Abstract
Sheep breeders requested that the U.S. Sheep Experiment Station (USSES) to participate in national genetic evaluation through the National Sheep Improvement Program (NSIP). The reasons included the need for (1) a comparison of the productivity of industry and United States Department of Agriculture (USDA) lines, (2) transparency of USDA flocks, (3) genetic ties for NSIP by sampling of industry flocks, and (4) development of premium genetic lines for public release. In response, USSES began to incorporate external sires from NSIP participating flocks into the USSES Targhee flock. Our objective, based on a pedigree analysis, was to test if introgression of external genetics into the flock was achieved. The pedigree included 13,189 animals with mean maximum generations, mean complete generations, and mean equivalent complete generations of 4.2, 1.8, and 2.6, respectively. The mean generation interval was 3.1 yr. The reference population was defined as lambs born from 2021 to 2023 (n = 792). Two additional populations were defined as the current mature ewe flock (n = 123) and the current mature rams (n = 14). The Genetic Conservation Index averaged 7.7 for the full population and 25.7 for the reference population. Overall inbreeding was 0.003 for the full population and 0.006 for the reference population. The rate of inbreeding was 0.0003 per generation. Average relatedness was 0.015 for the full population and 0.018 for the reference population. The effective number of founders, effective number of ancestors, and founder genome equivalents contributing to the reference population were 60, 39, and 19.1, respectively. The ratio of the effective number of founders to the effective number of ancestors was 1.5, indicating the presence of genetic bottlenecks. Measures of effective population size ranged from 102 to 547. Of the 704 offspring produced by external sires, 17 ram lambs and 132 ewe lambs were retained for breeding. The USSES sires produced 299 offspring with 2 ram lambs and 51 ewe lambs retained. Incorporating external sires resulted in a cumulative percentage of genetic variance of 48.8, 49.1, and 44.2 of external genetics for the reference population, current mature ewe flock, and current mature rams, respectively. Stakeholder needs were addressed by introgression of external sires and participation in NSIP, but future selection practices need to be modified to maintain a minimum of 50% USSES core genetics in the flock.
Keywords: genetic conservation index, genetic diversity, introgression, ovine, Targhee
This study provided an assessment of the genetic diversity present in the U.S. Sheep Experiment Station Targhee flock and an evaluation of the success of introgression of industry sires into the flock. The genetic impact of incorporating external sires into the flock was higher than expected; breeding managers can learn from our experiences to carefully manage the introgression of external genetics in their flock.
INTRODUCTION
The Targhee breed originated in 1926 at the U.S. Sheep Experiment Station (USSES) near Dubois, ID. The breed was developed as a dual-purpose for meat and wool to perform under extensive range conditions. The Targhee was developed from Rambouillet rams crossed with Corriedale × Lincoln-Rambouillet or Lincoln-Rambouillet ewes. The Targhee is polled and white-faced with a ewe body weight of 63 to 90 kg, a grease fleece weight of 4.5 to 5.4 kg, and a fiber diameter of 22 to 25 µm. Based on a molecular evaluation, Zhang et al. (2013) reported the Targhee was genetically similar to the Rambouillet (FST = 0.07) and had the highest within-breed variation of the breeds studied. The breed is primarily in large flocks in the Western United States, mainly in the state of Montana (Terrill, 1947; USTSA, 2023).
Participation in national genetic evaluation through the National Sheep Improvement Program (NSIP) is the primary means for sheep breeders to make genetic improvement in the U.S. Exchange of genetics among flocks is important to create genetic linkages to allow for across-flock evaluation. Genetic connectedness is needed to reduce bias and increase the reliability of estimates (Kuehn et al., 2009). Currently, NSIP needs more enrolled flocks with a greater diversity of external genetics compared to the currently enrolled flocks. Achieving this has become increasingly difficult, given the greater than 50% reduction in the U.S. breeding ewe inventory since the initiation of NSIP.
Formerly, United States Department of Agriculture (USDA) research sheep flocks were not enrolled in NSIP, due to general concern that USDA research agendas may not align with industry genetic goals. Regardless of the validity of such assumptions, which were never confirmed, stakeholders and USDA scientists later agreed that participation of USDA in NSIP would benefit NSIP and the U.S. sheep industry. Beginning in 2013, USSES began engaging U.S. sheep producers and associations about the necessity of enrolling their USDA flocks in NSIP. Over 3 yr, input was received from four breed associations, six industry associations, and nearly 100 seedstock and commercial sheep producers. The most common responses from producers and associations about the necessity of enrolling USDA flocks in NSIP were to
enable the public to compare productivity of industry lines with USDA founder lines (e.g., Polypay and Targhee breeds);
provide an “open view” of USDA flocks, enabling the public to assess the applicability of USDA research programs for industry genetic improvement;
increase genetic connectivity at a national scale for NSIP, requiring USDA flocks to sample from other NSIP flocks periodically; and
develop premium genetic lines for release to the public.
USSES and its collaborators moved forward with the first 3 responses, with (3) becoming the primary focus of past National Program project plans. Specifically, a core objective of these plans was “establish genetic linkages between experimental and industry flocks to support industry-wide genetic evaluations and development of comprehensive breeding objectives.” In accomplishing this objective, we began by sampling sires from external Targhee flocks.
