Abstract
The flavivirus genome is a positive-stranded ∼11-kb RNA including 5′ and 3′ noncoding regions (NCR) of approximately 100 and 400 to 600 nucleotides (nt), respectively. The 3′ NCR contains adjacent, thermodynamically stable, conserved short and long stem-and-loop structures (the 3′-SL), formed by the 3′-terminal ∼100 nt. The nucleotide sequences within the 3′-SL are not well conserved among species. We examined the requirement for the 3′-SL in the context of dengue virus type 2 (DEN2) replication by mutagenesis of an infectious cDNA copy of a DEN2 genome. Genomic full-length RNA was transcribed in vitro and used to transfect monkey kidney cells. A substitution mutation, in which the 3′-terminal 93 nt constituting the wild-type (wt) DEN2 3′-SL sequence were replaced by the 96-nt sequence of the West Nile virus (WN) 3′-SL, was sublethal for virus replication. An analysis of the growth phenotypes of additional mutant viruses derived from RNAs containing DEN2-WN chimeric 3′-SL structures suggested that the wt DEN2 nucleotide sequence forming the bottom half of the long stem and loop in the 3′-SL was required for viability. One 7-bp substitution mutation in this domain resulted in a mutant virus that grew well in monkey kidney cells but was severely restricted in cultured mosquito cells. In contrast, transpositions of and/or substitutions in the wt DEN2 nucleotide sequence in the top half of the long stem and in the short stem and loop were relatively well tolerated, provided the stem-loop secondary structure was conserved.
Dengue (DEN) viruses belong to the genus Flavivirus, within the family Flaviviridae. There are at least 70 flavivirus species, among which the most important human pathogens are the DEN viruses, yellow fever virus, and the Japanese encephalitis (JE) and tick-borne encephalitis viruses. The diseases caused by the four serotypes of DEN virus (DEN1 to -4), dengue fever (DF) and dengue hemorrhagic fever/shock syndrome (DHF/DSS), are endemic or epidemic in tropical and subtropical countries around the world. At present, there is no vaccine available to prevent DF and DHF/DSS (18).
The flavivirus genome is a single-stranded, positive-sense ∼11-kb RNA (22). It contains a single long open reading frame which includes 95% of the nucleotide sequence. The encoded polyprotein is processed to produce three structural proteins (capsid, premembrane, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (reviewed in reference 9). The 5′ and 3′ noncoding regions (NCR) of flavivirus genomes are approximately 100 and 400 to 600 nucleotides (nt) in length, respectively. These segments are expected to include promoter elements for full-length positive- and negative-sense RNA synthesis, since current evidence suggests that no subgenomic-size RNAs are synthesized during virus replication (5, 28).
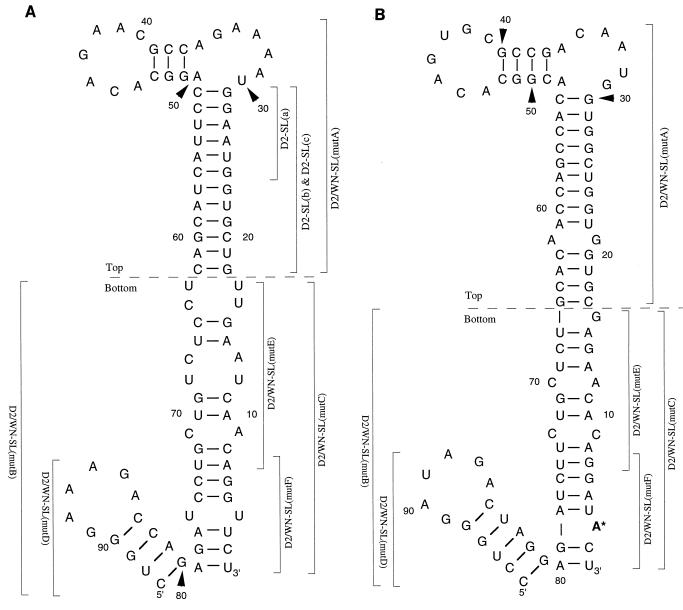
The terminal nucleotide sequences of both NCRs in flavivirus RNA are predicted to form stem-and-loop secondary structures (6, 7, 11, 17, 21, 29, 33). The 3′-terminal secondary structure (3, 33) includes a “short” stem and loop adjacent to a “long” stem and loop (the 3′-SL). For DEN 2, the predicted 3′-SL is formed by the 3′-terminal 93 nt of the genome (Fig. 1A). For West Nile virus (WN), it is formed by the 3′-terminal 96 nt (Fig. 1B). RNase probing confirmed the presence of the predicted 3′-SL in the WN genome and showed that interaction between the small loop and the lower portion of the adjacent long stem within the 3′-SL may result in a “pseudoknot” structure (24). Recent studies suggest a role for the 3′-SL in virus replication. (i) Three baby hamster kidney (BHK) cellular proteins were shown to bind specifically to an in-vitro-synthesized RNA containing the WN 3′-SL nucleotide sequence (3); one of these cellular proteins was subsequently identified as the translation elongation factor, eF1-α (4). It was proposed that the interaction of the 3′-SL with cellular proteins was related to the initiation of negative-strand RNA synthesis. (ii) RNA transcripts representing the JE virus 3′-SL were shown to bind the JE virus NS5 protein in vitro (10); NS5 contains RNA-dependent RNA polymerase activity (30). (iii) In an in vivo study, an internal deletion of 3′ NCR nucleotide sequences extending downstream into the small stem-and-loop nucleotide sequence within the 3′-SL were lethal for DEN4 virus replication (16).
FIG. 1.
(A) The proposed conformation and 93-nt sequence (12) of the DEN2 3′-SL. Nucleotides are numbered from the 3′ terminus of the DEN2 genome. For the purposes of this study, the DEN2 3′-SL was divided into top and bottom portions according to an approach taken for the 3′-SL of WN strain E101 (3). Segments of the DEN2 3′-SL that were mutagenized are indicated by brackets, labeled with the names of the respective mutant viruses. See Fig. 2 and 3 for the genotypes of mutant viruses. (B) The conformation and 96-nt sequence of the WN 3′-SL (4). Nucleotides are numbered beginning from the 3′ terminus of the genome. Top and bottom portions were previously defined (3). Segments of the WN 3′-SL nucleotide sequence that were substituted for the corresponding DEN2 nucleotide sequences in DEN2-WN chimeric RNAs are indicated by brackets labeled with the names of the resultant chimeric viruses (see Fig. 2).
Although the 3′-SL structure in flavivirus RNA is well conserved among species, the involved primary nucleotide sequences are at best semiconserved. Divergence of the nucleotide sequences is especially evident in the region of the long stem, while the nucleotide sequences of the loop segments are relatively well conserved (for an example, see Fig. 1). To study the required elements of the DEN2 3′-SL for virus replication, we created mutations in the relevant nucleotide sequence of a full-length infectious cDNA clone of the DEN2 RNA genome (20). Two strategies were adopted. (i) To study the requirement for the primary nucleotide sequence in the 3′-SL, nucleotide sequence elements of the WN 3′-SL were substituted for analogous nucleotide segments of the wild-type (wt) DEN2 3′-SL, resulting in a series of DEN2/WN hybrid genomes. (ii) To determine the relative importance of structure versus the primary nucleotide sequence of the long stem in the DEN2 3′-SL, additional mutants with transpositions of wt DEN2 nucleotide sequences within the long stem or an alteration of the wt nucleotide sequence to abrogate formation of the long stem were constructed.
wt and mutant DEN2 RNAs derived by in vitro transcription were transfected into monkey kidney (LLC-MK2) cells to determine their replication phenotypes. Results demonstrated that an 11-bp DEN2 nucleotide sequence constituting the uppermost portion of the “bottom” half of the long stem in the 3′-SL (Fig. 1) was essential for efficient DEN2 virus replication. In contrast, the structure, rather than the primary nucleotide sequence, of the “top” half of the 3′-SL was a determinant of virus growth. One viable mutant virus grew similarly to wt virus in monkey kidney cells but was markedly restricted for growth in cultured mosquito cells.
MATERIALS AND METHODS
Production of DEN2 infectious cDNAs containing mutations in the 3′-SL.
A full-length cDNA copy of a DEN2 genome (New Guinea C strain) had previously been cloned into yeast shuttle vector pRS424 (20). The recombinant plasmid was designated pRS424FLD2. We mutagenized the 3′ terminus of the cloned DEN2 genome by homologous recombination, according to a method previously described (26). Briefly, plasmid pRS424FLD2, containing unique restriction sites SacI and ApaI at the 3′ terminus and 181 nt upstream from the 3′ terminus of the DEN2 sequence, respectively, was digested with these two enzymes. A 181-nt fragment, including nucleotide sequences encoding the wt DEN2 3′-SL, was thus cleaved from pRS424FLD2 recombinant DNA. A PCR product which contained the desired mutations in the nucleotide sequence of the 3′-SL and which overlapped by 50 nt the 5′ and 3′ termini of the SacI- and ApaI-digested recombinant plasmid was cotransformed with the linear SacI- and ApaI-digested DNA into Saccharomyces cerevisiae YPH857 made competent with polyethylene glycol (PEG) (31). After transformation, yeast cells were plated on tryptophan-free medium (Trp−) (23) containing 2% agar and incubated at 30°C for 3 days.
