of anciently associated gut bacterial symbionts in a clade of stingless bees. ISME J. 15, 2813–2816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figueroa LL, Maccaro JJ, Krichilsky E, Yanega D & McFrederick QS Why did the bee eat the chicken? Symbiont gain, loss, and retention in the vulture bee microbiome. MBio 12, e0231721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kueneman JG, Bonadies E, Thomas D, Roubik DW & Wcislo WT Neotropical bee microbiomes point to a fragmented social core and strong species-level effects. Microbiome 11, 150 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holley J-AC, Jackson MN, Pham AT, Hatcher SC & Moran NA Carpenter bees (Xylocopa) harbor a distinctive gut microbiome related to that of honey bees and bumble bees. Appl. Environ. Microbiol 88, e0020322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engel P, Martinson VG & Moran NA Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U. S. A 109, 11002–11007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H et al. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. U. S. A 116, 25909–25916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Honeybee gut symbionts, including Bifidobacterium, Gilliamella and Lactobacillus strains, encode specific pectate lyases and glycoside hydrolases that are highly variable among strains and are involved in the degradation of recalcitrant components within pollen husks.
- 43.Zheng H et al. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. MBio 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee FJ, Miller KI, McKinlay JB & Newton ILG Differential carbohydrate utilization and organic acid production by honey bee symbionts. FEMS Microbiol. Ecol 94, (2018). [DOI] [PubMed] [Google Scholar]
- 45.Kwong WK, Mancenido AL & Moran NA Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci 4, 170003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horak RD, Leonard SP & Moran NA Symbionts shape host innate immunity in honeybees. Proc. Biol. Sci 287, 20201184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; The honeybee symbiont Snodgrassella alvi activates the expression of host antimicrobial peptides, such as apidaecin, defensin and hymenoptaecin, but live S. alvi cells appear to suppress some parts of innate immune signalling.
- 47.Raymann K & Moran NA The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci 26, 97–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steele MI, Motta EVS, Gattu T, Martinez D & Moran NA The gut microbiota protects bees from invasion by a bacterial pathogen. Microbiol. Spectr 9, e0039421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer-Young EC, Markowitz LM, Huang W-F & Evans JD High temperatures augment inhibition of parasites by a honey bee gut symbiont. Appl. Environ. Microbiol 89, e01023–23 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng H, Powell JE, Steele MI, Dietrich C & Moran NA Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U. S. A 114, 4775–4780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motta EVS, Powell JE, Leonard SP & Moran NA Prospects for probiotics in social bees. Philos. Trans. R. Soc. Lond. B Biol. Sci 377, 20210156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper summarizes recent research on probiotics for bees and presents new evidence that the use of defined native gut bacteria is a promising way to restore perturbed microbial communities in bees exposed to agrochemicals.
- 52.Wang X et al. High-fat diets with differential fatty acids induce obesity and perturb gut microbiota in honey bee. Int. J. Mol. Sci 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z et al. Honeybee gut Lactobacillus modulates host learning and memory behaviors via regulating tryptophan metabolism. Nat. Commun 13, 2037 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engel P et al. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. MBio 7, e02164–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emery O, Schmidt K & Engel P Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol 26, 2576–2590 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Engel P, Bartlett KD & Moran NA The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. MBio 6, e00193–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Leonard SP, Powell JE & Moran NA Species divergence in gut-restricted bacteria of social bees. Proc. Natl. Acad. Sci. U. S. A 119, e2115013119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used genome sequences from isolates of Snodgrassella alvi and GiHiamella spp. to identify ‘populations’ defined by evidence for homologous exchange and therefore representing biological species.