The USSES goals for participation in NSIP included (1) establish broad genetic connectivity at a national scale to enhance the utility of NSIP, thus positioning the USSES flock to be a national reference flock, (2) evaluate genetics from industry sires, and (3) incorporate genetics from NSIP Targhee flocks into the USSES flock. Accordingly, sires were selected from NSIP participating flocks with the intent to improve genetic ties for genetic evaluation and to evaluate their overall performance. Sires were sampled as widely as was feasible and were selected to represent sires that would be purchased by a commercial flock (i.e., not elite sires) and therefore not extreme in their EBV or in the accuracy of those EBV. Incorporating external sires allowed benchmarking of the performance of NSIP sires and the USSES flock, from which the breed originated. Sixteen sires were incorporated over a 3-yr period followed by an additional two sires 3 yr later. Offspring performance was recorded through 2023. Maintaining the core genetics of the original USSES flock while incorporating external genetics requires careful planning. The objectives of this study were to (1) evaluate the present pedigree-based genetic diversity in the USSES flock and 2) evaluate the introgression of the external sires into the USSES flock.
MATERIALS AND METHODS
Animal Care
The USSES Institutional Animal Care and Use Committee (IACUC) approved all husbandry practices and experimental procedures used in this study.
Data Structure
Sixteen external sires from 10 flocks were incorporated into the USSES flock during the 2015 to 2017 breeding seasons. Two additional sires from an additional flock were incorporated in 2020. There were 32 active Targhee flocks participating in NSIP in at least 1 yr during the study period of 2016 to 2023, including USSES. Sixteen rams originated in Montana from 10 flocks and 2 rams originated in Oregon from 1 flock. Six rams were used for a single breeding season, 11 rams for two breeding seasons, and 1 ram was used for three breeding seasons. Ewe lambs were mated to terminal sires; consequently, these rams were mated to the mature ewe flock which included yearlings through 7-yr-olds.
Pedigree records were obtained from the NSIP database and included all USSES-born sheep and their ancestors regardless of flock of origin. While USSES has only participated in NSIP for the past few years, production records as far back as 1999 were included when initially joining NSIP, with some pedigree records tracing as far back as 1991. Pedigree records were ordered so parents were placed before their offspring and the pedigree was recorded using the Animal Breeders Toolkit (Golden et al., 1992). The final pedigree included 13,189 animals with 6,267 males and 6,922 females. There were 341 sires and 2,689 dams. A reference population, representing the most recent generation interval, was defined as lambs born from 2021 to 2023 and included 792 lambs. Two additional populations were defined to understand the genetic contribution of ancestors to the current flock and included (1) the current mature ewe flock (n = 123) and (2) the mature ram flock (n = 14).
Relationship coefficients were computed among (1) external sires, (2) external sires and USSES sires, and (3) external sires and the current mature ewe flock (n = 123) using the ggroups package in R (Nilforooshan and Saavedra-Jiménez, 2020). The remaining genetic analyses were performed using ENDOG software (Gutiérrez and Goyache, 2005). The prepared file for the ENDOG analyses included the pedigree, birth date, sex, reference population designation, and external or USSES sire designation for sires evaluated in this study.
Genetic Diversity in the USSES Flock
Pedigree completeness was assessed by computing the mean maximum generations, mean complete generations, and mean equivalent complete generations. Equivalent complete generations were computed as the sum of the proportion of known ancestors over all generations (Maignel et al., 1996). The mean generation interval was computed using the four-path method, taking into account the age at which a parent is replaced by their offspring in a breeding population; the four paths are sire-son, sire-daughter, dam-son, and dam-daughter (James, 1977; Hill, 1979).
The genetic conservation index (GCI) was reported for all animals. According to Alderson (1992), capturing the genetic diversity of a population is best accomplished by retaining all the alleles in the original founder population. This is achieved by an animal receiving equal contributions from all the founder ancestors. Computationally, the GCI for an animal is the effective number of founders in its pedigree from , where Pi is the proportion of genes of founder animal i in the pedigree (Alderson, 1992). Animals can be ranked based on GCI, with a higher GCI value representing a higher level of genetic diversity. The maximum GCI value is equal to the number of founder ancestors in the breed. To evaluate the maximum potential GCI of the current USSES population, the current rams (n = 14) were pseudo mated to the current ewes (n = 123) and the maximum GCI for each ram was reported.
Genetic variance of the population was defined by Boichard et al. (1997) using the probability of gene origin approach which uses pedigree data to measure the presence of the alleles from the founder population in subsequent generations. Genetic variation is lost from one generation to the next from unbalanced contributions of parents and a loss of genes being passed from a parent to its progeny through Mendelian sampling. Measures of genetic variability in the population included the inbreeding coefficient (F), which is the probability that an individual has two identical alleles by descent; ENDOG computes F as described by Meuwissen and Luo (1992). The average relatedness coefficient (AR) was computed for each animal as the probability that a randomly chosen allele from the entire pedigreed population belongs to a specified animal (Gutiérrez and Goyache, 2005). The reference population was used to compute measures of the probability of gene origin, including the effective number of founders (fe), the effective number of ancestors (fa), and the founder genome equivalents (fg). The effective number of founders is the number of animals that would produce the genetic diversity in the population if all animals contributed equally and is computed as
where qk is the proportion of the genes of the reference population contributed by founder k, and f is the total number of founders (Lacy, 1989). The effective number of ancestors is defined as the minimum number of ancestors that explains the genetic diversity of the population, and takes into account genetic drift and population bottlenecks and is computed as
where qj is the marginal contribution of ancestor j, and a is the total number of ancestors (Boichard et al., 1997). The founder genome equivalents are the number of founders expected to produce the same genetic diversity present in the population if the founders were represented equally and all founder alleles are still present in the population and is computed as
where fk are gene frequencies of founder k, and f is the total number of founders (MacCluer et al., 1986; Lacy, 1989; Boichard et al., 1997). The ratio of fe/fa evaluates the presence of bottlenecks in the population (Boichard et al., 1997) where a ratio close to 1 indicates less influence of bottlenecks. The ratio of fe/fg determines the effect of genetic drift on the population (Lacy, 1989) where a higher value indicates a greater loss of genetic diversity due to genetic drift.