Resultant isolated yeast colonies were transferred to 3 ml of Trp− liquid medium and cultured for 16 to 18 h in a 30°C shaker (250 rpm). Yeast cells were pelleted and resuspended in 200 μl of lysis buffer (1% Triton X-100, 1% sodium dodecyl sulfate (SDS), 100 mM NaCl, 10 mM Tris-HCl [pH 8], 1 mM EDTA) and 200 μl of phenol-chloroform-isoamyl ethanol (25:24:1). The mixture was vortexed with a 200-μl volume of 425- to 600-μ-diameter glass beads (Sigma, St. Louis, Mo.) for 10 min and then centrifuged for 2 min. The supernatant was precipitated in ethanol and resuspended in 50 μl of Tris-EDTA buffer. One microliter of the resulting suspension was used to transform 50 μl of Escherichia coli STBL 2 competent cells (Life Technologies Inc., Bethesda, Md.) to ampicillin resistance. After 3 days of incubation at 30°C, colonies were cultured in 100 ml of superbroth (BioWhittaker; Walkersville, Md.) with ampicillin (100 μg/ml) for 16 h at the same temperature. Plasmid DNA was purified by Qiagen (Chatsworth, Calif.) column tip-100.
Construction of PCR products containing mutations in cDNA encoding the DEN2 3′-SL.
To construct the PCR products used as described above to generate mutations of the DEN2 3′-SL, we utilized a BstYI site or a HinfI site, 79 and 15 nt, respectively, from the 3′ terminus of the DEN2 nucleotide sequence (12). Both of these sites lie within nucleotide sequences constituting the 3′-SL. PCR fragments extending up- and downstream from either of these sites were generated separately, digested with the appropriate endonuclease, and then ligated together in vitro to form the required mutagenic fragment with sufficient overlap of adjacent sequences in pRS424FLD2 DNA. The ligated fragment was then amplified by PCR prior to the cotransformation of yeast. All PCR products were generated by 30 cycles of the following program: 94°C for 1 min, 55°C for 2 min, and 72°C for 1 min. Native Pfu DNA polymerase (Stratagene, La Jolla, Calif.) was used in all PCRs.
PCR products containing mutations D2-SL(c), D2/WN-SL(mutD), and D2/WN-SL(mutE) were constructed in the following manner (these designations are also used to refer to the corresponding mutant viruses) . To obtain the mutations located upstream of the BstYI site, genomic antisense primers containing the BstYI site, the corresponding mutant nucleotide sequence, and 18 3′-terminal complementary nucleotides were used for PCR amplification together with genomic sense primer 1 (5′-GCATGGCGTAGTGGACTAGCGG-3′), which begins 242 nt upstream from the DEN2 cDNA 3′ terminus. To obtain mutations located downstream of the BstYI site, genomic sense primers containing the BstYI site, the desired mutant nucleotide sequence, and 18 complementary nucleotides were used for PCR amplification together with antisense primer 2 (5′-ATGATTACGCCAAGCGCGC-3′) located 55 nt downstream from the DEN2 cDNA 3′ terminus, within the pRS424 vector nucleotide sequence. Two hundred micrograms of each of the PCR products was digested with BstYI and gel purified; then the respective products representing sequences up- and downstream from the BstYI site were ligated at room temperature for 16 h. A volume of 1.0 μl from this ligation reaction mixture was used as the template for further PCR amplification directed by primers 1 and 2. The final PCR products were ethanol precipitated prior to yeast transformation. In a similar manner, mutants D2/WN-SL(mutA), D2-SL(a), and D2-SL(b) were constructed by using the HinfI restriction site.
For mutants D2/WN-SL, D2/WN-SL(mutB), D2/WN-SL(mutC), and D2/WN-SL(mutF), each of the desired mutant fragments downstream from the BstYI site was first synthesized as a positive-sense 95-nt oligonucleotide, including the last 80 nt of either the DEN2 or the WN cDNA sequence and the 5′-proximal 15 nt of the downstream pRS424 vector sequence. Next, a 50-nt antisense oligonucleotide, complementary to vector DNA downstream from the 3′ terminus of the DEN2 cDNA and overlapping the positive-sense 95-nt mutagenic oligonucleotide by 15 nt at its 3′ terminus, was also synthesized. These pairs of oligonucleotides were annealed at the overlapping 15-nt termini and extended by PCR to create 130-bp mutant fragments representing the required mutant nucleotide sequences downstream from the BstYI site.
To generate revertants for the lethal and sublethal mutants D2/WN-SL, D2/WN-SL(mutB), D2/WN-SL(mutC), and D2/WN-SL(mutE), the corresponding wt DEN2 3′-SL cDNA sequence was amplified by PCR, with recombinant plasmid pRS424FLD2 as the template and with primers 1 and 2. This PCR product was used for homologous recombination with each of the respective mutant recombinant cDNAs, which had first been digested with ApaI and SacI to remove the nucleotide segment containing the mutant 3′-SL.
To verify the presence of desired mutations in the context of the pRS424 recombinant plasmids used to generate infectious RNA, all PCR-amplified regions were sequenced. Plasmids were also analyzed by restriction endonuclease digestion, by using the enzymes EcoRI, KpnI, and SacI in concert. Only recombinant plasmids that appeared to yield nine fragments of the correct predicted sizes were used to generate RNA for transfection.
RNA transcription, transfection of LLC-MK2 cells, and virus recovery.
wt or 3′-SL mutant recombinant plasmid DNA (2 μg) was linearized by digestion with the SacI restriction endonuclease and used as the template for RNA transcription catalyzed by SP6 RNA polymerase (Promega; Madison, Wis.), with an SP6 promoter that had been inserted upstream from the DEN2 cDNA insert in pRS424FLD2 (20). RNA transcripts (0.5 μg) were transfected into a continuous line of monkey kidney cells (LLC-MK2) by electroporation. Briefly, RNA was added to LLC-MK2 cells (106) suspended in 300 μl of phosphate-buffered saline. Cells and RNA were incubated on ice for 10 min prior to electroporation at 200 V and 950 μF using a Gene Pulser II with a capacitance extender (Bio-Rad, Hercules, Calif.). Transfected cells were then plated in one 35-mm-diameter well of a six-well tissue culture plate and fed with Eagle’s minimal essential medium containing 10% fetal bovine serum.
IFA to detect virus antigen production.
An indirect immunofluorescence assay (IFA) was performed on days 3 and 10 postelectroporation (p.e.) on cells that had been seeded to a 1-cm2 chamber on a slide (LabTek; Naperville, Ill.) on the day of electroporation. In a second type of experiment involving IFA, a transfected cell monolayer (one 35-mm-diameter well of a six-well plate) was trypsinized on days 5, 10, 15, and 20 p.e. On each of these days, 1/20 of the total cells were transferred to a 1-cm2 chamber slide and IFA was performed on this slide 16 h later. The remaining cells were replated in fresh medium prior to the next time point in each instance. A 1:50 dilution in phosphate-buffered saline of DEN2 hyperimmune mouse ascitic fluid (HMAF; American Type Culture Collection, Manassas, Va.) was used to detect viral antigens in acetone-fixed cells. Fluorescein-conjugated goat anti-mouse antibody (Kirkegaard and Perry Laboratories; Rockville, Md.) was used as a detector antibody at the same dilution. A Leitz Diaplan microscope fitted with a Leica/Wild MPS48 automated photographic system was used for all photomicrographs.
Virus growth curves and plaque morphology.
Each of the supernatants derived from transfected LLC-MK2 cells was harvested when about 70% of the cells were positive for viral antigens and passaged serially in a continuous line of Aedes albopictus cells (C6/36 cells) at 30°C or in LLC-MK2 cells at 37°C for mutant D2/WN-SL(mutF), in order to obtain sufficient titers of virus for further analysis. To determine plaque size, virus in media directly from transfected or from infected cells was serially diluted and used to infect LLC-MK2 cells in paired wells of six-well plates. Plates were incubated at 37°C for 8 or 20 days, and then the monolayer was stained with neutral red for 16 to 18 h. After the staining, plaques were counted and plaque size was measured. To determine a virus growth curve, wt DEN2 and each of the viable mutant viruses were used to infect both LLC-MK2 cells in six-well plates and C6/36 cells in T-25 flasks, at a multiplicity of infection (MOI) of 0.01 in each case. Then, 300 μl of supernatant from infected cells was harvested daily for 8 days. Virus titers for each day and each cell line were determined by plaque assay on LLC-MK2 cells by the method described above.
Verification of the sequences of the mutant viruses.
Viable mutant viruses D2/WN-SL(mutA) and -(mutF) and D2-SL(a) and -(b) were used to infect C6/36 cells [or LLC-MK2 cells for mutant D2/WN-SL(mutF)] in a T-75 flask after three passages each in the respective substrates. When widespread cytopathic effect was observed (7 to 14 days), infected cell media were harvested and clarified by low-speed centrifugation. Then, virus was precipitated with PEG-NaCl as described previously (20). For D2/WN-SL(mutF), the pellet was resuspended in TNE (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) and virus was further purified by being pelleted through an 8.5-ml cushion of 10% glycerol in TNE at 35,000 rpm in an SW40.1 Ti rotor for 4 h at 4°C. RNA was prepared from virus or PEG pellets with the RNEasy kit (Qiagen). The 5′ cap structure on virion RNA was removed by incubation at 37°C for 1 h in a reaction mixture containing 50 mM Na acetate (pH 6.0), 1 mM EDTA, 0.1% 2-mercaptoethanol, 0.01% Triton X-100, 0.2 mM ATP, and 10 to 25 U of tobacco acid pyrophosphatase (Epicentre Technologies) in a final volume of 50 μl. After extraction with phenol-chloroform and ethanol precipitation, “decapped” viral RNA was circularized by incubation overnight at 14°C in a 100-μl reaction volume containing 33 mM Tris-acetate (pH 7.8), 66 mM K acetate, 10 mM Mg acetate, 0.5 mM dithiothreitol, 1 mM ATP, 10% dimethylsulfoxide, 200 U of RNAsin (Promega), and 25 U of T4 RNA ligase (Epicentre Technologies). Circular RNA was used as the template for reverse transcription-PCR (RT-PCR) to amplify the joint containing the ligated 5′ and 3′ ends. The RT-PCR was primed with an oligonucleotide corresponding to antisense DEN2 nt 172 to 155 and a sense primer corresponding to DEN2 nt 10420 to 10437. Reaction conditions were essentially as described previously (20), except that in some cases Expand polymerase (Boehringer-Mannheim) was used instead of Pfu polymerase (Stratagene) for PCR. Amplified products were sequenced with the antisense primer described above.
