Abstract
Pyroptosis, a gasdermin-mediated lytic cell death, is a new hotspot topic in cancer research, and induction of tumor pyroptosis has emerged as a new target in cancer management. Quercetin (Que), a natural substance, demonstrates promising anticancer action. However, further information is required to fully comprehend the function and mechanism of Que in pyroptosis in colon cancer. This study revealed the underlying mechanism of Que-induced pyroptosis in colon cancer in vitro and in vivo. Que inhibited colon cancer cell growth through gasdermin D (GSDMD)-mediated pyroptosis. Depletion of GSDMD, rather than gasdermin E (GSDME), reversed the cytotoxic effects of Que on colon cancer cells. Que treatment upregulated NIMA-related kinase 7 (NEK7) protein expression, thus facilitating the assembly of the NLRP3 inflammasome and cleavage of GSDMD. NEK7 silencing resulted in colon cancer cell growth in vitro and in vivo. Mechanistically, NEK7 depression restrained the activation of the NLRP3 inflammasome-GSDMD pathway, thus attenuating pyroptosis triggered by Que in colon cancer cells. Furthermore, lower NEK7 and NLRP3 expression levels indicated colon cancer progression. Our results unveiled a novel pattern of anti-colon cancer activity of Que, and activation of NEK7-mediated pyroptosis is potentially a promising therapeutic target for colon cancer, which provides novel experimental proof for the clinical application of Que.
Keywords: Colon cancer, quercetin, pyroptosis, NEK7, NLRP3 inflammasome, GSDMD
Introduction
The most prevalent type of malignant tumor of the digestive system is colon cancer, ranking third in incidence and second in mortality among diverse types of cancers [1]. Despite recent and novel advances in treatment modalities, they have their limitations for clinical application [2-4]. Therefore, more emphasis should be placed on alternative therapies such as natural compounds with anticancer properties [5,6].
Pyroptosis, also known as gasdermin-mediated lytic cell death, is a novel form of programmed cell death (PCD) that occurs after the cytotoxic N-terminal cleavage of some gasdermins [7,8]. Recently, more comprehensive research suggests that pyroptosis affects tumor progression [9,10]. Determining the role and function of pyroptosis in colon cancer may provide novel perspectives and unravel potential therapeutic targets for colon cancer management. Gasdermins, the executor of pyroptosis, comprises six paralogous genes in humans [11]. Among the six proteins, the detection of GSDMD cleavage aids in identifying pyroptosis, as GSDMD can be cleaved by assembling inflammasomes [12]. In canonical signaling, the activated cytosolic inflammasome sensor NLRP3 recruits the adaptor apoptosis-associated speck-like protein containing a CARD (named, ASC adaptor) and Caspase1 (Cas1) to form the NLRP3 inflammasome that accelerates the proteolysis of Cleaved-caspase1 (C-Cas1) [13]. Subsequently, the mature C-Cas1 cleaves GSDMD into the N- and C-terminal fragments. The N-terminal fragment perforates the cell membrane resulting in pyroptosis [14]. Additionally, the mature C-Cas1 also converts the precursor form of IL-1β and IL-18 into their mature forms [15], which are released through the pores formed by GSDMD.
NEK7, a member of the NEK family, is a serine-threonine kinase that was primordially proven to drive cell cycle progression [16,17]. Yuan He et al [18] reported that NEK7 interacted with NLRP3 and that its deficiency specifically abrogated NLRP3 inflammasome activation. However, only less attention has been dedicated to the function of NEK7 in colon cancer progression and whether it can induce GSDMD-mediated pyroptosis in colon cancer cells by activating the NLRP3 inflammasome.
Quercetin (Que; C15H10O7) is part of the flavanol family that belongs to flavonoids, which are abundant in fruits, vegetables, and herbs [19]. Adverse effects following Que supplementation are rarely reported in clinical studies [20-22]. Mechanically, several studies have confirmed that Que exhibits anti-colon cancer activity both in vitro and in vivo by altering a variety of phenotypes [23-26]. Our previous study also demonstrated that Danggui Buxue decoction containing Que inhibited metastatic colon cancer development [27], implying that Que may be beneficial for the management of colon cancer. However, whether Que can trigger pyroptosis in colon cancer remains unknown and the underlying molecular mechanism remains unexplored.
In this study, we explored whether Que can induce pyroptosis in colon cancer by activating the NEK7-mediated NLRP3 inflammasome-GSDMD signaling pathway, which revealed a novel pattern of anti-colon cancer activity of Que and provided extra experimental proof for the clinical application of Que.
Materials and methods
Drugs and reagents
Que (Cat. #S2391, >98.93% purity) and GSK’872 (Cat. #S8465, 99.9% purity) were obtained from Selleck (Shanghai, China). McCoy’s 5A medium (Cat. #C3020), trypsin-EDTA (0.25%) (Cat. #C3530), and fetal bovine serum (Cat. #C04001) were purchased from VivaCell (Shanghai, China). Brdu (Cat. #B5002) was obtained from Sigma (Shanghai, China). Dimethyl sulfoxide (DMSO, Cat. #D8371) and Puromycin (Cat. #P8230) were purchased from Solarbio (Beijing, China). Enhanced BCA Protein Assay Kit (Cat. #P0010) were the products of Beyotime Biotech Inc. (Shanghai, China). CCK-8 kits (Cat. #C6005) were provided by US Everbright (Suzhou, China). 10% Giemsa solution (Cat. #DM0002) was purchased from Leagene Biotechnology Co., Ltd. (Beijing, China). LDH kits (Cat. #A020-2-2) were obtained from Nanjing Jiancheng Bioengineering Institute (Jiangsu, China). ELISA kits (IL-18, Cat. #JM-03294H1; IL-1β, Cat. #JM-03336H1) were purchased from Jingmei Biotechnology (Jiangsu, China). Primary antibodies specific for NEK7 (Cat. #DF4467), NLRP3 (Cat. #DF7438), ASC (Cat. #DF6304), Cas1 (Cat. #AF5418), C-Cas1 (Cat. #AF4005), GSDMD (Cat. #AF4012), RIPK3 (Cat. #AF4808), MLKL (Cat. #DF7412), p-MLKL (Cat. #AF7420), IL-1β (Cat. #AF5103), IL-18 (Cat. #DF6252), β-actin (Cat. #AF7018), and GAPDH (Cat. #AF7021) were produced by Affinity Biosciences (Cincinnati, USA). Antibodies against p-RIPK3 (Cat. #ab209384) and GSDME (Cat. #ab215191) were obtained from Abcam (Cambridge, MA, USA). Anti-Caspase3 (Cas3) antibody (Cat. #14220) was purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-Brdu antibody (Cat. #GB12051) was obtained from Servicebio Technology (Wuhan, China). Anti-Ki67 antibody (Cat. #AF10714) and 488-TSA solution (Cat. #AFIHCC023) were provided by AIFang Biological (Hunan, China). The secondary antibodies, goat anti-rabbit IgG (H+L) HRP (Cat. #S0001) and goat anti-mouse IgG (H+L) HRP (Cat. #S0002) were purchased form Affinity Biosciences. All antibodies can be searched in the CiteAb engine (http://www.citeab.com), an engine for ranking antibodies based on the number of times they have been cited [28].
Cell culture, and drug preparation
The third-generation human colon cancer cell lines authenticated using short tandem repeat (STR) DNA fingerprinting, namely, HCT116 and HT29 [29,30] (Cat. #CL-0096 and Cat. #CL-0118; Procell technology, Wuhan, China) were grown in McCoy’s 5A medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified environment of 5% CO2 at 37°C. For the intervention experiment, cells were preincubated with or without the RIPK3 inhibitor GSK’872 (12 µM) for 3 h before treatment with different concentrations of Que.