ENDOG provided several options for computing the effective population size (Ne), including the increase in inbreeding by maximum generation, complete generation, and equivalent generation (Gutiérrez and Goyache, 2005), individual increase in F (Gutiérrez et al., 2008), the regression on equivalent generations (Gutiérrez et al., 2003), the log regression on equivalent generations (Pérez‐Enciso, 1995), and individual increase in coancestry (Cervantes et al., 2011).
Genetic Impact of External Sires on the USSES Flock
The ancestors explaining the genetic variability of each population were identified in order of importance to the population in terms of genetic contribution; their marginal and cumulative contributions to the population were computed. Each additional ancestor was selected based on the proportion of genes it contributes that were not yet explained by previously selected ancestors until all genetic variability in the reference population was accounted for (Boichard et al., 1997). For the 18 external sires and the 12 USSES sires, the marginal contributions for each sire were determined. For each sire, the number of breeding years, number of offspring, number of retained offspring, and GCI were obtained.
To compare the two sire groups, total lambs produced and total contribution to retained lambs for the study period were computed. Population differentiation between the two sire groups was compared using Nei’s minimum distance (Nei, 1978) and Wright’s FST (Wright, 1951). Nei’s minimum distance (D) was computed as
where fii and fjj are the average coancestry within populations i and j and fij is the average coancestry between the two populations. Wright’s FST was computed as
where is the average coancestry for the subpopulation and is the mean coancestry for the entire population.
Results
Genetic Diversity in the USSES Flock
For the full pedigree (n = 13,189), the mean maximum generations traced was 4.2 with a range from 0 to 12 generations. The mean complete generations was 1.8 and ranged from 0 to 5. The mean equivalent generations traced was 2.6 with a range of 0 to 6.8 generations. For the reference population (n = 792), the mean maximum generations, mean complete generations, and mean equivalent generations were 9.8, 3.3, and 5.5, respectively. The overall generation interval was 3.1 yr, with the four paths of sire-son, sire-daughter, dam-son, and dam-daughter averaging 2.6, 2.6, 3.9, and 3.6 yr, respectively.
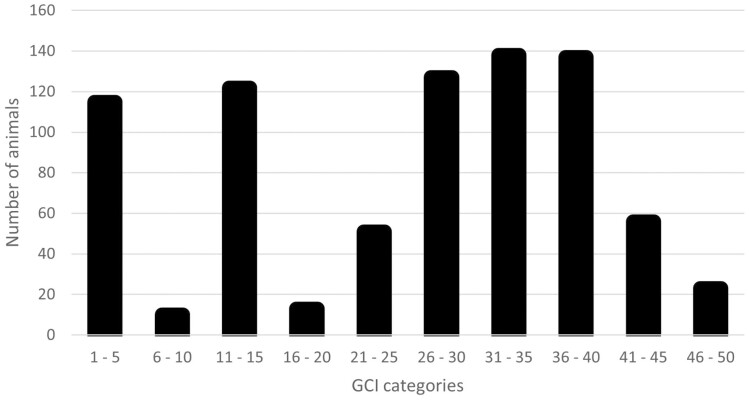
For the full pedigree, the GCI values ranged from 1 to 50.5 with a mean of 7.7. Animals in the reference population had a range of GCI values from 2 to 50.3 with a mean of 25.7. Reference population animals were summarized in GCI categories and plotted in Figure 1. There were 481 animals in the GCI categories of 26 and higher, indicating a large percentage of the reference population has many founder contributions and a high level of genetic diversity. Less than 15% of the reference population had a GCI of 5 or less, with few founders in the animal’s pedigree. The opportunity to increase GCI in the current population was evaluated by pseudo matings between the current rams and current ewes, resulting in a maximum GCI for each ram mating ranging from 13.0 to 65.3 (Table 1), which is an increase over the current maximum GCI in the pedigree (50.5).
Figure 1.
Targhee reference population by Genetic Conservation Index (GCI) category.
Table 1.