For mutants D2/WN-SL and D2/WN-SL(mutD), an RT-PCR product containing the 3′-SL nucleotides was derived by conventional methods from linear viral RNA, with a genomic antisense primer complementary to the expected 3′-terminal 18 nt of WN RNA (for D2/WN-SL) or the 3′-terminal 23 nt of DEN2 NGC RNA [for D2/WN-SL(mutD)] and a genomic sense primer representing the DEN2 genomic nucleotide sequence upstream from the 3′-SL. D2/WN-SL RNA was prepared from total cellular RNA after TRIzol extraction, whereas D2/WN-SL(mutD) RNA was prepared from PEG-precipitated virus. The nucleotide sequences of all PCR products were obtained by an automated method (ABI model 377 automated DNA sequencer and a Prism dye terminator cycle sequencing kit; ABI, Columbia, Md.).
Computer analysis of wt and mutant 3′-SL nucleotide sequences.
The predicted secondary structures of DEN2 and WN wt 3′-SL nucleotide sequences and of the corresponding mutant nucleotide sequences were ascertained by using the program DNAsis, version 2.0 on a Power Macintosh 9500 computer.
Viral protein and RNA studies.
Pairs of six-well plates containing confluent monolayers of LLC-MK2 cells were infected with wild-type DEN2 or each of the mutant viruses, at an MOI of 0.05. After 2 days, one such plate was starved for methionine and cysteine for 1 h and then labeled with [35S]methionine plus [35S]cysteine at a concentration of 100 μCi/ml (>3,000 Ci/mmol; Amersham, Arlington Heights, Ill.) for 4 h. Cells were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 100 mM Tris-HCl [pH 7.4], 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS), and DEN virus-specific proteins were immunoprecipitated with DEN2 HMAF at a 1:50 dilution. Immune complexes were collected on Pansorbin beads (Calbiochem; La Jolla, Calif.). Precipitated proteins were analyzed by electrophoresis on an SDS–12% polyacrylamide gel with a tricine-based buffer system (22). Total cellular RNA was prepared from the second plate of the pair with TRIzol reagent. Viral RNA was detected and quantified by slot blot hybridization on a Hybond-N nylon membrane (Amersham), as suggested by the supplier. Briefly, RNA samples were denatured at 65°C for 5 min with a mixture containing 50% formamide, 30% formaldehyde, and 1× MOPS (morpholinepropanesulfonic acid) buffer and then chilled on ice. SSC (20×; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was added to adjust the final concentration of the sample to 5× SSC. RNAs were applied to the nylon membrane and cross-linked to it with UV light. To generate the DEN2 cDNA probe, pRS424FLD2 DNA was digested with restriction enzymes SphI and StuI, followed by gel purification of a product cDNA containing nt 1379 to 7871 of the DEN2 sequence. This cDNA was radiolabeled with [32P]dCTP (Amersham) to a specific activity of 108 cpm/μg of DNA by using the Prime-It kit (Stratagene). Hybridization was performed at 50°C for 16 to 18 h in a buffer containing 5× SSC, salmon sperm DNA (100 μg/ml), 1% SDS, 1 mM EDTA, and radiolabeled DNA probe (2 × 105 cpm).
RESULTS
Impaired replication of a chimeric DEN2 virus containing the WN 3′-SL.
The 3′-SL in flavivirus RNA was defined as a short stem-and-loop structure together with an adjacent long stem-and-loop structure, formed with a predicted high level of thermal stability by the 3′-terminal nucleotide sequence (6). The 3′-SL is structurally conserved in flavivirus RNAs; however, the primary nucleotide sequences of the 3′-SLs of different species are only semiconserved, at best (reviewed in reference 9). For example, the primary nucleotide sequences of the 3′-SL in the DEN2 strain New Guinea C genome (12) (Fig. 1A) and in the WN strain E101 genome (4) (Fig. 1B) are quite divergent in the region of the long stem, whereas the nucleotide sequence of the short stem-and-loop structure is relatively well conserved between these species and among other flaviviruses.
To study the structural and nucleotide sequence requirement for the 3′-SL for virus replication, a DEN2-WN chimeric genome was first constructed, starting from a full-length cDNA copy of the genome of a mouse brain-adapted DEN2 virus cloned in a yeast shuttle vector, the recombinant plasmid pRS424FLD2 (20). The chimeric genome and other mutant genomes were constructed by homologous recombination of cleaved pRS424FLD2 DNA with the PCR product(s) containing the desired mutation(s) in yeast. The initial mutant construct (D2/WN-SL) contained the full-length wt DEN2 sequence, except that the last 96 nt of the WN genome (the WN 3′-SL) was substituted for the 3′-terminal 93 nt of the DEN2 sequence, comprising the wt DEN2 3′-SL (Fig. 1 and 2). The nucleotide sequence chosen to represent the WN 3′-SL had been determined from WN strain E101 viral RNA by Blackwell and Brinton (4).
FIG. 2.
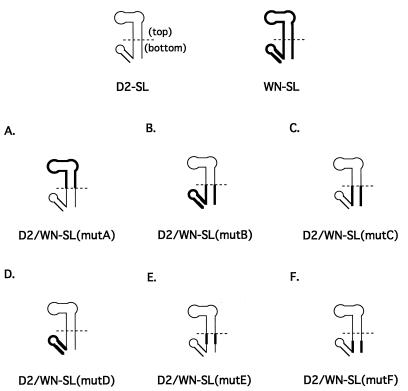
The composition of the 3′-SLs in mutant viruses are depicted. The DEN2 nucleotide sequence is shown as a thin line, and the WN nucleotide sequence is shown as a thicker line. (A) Mutant D2/WN-SL(mutA) contained a substitution of DEN2 nt 18 to 62 by WN nt 17 to 66, the top portion of the 3′-SL. (B) Mutant D2/WN-SL(mutB) contained a substitution of DEN2 nt 1 to 17 and 63 to 93 by WN nt 1 to 16 and 67 to 96, respectively, the bottom portion of the 3′-SL. (C) Mutant D2/WN-SL(mutC) contained a substitution of DEN2 nt 1 to 17 and 63 to 79 by WN nt 1 to 16 and 67 to 80, respectively, the bottom portion of the long stem. (D) Mutant D2/WN-SL(mutD) contained a substitution of DEN2 nt 80 to 93 by WN nt 81 to 96, the short stem and loop. (E) Mutant D2/WN-SL(mutE) contained a substitution of DEN2 nt 7 to 17 and 63 to 73 by WN nt 7 to 16 and 67 to 75, respectively. (F) Mutant D2/WN-SL(mutF) contained a substitution of DEN2 nt 1 to 7 and 73 to 79 by WN nt 1 to 7 and 75 to 80, respectively.
Initially, RNAs derived from transcription of the linearized DEN2 wt and D2/WN-SL mutant recombinant plasmids were electroporated into LLC-MK2 cells. The infectivity of mutant RNA compared to wt RNA was assessed by IFA for DEN2 antigens in transfected cells, by using murine anti-DEN2 antibodies. The D2/WN-SL chimera was negative for DEN2 antigens on day 3 (Table 1), and less than 10% of transfected cells were DEN2 antigen positive by day 10. In contrast, DEN2 wt RNA-transfected cells were positive for DEN2 antigens by IFA after 24 h and nearly 100% of cells in the monolayer were positive by day 6 or 7. Assuming that the efficiencies of transfection of cells by wt and mutant RNAs were approximately equal and that cells constituting the monolayer were homogeneous with respect to their ability to support virus replication, this result suggested that the D2/WN-SL mutant virus did replicate in the transfected cells but that replication was markedly impaired in comparison to that of wt virus. Thus, we interpreted the relatively late, delayed spread of positive fluorescence in the monolayer as evidence that progeny virions resulting from transfection were infectious. We termed this phenotype sublethal.
TABLE 1.
Properties of wt DEN2 and 3′-SL mutant viruses derived by transfection of LLC-MK2 cells
| Virus | Result of IFAa at dayb:
|
Plaque size (mm) (day)c | |
|---|---|---|---|
| 3 | 10 | ||
| wt DEN2 | ++ | ++++ | 2.0 (8) |
| D2/WN-SL | − | + | 1.5 (20) |
| D2/WN-SL(mutA) | + | +++ | 2.0 (20) |
| D2/WN-SL(mutB) | − | − | NA |
| D2/WN-SL(mutC) | − | − | NA |
| D2/WN-SL(mutD) | + | ++ | 3.0 (20) |
| D2/WN-SL(mutE) | − | + | 1.5 (20) |
| D2/WN-SL(mutF) | + | ++ | 4.0 (20) |
| D2-SL(a) | + | +++ | 4.0 (20) |
| D2-SL(b) | + | ++ | 1.5 (20) |
| D2-SL(c) | − | − | NA |
Cells were stained on the days indicated with DEN2 HMAF (see Materials and Methods). The percentage of DEN antigen-positive cells was determined by examination of at least 100 cells. −, no positive cells; +, <10% positive cells; ++, 10 to 40% positive cells; +++, 40 to 70% positive cells; ++++, 70 to 100% positive cells.