To generate a 200 mM stock solution, Que powder was dissolved in DMSO. The final concentration of DMSO in the solution was limited to 0.1%. The stock solution was kept at -80°C and was defrosted right before each experiment.
Cell viability assay
Cells in the logarithmic growth phase were harvested and planted in a 96-well plate (5000 cells per well) before being treated for 24 or 48 h with the appropriate doses of Que. Cell viability was then assessed using the CCK-8 kits according to the manufacturer’s protocol. IC50 values were calculated using GraphPad Prism 9.0 software (GraphPad Software, CA, USA).
Colony formation assay
Cells subjected to Que for 48 h (1000 cells) were maintained in 6-cm culture dishes. The medium was replenished every 3 days. After 2-week incubation, colonies were fixed and stained with 10% Giemsa solution for 45 min, after which the dishes were allowed to dry at room temperature. Cell colonies (>50 cells) were counted and assessed using ImageJ software, version 11.53k (National Institutes of Health, USA).
LDH assay
Cells were seeded into 96-well plates for 12 h prior to being exposed to Que for 48 h. Cell membrane rupture was identified using LDH kits corresponding to the company’s procedure.
Morphological assay
Pyroptotic morphological alterations in HCT116 and HT29 cells subjected to Que were observed under an IX71 microscope (Olympus Corporation). In brief, cells sown in 6-well culture dishes were treated with 100 μM of Que for 48 h and observed under the light.
ELISA assay
Cells were seeded in 96-well plates with or without Que for 48 h, after which the culture supernatant was collected and centrifuged at 800×g for 20 min at 4°C. IL-1β and IL-18 levels were quantified using ELISA kits following the manufacturer’s instructions. A Varioskan LUX microplate reader (Thermo Fisher Scientific, Inc.) was used to measure the absorbance of the sample at 450 nm.
Cell transduction assay
GeneChem Co., Ltd. (Shanghai, China) designed and provided the recombinant len-tiviral shRNA specific for human NEK7 (NM_133494), NLRP3 (NM_004895), Cas1 (NM_033292), GSDMD (NM_024736), Cas3 (NM_004346), and GSDME (NM_004403), as well as their negative control vectors. All transduction procedures were executed according to the company’s guidelines. Briefly, HCT116 and HT29 cells were cultivated in 6-well plates (4×104 per well) for 24 h. After that, cells were infected for 14 h with the RNAi mixtures, and rinsed with sterile PBS and grown in McCoy’s 5A medium with 10% FBS for 72 h. Puromycin (2 ug/ml and 8 ug/ml for HCT 116 and HT29 cells, respectively) in growth media was employed for 7 days to establish stable cell lines. The remaining cells were frozen, and total protein was extracted for western blotting analysis.
Bioinformatics analysis
Data on correlation between NEK7 and NLRP3 gene expression in colon cancer was analyzed using Spearman rank correlation in the TIMER2.0 [31] database (http://timer.cistrome.org/). The TCGA Gene module of The University of Alabama at Birmingham CANCER data analysis portal (UALCAN; https://ualcan.path.uab.edu/) [32] was utilized to assess the expression of NEK7 and NLRP3, and to investigate the association between their expression levels and colon cancer progression. All samples used for online analysis were chosen from The Cancer Genome Atlas (TCGA)-Colon Adenocarcinoma (COAD) dataset.
Colon cancer tissue collection
Colon cancer and paired adjacent noncancerous paraffin specimens were collected through surgical resection at Yong Chuan Hospital of Chongqing Medical University (Chongqing, China) from January 2018 to September 2018. Eighty patients with colon cancer who did not undergo any treatment before the surgery were included. No ethical issues occurred during this experiment. The use of specimens was approved by the Ethics Committee of Yongchuan Hospital, Chongqing Medical University on 25 February, 2017, in accordance with the Declaration of Helsinki (No. YCHCQMU2017022522). In accordance with the institutional rules, informed consent was collected from all participants.
Immunofluorescence staining assay
The human colon cancer tissue microarray was constructed by AIFang Biological (Hunan, China) with a 1.5 mm diameter for each sample. To verify whether co-expression existed between NEK7 and NLRP3 in cancer specimens, tyramide signal amplification (TSA) fluorescence double-labeled staining was performed. Briefly, the tissue microarray slide was deparaffinized, rehydrated, retrieved, and blocked with 3% hydrogen peroxide and then with 3% bovine serum albumin to inhibit nonspecific binding. Then, the slide was incubated with rabbit anti-NEK7 antibody (1:100 dilution) overnight at 4°C and then with goat anti-rabbit horseradish peroxidase (HRP) - conjugated secondary antibody (Cat. #AFIHC004, AIFang Biological). Subsequently, the slide was treated with a 488-TSA solution, followed by microwave retrieval to remove noncovalently bound antibodies. The slide was again incubated with rabbit anti-NLRP3 antibody (1:250 dilution) overnight at 4°C. Fluorescence emerged by exposing the slide for 1 h at 37°C with Cy3-labeled goat antirabbit IgG antibody (Cat. #AFSA006, AIFang Biological). Finally, the nucleus was counterstained with DAPI before visualization of the slide with a fluorescent microscope (NIKON ECLIPSE C1, Japan). The staining intensity and proportion of positively stained cells were independently determined by two expert pathologists who were blinded to the clinicopathological details of the patients involved in this study. The staining intensity was graded on a scale of 0 to 3, which respectively indicates no, mild, moderate, and strong staining [33]. The proportion of positive cells was assessed on a scale of 0 to 3, which respectively represents 5%, 5-25%, 26-50%, and >50% positive cells [33]. The staining intensity and staining cell scores were added together to obtain a total score. All patients were separated into two groups based on a total score: 0-2, negative expression, and 3-6, positive expression.
For animal xenograft tumor biospecimens, fixed, paraffin-embedded sections (1.5-μm thick) were deparaffinized, rehydrated, and blocked with 3% bovine serum albumin. Subsequently, these sections were incubated overnight with primary antibodies against NEK7, NLRP3, ASC, Cas1, and GSDMD (all 1:200 dilution) and against Brdu and Ki67 (both 1:300 dilution) at 4°C, followed by incubation with fluorescent-labeled secondary antibody (Cy3-labeled goat antirabbit IgG). After staining with DAPI, a fluorescence microscope (NIKON ECLIPSE C1, Japan) was applied to view sections.
Animal experiments
All protocols involving animals were approved by the Institutional Animal Care and Use Committee of Chongqing Medical University on 11 May, 2023, in compliance with the code of practice for the care and use of animals for scientific purposes (No. IACUC-CQMU-2023-0089). Forty female BABL/c-nu mice aged 4 weeks (weighing 16±2 g) were purchased from Hunan Slack Jingda Experimental Animal Co., Ltd. (Hunan, China; SCXK-(xiang)2019-0004). During the experiment, all mice were housed in regular husbandry conditions (22±2°C, 60%-80% relative humidity, and a 12-h photoperiod) and had free access to food and water. After 1 week of adaptation, 100 µl of 3×106 single-cell suspension (HT29-NC and HT29-shNEK7 cells) was subcutaneously injected into the right axilla. Four days later, the mice were randomly divided into four groups (n=10 per group) and allowed an intraperitoneal injection of Que (40 mg/kg body weight [34,35], in 2% DMSO+30% PEG300+2% Tween 80+66% ddH2O; Cat. #S1102; Selleck) for 15 consecutive days. Tumor volume was monitored every 2 days and calculated using the following formula: Tumor volume (mm3) = ½ (width2 × length). Mice were given a 100 mg/kg Brdu solution intraperitoneally before being euthanized with sodium pentobarbital. Xenograft tumors and blood samples were collected two hours later. The serum was centrifuged at 1500×g for 10 min at 4°C for LDH and ELISA tests.