Maximum GCI for pseudo matings between current rams (n = 14) and current ewes (n = 123)
| Current ram | GCI |
|---|---|
| Ram 1 | 52.9 |
| Ram 2 | 13.4 |
| Ram 3 | 13.3 |
| Ram 4 | 65.3 |
| Ram 5 | 50.3 |
| Ram 6 | 59.1 |
| Ram 7 | 56.8 |
| Ram 8 | 33.7 |
| Ram 9 | 60.9 |
| Ram 10 | 59.9 |
| Ram 11 | 13.0 |
| Ram 12 | 61.1 |
| Ram 13 | 54.9 |
| Ram 14 | 57.4 |
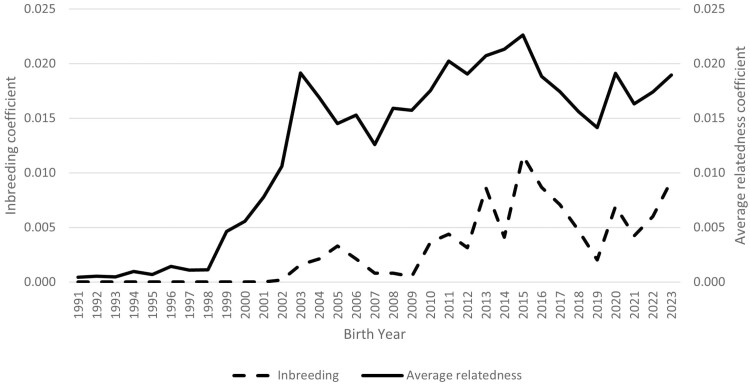
The average inbreeding and average relatedness coefficients were computed by birth year (Figure 2). Inbreeding was low, with fluctuations prior to 2015 likely due to incomplete pedigree records and the decrease in inbreeding after 2015 attributed to the introduction of external sires. The rate of inbreeding was 0.0003 per generation and did not differ across generations (P = 0.91). The simple linear regression from 2003 to 2023 was 0.0004. Overall inbreeding was 0.003 for the full population with a range from 0 to 0.250. The average inbreeding for the reference population was 0.006 with a range of 0 to 0.036. The average relatedness and range for the full population and reference population were 0.015 (0 to 0.052) and 0.018 (0.001 to 0.026), respectively. Founder, ancestor, and effective population size statistics were summarized in Table 2. Since the ratio of fe to fa is greater than 1, there were bottlenecks at some time in the history of the flock. The ratio of fe to fg of 3.1 is indicative of genetic drift in the flock.
Figure 2.
Targhee inbreeding and average relatedness coefficient by birth year.
Table 2.
Measures of probability of gene origin for the Targhee reference population
| Gene origin parameter | Value |
|---|---|
| Reference population size | 792 |
| Ancestors contributing to the reference population | 167 |
| Effective number of founders (fe) | 60 |
| Effective number of ancestors (fa) | 39 |
| Founder genome equivalents (fg) | 19.1 |
| f e /fa | 1.5 |
| f e /fg | 3.1 |
| Ancestors explaining 50% of the gene pool | 13 |
Estimates of Ne ranged from 102.2 for the individual increase in coancestry to 546.9 for the increase in inbreeding by maximum generation (Table 3). Across a breed, a minimum Ne value of 50 to 100 is generally recommended to maintain genetic diversity in a population (FAO, 1998; Meuwissen, 2009). The Ne estimates may be inflated due to incomplete pedigree records; however, given this is a single flock within a breed, this measure of genetic diversity is quite large.
Table 3.
Effective population size (Ne) measures for the USSES Targhee flock
| Method | N e estimate |
|---|---|
| Increase in F by maximum generation | 546.9 |
| Increase in F by complete generation | 194.5 |
| Increase in F by equivalent generation | 259.9 |
| Individual increase in F | 449.6 |
| Regression on equivalent generations | 170.2 |
| Log regression on equivalent generations | 170.3 |
| Individual increase in coancestry | 102.2 |
Genetic Impact of External Sires on the USSES Flock
The external sires had a coefficient of relationship with each other ranging from 0 to 0.20 with an average relationship of 0.02. External rams were related to between 0 and 13 other external rams. In contrast, there were no relationships between the external and USSES rams. By 2023, the external rams had an average coefficient of relationship with the ewe flock ranging from 0 to 0.06 with a maximum relationship of 0.5, indicating a retained daughter. The external sires were related to between 0 and 109 ewes of the 123 mature ewe flock (Table 4).
Table 4.
Mean, maximum (Max), and count (N) greater than 0 of the coefficient of relationships among each external sire (n = 18) and each external sire with the current mature ewe flock (n = 123)
| Sire | Coefficient of relationship with other external sires | Coefficient of relationship with current mature ewe flock | ||||
|---|---|---|---|---|---|---|
| Mean | Max | N > 0 | Mean | Max | N > 0 | |
| E1 | 0.00 | 0.00 | 0 | 0.02 | 0.50 | 4 |
| E2 | 0.03 | 0.20 | 9 | 0.06 | 0.50 | 101 |
| E3 | 0.00 | 0.00 | 0 | 0.06 | 0.50 | 14 |
| E4 | 0.04 | 0.16 | 11 | 0.04 | 0.50 | 103 |
| E5 | 0.01 | 0.06 | 4 | 0.02 | 0.50 | 39 |
| E6 | 0.00 | 0.00 | 0 | 0.04 | 0.50 | 24 |
| E7 | 0.02 | 0.18 | 13 | 0.03 | 0.50 | 109 |
| E8 | 0.02 | 0.03 | 12 | 0.02 | 0.50 | 108 |
| E9 | 0.03 | 0.08 | 12 | 0.03 | 0.50 | 105 |
| E10 | 0.02 | 0.20 | 9 | 0.04 | 0.50 | 101 |
| E11 | 0.02 | 0.06 | 9 | 0.02 | 0.50 | 68 |
| E12 | 0.03 | 0.16 | 11 | 0.02 | 0.25 | 103 |
| E13 | 0.02 | 0.08 | 9 | 0.03 | 0.27 | 101 |
| E14 | 0.01 | 0.05 | 5 | 0.01 | 0.26 | 55 |
| E15 | 0.02 | 0.13 | 6 | 0.01 | 0.50 | 50 |
| E16 | 0.01 | 0.03 | 9 | 0.04 | 0.26 | 101 |
| E17 | 0.00 | 0.00 | 0 | 0.00 | 0.00 | 0 |
| E18 | 0.03 | 0.18 | 11 | 0.01 | 0.09 | 103 |
Of the 167 ancestors contributing genetic variation to the reference population, 48.8% of the variation was contributed by external genetics and 51.1% by USSES genetics. The current mature ewe flock is comprised of 49.1% external genetics compared to 50.9% USSES genetics. The current mature rams are 44.2% external genetics and 55.8% USSES genetics (Table 5).