Days p.e. of LLC-MK2 cells. One-half milligram of RNA was transfected into 106 cells. On day 0, 105 transfected cells were seeded to a 1-cm2 chamber for IFA.
Viable mutant viruses were passaged three times in C6/36 cells or in LLC-MK2 cells [D2/WN-SL(mutE)]. The sequence of the 3′-terminal 242 nt in viral RNA was then verified, and the diameter of plaques was determined in LLC-MK2 cells. For all viable mutant viruses, plaques were not evident at day 8 postinfection. NA, not applicable; the mutation was lethal.
Because transfected cell monolayers did not always remain viable for sufficient periods of time to observe the growth of D2/WN-SL and other mutant viruses with the sublethal phenotype, a second type of assay for infectivity after transfection was performed. Here, IFA was carried out on days 5, 10, 15, 20, and, in some instances, day 25 p.e., but the transfected cell monolayer was reseeded in fresh medium at each time point. In this assay also, D2/WN-SL virus exhibited poor growth compared to wt (Fig. 3). Only about 40% of cells were positive by day 25 (data not shown). Thus, the WN 3′-SL could not efficiently substitute for the DEN2 3′-SL to support DEN2 replication, despite the nearly identical predicted secondary structures of the two nucleotide sequences.
FIG. 3.
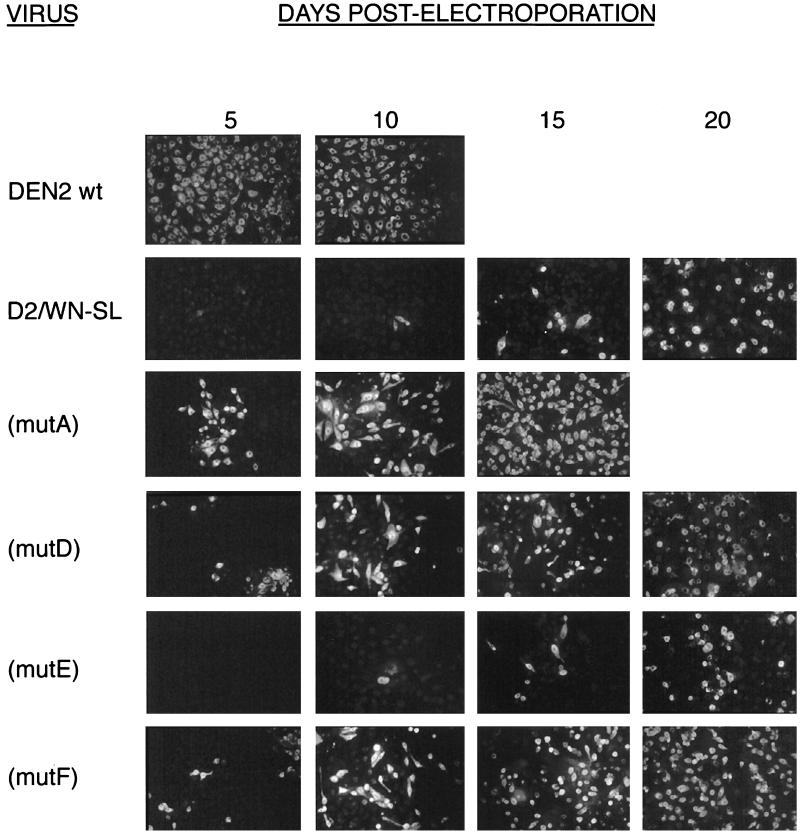
IFA for growth of DEN2 wt and DEN2-WN chimeric mutant viruses. A total of 106 LLC-MK2 cells were transfected by electroporation with 500 ng of wt or mutant viral RNAs that had been transcribed in vitro. Cells were then seeded to one 35-mm-diameter well of a six-well tissue culture plate. On days 5, 10, 15, and 20 p.e., the monolayer was disrupted by trypsinization and 5% of the total cells were seeded to a coverslip for IFA. The remaining cells were recultured in fresh medium at each time point. DEN2 murine hyperimmune ascitic fluid was used in the assay at a 1:50 dilution to detect DEN2 antigen-positive cells. Cells were fixed in acetone. See Fig. 2 for genotypes of mutant viruses.
Nucleotide sequence elements of the DEN2 3′-SL required for virus replication.
A series of DEN2-WN 3′-SL chimeric genomes was next constructed in order to determine which DEN2 nucleotide sequence elements within the 3′-SL were required for efficient virus replication. In this study, specific regions of the DEN2 3′-SL were substituted for by the structurally analogous specific regions of the WN 3′-SL (Fig. 2). Initially, two such genomes were constructed, and their infectivity was assessed. D2/WN-SL(mutA) (Fig. 2A) contained a substitution of the top half of the DEN2 3′-SL (nt 18 to 62, numbering in the upstream direction from the 3′-terminal nucleotide of the genome; Fig. 1A) with that of WN (nt 17 to 66; Fig. 1B). D2/WN-SL(mutB) contained the converse substitution (Fig. 2B); the bottom half of the DEN2 3′-SL sequence, nt 1 to 17 and 63 to 93, was swapped for WN nt 1 to 16 and 67 to 96, respectively. The bottom half of the WN 3′-SL alone and the analogous segment of the DEN3 3′-SL had previously been shown to contain the binding site for an unidentified 84-kDa BHK cell protein in vitro, whereas the bottom half plus the next 5 bp of the top half of the WN structure were required to bind a specific 105-kDa BHK cell protein (Fig. 1B) (3). The 50-kDa translation elongation factor, eF1-α, bound to a 3-nt linear site in the top half of the WN long stem (4).
For D2/WN-SL(mutA), IFA was positive by day 3 after transfection of LLC-MK2 cells, and 40 to 70% of cells were positive by day 10, when the monolayer was maintained continuously after transfection (Table 1). In the second IFA, when monolayers were reseeded at 5-day intervals, cells became ∼100% antigen positive by day 15, whereas wt RNA-transfected cells were 100% IFA positive between days 5 and 10 (Fig. 3). Thus, this mutant was “viable” but appeared to replicate less efficiently than wt. In marked contrast, the mutation of the DEN2 genome in D2/WN-SL(mutB) was lethal; IFA for DEN2 antigens in transfected cells was negative, even after 25 days (data not shown). This suggested that substitution of analogous WN 3′-SL nucleotide sequences was tolerated for the top half of the DEN2 3′-SL but that there was a specific requirement for DEN2 nucleotide sequences comprising the bottom half of the structure.
To define which portion of the DEN2 nucleotide sequence in the bottom half of the 3′-SL was absolutely required for viral replication, four additional mutants were generated. D2/WN-SL(mutC) contained the bottom half of the WN 3′-SL long stem in place of the analogous domain of the wt DEN2 3′-SL (Fig. 2C); WN nt 1 to 16 and 67 to 80 replaced, respectively, DEN2 nt 1 to 17 and 63 to 79 (Fig. 1). Thus, it retained the DEN2 nucleotide sequence of the short stem-and-loop structure. Conversely, only the WN short stem-and-loop nucleotide sequence (WN nt 81 to 96) was substituted for analogous DEN2 nucleotide sequences (DEN2 nt 80 to 93) in D2/WN-SL(mutD) (Fig. 2D). Mutation D2/WN-SL(mutC) was lethal; IFA of the transfected LLC-MK2 cell monolayer for DEN2 antigens remained negative at all times p.e. up to 20 days. In contrast, mutant virus D2/WN-SL(mutD) replicated efficiently; a small percentage of cells were DEN2 antigen positive by day 3 (Table 1), and essentially all cells were displaying viral antigen by day 20 in the discontinuous culture assay (Fig. 3). As for the other viable mutant viruses, replication of D2/WN-SL(mutD) was slightly less vigorous than that of wt DEN2 virus, as judged by the spread of fluorescence in the transfected monolayer (Table 1; Fig. 3). Thus, the incompatibility between WN and DEN2 nucleotide sequences in the hybrid 3′-SL structures appeared to be much more related to the presence of WN nucleotide sequences in the bottom half of the long stem than to the presence of such sequences in the short stem and loop or in the top half of the long stem and loop.
Mutation D2/WN-SL(mutE) and D2/WN-SL(mutF) were next constructed to define which WN nucleotide sequences in the bottom half of the long stem were not compatible with efficient DEN2 virus replication (Fig. 2E and F). D2/WN-SL(mutE) contained an 11-bp substitution of the upper portion of the bottom of the long stem sequence, nt 7 to 17 and 63 to 73, by that of WN nt 7 to 16 and 67 to 75, respectively, whereas, in D2/WN-SL(mutF), the bottom portion of the same region of the long stem (nt 1 to 7 and 73 to 79) was exchanged for the WN counterpart (nt 1 to 7 and 75 to 80).
The D2/WN-SL(mutE) mutation was sublethal; IFA was negative at day 3, and less than 10% of cells were positive on day 10 (Table 1). As assessed by the IFA on discontinuously cultured transfected cells (Fig. 3), replication of D2/WN-SL(mutE) virus could be seen to parallel that of the parent mutant, D2/WN-SL. Virus derived from construct D2/WN-SL(mutF) was obviously more viable; about 10% of cells were positive on day 3, and about 40% of cells were positive by day 10 (Table 1). Essentially all cells were positive by day 20 in the discontinuous culture assay (Fig. 3). Therefore, the DEN2 nucleotide sequence of the uppermost portion of the bottom half of the long stem in the 3′-SL was not dispensable for efficient virus replication; replacement of that segment by the analogous WN nucleotide sequence was lethal or sublethal in all mutant constructs (top portion of Fig. 2; Fig. 2B, C, and E). In contrast, other DEN2 nucleotide sequences within the 3′-SL (Fig. 2A, D, and F) appeared to be exchangeable for analogous WN nucleotide sequences with much less significant loss of replication efficiency.