Hematoxylin-eosin staining
Tumor specimens of mice were fixed in 4% paraformaldehyde before being embedded in paraffin and sliced into 5-μm-thick sections, which were then stained with hematoxylin and eosin. Histopathological changes in the tumor tissues were evaluated using a light microscope (IX71 microscope, Olympus Corporation).
Protein extraction and western blot analysis
The samples were lysed on ice with radioimmunoprecipitation assay lysis buffer containing 1% phenylmethylsulfonyl fluoride and phosphatase inhibitor cocktails to extract total protein. Protein concentration was quantified with an Enhanced BCA Protein Assay Kit. Then, 25 µg of the sample containing the total protein from each group was separated using SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes (Millipore; Billerica, MA, USA). After blocking with 5% nonfat milk, the membranes were incubated at 4°C overnight with primary antibodies against NEK7, NLRP3, ASC, Cas1, C-Cas1, GSDMD, RIPK3, p-RIPK3, MLKL, p-MLKL, IL-1β, IL-18, Cas3, GSDME, β-actin, and GAPDH. All antibodies were used in 1:1000 dilution except for β-actin and GAPDH (1:5000 dilution). Subsequently, the membranes were washed and incubated for 1 h at room temperature with HRP-conjugated secondary antibodies. Protein bands were detected with the Electrochemiluminescence Western Blotting Detection Kit (Cat. #BMU102-CN; Abbkine, Wuhan, China) and quantified using ImageJ software (version 11.53k, National Institutes of Health, USA).
Statistical analysis
Statistical analysis was performed using Prism 9.0 (GraphPad Software, CA, USA). Data of quantitative variables were expressed as mean ± standard deviation (SD) and analyzed using Student’s t-test for two groups or one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test for multiple groups. The Wilcoxon matched-pairs signed rank test was used to analyze the immunofluorescence scores of NEK7 and NLRP3 in paired colon cancer tissues. Data on protein expression levels and clinicopathological features in patients with colon cancer were assessed by Chi-square tests (χ2 test). All numeric data were obtained from at least three separate tests. P<0.05 indicated statistical significance.
Results
Que inhibits cell growth and induces pyroptosis in colon cancer cells
The CCK-8 assay showed that Que restrained cell viability in a dose- and time-dependent manner. However, HCT116 cells are more sensitive to Que (Figure 1A, 1B), which is reflected by the lower IC50 values in HCT116 cells than in HT29 cells. Additionally, cell confluency reduced with Que treatment in a dose-dependent manner (Figure 1C). As the CCK-8 assay result is an endpoint detection method, we initially treated the cells with Que for 48 h, after which colony formation tests were performed to assess cancer cell proliferation during the interval without Que treatment. Consistent with the CCK-8 assay results, the number of colonies markedly decreased in the intervention group, as compared with that in the vehicle group (Figure 1D).
Figure 1.

Que inhibits cell growth in colon cancer cells. A, B. HCT116 and HT29 cells were exposed to different concentrations of Que (0, 25, 50, 100, and 200 μM) for 24 h or 48 h. Cell viability was then measured using the CCK-8 assay, with six technical replicates per group, and the mean value utilized for quantitative analysis (n=10, where “n” represents the number of independent repeated experiments). C. Bright-field images of confluency in HCT116 and HT29 cells after Que treatment for 48 h (n=3, where “n” represents the number of independent repeated experiments. Scale bar: 200 μm, 40×). D. Colony formation assay. Colonies were visualized by Giemsa staining (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. **P<0.01, ***P<0.001, ****P<0.0001, compared with the vehicle group.
Based on the IC50 value of Que treatment for 48 h, Que concentrations of 50 and 100 μM were chosen for further study. Loss of membrane integrity is one of the characteristic features of pyroptosis cells. Along with membrane rupture, intracellular LDH is released to the extracellular compartment. After incubating the cells with Que for 48 h, the extracellular level of LDH elevated in a concentration-dependent manner (Figure 2A). Morphologically, cytoplasmic swelling and the formation of large bubbles were observed in HCT116 and HT29 cells treated with 100 μM of Que (Figure 2B), which highly resembled pyroptosis. The full and N-terminal expression of GSDMD was higher in the treatment group than in the vehicle group (Figure 2C, 2D). The intracellular levels of IL-1β and IL-18 (pro and mature) also increased in HCT116 and HT29 cells (Figure 2E, 2F). Moreover, the IL-1β and IL-18 released to the extracellular compartments were elevated in concentration (Figure 2G). Taken together, alterations in morphology and type of molecules indicated that Que induces pyroptosis in colon cancer cells, which may correlate with weakened cell viability.
Figure 2.
Que induces pyroptosis in colon cancer cells. A. Release of LDH from Que-treated HCT116 and HT29 cells measured using LDH kits, with three technical replicates per group, and the mean value utilized for quantitative analysis (n=4, where “n” represents the number of independent repeated experiments). B. Morphology images of HCT116 and HT29 cells treated with 100 μM of Que for 48 h. Black arrowheads indicate the large bubbles emerging from the plasma membrane and cytoplasmic swelling (n=3, where “n” represents the number of independent repeated experiments. Scale bar: 100 μm, 200×). C-F. Protein expression of GSDMD, N-GSDMD, IL-1β, and IL-18 (pro and mature) in Que-treated HCT116 and HT29 cells was measured using western blotting (n=3, where “n” represents the number of independent repeated experiments). G. Release of IL-1β and IL-18 from Que-treated HCT116 and HT29 cells was measured using ELISA kits, with three technical replicates per group, and the mean value utilized for quantitative analysis (n=5, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, compared with the vehicle group.
Que activates the Caspase3-GSDME axis
GSDME, another member of the gasdermin family, contains a two-domain architecture, similar to that of GSDMD [8]. GSDME can be cleaved by Cleaved-caspase3 (C-Cas3), followed by pyroptosis occurrence [36]. To assess the effect of Que on the N-terminal cleavage of GSDME, a western blot was performed. The results revealed that C-Cas3 expression was significantly higher in the treatment group than in the vehicle group, which subsequently enhanced the N-terminal cleavage of GSDME in a dose-dependent manner (Figure 3). These findings suggested that the Cas3-GSDME axis may also be associated with Que-induced cell death.
Figure 3.
Que actives the Cas3-GSDME axis. A, B. Effect of Que on the activation of the Cas3-GSDME pathway was detected by western blot (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, compared with the vehicle group.
Que-induced cell death independent of necroptosis pathway activation
Phosphorylated MLKL in the necroptosis pathway also contributes to membrane rupture [37]. To exclude necroptosis signal transduction induced by Que that resulted in decreased cell viability, various concentrations of the inhibitor GSK’872 were applied to block the necroptosis pathway (Figure 4A). Treatment with a 12 μM dose of GSK’872 did not reverse decreased cell viability triggered by the 100 μM dose of Que (Figure 4B), suggesting that cell death triggered by Que is independent of necroptosis activation. To further verify the results, we measured the expression of proteins implicated in the necroptosis pathway. 100 μM of Que downregulated the protein expression of RIPK3 and MLKL, and inhibited the activation of p-RIPK3 and p-MLKL as determined by Western blotting, which restrained the activation of the necroptosis pathway (Figure 4C-E). Next, we assessed whether the inhibition of the necroptosis pathway affected cell pyroptosis. Western blotting results showed that GSK’872 remarkably enhanced the N-terminal cleavage of GSDMD in both colon cancer cells compared to the vehicle group. Although elevated N-GSDME was observed, there was no statistical significance (Figure 4F, 4G). Additionally, when co-incubated, GSK’872 showed a synergistic effect with Que on the cleavage of pyroptosis-related protein, which may have resulted in marked cell death in the co-incubated group (Figure 4B). These results indicated that Que-induced colon cancer cell death is independent of necroptosis pathway activation.
Figure 4.