Table 5.
Marginal contributions of external and USSES ancestors to the current generation interval (lambs born from 2021 to 2023), the current mature ewe flock, and the current mature rams
| Population | Number of ancestors | External marginal contributions | USSES marginal contributions |
|---|---|---|---|
| Current generation interval | 167 | 48.8 | 51.1 |
| Current mature ewe flock | 115 | 49.1 | 50.9 |
| Current mature rams | 18 | 44.2 | 55.8 |
The two external sires with the largest marginal contributions to the reference population each defined 4.9% of the genetic variance. One sire had 1 son and 11 daughters retained in the flock while the other had 4 sons and 13 daughters retained in the flock (Table 6). The USSES sires had a higher average GCI of 13.9 (range of 10.4 to 18.3) compared to the external sires with an average of 6.9 (range of 1.0 to 10.7). The 18 external sires had 704 offspring compared to 299 for the 12 USSES sires. External sires had 17 ram lambs and 132 ewe lambs retained for the breeding flock while the USSES sires had 2 ram lambs and 51 ewe lambs retained. Because of the disproportionate number of ram lambs retained from external sires, by 2023 there were no 100% USSES offspring in the flock. Population differentiation between the external sires and USSES sires was computed as 0.05 and 0.03 using Nei’s minimum distance and Wright’s FST, respectively.
Table 6.
Marginal contributions of ancestors, offspring, and GCI for the externally purchased and USSES sires
| Sire | Sire group | Marginal contributions | Number of breeding years | Number of offspring | Retained sons | Retained daughters | GCI |
|---|---|---|---|---|---|---|---|
| E1 | External | 0.049 | 2 | 54 | 1 | 11 | 1.0 |
| E2 | External | 0.049 | 1 | 26 | 4 | 13 | 6.4 |
| E3 | External | 0.044 | 2 | 44 | 1 | 13 | 1.0 |
| E4 | External | 0.038 | 3 | 78 | 1 | 15 | 9.1 |
| E5 | External | 0.032 | 2 | 56 | 2 | 5 | 8.0 |
| E6 | External | 0.028 | 2 | 47 | 0 | 11 | 1.0 |
| E7 | External | 0.018 | 2 | 46 | 1 | 10 | 8.5 |
| E8 | External | 0.015 | 2 | 42 | 0 | 9 | 6.7 |
| E9 | External | 0.010 | 1 | 27 | 1 | 6 | 9.1 |
| E10 | External | 0.008 | 2 | 55 | 1 | 9 | 9.1 |
| E11 | External | 0.008 | 1 | 23 | 1 | 6 | 9.1 |
| E12 | External | 0.006 | 2 | 35 | 0 | 7 | 10.7 |
| E13 | External | 0.005 | 2 | 35 | 1 | 3 | 8.0 |
| E14 | External | 0.005 | 1 | 7 | 1 | 2 | 8.0 |
| E15 | External | 0.005 | 2 | 47 | 0 | 5 | 9.1 |
| E16 | External | 0.000 | 2 | 49 | 2 | 5 | 8.5 |
| E17 | External | 0.000 | 1 | 17 | 0 | 1 | 1.0 |
| E18 | External | 0.000 | 1 | 16 | 0 | 1 | 10.0 |
| U1 | USSES | 0.0001 | 1 | 23 | 0 | 2 | 17.8 |
| U2 | USSES | 0.000 | 1 | 31 | 0 | 8 | 10.4 |
| U3 | USSES | 0.000 | 1 | 26 | 0 | 6 | 14.2 |
| U4 | USSES | 0.000 | 2 | 52 | 0 | 8 | 12.3 |
| U5 | USSES | 0.000 | 1 | 17 | 0 | 3 | 13.1 |
| U6 | USSES | 0.000 | 1 | 5 | 1 | 0 | 11.8 |
| U7 | USSES | 0.000 | 1 | 27 | 0 | 6 | 11.4 |
| U8 | USSES | 0.000 | 1 | 26 | 0 | 5 | 12.8 |
| U9 | USSES | 0.000 | 1 | 27 | 0 | 6 | 18.3 |
| U10 | USSES | 0.000 | 1 | 28 | 0 | 3 | 12.9 |
| U11 | USSES | 0.000 | 1 | 16 | 0 | 3 | 14.5 |
| U12 | USSES | 0.000 | 1 | 21 | 1 | 1 | 17.6 |
Discussion
Genetic Diversity in the USSES Flock
The generation interval for sires was more than a year shorter than for dams. This practice is typical of that in the sheep industry and of management practices at USSES. Rapid turnover of sires helps to achieve genetic gains while maintaining productive ewes in the flock for as long as possible improves production efficiency. The average generation interval of 3.1 yr was lower than Santa Inês sheep in Brazil (3.7 yr), Morada Nova hair sheep in Brazil (3.6 yr), Dorset in Canada (3.9 yr), Polypay in Canada (3.4 yr), and Suffolk in Canada (3.3 yr), but higher than Suffolk in the U.S. (2.9 yr) and Romanov in Canada (2.9 yr; Pedrosa et al., 2010; Stachowicz et al., 2018; McManus et al., 2019; Wilson et al., 2022).