Additional mutations of the long stem in the DEN2 3′-SL.
An additional group of mutants was constructed to verify the suggestion that the conformation of the upper half of the long stem in the DEN2 3′-SL, rather than its nucleotide sequence, was of primary importance for virus replication. In mutant D2-SL(a), the wt nucleotide sequences comprising the uppermost 6 bp of the top half of the long stem were transposed (Fig. 1A and 4A). DEN2 nt 24 to 29 in the “right” strand of the stem were substituted for by nt 51 to 56. Conversely, nt 51 to 56 in the “left” strand were substituted for by nt 24 to 29. In mutant D2-SL(b), the 12-nt complementary sequences of the right and left strands of the entire top half of the long stem (nt 18 to 29 and 51 to 62, respectively) were similarly transposed (Fig. 1A and 4B). For both these mutants, only the positions of portions of the wt long stem nucleotide sequence were altered; both constructs would be predicted to retain a double-stranded configuration with free energy identical to that of the wt DEN2 3′-SL. In mutant D2-SL(c), base pairing in the upper portion of the long stem was disrupted; nt 18 to 29 were substituted for by a repeat of the complementary sequence of the opposite strand, nt 51 to 62 (Fig. 1A and 4C). These predicted effects of mutations D2-SL(a), -(b), and -(c) on the secondary structure of the 3′-SL were confirmed by computer analysis of the respective mutant nucleotide sequences.
FIG. 4.
Mutations in the context of the all-DEN2 3′-SL and their putative conformations are depicted. (A) The 6-bp segment comprising the uppermost portion of the long stem was transposed; DEN2 nt 24 to 29 and 51 to 56, indicated by crosshatched rectangles, were exchanged between strands. (B) The 12-bp segment comprising the top half of the long stem was transposed; DEN2 nt 18 to 29 and 51 to 62, indicated by crosshatched rectangles, were exchanged between strands. (C) Base pairing in the top half of the long stem was disrupted. nt 18 to 29 were deleted and replaced by a repeat of the sequence of the complementary strand, nt 51 to 62.
Mutant RNAs D2-SL(a) and -(b) yielded viable virus after electroporation into LLC-MK2 cells (Table 1). Both gave positive fluorescence in up to 10% of cells by 3 days. Cells transfected with mutant D2-SL(a) RNA were up to 70% positive by IFA for DEN2 antigens after 10 days, and essentially 100% of cells in the monolayer were positive by day 15 in the discontinuous culture IFA (Fig. 5). For mutant D2-SL(b), IFA was positive in up to 40% of cells in 10 days and in nearly all cells by day 20 for the same assay. In contrast, mutation D2-SL(c) was lethal. No DEN2 antigen-producing cells were detected at any time after electroporation up to day 20. These results were consistent with those obtained for the DEN2-WN chimeric viruses: Repositioning of wt DEN2 nucleotide sequences within the top half of the long stem in the 3′-SL did not have severe effects on virus replication, as long as the double-stranded configuration of the structure was conserved. However, disruption of base pairing in the top half of the long stem was lethal.
FIG. 5.
IFA for growth of DEN2 wt and DEN2 3′-SL mutant viruses after transfection of LLC-MK2 cells. The assay was conducted as described in the legend to Fig. 3. See Fig. 4 for genotypes of mutant viruses.
Construction of wt revertants from genomes of viruses with lethal and sublethal phenotypes and computer analysis of mutant 3′-SLs.
To reduce the possibility that a technical or procedural error could account for the observed lethal or sublethal phenotypes of the D2/WN-SL, D2/WN-SL(mutB), D2/WN-SL(mutC), D2/WN-SL(mutE), and D2-SL(c) mutations, each transfection experiment was repeated four times. In addition, to reduce the possibility that we had introduced an occult lethal mutation into the wt DEN2 cDNA during mutagenesis, we “rescued” mutants by replacing the 3′-terminal 242-nt sequence of the respective mutant cDNAs with that of wt DEN2. The resulting viruses consistently had growth properties indistinguishable from those of wt DEN2 (data not shown), indicating that the growth phenotypes of the mutant viruses were likely due to the intentional introduction of mutations into the 3′-SL nucleotide sequence.
In addition to rescuing the lethal and sublethal mutations, we also analyzed all mutations by computer to determine the predicted secondary structures of mutant 3′-SLs. All mutant viral RNAs, except mutant RNA D2-SL(c), were predicted to contain 3′-SL secondary structures with base pairing identical to that predicted for the corresponding wt DEN2 or WN 3′-SL nucleotide sequences. Therefore, disruption or alteration of the 3′-SL secondary structure per se was unlikely to account for the observed differences in the viability of mutants, except as intended in the case of mutant D2-SL(c).
Kinetics of replication of mutant viruses in LLC-MK2 and C6/36 cells.
Supernatant from cells electroporated with the “parent” mutant RNA, that of D2/WN-SL, was initially used to infect both LLC-MK2 cells and C6/36 cells. Both of these cell lines are permissive for WN and DEN2 replication. After incubation periods of up to 3 weeks, virus released into the medium was quantified by plaquing in LLC-MK2 monolayers. The highest titer achieved in medium from either cell line, even after several passages, was ∼60 PFU/ml (data not shown). Therefore, D2/WN-SL virus could not be included in the analysis of growth kinetics. For similar reasons, mutants D2/WN-SL(mutB), -(mutC), and -(mutE) were also excluded. The viable mutant viruses were passaged three times in C6/36 cells in order to obtain titers sufficient for determining growth curves, with the exception of mutant D2/WN-SL(mutF), which replicated very poorly in C6/36 cells and was therefore passaged in LLC-MK2 cells prior to titration in both cell lines.
The growth rates of the viable viruses in both LLC-MK2 and C6/36 cells were determined. Cells were infected with each of the mutants or wt DEN2 at an MOI of 0.01. Virus secreted into the medium was then titrated daily for 8 days. The peak titer for wt DEN2 in LLC-MK2 cells was between 105 and 106 PFU/ml, achieved on day 6 postinfection (Fig. 6A). Mutants D2-SL(a), D2-SL(b), and D2/WN-SL(mutA) were about 10-fold reduced in their peak titers compared to the wt on day 6. However, two of the mutants, D2/WN-SL(mutD) and D2/WN-SL(mutF), achieved titers of about 105 PFU/ml by day 8 postinfection, nearly comparable to the day 6 peak titers for wt DEN2 (Table 2). We noted that titers of D2/WN-SL(mutF) were 100- to 1,000-fold reduced compared to those of the wt on days 2 through 4 after infection (Fig. 6A). Thus, it was possible that this mutant further adapted to growth in LLC-MK2 cells during the course of the experiment by an occult mutation (in addition to that noted below). This seemed unlikely, however, since the virus had already been passaged three times in LLC-MK2 cells prior to the growth assay.
FIG. 6.
Growth curves for wt and viable mutant DEN2 viruses. The supernatant of LLC-MK2 cells that had been transfected with in-vitro-transcribed wt and mutant DEN2 viral RNAs was used to infect C6/36 cells [or LLC-MK2 cells for mutant D2/WN-SL(mutF)], when the transfected cell monolayer was about 70% positive for DEN2 antigens by IFA. wt and viable mutant viruses were harvested and passaged three times in either of the substrates, and then the 3′-terminal 242 nt of each of the viral genomes were sequenced to confirm the stability of the respective mutations. Virus stocks were then used to infect LLC-MK2 cells (A) and C6/36 cells (B) at an MOI of 0.01 in each case, and growth was monitored by plaque titration (in LLC-MK2 cell monolayers; see Materials and Methods) of virus in supernatants on the days indicated.
TABLE 2.
Peak titers of wt and viable chimeric mutant viruses in LLC-MK2 and C6/36 cellsa
| Virusb | Peak titer (104 PFU/ml) (day postinfection) in:
|
|
|---|---|---|
| LLC-MK2 cells | C6/36 cells | |
| DEN2 wt | >10 (6) | >1,000 (3) |
| D2/WN-SL(mutA) | 1 (5) | >100 (4) |
| D2/WN-SL(mutD) | 10 (6–8) | >10 (5) |
| D2/WN-SL(mutF) | >10 (8) | 10−2 (7) |
| D2-SL(a) | >1 (5) | >100 (6) |
| D2-SL(b) | 1 (5) | >10 (5) |
All viruses tested yielded higher titers in C6/36 cells than in LLC-MK2 cells, except D2/WN-SL(mutF) (Table 2). In addition, all mutant viruses were at least 10-fold reduced in peak titer compared to wt DEN2 (>107 PFU/ml on days 3 to 5 [Fig. 6B]). Mutants D2-SL(a) and D2/WN-SL(mutA) attained peak titers of >106 PFU/ml, and mutants D2-SL(b) and D2/WN-SL(mutD) attained peak titers of >105 PFU/ml. Peak titers for all mutant viruses were achieved from 1 to 3 days later after infection than the corresponding time for wt virus. Surprisingly, D2/WN-SL(mutF) grew very poorly in C6/36 cells. The peak titer was 102 PFU/ml on day 7, 105-fold reduced compared to that for wt virus in C6/36 cells. However, this mutant grew at late times after infection to titers similar to that for wt virus in LLC-MK2 cells. D2/WN-SL(mutF) virus was significantly and specifically restricted for growth in C6/36 cells compared to all other viable mutant viruses.