Que-induced cell death independent of necroptosis pathway activation. A. Protein levels of MLKL and p-MLKL in HCT116 and HT29 cells treated with various doses of GSK’872 were measured using western blotting assay (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, compared with the vehicle group. B. Effect of the RIPK3-specific inhibitor GSK’872 on the viability of HCT116 and HT29 cells in the presence of 100 μM of Que treatment for 48 h assessed using the CCK-8 assay, with six technical replicates per group, and the mean value utilized for quantitative analysis (n=6, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. ****P<0.0001. C-E. Influence of 100 μM of Que on the initiation of the necroptosis pathway measured using western blotting (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. &P<0.05, &&P<0.01 (vs the vehicle group); *P<0.05, **P<0.01, ***P<0.001 (vs the vehicle group); #P<0.05 (vs the Que group). F, G. Effects of GSK’872 on the cleavage of GSDMD and GSDME were measured using western blot (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. &P<0.05 (vs the vehicle group); **P<0.01, ***P<0.001 (vs the vehicle group); #P<0.05 (vs the Que group).
Que initiates GSDMD-mediated pyroptosis in colon cancer cells by activating the NLRP3 inflammasome
We evaluated the expression level of NLRP3 inflammasome-related proteins. As expected, Que treatment upregulated the protein level of NLRP3 and ASC, significantly enhancing C-Cas1 cleavage (Figure 5A). To further determine whether Que-induced pyroptosis depended on NLRP3 inflammasome activation and to clarify the effect of the NLRP3 inflammasome-GSDMD signaling pathway in colon cancer, RNAi technology was used to knock down the expression of NLRP3/Cas1/GSDMD in HCT116 and HT29 cells (Figure 5B). Interestingly, CCK-8 assay results showed that depletion of NLRP3, Cas1, and GSDMD promoted cell survival (Figure 5C), and the decreased viability in HCT116 and HT29 cells upon Que treatment was rescued by interfering with the expression of the aforementioned proteins (Figure 5C). Mechanistically, the expression level of downstream proteins, namely, ASC, C-Cas1, and N-GSDMD were decreased with NLRP3 knockdown. Elevated levels of NLRP3, ASC, and C-Cas1 induced by Que were reduced with NLRP3 depletion, which further inhibited the formation of the N-terminal fragment of GSDMD (Figure 5D, 5E). Que treatment also increased the protein levels of Cas3 and GSDME (Figure 3). To distinguish the pyroptosis triggered by NLRP3 inflammasome-GSDMD activation from Cas3-GSDME signal transduction, we transfected lentiviral vectors of shCas3 and shGSDME into HCT116 and HT29 cells (Figure 6A). Silencing of Cas3 and GSDME failed to prevent the weakening of cell viability in HCT116 and HT29 cells triggered by Que (Figure 6B). Thus, impaired cell viability induced by Que treatment depends on NLRP3 inflammasome-mediated GSDMD activation rather than Cas3-GSDME signal transduction. These results indicated that Que-induced GSDMD-mediated pyroptosis in colon cancer cells depends on NLRP3 inflammasome activation, which resulted in weakened cell viability in colon cancer cells.
Figure 5.
Que induces GSDMD-mediated pyroptosis in colon cancer cells through NLRP3 inflammasome activation. A. Expression of NLRP3 inflammasome-related proteins in Que-treated HCT116 and HT29 cells detected using western blot (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001 (vs the vehicle group). B. Protein levels of NLRP3, Cas1, and GSDMD in short hairpin RNA-transfected cells were determined using western blot (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01 (vs the vector group). C. Effect of depression of NLRP3, Cas1, and GSDMD on cell viability in the presence of 100 μM of Que treatment for 48 h were assessed using the CCK-8 assay, with six technical replicates per group, and the mean value utilized for quantitative analysis (n=4, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. D, E. Effects of shNLRP3 on the Que-triggered activation of ASC and cleavage of C-Cas1 and N-GSDMD were detected using western blotting (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. &P<0.05, &&P<0.01 (vs the vector group); *P<0.05, **P<0.01 (vs the vector group); #P<0.05, ##P<0.01, ###P<0.001 (vs the Q-100 group).
Figure 6.

Que-induced impaired cell viability in colon cancer cells does not depend on the Cas3-GSDME axis. A. Western blotting analysis to examine the protein levels of Cas3 and GSDME in short hairpin RNA-transfected cells (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, ***P<0.001 (vs the vector group). B. CCK-8 assay to determine the influence of depletion of Cas3 and GSDME on decreased cell viability triggered by Que, with six technical replicates per group, and the mean value utilized for quantitative analysis (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. ****P<0.0001.
NEK7 expression is downregulated in human colon cancer tissues and is correlated with NLRP3 expression
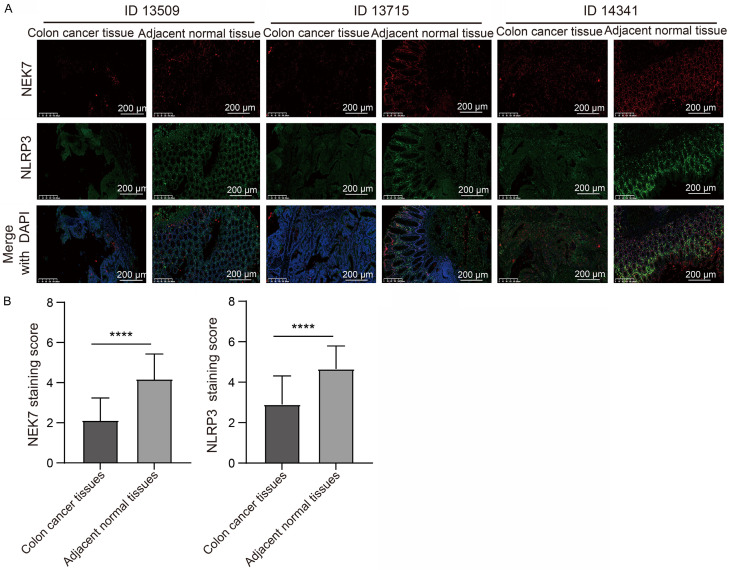
NEK7 was reported to interact with NLRP3 and that its deficiency specifically abrogated NLRP3 inflammasome activation [18]. We then speculated that Que induces GSDMD-mediated pyroptosis in colon cancer through the activation of the NLRP3 inflammasome, which is related to NEK7 expression. To confirm our hypothesis, we first searched in the TIMER2.0 database for information on gene expression association. We found that the gene expression of NLRP3 was positively correlated with NEK7 expression in colon cancer tissues (Figure 7A), implying that NEK7 may regulate NLRP3 expression. We interfered with NEK7 expression in both colon cancer cells. As expected, NEK7 is an upstream modulator of NLRP3 in HCT116 and HT29 cells, as evidenced by the result that loss of NEK7 downregulated NLRP3 protein expression (Figure 7B). The result from the UALCAN database showed that both NEK7 and NLRP3 expression are downregulated in colon cancer tissues when compared with that in normal tissues (Figure 7C, 7D). Further analysis indicated that decreased NEK7 and NLRP3 expression in colon cancer tissues are reflected in advanced cancer stages and more metastasis to lymph nodes (Figure 7C, 7D). Considering the data collected from public databases were based on transcriptional levels, we subsequently performed TSA fluorescence staining assay in tissue microarrays to disclose the effect of NEK7 protein expression on colon cancer progression. The results revealed that NEK7 and NLRP3 were found to be co-expressed in colon cancer tissues. NEK7 and NLRP3 protein expression levels were lower in colon cancer tissues compared to adjacent normal tissues (Figure 8A, 8B). In line with the results of the public database analysis, the decreased protein expression level of NEK7 was associated with poor stage and more distant metastasis (Table 1) in colon cancer samples. Similarly, we investigated the association between NLRP3 protein expression level and clinical features in colon cancer tissue samples. Intriguingly, the lower expression level of NLRP3 was significantly associated with an advanced stage and more lymph node metastasis (Table 1). Taken together, these data implied that the lower expression levels of NEK7 and NLRP3 were both closely related to malignant progression of colon cancer.