If conservation of the genetic variation present in the original population is the breeding objective, animals retained for breeding should be selected for a large GCI. This is rarely the objective of a production flock but can still be used to evaluate the representation of founders in the current population. There were 167 ancestors contributing to the reference population. The average GCI of the reference population was 25.7, which was higher than the overall flock average of 7.7. Thus, GCI in the reference population is high, without any selection pressure for genetic diversity. The reference population has a small group of low GCI animals and a large group of higher GCI animals (Figure 1).
Minimizing closely related matings is a priority for the USSES flock and is reflected in the low overall average inbreeding of 0.003 for the full population and 0.006 for the reference population. In the most recent year, the average inbreeding was 0.009. For the most recent year reported, inbreeding was 0.055, 0.027, 0.029, 0.030, 0.035, 0.048, 0.009, and 0.044 for U.S. Suffolk, Canadian Dorset, Canadian Suffolk, Canadian Rideau-Arcott, Canadian Polypay, Canadian Romanov, Irish Charollais, and Irish Belclare, respectively (Stachowicz et al., 2018; Rafter et al., 2022; Wilson et al., 2022). While all inbreeding levels reported were relatively low, the USSES Targhee flock was on the low end of this range. A rate of inbreeding less than 1% per generation is recommended to maintain genetic diversity in a population (FAO, 1998); the Targhee population is well below this rate, even when considering incomplete pedigree records. For the Targhee reference population, the average relatedness value was 0.018. Average relatedness helps to identify unrelated animals for mating, which has a long-term impact on the development of inbreeding (Goyache et al., 2003; Machová et al., 2021). Formal methods to maximize genetic gain while minimizing kinship are available through optimal contribution selection (OCS) methods (Meuwissen, 2009).
Estimates of effective number of founders, ancestors, founder genome equivalents, and their ratios are informative about the current status of the flock relative to its original population. For the Targhee reference population, the fe, fa, and fg were 60, 39, and 19, respectively. In comparison, a subset of the U.S. Suffolk breed had a fe, fa, and fg of 255, 107, and 50, respectively (Wilson et al., 2022). Six Irish sheep breeds had a range of 51 to 303, 26 to 125, and 12 to 62 for fe, fa, and fg, respectively. In this study, the ratio of fe to fa was 1.5 for Targhee and ranged from 1.6 to 3.4 for six Irish sheep breeds (Rafter et al., 2022), all indicative of the presence of bottlenecks in the history of the breed. In fact, most sheep breeds reported in the literature indicated some evidence of bottlenecks (Pedrosa et al., 2010; Sheikhlou and Abbasi, 2016; Stachowicz et al., 2018; Machová et al., 2021). Unequal contributions of breeding animals to the next generation are the primary source of these bottlenecks and are expected in a breed (or flock) undergoing selection. Thirteen ancestors explained 50% of the gene pool of the Targhee reference population. This is similar to reports from other breeds: Iranian Lori-Bakhtiari (15), Canadian Romanov (10), Canadian Rideau-Arcott (14), and Canadian Polypay (14), but lower than Canadian Dorset (49) and Canadian Suffolk (52; Sheikhlou and Abbasi, 2016; Stachowicz et al., 2018).
Effective population size ranged from 102 to 547 for the Targhee flock depending on the computation method. In comparison, Ne for other breeds reported were 28 to 244 for U.S. Suffolk sheep, 55 to 99 for five Canadian sheep breeds, and 116 to 315 for six Irish sheep breeds (Stachowicz et al., 2018; Rafter et al., 2022; Wilson et al., 2022). The Targhee flock was on the high end of reported Ne and sheep breeds in general reported higher Ne than other livestock species (Welsh et al., 2010; Kijas et al., 2012; Faria et al., 2019; Makanjuola et al., 2020). Even if the Ne values are inflated due to incomplete pedigree records, there appears to be significant genetic diversity in the Targhee flock for long-term sustainability. Further analysis comparing external and USSES sires using molecular methods is warranted.
Genetic Impact of External Sires on the USSES Flock
While inbreeding in the Targhee flock was low, a decrease in the inbreeding trend was observed after external sires were introduced from 2015 to 2017 for offspring born from 2016 to 2019 (Figure 2). This was expected as the external sires were expected to be less related to the flock than the sires selected from within the flock. The USSES sires from this time period had an average GCI of 13.9 compared to 6.9 for the external sires. This confirms the external sires were less representative of the founders of the USSES flock, which would lead to a reduction in inbreeding. It may also be an artifact of the pedigree structure, where four external sires were of unknown parentage and had a subsequent GCI of 1. Population differentiation between the external sires and the USSES sires was low, indicating the populations have not experienced a major genetic shift due to geographical distance or differing selection objectives.
Early observations on the performance and phenotypes of the progeny of external and USSES sires resulted in the decision to proceed with integration of the external genetics into the USSES Targhee flock. As stated by Machová et al. (2021), even a single excessively used sire can significantly decrease the genetic diversity of a breed if the offspring of that sire are used excessively. This was the case specifically with the top five external sires with disproportionate representation of these sires for both retained ram lambs and ewe lambs, and generally for the external sires when compared to the USSES sires. Over the study period, the external sires produced more than twice as many offspring as the USSES sires. By 2023, none of the lambs born were of 100% USSES genetics.