- 58.Bobay L-M, Wissel EF & Raymann K Strain structure and dynamics revealed by targeted deep sequencing of the honey bee gut microbiome. mSphere 5, e11694–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raymann K, Bobay L-M & Moran NA Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol 27, 2057–2066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steele MI, Kwong WK, Whiteley M & Moran NA Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. MBio 8, e01630–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brochet S et al. Niche partitioning facilitates coexistence of closely related honey bee gut bacteria. Elife 10, e68583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellegaard KM et al. Genomic changes underlying host specialization in the bee gut symbiont Lactobacillus Firm5. Mol. Ecol 28, 22.24–2231 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Motta EVS et al. Host-microbiome metabolism of a plant toxin in bees. Elife 11, e82595 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kešnerová L et al. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol 15, e2003467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study combined metabolomics and gnotobiotic bees to investigate the metabolic contributions of individual members of the bee gut microbiota.
- 65.Koch H et al. Host and gut microbiome modulate the antiparasitic activity of nectar metabolites in a bumblebee pollinator. Philos. Trans. R. Soc. Lond. B Biol. Sci 377, 20210162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study disentangled the contributions of host and gut microbial enzymes to the metabolism of some plant secondary metabolites.
- 66.Tauber JP et al. Colony-level effects of amygdalin on honeybees and their microbes. Insects 11,783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwong WK, Engel P, Koch H & Moran NA Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. U. S. A 111, 11509–11514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinn A et al. Foraging on host synthesized metabolites enables the bacterial symbiont Snodgrassella alvi to colonize the honey bee gut. bioRxiv 2023.01.23.524906 (2023) doi: 10.1101/2023.01.23.524906. [DOI] [Google Scholar]
- 69.Powell JE, Leonard SP, Kwong WK, Engel P & Moran NA Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. U. S. A 113, 13887–13892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cabirol A et al. A defined community of core gut microbiota members promotes cognitive performance in honey bees. bioRxiv 2023.01.03.522593 (2023) doi: 10.1101/2023.01.03.522593. [DOI] [Google Scholar]
- 71.Steele MI & Moran NA Evolution of interbacterial antagonism in bee gut microbiota reflects host and symbiont diversification. mSystems 6, e00063–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen C, Yang X & Shen X Confirmed and potential roles of bacterial T6SSs in the intestinal ecosystem. Front. Microbiol 10, 1484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt K et al. Integration host factor regulates colonization factors in the bee gut symbiont Frischella perrara. Elife 12, e76182 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le N-H, Pinedo V, Lopez J, Cava F & Feldman MF Killing of Gram-negative and Gram-positive bacteria by a bifunctional cell wall-targeting T6SS effector. Proc. Natl. Acad. Sci. U. S. A 118, e2106555118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coyte KZ, Schluter J & Foster KR The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Heilbronner S, Krismer B, Brötz-Oesterhelt H & Peschel A The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol 19, 726–739 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Zendo T et al. Kunkecin A, a new nisin variant bacteriocin produced by the fructophilic lactic acid bacterium, Apilactobacillus kunkeei FF30-6 isolated from honey bees. Front. Microbiol 11, 571903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shkoporov AN, Turkington CJ & Hill C Mutualistic interplay between bacteriophages and bacteria in the human gut. Nat. Rev. Microbiol 20, 737–749 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Bonilla-Rosso G, Steiner T, Wichmann F, Bexkens E & Engel P Honey bees harbor a diverse gut virome engaging in nested strain-level interactions with the microbiota. Proc. Natl. Acad. Sci. U. S. A 117, 7355–7362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that honeybee guts harbour bacteriophage communities that have coevolved with core bacterial symbionts and may play critical roles in mediating antagonistic and beneficial interactions within the bee gut microbiota.
- 80.Busby TJ, Miller CR, Moran NA & Van Leuven JT Global composition of the bacteriophage community in honey bees. mSystems 7, e0119521 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deboutte W. et al. Honey-bee–associated prokaryotic viral communities reveal wide viral diversity and a profound metabolic coding potential. Proc. Natl. Acad. Sci. U. S. A 117, 10511–10519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J. et al. Stably transmitted defined microbial community in honeybees preserves Hafnia alvei inhibition by regulating the immune system. Front. Microbiol 13, 1074153 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller DL, Smith EA & Newton ILG A bacterial symbiont protects honey bees from fungal disease. MBio 12, e0050321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that the symbiont Bombella apis protects honeybee larvae against the fungal pathogen Aspergillus flavus by producing specific antifungal molecules.