Nucleotide sequence analysis of the genomes of viable mutant viruses and of mutant D2/WN-SL.
Initially, after three passages in C6/36 cells [or three passages in LLC-MK2 cells for D2/WN-SL(mutF)], the nucleotide sequence of the 3′ terminus of each viable mutant virus genome was wholly or partially verified. For mutants D2/WN-SL(mutA) and -(mutF) and D2-SL(a) and -(b), purified viral RNA was decapped and circularized by ligation with T4 RNA ligase. Then RT-PCR was performed to derive a cDNA that spanned the 5′-to-3′ junction of the viral RNA and that included the entire 3′-SL nucleotide sequence, and these PCR products were sequenced.
By this analysis, mutant D2-SL(a) was shown to have sustained no spontaneous mutations within the 3′-SL during virus replication. However, mutant D2-SL(b) and D2/WN-SL(mutA) RNAs each contained an identical spontaneous mutation of nt G5 to U, in the context of the DEN2 nucleotide sequence forming the bottom half of the 3′-SL in each of these mutant constructs (Fig. 1A). This mutation had the effect of abrogating a G-C base pairing in the DEN2 long stem. The significance of its presence in the passaged viable mutant viruses is currently the subject of further investigation.
D2/WN-SL(mutF) RNA also contained a spontaneous point mutation, a deletion of nt A3 (see the WN 3′-SL nucleotide sequence; Fig. 1B). This nucleotide is unpaired in the WN 3′-SL sequence determined by Blackwell and Brinton (3, 4) (Fig. 7A), and its deletion alters the 3′-terminal 7-nt sequence of the D2/WN-SL(mutF) RNA from 3′-UCA3UAGG to 3′-UCUAGGA (in which all nucleotides are hydrogen bonded to the opposite strand of the long stem; Fig. 7B). For comparison, the wt DEN2 3′-terminal 7-nt sequence is 3′-UCUUGGA, where U4 is part of a U-U unbonded “bulge” in the long stem (Fig. 1A and 7C). The two nucleotide differences in the 3′-SL of RNA from replicating D2/WN-SL(mutF) virus compared to that of wt DEN2 RNA, A versus U at nt 4 and U versus C at nt 74 in the 3′-SL, were apparently sufficient to abrogate replication when D2/WN-SL(mutF) virus derived in monkey kidney cells was used to infect mosquito cells. This result raised the possibility that the lethal or sublethal phenotype of mutants containing the bottommost segment of the WN 3′-SL was related to the presence of nt A3 in genomic RNA. For mutant D2/WN-SL(mutD), no spontaneous mutations in the 3′-SL were detected by a technique that excluded sequence analysis of the 3′-terminal 23 nt (see Materials and Methods).
FIG. 7.
Locus of a spontaneous mutation in the 3′-SL of D2/WN-SL(mutF) RNA associated with replication in monkey kidney cells and its effect on 3′-SL structure. Nucleotides are numbered from the 3′ terminus of the RNA genome, as in Fig. 1A and B. (A) Nucleotide sequence of the bottommost segment of the long stem in the 3′-SL of input D2/WN-SL(mutF) virion RNA, representing the substitution mutation introduced into D2/WN-SL(mutF) cDNA (see also Fig. 1A, 1B, and 2F). The substituted nucleotides were derived from the WN strain E101 nucleotide sequence (4) (Fig. 1B). Broad dashed lines indicate that the remainder of the 3′-SL nucleotide sequence is the same as that of wt DEN2 NGC RNA. Unpaired nt A3 (shown in boldface) was deleted (dl A3) in RNA isolated from D2/WN-SL(mutF) virus replicating in LLC-MK2 cells after transfection, as shown in panel B and as suggested by an arrow. (Note that the nucleotide numbering has been altered in panel B to reflect the deletion mutation.) (C) The analogous segment in the 3′-SL of wt DEN2 strain NGC virus RNA. Compared to the D2/WN-SL(mutF) nucleotide sequence, U74 is replaced by C74 (boldface) and the hydrogen-bonded U76-A4 base pair is replaced by unbonded U76/U4, forming a bulge in the long stem. (D) The nucleotide sequence of the analogous segment in the 3′-SL of an unspecified strain of WN virus, as determined by Wengler and Castle (33). The Wengler and Castle WN nucleotide sequence does not contain an unpaired A in the position analogous to that found in the Blackwell and Brinton nucleotide sequence. In panel D only, the fine dashed lines signify that the remainder of the 3′-SL nucleotide sequence is that of WN virus RNA, as determined by Wengler and Castle (data not shown).
The sequencing procedure obviously did not rule out the possibility that viable mutant viruses might have sustained additional spontaneous mutations upstream from the sequenced 3′ terminus. This is the subject of further study. However, a partial genomic nucleotide sequence upstream from the 3′-SL was obtained from D2/WN-SL virus RNA prepared 20 days after infection of LLC-MK2 cells, to determine whether the spread of infection by this sublethal mutant virus at very late times after infection was related to the occurrence of a second-site mutation. No such mutation was detected within a domain that included the entire NS5 gene sequence, as well as the entire 3′ NCR of the D2/WN-SL genome, except for the 3′-terminal 18 nt.
Plaque morphology in LLC-MK2 cells.
The size of plaques formed by the viruses bearing the sublethal mutations D2/WN-SL and D2/WN-SL(mutE) was assessed by using virus harvested directly from transfected LLC-MK2 cells. Plaque size for the viable mutant viruses was assessed by using virus passaged in C6/36 cells [or in LLC-MK2 cells for mutant D2/WN(mutF)], as well as virus derived directly from transfected cells. wt DEN2 virus produced plaques with a diameter of 2 mm after 8 days of infection, while all the mutant viruses required 20 days to produce easily detectable plaques (Table 1). After 20 days, mutants D2/WN-SL(mutF) and D2-SL(a) produced 4-mm-diameter plaques. Mutants D2/WN-SL(mutA) and D2/WN-SL(mutD) produced 2- and 3-mm-diameter plaques, respectively, and mutants D2/WN-SL, D2/WN-SL(mutE), and D2-SL(b) produced plaques 1.5 mm in diameter. In general, plaque size correlated with results of IFA; viruses that were seen to spread from cell to cell most rapidly by that assay also made the largest plaques, with the exception of the relatively small plaque size seen for mutant D2-SL(b). For the viable mutants, plaques formed by virus derived directly from transfection were not different in size from plaques formed by passaged virus.
Analyses of viral RNA and protein synthesis.
Viable mutants D2-SL(a), D2-SL(b), D2/WN-SL(mutA), D2/WN-SL(mutD), and D2/WN-SL(mutF) were used to infect LLC-MK2 cells and C6/36 cells at an MOI of 0.05. Total cellular RNA was extracted after 2 days. Slot blot hybridization was performed with a [32P]dCTP-labeled DEN2 cDNA probe representing nt 1379 to 7871 of the DEN2 nucleotide sequence (12). Since existing evidence suggests that subgenomic-sized RNAs are not produced during flavivirus replication (5, 28), this assay was expected to detect all positive- and negative-sense DEN2 RNAs. The amount of viral RNA detected correlated with the titers of viruses in the growth curves at day 2 (Fig. 8; see also Fig. 6A and B). wt DEN2 viral RNA was more abundant than that of any of the mutant viruses in both cell lines. Viral RNA in D2/WN-SL(mutA)-infected cells was next most abundant, and the titer of this virus on day 2 was about 10-fold higher than those for the other mutants, in both cell lines. As would be expected based on its growth characteristics, D2/WN-SL(mutF) RNA was the least abundant of the viral RNAs in LLC-MK2 cells at day 2 and was undetectable in C6/36 cells.
FIG. 8.
Viral RNAs in infected LLC-MK2 and C6/36 cells. Viruses derived by transfection were passaged in C6/36 or in LLC-MK2 cells, as described in the legend to Fig. 6, and then used to infect cell monolayers at an MOI of 0.05. After 2 days, cells were lysed and RNA derived from one 35-mm-diameter well of a six-well plate was recovered and applied to a nylon membrane. Virus-specific RNA was detected with a 32P-labeled cDNA probe representing nt 1379 to 7871 of the DEN2 genome.
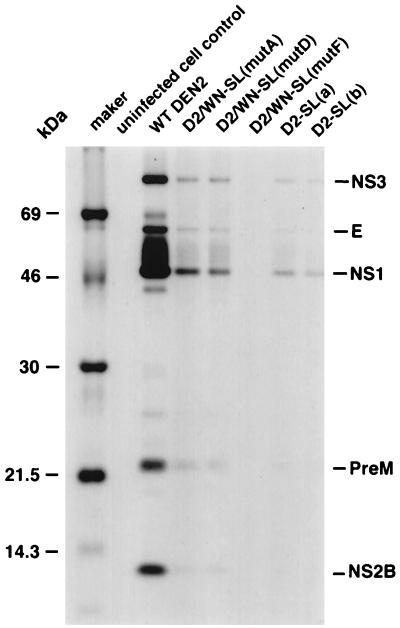
Viral protein synthesis in infected LLC-MK2 cells was also estimated (Fig. 9). For this experiment, LLC-MK2 cells were infected and processed in parallel with cells used for viral RNA analysis. Two days postinfection, cells were labeled for 4 h with [35S]methionine and [35S]cysteine and then lysed. Proteins were immunoprecipitated with DEN2 HMAF and analyzed by SDS-polyacrylamide gel electrophoresis. The DEN2 viral proteins prM, E, NS1, NS2B, and NS3 were then identified by size. The relative amounts of viral proteins detected paralleled the amounts of viral RNA detected at the chosen time point. Cells infected with mutant viruses yielded much less radiolabeled viral protein than did cells infected with wt DEN2. However, the ratios of each of the identifiable proteins were not obviously different for any of the mutants, compared to those for wt DEN2. In keeping with the results of the assay for virus-specific RNA, D2/WN-SL(mutF)-specific proteins were barely detectable on day 2, when the titer of this virus was 10- to 100-fold reduced compared to those of the other viable mutant viruses and to that of the wt (Fig. 6A). Since there was a reasonable correlation between amounts of viral proteins and amounts of viral RNAs detected on day 2, we inferred that the defects in the replication of the mutant viruses were likely to be at the level of RNA synthesis, rather than at the level of translation. The assays for RNA and protein were repeated at day 4 after infection, with completely analogous results (data not shown).