When evaluating the introgression of external sires into the USSES flock, the measures of success are multifaceted. Clearly, stakeholder objectives were met with the USSES participation in NSIP, the introgression of external genetics into the flock, and the evaluation of industry sire performance (results not reported here). The number of external sires incorporated into the flock was well designed, but the preferential selection of retained offspring for the breeding flock from each sire was not evaluated at each selection cycle, resulting in the potential for genetic replacement rather than introgression of genetics. Currently, the flock is split almost evenly between external and USSES genetics (Table 5). It is useful to recognize that this is not a breed-level introgression. Instead, genetic material from flocks derived, and subsequently largely isolated, from the original Targhee source flock was reintroduced into the USSES flock. This introduction was anticipated to restore potentially useful alleles that may have been lost or kept at low frequencies due to genetic drift in the USSES flock.
Moving forward, to conserve the USSES foundation genetics at a minimum of 50%, OCS methods will be applied in conjunction with maximizing genetic gain through the use of the NSIP Western Range Index (Borg et al., 2007). The breeding objective for the maternal breeds at USSES, which includes the Targhee, is to improve maternal traits, including future selection for lamb survivability, ewe longevity, and twinning rate. Maintaining the core USSES genetics while exploiting the added genetic variation from the introgression of external genetics provides the opportunity for the USSES Targhee flock to serve as a national genetic reference flock. Maintaining a balance of 50% each USSES and external genetics will help achieve the goals set forth by stakeholders of introducing industry genetics to the USSES flock and comparing the performance of these sires.
When making the decision to introgress external genetics into an established flock, breeding managers should consider their end goal before they begin. Is the goal to replace the flock with superior genetics? Is the objective to increase the genetic diversity of the flock? Should the original genetics be maintained as a separate genetic line? How many offspring should be retained from each sire? The answers to these questions should guide decisions and be reevaluated at each breeding cycle. Successful introgression can be achieved through the use of carefully designed selection and mating plans, which can be made easier with the application of OCS methods.
Conclusion
This study provided an assessment of the genetic diversity present in the USSES Targhee flock and an evaluation of the success of the introgression of industry sires into the flock. While acknowledging the limited depth of pedigree records available for the flock, the genetic diversity present in the USSES Targhee flock is high based on all measured parameters, including low levels of inbreeding and average relatedness, high GCI in the reference population, and high effective population size. The long-term genetic variability in the Targhee flock is not of concern. The genetic impact of incorporating external sires into the flock was higher than expected, resulting in a cumulative percentage of genetic variance of 48.8, 49.1, and 44.2 for the reference population, current mature ewe flock, and current mature rams, respectively. Over an 8-yr period, the external sires replaced almost 50% of the core genetics of the original flock with no 2023 born lambs being of 100% USSES genetics. Stakeholder requests for NSIP participation and incorporating industry sires into the USSES flock were met. Future breeding objectives at USSES will benefit from the inclusion of external genetics in the flock. Breeding managers can learn from our experiences and apply OCS methods for each breeding cycle to carefully manage the introgression of external genetics in their flock.
Acknowledgments
We would like to thank Natalie Cherry for meticulous record keeping of the USSES sheep production data. The USDA is an equal opportunity provider and employer. The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Contributor Information
Carrie S Wilson, Range Sheep Production Efficiency Research Unit, U.S. Sheep Experiment Station, ARS, USDA, Dubois, ID, 83423, USA.
J Bret Taylor, Range Sheep Production Efficiency Research Unit, U.S. Sheep Experiment Station, ARS, USDA, Dubois, ID, 83423, USA.
Ronald M Lewis, Department of Animal Science, University of Nebraska-Lincoln, Lincoln, NE, 68583, USA.
David R Notter, School of Animal Sciences, Virginia Tech, Blacksburg, VA, 24061, USA.
Conflict of Interest Statement
The authors declare no conflict of interest.