- 84.Lang H. et al. Engineered symbiotic bacteria interfering Nosema redox system inhibit microsporidia parasitism in honeybees. Nat. Commun 14, 2778 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang Q, Lariviere PJ, Powell JE & Moran NA Engineered gut symbiont inhibits microsporidian parasite and improves honey bee survival. Proc. Natl. Acad. Sci. U. S. A 120, e2220922120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li JH et al. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS One 12, e0187505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dosch C. et al. The gut microbiota can provide viral tolerance in the honey bee. Microorganisms 9, 871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lang H. et al. Specific strains of honeybee gut Lactobacillus stimulate host immune system to protect against pathogenic Hafnia alvei. Microbiol Spectr 10, e0189621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Evans JD et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol 15, 645–656 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nappi AJ & Christensen BM Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem. Mol. Biol 35, 443–459 (2005). [DOI] [PubMed] [Google Scholar]
- 91.Raymann K, Coon KL, Shaffer Z, Salisbury S & Moran NA Pathogenicity of Serratia marcescens strains in honey bees. MBio 9, e01649–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burritt NL et al. Sepsis and hemocyte loss in honey bees (Apis mellifera) infected with Serratia marcescens strain Sicaria. PLoS One 11, e0167752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.El Sanousi SM, El Sarag MSA & Mohamed SE Properties of Serratia marcescens isolated from diseased honeybee (Apis mellifera) larvae. Microbiology 133, 215–219 (1987). [Google Scholar]
- 94.Evans JD & Spivak M Socialized medicine: individual and communal disease barriers in honey bees. J. Invertebr. Pathol 103 Suppl 1, S62–72 (2010). [DOI] [PubMed] [Google Scholar]
- 95.Baracchi D, Fadda A & Turillazzi S Evidence for antiseptic behaviour towards sick adult bees in honey bee colonies. J. Insect Physiol 58, 1589–1596 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Raymann K, Shaffer Z & Moran NA Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15, e2001861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Forsgren E. European foulbrood in honey bees. J. Invertebr. Pathol 103 Suppl 1, S5–9 (2010). [DOI] [PubMed] [Google Scholar]
- 98.Genersch E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol 103 Suppl 1, S10–9 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Kačániová M, Gasper J & Terentjeva M Antagonistic effect of gut microbiota of honeybee (Apis mellifera) against causative agent of American foulbrood Paenibacillus larvae. J. Microbiol. Biotechnol. Food Sci 9, 478–481 (2019). [Google Scholar]
- 100.Al-Ghamdi A. et al. In vitro antagonistic potential of gut bacteria isolated from indigenous honey bee race of Saudi Arabia against Paenibacillus larvae. J. Apic. Res 59, 825–833 (2020). [Google Scholar]
- 101.Koch H & Schmid-Hempel P Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U. S. A 108, 19288–19292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koch H & Schmid-Hempel P Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett 15, 1095–1103 (2012). [DOI] [PubMed] [Google Scholar]
- 103.Mockler BK, Kwong WK, Moran NA & Koch H Microbiome structure influences infection by the parasite Crithidia bombi in bumble bees. Appl. Environ. Microbiol 84, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koch H, Woodward J, Langat MK, Brown MJF & Stevenson PC Flagellum removal by a nectar metabolite inhibits infectivity of a bumblebee parasite. Curr. Biol 29, 3494–3500.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Palmer-Young EC, Raffel TR & McFrederick QS pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology 146, 380–388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Floyd AS et al. Microbial ecology of European foul brood disease in the honey bee (Apis mellifera): Towards a microbiome understanding of disease susceptibility. Insects 11, 555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rubanov A, Russell KA, Rothman JA, Nieh JC & McFrederick QS Intensity of Nosema ceranae infection is associated with specific honey bee gut bacteria and weakly associated with gut microbiome structure. Sci. Rep 9, 3820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo J. et al. Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis cerana. J. Invertebr. Pathol 127, 110–114 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Kim C. et al. Analysis of the gut microbiome of susceptible and resistant honeybees (Apis cerana) against sacbrood virus disease. J. Appl. Entomol 146, 1078–1086 (2022). [Google Scholar]