Figure 7.

Downregulated expression of NEK7 in human colon cancer tissues and correlated with NLRP3 expression. A. Gene correlation between NLRP3 and NEK7 in colon cancer investigated using the TIMER2.0 database based on Spearman’s correlation analysis. Rho represents the correlation index. B. Influence of NEK7 knockdown on NLRP3 protein expression determined using western blotting (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01 (vs the vector group). C, D. The correlation between lower expression of NEK7 and NLRP3 and colon cancer progression was detected using the UALCAN database (*P<0.05, **P<0.01, ****P<0.0001) vs normal tissues.
Figure 8.
Representative immunofluorescence staining images of NEK7 and NLRP3 expression. A. Representative immunofluorescence staining images of NEK7 and NLRP3 expression on colon cancer tissues and adjacent normal tissues (n=80, where “n” represents the amount of sample. Scale bar: 200 μm, 200×). B. Immunofluorescence staining scores of NEK7 and NLRP3 on colon cancer tissues and adjacent normal tissues (n=80, where “n” represents the number of samples). Values are presented as mean ± SD. ****P<0.0001.
Table 1.
Correlation between NEK7, NLRP3 expression and clinicopathologic parameters of colon cancer patients (χ2 test)
| Variables | Case (n=80) | NEK7 expression | NLRP3 expression | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Positive (n=49) | Negative (n=31) | P value | Positive (n=48) | Negative (n=32) | P value | ||
| Age | 0.349 | 0.454 | |||||
| <65 | 49 | 32 (65.3%) | 17 (54.8%) | 31 (64.6%) | 18 (56.3%) | ||
| ≥65 | 31 | 17 (34.7%) | 14 (45.2%) | 17 (35.4%) | 14 (43.8%) | ||
| Gender | 0.159 | 0.783 | |||||
| Female | 44 | 30 (61.2%) | 14 (45.2%) | 27 (56.3%) | 17 (53.1%) | ||
| Male | 36 | 19 (38.8%) | 17 (54.8%) | 21 (43.7%) | 15 (46.9%) | ||
| Tumor Size (cm) | 0.301 | 0.566 | |||||
| <5 | 28 | 15 (30.6%) | 13 (41.9%) | 18 (37.5%) | 10 (31.3%) | ||
| ≥5 | 52 | 34 (69.4%) | 18 (58.1%) | 30 (62.5%) | 22 (68.8%) | ||
| AJCC stage | 0.002** | 0.001** | |||||
| I/II | 41 | 32 (65.3%) | 9 (29.0%) | 32 (66.7%) | 9 (28.1%) | ||
| III/IV | 39 | 17 (34.7%) | 22 (71.0%) | 16 (33.3%) | 23 (71.9%) | ||
| Lymphatic metastasis | 0.050 | 0.026* | |||||
| N0 | 47 | 33 (67.3%) | 14 (45.2%) | 33 (68.8%) | 14 (43.8%) | ||
| N1/N2 | 33 | 16 (32.7%) | 17 (54.8%) | 15 (31.2%) | 18 (56.2%) | ||
| Distant metastasis | 0.005** | 0.947 | |||||
| M0 | 69 | 47 (95.9%) | 22 (71.0%) | 42 (87.5%) | 27 (84.4%) | ||
| M1 | 11 | 2 (4.1%) | 9 (11.3%) | 6 (12.5%) | 5 (15.6%) | ||
| Differentiation | 0.176 | 0.911 | |||||
| Poor | 17 | 8 (16.3%) | 9 (29.0%) | 10 (20.8%) | 7 (21.9%) | ||
| Moderate/Well | 63 | 41 (83.7%) | 22 (71.0%) | 38 (79.2%) | 25 (78.1%) | ||
P<0.05, significant difference.
P<0.01, significant difference.
Que activates the NLRP3 inflammasome-GSDMD signaling pathway by upregulating NEK7 expression in colon cancer cells
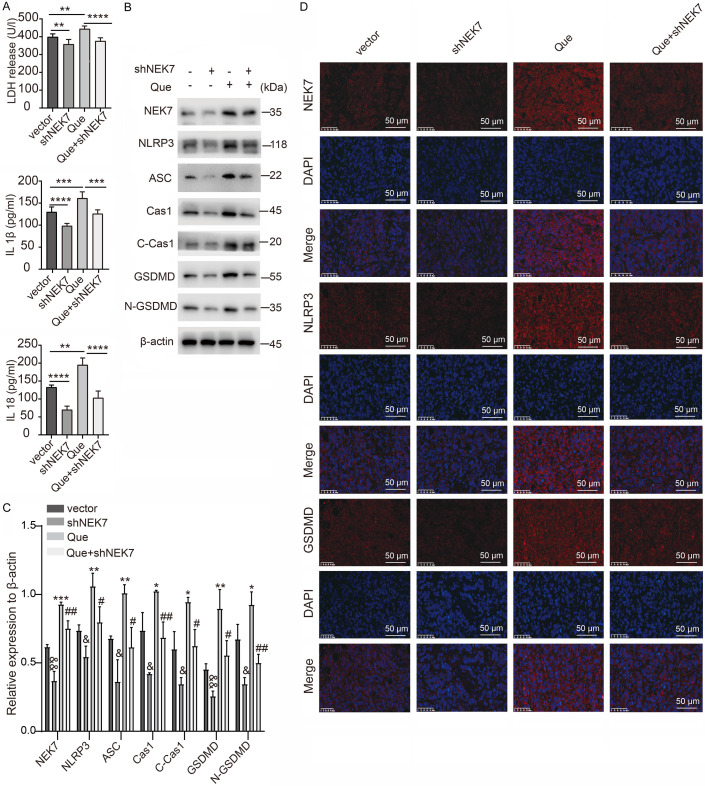
Although our results revealed that NEK7 depression decreased NLRP3 expression, whether Que can regulate NEK7 expression remains unclear. Surprisingly, Que treatment upregulated NEK7 expression in a dose-dependent manner in HCT116 and HT29 cells (Figure 9A). To clarify whether NEK7 was the key molecule in Que-induced pyroptosis, HCT116 and HT29 cells with NEK7 depletion were exposed to Que for 48 h. Silencing of NEK7 resulted in a decreased release of LDH, IL-1β, and IL-18, which restrained the effect of Que-triggered pyroptosis in colon cancer cells (Figure 9B). Interestingly, the reduction of NEK7 levels contributed to remarkably elevated cell viability in HCT116 and HT29 cells. By contrast, the inhibitory effect of Que on cell viability was neutralized with NEK7 knockdown (Figure 9D). Consistent with the CCK-8 analysis results, NEK7 knockdown significantly promoted colony formation and reversed fewer colonies treatment with Que in HCT116 and HT29 cells (Figure 9C, 9D). Mechanically, NEK7 knockdown downregulated the expression of NLRP3 and ASC, further leading to the blockade of Cas1 and GSDMD cleavage (Figure 9E-G). Thus, NEK7 is an upstream regulator of NLRP3 inflammasome-GSDMD signaling-dependent pyroptosis in colon cancer. The higher expression of NEK7, NLRP3, ASC, and the increased generation of C-Cas1 and N-GSDMD induced by Que were rescued by shNEK7 (Figure 9E-G). Altogether, Que induces pyroptosis in colon cancer by upregulating NEK7 to activate the NLRP3 inflammasome-GSDMD signaling pathway.
Figure 9.