FIG. 9.
Viral proteins in infected LLC-MK2 cells. Viruses derived by transfection were passaged in C6/36 or in LLC-MK2 cells, as described in the legend to Fig. 6, and then used to infect LLC-MK2 cell monolayers at an MOI of 0.05, in parallel with cells used in the analysis of viral RNA synthesis shown in Fig. 7. After 2 days, cells were radiolabeled for 4 h with [35S]promix (methionine plus cysteine). Then cell monolayers were disrupted by trypsinization, and cells were lysed in radioimmunoprecipitation assay buffer. Proteins in the lysate from one 35-mm-diameter well of a six-well plate were immunoprecipitated with DEN2 HMAF, and precipitates were collected with Pansorbin beads. Radiolabeled proteins were analyzed on a tricine-buffered SDS–12% polyacrylamide gel. DEN2 proteins were presumptively identified by size.
DISCUSSION
Flavivirus genomic RNAs contain 5′ and 3′ NCRs with lengths of approximately 100 and 400 to 600 nt, respectively. The 3′-terminal 90 to 100 nt of the 3′ NCR is predicted to form thermodynamically stable, adjacent stem-loop structures, collectively referred to here as the 3′-SL (5, 6, 11, 17, 21, 33). Although the 3′-SL structure is conserved among flaviviruses, the primary nucleotide sequences in this region of the genome are quite heterogeneous (9). For example, the WN and DEN2 nucleotide sequences are only 37% homologous (data not shown). However, higher levels of homology exist over localized areas within the 3′-SL, such as in the small stem-loop structure (Fig. 1).
Accumulated evidence suggests that the 3′-SL and an analogous conserved structure in the 5′ NCR play a crucial role in flavivirus replication. In vivo, deletions of 3′-NCR nucleotide sequences upstream from the 3′-SL in a DEN4 infectious cDNA were relatively well tolerated, whereas a deletion that extended into the nucleotide sequence required to form the small stem-loop structure in the 3′-SL was lethal for DEN4 replication (16). In vitro, RNA transcripts containing all or only the bottom portion of the nucleotide sequence of the WN 3′-SL (Fig. 1B) bound specifically to 56-, 84-, and 105-kDa proteins in uninfected BHK cellular extracts (3), and the “56-kDa” protein was subsequently identified as the 50-kDa translation elongation factor, eF1-α (4). The binding of eF1-α to the WN 3′-SL was dependent upon its phosphorylation. Another recent study (10) showed that the 3′-terminal 83 nt of the JE virus genome (the long stem and loop within the 3′-SL) could compete with full-length JE virus RNA for binding to the viral RNA-dependent RNA polymerase, NS5 (30), and that longer portions of 3′-terminal sequences also bound the virus-encoded helicase, NS3, to form a “replication complex”. Preliminary data from another study suggested a requirement for the flavivirus 3′-SL in translation (15). Stable stem-and-loop secondary structures at the 5′ and 3′ termini of rubella virus genome RNA bound La protein and calreticulin, respectively (1, 19). Phosphorylation-dependent binding of the rubella 3′-SL by calreticulin was linked to initiation of negative-strand RNA synthesis and to an effect of virus infection on the arrest of the cell cycle.
A study of the binding of the WN 3′-SL by eF1-α and studies of the binding of other cellular proteins to viral RNA secondary structures support the proposition that highly specific nucleotide sequence elements within such structures may be important for the binding of regulatory proteins (2–4, 27, 32, 34). Experiments described here were designed to study the structural and nucleotide sequence requirements for the 3′-SL in vivo in the context of the replication of DEN2 virus. The chimeric virus D2/WN-SL, which contained the 96-nt sequence of the WN 3′-SL as a substitute for the 93-nt DEN2 3′-SL, was greatly impaired for viral replication. This defect could occur by two mechanisms. (i) Some essential RNA-RNA or protein-RNA interaction is reduced in efficiency or abrogated for the majority of transfected molecules; virus replication occurs at a normal rate from a reduced number of substrate genomes in a reduced number of cells. (ii) Replication of all transfected genomes occurs at a slower-than-normal rate in all transfected cells, due to a series of impaired interactions that must occur at successive points in the replication process. In either case, the 3′-SL conformation alone was not sufficient to support replication; specific DEN2 nucleotide sequence elements within the 3′-SL were required for interaction either with viral proteins or with other regions of the viral genome. Possibly, specific nucleotide sequences of the DEN2 3′-SL are required to bind the DEN2 NS5 and/or NS3 to form the putative replication complex recently described for JE virus (10) or to interact with a specific sequence(s) in the DEN2 5′ NCR. It seemed less likely that the defect in replication was related to a reduction in binding of cellular proteins by the 3′-SL, assuming that the DEN2 3′-SL binds the same set of proteins as does the WN 3′-SL. Truncated DEN3 3′-SL RNAs efficiently competed with analogous WN 3′-SL segments for binding two (as yet unidentified) BHK cell proteins of the three specifically bound by the WN sequence (the 84- and 105-kDa species [3]), and the DEN2 3′-SL long stem contains nucleotide sequence (C62-U63-C64; Fig. 1A) in a position analogous to that shown to be the major binding site for the third 3′-SL binding protein, eF1-α, in the WN 3′-SL (C63-A64-C65; Fig. 1B [4]). The phenotype was probably not due to the accidental introduction of other occult mutations into the DEN2 genome during the mutagenesis procedure, since this and all other lethal and sublethal mutations (see below) could be rescued by replacement of the respective chimeric 3′-SL structures with the wt DEN2 3′-SL nucleotide sequence.
We next constructed mutant DEN2 cDNAs in which various segments of the DEN2 3′-SL were substituted for by analogous segments within the WN 3′-SL to determine which elements of the DEN2 nucleotide sequence in the 3′-SL were required for efficient virus replication. We defined top and bottom portions of the DEN2 and WN 3′-SL structures, since information regarding the in vitro cell protein-binding properties of the bottom portion of the WN 3′-SL had previously been defined (3, 4). We also constructed a set of mutations in the context of the “all-DEN2” 3′-SL. All of the D2/WN substitution mutant 3′-SL nucleotide sequences and the all-DEN2 mutants D2-SL(a) and -(b) were predicted by computer analysis to form 3′-SL structures in which the base pairing predicted for the wt parental 3′-SLs was preserved, allowing us to infer that modulation of the efficiency of replication of mutant viruses with respect to that of the wt was largely due to alterations of the wt DEN2 nucleotide sequence comprising the 3′-SL.
Phenotypes of the mutant viruses fell into three categories. (i) Viable, but slightly impaired for replication in LLC-MK2 and/or C6/36 cells compared to DEN2 wt. Cells transfected with these mutant RNAs were typically negative by IFA for DEN virus antigens at 24 h after transfection (in contrast to cells transfected with wt RNA) but positive after 3 days. A comparison of viral RNA and protein synthesis between wt and viable mutant viruses showed no obvious lesion at the level of translation, and we inferred that viable mutants were more or less defective compared to the wt at the level of RNA replication. (ii) Sublethal. IFA for DEN2 antigens was negative at 3 days and positive by day 10 after transfection. The spread of IFA positivity in the transfected monolayer indicated that infectious virus was produced. However, growth curves for these mutants in LLC-MK2 cells and C6/36 cells could not be obtained. The parent mutant, D2/WN-SL, was in this category. (iii) Lethal. IFA of the transfected monolayer for DEN2 antigens remained negative at all times up to 25 days.
Three of the DEN2-WN chimeric substitution mutations, D2/WN-SL(mutB), -(mutC), and -(mutE), (Fig. 2), were lethal or sublethal for DEN2 replication. Each of the associated constructs contained substitution mutations involving all or part of the bottom half of the long stem. Mutations D2/WN-SL(mutB) and D2/WN-SL(mutC), which substituted the entire bottom half of the WN 3′-SL, or only the bottom half of the long stem within the WN 3′-SL, for the respective analogous DEN2 nucleotide sequences, were lethal. D2/WN-SL(mutE) contained the most minimal exchange of DEN2 for WN nucleotide sequences, involving only the uppermost 11-bp of the bottom half of the long stem (DEN2 nt 7 to 17 and 63 to 73) and had the sublethal phenotype of the parent mutant, D2/WN-SL. This result suggested that DEN2 nt 7 to 17 and 63 to 73 were required for the viable phenotype of mutant viruses. However, the present data do not permit a simple explanation of the finding that some of the mutations involving the bottom half of the long stem were sublethal and some were lethal. We speculate that the two lethal mutations [D2/WN-SL(mutB) and -(mutC)] must have induced an additional defect in RNA replication or translation that was not conferred by the sublethal mutations D2/WN-SL and D2/WN-SL(mutE), related to the specific composition of the respective chimeric 3′-SL nucleotide sequences. For example, in the lethal mutations, the bottommost 7-bp segment of the long stem was derived from the WN nucleotide sequence and the entire top of the long stem was derived from the DEN2 nucleotide sequence. However, for the sublethal mutations those respective nucleotide sequences in the 3′-SL were derived either entirely from the WN sequence (D2/WN-SL) or entirely from the DEN2 sequence [D2/WN-SL(mutE)].