- 110.Sullivan JP, Fahrbach SE & Robinson GE Juvenile hormone paces behavioral development in the adult worker honey bee. Horm. Behav 37, 1–14 (2000). [DOI] [PubMed] [Google Scholar]
- 111.Toth AL, Kantarovich S, Meisel AF & Robinson GE Nutritional status influences socially regulated foraging ontogeny in honey bees. J. Exp. Biol 208, 4641–4649 (2005). [DOI] [PubMed] [Google Scholar]
- 112.Bajgar A, Jindra M & Dolezel D Autonomous regulation of the insect gut by circadian genes acting downstream of juvenile hormone signaling. Proc. Natl. Acad. Sci. U. S. A 110, 4416–4421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Z, Mu X, Shi Y & Zheng H Distinct roles of honeybee gut bacteria on host metabolism and neurological processes. Microbiol Spectr 10, e0243821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li L. et al. Gut microbiome drives individual memory variation in bumblebees. Nat. Commun 12, 6588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liberti J. et al. The gut microbiota affects the social network of honeybees. Nat Ecol Evol 6, 1471–1479 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that microbiota-colonized bees, when compared to microbiota-deprived bees, exhibit increased levels of brain metabolites, such as serine and ornithine, and increased specialized head-to-head interactions between nestmates.
- 116.Cabirol A, Moriano-Gutierrez S & Engel P Neuroactive metabolites modulated by the gut microbiota in honey bees. Mol. Microbiol (2023) doi: 10.1111/mmi.15167. [DOI] [PubMed] [Google Scholar]
- 117.Billard J-M D-Amino acids in brain neurotransmission and synaptic plasticity. Amino Acids 43, 1851–1860 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Scofield HN & Mattila HR Honey bee workers that are pollen stressed as larvae become poor foragers and waggle dancers as adults. PLoS One 10, e0121731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brodschneider R & Crailsheim K Nutrition and health in honey bees. Apidologie 41, 278–294 (2010). [Google Scholar]
- 120.Wu Y. et al. Honey bee (Apis mellifera) gut microbiota promotes host endogenous detoxification capability via regulation of P450 gene expression in the digestive tract. Microb. Biotechnol 13, 1201–1212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tian B, Fadhil NH, Powell JE, Kwong WK & Moran NA Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. MBio 3, 00377–00312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Soares KO et al. Tetracycline exposure alters key gut microbiota in africanized honey bees (Apis mellifera scutellata x spp.). Front. Ecol. Evol 9, 716660 (2021). [Google Scholar]
- 123.Baffoni L. et al. Honeybee exposure to veterinary drugs: How is the gut microbiota affected? Microbiol Spectr 9, e0017621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Motta EVS & Moran NA Impact of glyphosate on the honey bee gut microbiota: Effects of intensity, duration, and timing of exposure. mSystems 5, 00268–00220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Powell JE, Carver Z, Leonard SP & Moran NA Field-realistic tylosin exposure impacts honey bee microbiota and pathogen susceptibility, which is ameliorated by native gut probiotics. Microbiol. Spectr 9, e0010321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ortiz-Alvarado Y. et al. Antibiotics in hives and their effects on honey bee physiology and behavioral development. Biol. Open 9, bio053884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Duan X. et al. Antibiotic treatment decrease the fitness of honeybee (Apis mellifera) larvae. Insects 12, 301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Raymann K Honey bee microbiota and the physiology of antimicrobial resistance. in Honey Bee Medicine for the Veterinary Practitioner 125–134 (Wiley, 2021). [Google Scholar]