Que induces pyroptosis via upregulating NEK7 to activate the NLRP3 inflammasome-GSDMD pathway. A. Protein levels of NEK7 in Que-treated HCT116 and HT29 cells (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001, compared with the vehicle group. B. Release of LDH, IL-1β, and IL-18 in HCT116 and HT29 cells with depletion of NEK7 in the presence of 100 μM of Que treatment for 48 h, with three technical replicates per group, and the mean value utilized for quantitative analysis (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. C, D. The number of colonies and cell viability measured using colony formation (n=3, where “n” represents the number of independent repeated experiments) and CCK-8 assays in HCT116 and HT29 cells with NEK7 depletion in the presence of 100 μM of Que for 48 h. Six technical were performed per group, and the mean value was used for quantitative analysis (n=4, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. **P<0.01, ***P<0.001, ****P<0.0001. E-G. Effect of NEK7 knockdown on the activation of NLRP3 inflammasome-GSDMD signaling pathway and Que-induced pyroptosis in HCT116 and HT29 cells detected using western blotting (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. &P<0.05, &&P<0.01, &&&P<0.001 (vs the vector group); *P<0.05, **P<0.01, ***P<0.001 (vs the vector group); #P<0.05, ##P<0.01 (vs the Q-100 group).
Que-induced pyroptosis in vivo depends on the activation of the NEK7-mediated NLRP3 inflammasome-GSDMD signaling pathway
To further evaluate the function of NEK7-mediated pyroptosis in cancer growth on Que treatment, HT29 cells with deficient NEK7 expression as well as their control cells were subcutaneously injected into nude mice. When the tumor size reached approximately 100 mm3, mice received an intraperitoneal injection of Que. NEK7-knockdown accelerated tumor growth and resulted in elevated tumor weight when compared to the vector group (Figure 10A-C). Treatment with 40 mg/kg Que significantly restrained tumor growth and reduced tumor weight. Conversely, the inhibitory impact of Que on tumor xenografts was neutralized with NEK7 knockdown. Pathological staining showed higher tumor cell density in the interference group compared to the control group. A larger necrotic lesion and infiltration of a greater number of inflammatory cells were observed with Que treatment than those of the vector group, whereas a smaller necrotic lesion and infiltration of fewer inflammatory cells were found in the co-treatment group (Figure 10D). In addition, the number of Brdu- and Ki67-stained cells in the nucleus significantly increased in the shNEK7 group compared to the vector group (Figure 10E). The number of deceased positive cells in response to Que was reduced with knockdown of NEK7 expression (Figure 10E). These results demonstrated that NEK7 knockdown restrains Que-inhibited tumor growth. We further evaluated whether the inhibitory effect of Que on tumor growth is related to pyroptosis. We observed that the serum level of LDH, IL-1β, and IL-18 in response to Que was remarkably elevated. Moreover, Que-induced alteration was inhibited by depressing NEK7 expression (Figure 11A). To reveal the potential mechanisms in vivo, western blot assays were conducted in xenograft tumors. The findings indicated that NEK7 knockdown lowered the expression of NLRP3 and ASC, further leading to the blockade of Cas1 and GSDMD cleavage (Figure 11B, 11C). Furthermore, Que did indeed upregulate NEK7 expression, which then activated the NLRP3 inflammasome, contributing to the enhanced cleavage of GSDMD (Figure 11B, 11C). However, the elevated level of NEK7-NLRP3 inflammasome-GSDMD pathway-related proteins after Que treatment was rescued by NEK7 inhibition (Figure 11B, 11C). Immunofluorescence staining of the proteins related to the NEK7-NLRP3 inflammasome-GSDMD pathway demonstrated similar results (Figures 11D and 12). Altogether, these results indicated that Que-induced pyroptosis in vivo depends on the activation of the NEK7-mediated NLRP3 inflammasome-GSDMD signaling pathway, which is consistent with the results obtained in vitro.
Figure 10.
NEK7 knockdown neutralized Que-inhibited tumor growth in colon cancer. A. Image of xenograft tumors formed by HT29 cells treated with 40 mg/kg Que in the presence or absence of NEK7 knockdown. B. Xenograft tumor growth curve for the group of mice treated with 40 mg/kg Que in the presence or absence of NEK7 knockdown (n=10 per group). C. Tumor weight in different groups (n=10 per group). D. Representative images of hematoxylin-eosin staining of xenograft tumor sections. The area marked with a golden line indicates a necrotic lesion (n=5, where “n” represents the amount of sample. Scale bar: 100 μm, 200×). E. Representative images of immunofluorescence assays of Brdu and ki-67 proteins of the xenograft tumor (n=5, where “n” represents the amount of sample. Scale bar: 50 μm, 400×). Values are presented as mean ± SD. *P<0.5, **P<0.01, ***P<0.001, ****P<0.0001.
Figure 11.
Que induces proptosis in vivo depends on the activation of the NEK7-mediated NLRP3 inflammasome-GSDMD signaling pathway. A. Serum levels of LDH, IL-1β, and IL-18 in xenograft tumor-bearing mice treated with 40 mg/kg Que in the presence or absence of NEK7 knockdown detected using LDH kits and ELISA kits, with three technical replicates for each sample, and the mean value utilized for quantitative analysis (n=6, where “n” represents the amount of sample). Values are presented as mean ± SD. **P<0.01, ***P<0.001, ****P<0.0001. B, C. Effect of NEK7 knockdown on the activation of the NLRP3 inflammasome-GSDMD signaling pathway and Que-induced pyroptosis in vivo detected using western blotting (n=3, where “n” represents the number of independent repeated experiments). Values are presented as mean ± SD. &P<0.05, &&P<0.01 (vs the vector group); *P<0.05, **P<0.01, ***P<0.001 (vs the vector group); #P<0.05, ##P<0.01 (vs the Q-100 group). D. Representative images of immunofluorescence assays of NEK7, NLRP3, and GSDMD proteins of the xenograft tumor (n=5, where “n” represents the amount of sample. Scale bar: 50 μm, 400×).
Figure 12.
Representative images of immunofluorescence staining. Fluorescence images of ASC and Cas1 expression of the xenograft tumor (n=5, where “n” represents the amount of sample. Scale bar: 50 μm, 400×).
Discussion
The following were the primary findings of the current study: On the one hand, the expression of NEK7 is decreased in colon cancer, and this reduced expression accelerates tumor growth and correlates with colon cancer progression. On the other hand, silencing NEK7 restrains the activation of the NLRP3 inflammasome-GSDMD pathway, thus attenuating pyroptosis triggered by Que in colon cancer cells. Finally, whereas Que can facilitate the cleavage of both GSDMD and GSDME, GSDMD cleavage is required for Que-induced cell death.
Que is a significant flavonoid abundantly found in various plant sources, such as garden onion, capers, angelica, and black elderberry [19]. The anticancer effect of Que has been widely reported in this decade [38], whether GSDMD-mediated pyroptosis involves the anti-colon cancer effect of Que remains unknown. Our study showed that Que promoted the N-terminus release of GSDMD, accompanied by elevated levels of LDH, IL-1β, and IL-18 in vitro and in vivo. In colon cancer cells, the stimulation effect of Que on Cas3 has been reported [39]. Recently, several studies confirmed that C-Cas3 activation can switch apoptosis to pyroptosis in cells with GSDME expression [40,41]. In our study, Que treatment promoted the cleavage of C-Cas3 and GSDME. Although Que can simultaneously facilitate the cleavage of GSDMD and GSDME, the elevated cell death triggered by Que could be rescued by shGSDMD rather than shGSDME. This finding indicated that Que-induced cell death in colon cancer cells relies on GSDMD cleavage, and it is reasonable to speculate that Que may possess a superior potential to initiate pyroptosis in colon cancer cells. Consistent with our study, Que has been reported to induce pyroptosis in triple-negative breast cancer [42] and gastric cancer cells [43]. More recently, pyroptosis exerts a tumor-suppressive role, which correlates with the evocation of antitumor immunity, facilitated by the release of IL-1β and IL-18 [44-46]. Que-induced pyroptosis may initiate antitumor immunity in colon cancer, which needs further exploration in studies conducted in the future.