Mutant D2/WN-SL(mutA) contained the entire top portion of the WN 3′-SL, and the virus replicated efficiently, to only about a 10-fold-lower peak titer than did DEN2 wt in both LLC-MK2 and C6/36 cells. Since the nucleotide sequence of the top half of the WN 3′-SL diverges from that of the DEN2 3′-SL, the viability of the mutant suggested that the conformation of this domain, rather than its primary nucleotide sequence, was the more critical factor for virus replication. As a second test of this hypothesis, we constructed additional mutations of the top half of the 3′-SL in an all-DEN2 context (Fig. 3). In two of these mutants, part [D2-SL(a)] or all [D2-SL(b)] of the nucleotide sequences comprising the complementary strands of the top half of the long stem were transposed, thus repositioning the respective nucleotide sequence elements in the DEN2 3′-SL, while not altering its predicted stability compared to wt. In the third mutant, D2-SL(c), the double-stranded configuration of the top half of the long stem was completely disrupted by the substitution of the nucleotide sequence of one strand with a repeat of the nucleotide sequence of its opposite strand. Mutations D2-SL(a) and D2-SL(b) yielded viable virus in both cell lines, whereas the D2-SL(c) mutant had the lethal phenotype. Thus, the conformation of the top half of the 3′-SL, rather than its primary nucleotide sequence, was the more critical factor for viability.
Mutants that contained substitutions of WN nucleotide sequences for DEN2 nucleotide sequences in the small stem and loop [D2/WN-SL(mutD)] and in the bottommost portion of the long stem [D2/WN-SL(mutF)] also were viable. The homology between the DEN2 and WN 3′-SL nucleotide sequences is greatest in the small stem-and-loop domain: the 6 nt that constitute the loop region (Fig. 1), the sequences 5′-GAAAGA-3′ for DEN2 (nt 89 to 84) and 5′-GAUAGA-3′ for WN (nt 91 to 86), differ by only 1 nt, and the stem of the WN structure is longer than that of DEN2 by one G-C base pair. Shi et al. have suggested that 4 of the first 6 nt of the WN small-loop sequence may form a pseudoknot by hydrogen bonding with nt 71 to 74 in the adjacent long stem (24), and a similar structure in poliovirus genomic RNA has been implicated in RNA amplification (13). An inspection of the nucleotide sequence of the long stem for DEN2 suggests that formation of a pseudoknot might also be possible for the chimeric structure formed by D2/WN-SL(mutD) RNA.
Replication-competent D2/WN-SL(mutF) virus derived in LLC-MK2 cells was shown to contain a spontaneous deletion mutation within the substituted WN segment of the 3′-SL (Fig. 7). The 3′-SL of the resultant mutant genome thus resembled that of wt DEN2 more closely than did the original mutF construction, save for a U-to-C change in mutF RNA versus wt DEN2 RNA at nt 74 and the absence in the mutant genome of a bulge in the long stem created by the alignment of U4 with U76 in the wt DEN2 sequence (Fig. 7). This difference between the mutant genome and the wt nucleotide sequence apparently accounted for the observed failure of D2/WN-SL(mutF) virus to replicate in C6/36 cells. Spontaneous deletion of WN nt A3 in replicating D2/WN-SL(mutF) viral RNA may provide a clue to the lethal or sublethal phenotypes of other mutants that contained the bottommost portion of the WN 3′-SL; failure of those mutants to replicate efficiently in monkey cells may have been related to a deleterious effect of nt A3 on DEN2 replication.
D2/WN-SL(mutF) virus was uniquely defective with regard to its host cell-specific interactions, a phenotype that could be related to binding of cellular proteins to the 3′-SL. For example, mosquito cell proteins putatively required for binding to the 3′-SL may have binding specificities different from those of the analogous mammalian cell proteins. This hypothesis is currently under test in our laboratory. A similar one was advanced to explain the phenotypes of Sindbis virus host range mutants with deletions in the 5′ or 3′ NCR (14). Also, a DEN4 host range mutant that had sustained a 6-nt deletion in the 5′ NCR was similarly restricted for growth in mosquito cells but grew well in monkey kidney cells (8). The complement of the genomic 5′ NCR, the 3′ NCR in negative-stranded flavivirus RNA, is also predicted to form a stable stem-loop structure and also binds specific cellular proteins (25). It is possible that positive- and negative-strand RNA synthesis may in part be regulated by the analogous interactions of the two different stem-loop structures with cellular proteins, as others have suggested (3, 4).
ACKNOWLEDGMENTS
We thank Margo Brinton for helpful discussions.
This research was supported in part by an appointment to the Postgraduate Research Program at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
REFERENCES
- 1.Atreya C D, Singh N K, Nakhasi H L. The rubella virus RNA binding activity of human calreticulin is localized to the N-terminal domain. J Virol. 1995;69:3848–3851. doi: 10.1128/jvi.69.6.3848-3851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in the viral RNA. Cell. 1991;17:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinton M A. Replication of flaviviruses. In: Schlesinger S, Schlesinger M, editors. The Togaviridae and the Flaviviridae. New York, N.Y: Plenum Press; 1986. pp. 327–374. [Google Scholar]
- 6.Brinton M A, Fernandez A V, Dispoto J H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 7.Brinton M A, Dispoto J H. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1987;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 8.Cahour A, Pletnev A, Vazeille-Falcoz M, Rosen L, Lai C-J. Growth-restricted dengue virus mutants containing deletions in the 5′-noncoding region of the dengue virus genome. Virology. 1995;207:68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- 9.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 10.Chen C-J, Kuo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grange T, Bouloy M, Girard M. Stable secondary structure at the 3′ end of the genome of yellow fever virus (17D vaccine strain) FEBS Lett. 1985;188:159–163. doi: 10.1016/0014-5793(85)80895-4. [DOI] [PubMed] [Google Scholar]
- 12.Irie K, Mohan P M, Sasaguri Y, Putnak R, Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea C strain) Gene. 1989;75:197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn R J, Griffin D E, Zhang H, Niesters H G, Strauss J H. Attenuation of Sindbis virus neurovirulence by using defined mutations in nontranslated regions of the genome RNA. J Virol. 1992;66:7121–7127. doi: 10.1128/jvi.66.12.7121-7127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Brinton M A. Abstracts of the American Society for Virology 15th Annual Meeting. 1996. Effect of West Nile virus (WNV) 5′ and 3′ regions on translation, abstr. W2-1; p. 85. [Google Scholar]
- 16.Men R, Bray M, Clark D, Chanock R M, Lai C-J. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan P M, Padmanabhan R. Detection of stable secondary structure at the 3′ terminus of dengue virus type 2 RNA. Gene. 1991;108:185–191. doi: 10.1016/0378-1119(91)90433-c. [DOI] [PubMed] [Google Scholar]
- 18.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pogue G P, Hofmann J, Duncan R, Best J M, Etherington J, Sontheimer R D, Nakhasi H L. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polo S, Ketner G, Levis R, Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J Virol. 1997;71:5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice C M, Lenches E M, Eddy S R, Shin S J, Sheets R L, Strauss J H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–735. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 22.Schagger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 23.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 24.Shi P-Y, Brinton M, Veal J M, Zhong Y Y, Wilson W D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- 25.Shi P-Y, Li W, Brinton M A. Cell proteins bind specifically to West Nile virus minus-strand 3′ stem-loop RNA. J Virol. 1996;70:6278–6287. doi: 10.1128/jvi.70.9.6278-6287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer F, Ketner G, Connelly C, Hieter P. Targeted recombination-based cloning and manipulation of large DNA fragments in yeast. Methods Companion Methods Enzymol. 1993;5:161–175. [Google Scholar]
- 27.Stern S, Wilson R C, Noller H F. Location of the binding site for protein S4 on 16 Ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 28.Stollar V, Schlesinger R W, Steven T M. Studies of the nature of dengue viruses. III. RNA synthesis in cells infected with type 2 dengue virus. Virology. 1967;33:650–658. doi: 10.1016/0042-6822(67)90065-7. [DOI] [PubMed] [Google Scholar]
- 29.Takegami T, Washizu M, Yasui K. Nucleotide sequence at the 3′ end of Japanese encephalitis virus genome RNA. Virology. 1986;152:483–486. doi: 10.1016/0042-6822(86)90152-2. [DOI] [PubMed] [Google Scholar]
- 30.Tan B H, Fu J, Sugrue R J, Yap E H, Chan Y C, Tan Y H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 31.Valle R P C, Wickner R B. Elimination of L-A double-stranded RNA virus of Saccharomyces cerevisiae by expression of gag and gag-pol from an L-A cDNA clone. J Virol. 1993;67:2764–2771. doi: 10.1128/jvi.67.5.2764-2771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weeks K M, Crowthers D M. RNA recognition by Tat derived peptides: interaction with the major groove? Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- 33.Wengler G, Castle E. Analysis of structure properties which possibly are characteristic for the 3′-terminal sequence of the genome RNA of flaviviruses. J Gen Virol. 1986;67:1183–1188. doi: 10.1099/0022-1317-67-6-1183. [DOI] [PubMed] [Google Scholar]
- 34.Wu H-N, Uhlenbeck O C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987;26:8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]