- 129.Ramsey SD et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. U. S. A 116, 1792–1801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu L. et al. Honey bee Apis mellifera larvae gut microbial and immune, detoxication responses towards flumethrin stress. Environ. Pollut 290, 118107 (2021). [DOI] [PubMed] [Google Scholar]
- 131.Qi S. et al. Acaricide flumethrin-induced sublethal risks in honeybees are associated with gut symbiotic bacterium Gilliamella apicola through microbe-host metabolic interactions. Chemosphere 307, 136030 (2022). [DOI] [PubMed] [Google Scholar]
- 132.Kakumanu ML, Reeves AM, Anderson TD, Rodrigues RR & Williams MA Honey bee gut microbiome is altered by in-hive pesticide exposures. Front. Microbiol 7, 1255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rouzé R, Moné A, Delbac F, Belzunces L & Blot N The honeybee gut microbiota is altered after chronic exposure to different families of insecticides and infection by Nosema ceranae. Microbes Environ. 34, 226–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Diaz T, Del-Val E, Ayala R & Larsen J Alterations in honey bee gut microorganisms caused by Nosema spp. and pest control methods. Pest Manag. Sci 75, 835–843 (2019). [DOI] [PubMed] [Google Scholar]
- 135.Cuesta-Maté A. et al. Resistance and vulnerability of honeybee (Apis mellifera) gut bacteria to commonly used pesticides. Front. Microbiol 12, 717990 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hladik ML et al. Year-round presence of neonicotinoid insecticides in tributaries to the Great Lakes, USA. Environ. Pollut 235, 1022–1029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Buszewski B, Bukowska M, Ligor M & Staneczko-Baranowska I A holistic study of neonicotinoids neuroactive insecticides-properties, applications, occurrence, and analysis. Environ. Sci. Pollut. Res. Int 26, 34723–34740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Y-J et al. Thiacloprid exposure perturbs the gut microbiota and reduces the survival status in honeybees. J. Hazard. Mater 389, 121818 (2020). [DOI] [PubMed] [Google Scholar]
- 139.Castelli L, Branchiccela B, Zunino P & Antúnez K Insights into the effects of sublethal doses of pesticides glufosinate-ammonium and sulfoxaflor on honey bee health. Sci. Total Environ 868, 161331 (2023). [DOI] [PubMed] [Google Scholar]
- 140.Raymann K. et al. Imidacloprid decreases honey bee survival rates but does not affect the gut microbiome. Appl. Environ. Microbiol 84, e00545–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rothman JA, Russell KA, Leger L, McFrederick QS & Graystock P The direct and indirect effects of environmental toxicants on the health of bumblebees and their microbiomes. Proc. Biol. Sci 287, 20200980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blot N, Veillat L, Rouzé R & Delatte H Glyphosate, but not its metabolite AMPA, alters the honeybee gut microbiota. PLoS One 14, e0215466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Motta EVS, Raymann K & Moran NA Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. U. S. A 115, 10305–10310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Castelli L. et al. Impact of chronic exposure to sublethal doses of glyphosate on honey bee immunity, gut microbiota and infection by pathogens. Microorganisms 9, 845 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Almasri H, Liberti J, Brunet J-L, Engel P & Belzunces LP Mild chronic exposure to pesticides alters physiological markers of honey bee health without perturbing the core gut microbiota. Sci. Rep 12, 4281 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thompson HM et al. Evaluating exposure and potential effects on honeybee brood (Apis mellifera) development using glyphosate as an example. Integr. Environ. Assess. Manag 10, 463–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Motta EVS & Moran NA The effects of glyphosate, pure or in herbicide formulation, on bumble bees and their gut microbial communities. Sci. Total Environ 872, 162102 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cullen MG, Bliss L, Stanley DA & Carolan JC Investigating the effects of glyphosate on the bumblebee proteome and microbiota. Sci. Total Environ 864, 161074 (2023). [DOI] [PubMed] [Google Scholar]