Necroptosis, another important mechanism of PCD also causes cell membrane rupture, cell swelling, and lysis. The molecular mechanisms underlying necroptosis are reported to depend critically on receptor-interacting serine-threonine kinase 1 (RIPK1), RIPK3, and MLKL, regardless of the upstream trigger [47]. Que induced cell death through pyroptosis, not through necroptosis activation, based on the following evidence: first, the RIPK3 inhibitor GSK’872 accelerated cell death triggered by Que in colon cancer cells, instead of neutralizing the cytotoxicity effect; second, Que inhibited the activation of the RIPK3-MLKL axis; moreover, the protein expression level related to the NLRP3 inflammasome-GSDMD signaling pathway had increased, which was attributed to pyroptosis induction, and the shRNA of the NLRP3 inflammasome-GSDMD signaling pathway markedly rescued Que-induced cell death. Surprisingly, we found that GSK’872 could promote the N-terminal cleavage of both GSDMD and GSDME. It showed a synergistic effect with Que on the cleavage of pyroptosis-related proteins, which may have resulted in marked cell death in the co-incubated group. These results suggest that crosstalk exists between necroptosis and pyroptosis, although the underlying mechanism remains to be investigated.
GSDMD-mediated pyroptosis triggered by the assembly of the NLRP3 inflammasome has been widely elaborated in autoimmune diseases, inflammatory diseases, and microbial infection [48]. NLRP3 is expressed in both immune cells and colonic epithelial cells [49], indicating a possible association between NLRP3 and colon cancer. Mice deficient in ASC, Cas1, and NLRP3 are prone to inflammation associated colon cancer [50]. Similar to the previous study [50], our work showed that depletion of NLRP3 and Cas1 enhances cell viability in both HCT116 and HT29 cells. Lower NLRP3 expression is reflected in advanced colon cancer stages and more metastasis to lymph nodes, suggesting that NLRP3 inflammasome activation may restrain colon cancer development. Crohn’s disease is a well-known risk factor for colorectal cancer. Zaki et al [51] observed that mice deficient in NLRP3 or ASC and Cas1 were highly susceptible to dextran sodium sulfate-induced colitis. The NLRP3 inflammasome activation may play an important role in colon cancer prevention and treatment. Surprisingly, our results demonstrated that Que treatment significantly activated the NLRP3 inflammasome, thus facilitating the cleavage of GSDMD, which may be a candidate therapeutic drug for colon cancer management.
The cytosolic inflammasome sensor NLRP3 can be initiated by a variety of stimuli [52]. In macrophages, NEK7 is an NLRP3-binding protein that regulates NLRP3 oligomerization and activation [18]. We speculated that NEK7 is associated with Que-induced pyroptosis in colon cancer. Our study demonstrated that Que upregulated NEK7 expression and NEK7 knockdown restrained elevation in the protein expression related to the NLRP3 inflammasome-GSDMD pathway, which was activated by Que. Moreover, the release of LDH, IL-1β, and IL-18 activated by Que was inhibited with NEK7 depletion. Those results implied that Que induces GSDMD-mediated pyroptosis in colon cancer by activating the NLRP3 inflammasome signaling pathway through the upregulation of NEK7. Furthermore, the transcriptional and protein level of NEK7 expression was both lower in cancer tissues than in normal and adjacent tissues, and the decreased NEK7 expression in colon cancer tissues is related to advanced progression. The lack of NEK7 markedly enhanced cell viability and colony formation in vitro. An in vivo experiment showed that NEK7 deficiency accelerated xenograft tumor growth, and more tumor cells stained by Ki67 and Brdu were detected with the use of shNEK7. Briefly, the high expression of NEK7 may be a protective factor for colon cancer, and the activation of the NEK7-mediated NLRP3 inflammasome-GSDMD signaling pathway may expect to become a new therapeutic target for colon cancer management. However, the influence of the expression of NEK7 on the survival of patients with colon cancer should be clarified through clinical trials in the future.
To the best of our knowledge, this study is the first to reveal that upregulating NEK7 expression may serve as a protective factor in colon cancer management, highlighting its previously undisclosed role in inducing pyroptosis in colon cancer. Additionally, our findings uncover a novel anti-colon cancer profile for Que, suggesting its potential clinical application as a targeted medication for NEK7-mediated pyroptosis. Future research is needed to determine whether Que can target NEK7 and elucidate the role of NEK7 in colon cancer diagnosis and prognosis. Furthermore, Que, which is abundant in fruits and vegetables, holds profound significance in providing dietary guidelines for the prevention of colon cancer due to the association between diet and colon cancer occurrence.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81704033) and Xinglin program of Chongqing TCM/TCM-integrated Key discipline (No. 2021-ZDXK-DB04) granted by Chongqing Key Laboratory of Traditional Chinese Medicine for Prevention and Cure of Metabolic Diseases.
Disclosure of conflict of interest
None.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chakrabarti S, Peterson CY, Sriram D, Mahipal A. Early stage colon cancer: current treatment standards, evolving paradigms, and future directions. World J Gastrointest Oncol. 2020;12:808–832. doi: 10.4251/wjgo.v12.i8.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taieb J, André T, Auclin E. Refining adjuvant therapy for non-metastatic colon cancer, new standards and perspectives. Cancer Treat Rev. 2019;75:1–11. doi: 10.1016/j.ctrv.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Tolba MF. Revolutionizing the landscape of colorectal cancer treatment: the potential role of immune checkpoint inhibitors. Int J Cancer. 2020;147:2996–3006. doi: 10.1002/ijc.33056. [DOI] [PubMed] [Google Scholar]
- 5.Esmeeta A, Adhikary S, Dharshnaa V, Swarnamughi P, Ummul Maqsummiya Z, Banerjee A, Pathak S, Duttaroy AK. Plant-derived bioactive compounds in colon cancer treatment: an updated review. Biomed Pharmacother. 2022;153:113384. doi: 10.1016/j.biopha.2022.113384. [DOI] [PubMed] [Google Scholar]
- 6.Macharia JM, Zhang L, Mwangi RW, Rozmann N, Kaposztas Z, Varjas T, Sugár M, Alfatafta H, Pintér M, Bence RL. Are chemical compounds in medical mushrooms potent against colorectal cancer carcinogenesis and antimicrobial growth? Cancer Cell Int. 2022;22:379. doi: 10.1186/s12935-022-02798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broz P, Pelegrín P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 8.Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42:245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Pezuk JA. Pyroptosis in combinatorial treatment to improve cancer patients’ outcome, is that what we want? EBioMedicine. 2019;41:17–18. doi: 10.1016/j.ebiom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6:128. doi: 10.1038/s41392-021-00507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura M, Tanaka S, Fujii T, Aoki A, Komiyama H, Ezawa K, Sumiyama K, Sagai T, Shiroishi T. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics. 2007;89:618–629. doi: 10.1016/j.ygeno.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 13.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 15.Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in cell death, inflammation, and pyroptosis. Annu Rev Immunol. 2020;38:567–595. doi: 10.1146/annurev-immunol-073119-095439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell MJ, Krien MJ, Hunter T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003;13:221–228. doi: 10.1016/s0962-8924(03)00056-4. [DOI] [PubMed] [Google Scholar]
- 17.O’Regan L, Blot J, Fry AM. Mitotic regulation by NIMA-related kinases. Cell Div. 2007;2:25. doi: 10.1186/1747-1028-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Zeng MY, Yang D, Motro B, Núñez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai WF, Wong WT. Design and optimization of quercetin-based functional foods. Crit Rev Food Sci Nutr. 2022;62:7319–7335. doi: 10.1080/10408398.2021.1913569. [DOI] [PubMed] [Google Scholar]
- 20.Brüll V, Burak C, Stoffel-Wagner B, Wolffram S, Nickenig G, Müller C, Langguth P, Alteheld B, Fimmers R, Stehle P, Egert S. No effects of quercetin from onion skin extract on serum leptin and adiponectin concentrations in overweight-to-obese patients with (pre-)hypertension: a randomized double-blinded, placebo-controlled crossover trial. Eur J Nutr. 2017;56:2265–2275. doi: 10.1007/s00394-016-1267-0. [DOI] [PubMed] [Google Scholar]
- 21.Andres S, Pevny S, Ziegenhagen R, Bakhiya N, Schafer B, Hirsch-Ernst KI, Lampen A. Safety aspects of the use of quercetin as a dietary supplement. Mol Nutr Food Res. 2018;62:10. doi: 10.1002/mnfr.201700447. [DOI] [PubMed] [Google Scholar]
- 22.Han MK, Barreto TA, Martinez FJ, Comstock AT, Sajjan US. Randomised clinical trial to determine the safety of quercetin supplementation in patients with chronic obstructive pulmonary disease. BMJ Open Respir Res. 2020;7:e000392. doi: 10.1136/bmjresp-2018-000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatiya M, Pathak S, Jothimani G, Duttaroy AK, Banerjee A. A comprehensive study on the anti-cancer effects of quercetin and its epigenetic modifications in arresting progression of colon cancer cell proliferation. Arch Immunol Ther Exp (Warsz) 2023;71:6. doi: 10.1007/s00005-023-00669-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin R, Piao M, Song Y, Liu C. Quercetin suppresses AOM/DSS-induced colon carcinogenesis through its anti-inflammation effects in mice. J Immunol Res. 2020;2020:9242601. doi: 10.1155/2020/9242601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Özsoy S, Becer E, Kabadayı H, Vatansever HS, Yücecan S. Quercetin-mediated apoptosis and cellular senescence in human colon cancer. Anticancer Agents Med Chem. 2020;20:1387–1396. doi: 10.2174/1871520620666200408082026. [DOI] [PubMed] [Google Scholar]
- 26.Trinh NT, Nguyen TMN, Yook JI, Ahn SG, Kim SA. Quercetin and quercitrin from agrimonia pilosa ledeb inhibit the migration and invasion of colon cancer cells through the JNK signaling pathway. Pharmaceuticals (Basel) 2022;15:364. doi: 10.3390/ph15030364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng SH, Zhao B, Zhan X, Motanyane R, Wang SM, Li A. Danggui Buxue Decoction in the treatment of metastatic colon cancer: network pharmacology analysis and experimental validation. Drug Des Devel Ther. 2021;15:705–720. doi: 10.2147/DDDT.S293046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helsby MA, Leader PM, Fenn JR, Gulsen T, Bryant C, Doughton G, Sharpe B, Whitley P, Caunt CJ, James K, Pope AD, Kelly DH, Chalmers AD. CiteAb: a searchable antibody database that ranks antibodies by the number of times they have been cited. BMC Cell Biol. 2014;15:6. doi: 10.1186/1471-2121-15-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie W, Peng M, Liu Y, Zhang B, Yi L, Long Y. Simvastatin induces pyroptosis via ROS/caspase-1/GSDMD pathway in colon cancer. Cell Commun Signal. 2023;21:329. doi: 10.1186/s12964-023-01359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Xu T, Tian H, Wu J, Yu X, Zeng L, Liu F, Liu Q, Huang X. Coxsackievirus group B3 has oncolytic activity against colon cancer through gasdermin E-mediated pyroptosis. Cancers (Basel) 2022;14:6206. doi: 10.3390/cancers14246206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Zhang T, Cheng M, Hong L, Zhang C, Xie M, Sun P, Fan R, Wang Z, Wang L, Zhong J. TRIM29 facilitates the epithelial-to-mesenchymal transition and the progression of colorectal cancer via the activation of the Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res. 2019;38:104. doi: 10.1186/s13046-019-1098-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Hashemzaei M, Delarami Far A, Yari A, Heravi RE, Tabrizian K, Taghdisi SM, Sadegh SE, Tsarouhas K, Kouretas D, Tzanakakis G, Nikitovic D, Anisimov NY, Spandidos DA, Tsatsakis AM, Rezaee R. Anticancer and apoptosis-inducing effects of quercetin in vitro and in vivo. Oncol Rep. 2017;38:819–828. doi: 10.3892/or.2017.5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kee JY, Han YH, Kim DS, Mun JG, Park J, Jeong MY, Um JY, Hong SH. Inhibitory effect of quercetin on colorectal lung metastasis through inducing apoptosis, and suppression of metastatic ability. Phytomedicine. 2016;23:1680–1690. doi: 10.1016/j.phymed.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 37.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 38.Zang X, Cheng M, Zhang X, Chen X. Quercetin nanoformulations: a promising strategy for tumor therapy. Food Funct. 2021;12:6664–6681. doi: 10.1039/d1fo00851j. [DOI] [PubMed] [Google Scholar]
- 39.Tezerji S, Nazari Robati F, Abdolazimi H, Fallah A, Talaei B. Quercetin’s effects on colon cancer cells apoptosis and proliferation in a rat model of disease. Clin Nutr ESPEN. 2022;48:441–445. doi: 10.1016/j.clnesp.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Yu J, Li S, Qi J, Chen Z, Wu Y, Guo J, Wang K, Sun X, Zheng J. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis. 2019;10:193. doi: 10.1038/s41419-019-1441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahoo P, Jana P, Kundu S, Mishra S, Chattopadhyay K, Mukherjee A, Ghosh CK. Quercetin@Gd(3+) doped Prussian blue nanocubes induce the pyroptotic death of MDA-MB-231 cells: combinational targeted multimodal therapy, dual modal MRI, intuitive modelling of r(1)-r(2) relaxivities. J Mater Chem B. 2023;11:6646–6663. doi: 10.1039/d3tb00316g. [DOI] [PubMed] [Google Scholar]
- 43.Rong Y, Liu SH, Tang MZ, Yang XJ. Quercetin inhibits the proliferative effect of gastric cancer cells by activating the pyroptosis pathway. Asian J Surg. 2023;46:5286–5288. doi: 10.1016/j.asjsur.2023.07.051. [DOI] [PubMed] [Google Scholar]
- 44.Hsu SK, Li CY, Lin IL, Syue WJ, Chen YF, Cheng KC, Teng YN, Lin YH, Yen CH, Chiu CC. Inflammation-related pyroptosis, a novel programmed cell death pathway, and its crosstalk with immune therapy in cancer treatment. Theranostics. 2021;11:8813–8835. doi: 10.7150/thno.62521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakanishi K. Unique action of interleukin-18 on T cells and other immune cells. Front Immunol. 2018;9:763. doi: 10.3389/fimmu.2018.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity. 2018;48:35–44. e6. doi: 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu B, Gao J, Shi J, Zhang F, Shi C, Wen P, Wang Z, Guo W, Zhang S. Necroptosis throws novel insights on patient classification and treatment strategies for hepatocellular carcinoma. Front Immunol. 2022;13:970117. doi: 10.3389/fimmu.2022.970117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You R, He X, Zeng Z, Zhan Y, Xiao Y, Xiao R. Pyroptosis and its role in autoimmune disease: a potential therapeutic target. Front Immunol. 2022;13:841732. doi: 10.3389/fimmu.2022.841732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kummer JA, Broekhuizen R, Everett H, Agostini L, Kuijk L, Martinon F, van Bruggen R, Tschopp J. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem. 2007;55:443–452. doi: 10.1369/jhc.6A7101.2006. [DOI] [PubMed] [Google Scholar]
- 50.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y, Xu W, Zhou R. NLRP3 inflammasome activation and cell death. Cell Mol Immunol. 2021;18:2114–2127. doi: 10.1038/s41423-021-00740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]