- 149.Helander M. et al. Glyphosate and a glyphosate-based herbicide affect bumblebee gut microbiota. FEMS Microbiol. Ecol 99, fiad065 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Motta EVS, Powell JE & Moran NA Glyphosate induces immune dysregulation in honey bees. Anim. Microbiome 4, 16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Motta EVS et al. Oral or topical exposure to glyphosate in herbicide formulation impacts the gut microbiota and survival rates of honey bees. Appl. Environ. Microbiol 86, 116–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tan S. et al. Effects of glyphosate exposure on honeybees. Environ. Toxicol. Pharmacol 90, 103792 (2022). [DOI] [PubMed] [Google Scholar]
- 153.Pettis JS et al. Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS One 8, e70182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Al Naggar Y, Singavarapu B, Paxton RJ & Wubet T Bees under interactive stressors: the novel insecticides flupyradifurone and sulfoxaflor along with the fungicide azoxystrobin disrupt the gut microbiota of honey bees and increase opportunistic bacterial pathogens. Sci. Total Environ 849, 157941 (2022). [DOI] [PubMed] [Google Scholar]
- 155.Ludvigsen J, Amdam GV, Rudi K & L’Abée-Lund TM Detection and characterization of streptomycin resistance (strA-strB) in a honeybee gut symbiont (Snodgrassella alvi) and the associated risk of antibiotic resistance transfer. Microb. Ecol 76, 588–591 (2018). [DOI] [PubMed] [Google Scholar]
- 156.Huang SK et al. Influence of feeding type and Nosema ceranae infection on the gut microbiota of Apis cerana workers. mSystems 3, e00177–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Castelli L. et al. Impact of nutritional stress on honeybee gut microbiota, immunity, and Nosema ceranae Infection. Microb. Ecol 80, 908–919 (2020). [DOI] [PubMed] [Google Scholar]
- 158.Powell JE et al. The microbiome and gene expression of honey bee workers are affected by a diet containing pollen substitutes. PLoS One 18, e0286070 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chmiel JA, Pitek AP, Burton JP, Thompson GJ & Reid G Meta-analysis on the effect of bacterial interventions on honey bee productivity and the treatment of infection. Apidologie 52, 960–972 (2021). [Google Scholar]
- 160.Damico ME, Beasley B, Greenstein D & Raymann K A need for stronger regulation: Commercially sold probiotics for honey bees do not live up to their claims. bioRxiv 2023.09.13.557574 (2023) doi: 10.1101/2023.09.13.557574. [DOI] [Google Scholar]
- 161.Daisley BA et al. Novel probiotic approach to counter Paenibacillus larvae infection in honey bees. ISME J. 14, 476–491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stephan JG et al. Honeybee-specific lactic acid bacterium supplements have no effect on American foulbrood-infected honeybee colonies. Appl. Environ. Microbiol 85, e00606–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lamei S. et al. Feeding honeybee colonies with honeybee-specific lactic acid bacteria (Hbs-LAB) does not affect colony-level Hbs-LAB composition or Paenibacillus larvae spore levels, although American foulbrood affected colonies harbor a more diverse Hbs-LAB community. Microb. Ecol 79, 743–755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Daisley BA et al. Delivery mechanism can enhance probiotic activity against honey bee pathogens. ISME J. (2023) doi: 10.1038/s41396-023-01422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Leonard SP et al. Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol 7, 1279–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lariviere PJ, Leonard SP, Horak RD, Powell JE & Barrick JE Honey bee functional genomics using symbiont-mediated RNAi. Nat. Protoc 18, 902–928 (2023). [DOI] [PubMed] [Google Scholar]
- 167.Leonard SP et al. Engineered symbionts activate honey bee immunity and limit pathogens. Science 367, 573–576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang Y, Kaftanoglu O, Brent CS, Page RE Jr & Amdam GV Starvation stress during larval development facilitates an adaptive response in adult worker honey bees (Apis mellifera L.). J. Exp. Biol 219, 949–959 (2016). [DOI] [PubMed] [Google Scholar]
- 169.Faita MR, Cardozo MM, Amandio DTT, Orth AI & Nodari RO Glyphosate-based herbicides and Nosema sp. microsporidia reduce honey bee (Apis mellifera L.) survivability under laboratory conditions. J. Apic. Res 59, 332–342 (2020). [Google Scholar]
- 170.Kwong WK, Zheng H & Moran NA Convergent evolution of a modified, acetate-driven TCA cycle in bacteria. Nat. Microbiol 2, 17067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