Literature Cited
- Alderson, G. L. H. 1992. A system to maximize the maintenance of genetic variability in small populations. In: Alderson, G. L. H., and Bodo I., editors. Genetic conservation of domestic livestock No. 2. United Kindom: CAB International; p. 18–29. [Google Scholar]
- Boichard, D., Maignel L., and Verrier E... 1997. The value of using probabilities of gene origin to measure genetic variability in a population. Genet. Sel. Evol. 29:5–5. doi: 10.1186/1297-9686-29-1-5 [DOI] [Google Scholar]
- Borg, R., Notter D., Kuehn L., and Kott R... 2007. Breeding objectives for Targhee sheep. J. Anim. Sci. 85:2815–2829. doi:10.2527/jas.2006-064. [DOI] [PubMed] [Google Scholar]
- Cervantes, I., Goyache F., Molina A., Valera M., and Gutiérrez J. P... 2011. Estimation of effective population size from the rate of coancestry in pedigreed populations. J. Anim. Breedg. Genet. 128:56–63. doi: 10.1111/j.1439-0388.2010.00881.x [DOI] [PubMed] [Google Scholar]
- FAO. 1998. FAO Secondary Guidelines for development of national farm animal genetic resources management plans. Rome, Italy: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Faria, D. A., Wilson C., Paiva S., and Blackburn H. D... 2019. Assessing Sus scrofa diversity among continental United States, and Pacific islands populations using molecular markers from a gene banks collection. Sci. Rep. 9:3173. doi: 10.1038/s41598-019-39309-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, B. L., Snelling W. M., and Mallinckrodt C. H... 1992. Animal breeders tool-kit: user’s guide. Colorado State University. (Exp. Sta. Tech. Bull., LTB92-2). [Google Scholar]
- Goyache, F., Gutiérrez J. P., Fernández I., Gomez E., Álvarez I., Díez J., and Royo L... 2003. Using pedigree information to monitor genetic variability of endangered populations: the Xalda sheep breed of Asturias as an example. J. Anim. Breed. Genet. 120:95–105. doi:10.1046/j.1439-0388.2003.00378.x. [Google Scholar]
- Gutiérrez, J. P., Altarriba J., Díaz C., Quintanilla R., Cañón J., and Piedrafita J... 2003. Pedigree analysis of eight Spanish beef cattle breeds. Genet. Sel. Evol. 35:43–63. doi: 10.1186/1297-9686-35-1-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, J. P., Cervantes I., Molina A., Valera M., and Goyache F... 2008. Individual increase in inbreeding allows estimating effective sizes from pedigrees. Genet. Sel. Evol. 40:359–378. doi: 10.1186/1297-9686-40-4-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, J. P., and Goyache F... 2005. A note on ENDOG: a computer program for analysing pedigree information. J. Anim. Breedg. Genet. 122:172–176. doi: 10.1111/j.1439-0388.2005.00512.x [DOI] [PubMed] [Google Scholar]
- Hill, W. G. 1979. A note on effective population size with overlapping generations. Genetics. 92:317–322. doi: 10.1093/genetics/92.1.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, J. W. 1977. A note on selection differential and generation length when generations overlap. Anim. Sci. 24:109–112. doi: 10.1017/s0003356100039271 [DOI] [Google Scholar]
- Kijas, J. W., Lenstra J. A., Hayes B., Boitard S., Neto L. R., Cristobal M. S., Servin B., McCulloch R., Whan V., Gietzen K.,. et al. 2012. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10:e1001258. doi: 10.1371/journal.pbio.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn, L. A., Lewis R. M., and Notter D. R... 2009. Connectedness in Targhee and Suffolk flocks participating in the United States national sheep improvement program. J. Anim. Sci. 87:507–515. doi: 10.2527/jas.2008-1092 [DOI] [PubMed] [Google Scholar]
- Lacy, R. C. 1989. Analysis of founder representation in pedigrees: founder equivalents and founder genome equivalents. Zoo Biol. 8:111–123. doi: 10.1002/zoo.1430080203 [DOI] [Google Scholar]
- MacCluer, J. W., VandeBerg J. L., Read B., and Ryder O. A... 1986. Pedigree analysis by computer simulation. Zoo Biol. 5:147–160. doi: 10.1002/zoo.1430050209 [DOI] [Google Scholar]
- Machová, K., Milerski M., Rychtářová J., Hofmanová B., Vostrá-Vydrová H., Moravčíková N., Kasarda R., and Vostrý L... 2021. Assessment of the genetic diversity of two Czech autochthonous sheep breeds. Small Ruminant Res. 195:106301. doi: 10.1016/j.smallrumres.2020.106301 [DOI] [Google Scholar]
- Maignel, L., Boichard D., and Verrier E... 1996. Genetic variability of French dairy breeds estimated from pedigree information. Interbull Bull. 14:49–49. [Google Scholar]
- Makanjuola, B. O., Miglior F., Abdalla E. A., Maltecca C., Schenkel F. S., and Baes C. F... 2020. Effect of genomic selection on rate of inbreeding and coancestry and effective population size of Holstein and Jersey cattle populations. J. Dairy Sci. 103:5183–5199. doi: 10.3168/jds.2019-18013 [DOI] [PubMed] [Google Scholar]
- McManus, C., Facó O., Shiotsuki L., Rolo J. L. J. P., and Peripolli V... 2019. Pedigree analysis of Brazilian Morada Nova hair sheep. Small Ruminant Res. 170:37–42. 10.1016/j.smallrumres.2018.11.012. [DOI] [Google Scholar]
- Meuwissen, T. 2009. Genetic management of small populations: a review. Acta Agric. Scand. Sect. A. 59:71–79. doi: 10.1080/09064700903118148 [DOI] [Google Scholar]
- Meuwissen, T. H. E., and Luo Z... 1992. Computing inbreeding coefficients in large populations. Genet. Sel. Evol. 24:305–313. doi: 10.1186/1297-9686-24-4-305. [DOI] [Google Scholar]
- Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 89:583–590. doi: 10.1093/genetics/89.3.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilforooshan, M. A., and Saavedra-Jiménez L. A... 2020. ggroups: an R package for pedigree and genetic groups data. Hereditas. 157:17. doi: 10.1186/s41065-020-00124-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa, V. B., M.Santana, Jr, Oliveira P. S., Eler J. P., and Ferraz J. B. S... 2010. Population structure and inbreeding effects on growth traits of Santa Inês sheep in Brazil. Small Ruminant Res. 93:135–139. doi: 10.1016/j.smallrumres.2010.05.012 [DOI] [Google Scholar]
- Pérez‐Enciso, M. 1995. Use of the uncertain relationship matrix to compute effective population size. J. Anim. Breed. Genet. 112:327–332. doi: 10.1111/J.1439-0388.1995.TB00574.X [DOI] [Google Scholar]
- Rafter, P., McHugh N., Pabiou T., and Berry D... 2022. Inbreeding trends and genetic diversity in purebred sheep populations. Animal. 16:100604. doi: 10.1016/j.animal.2022.100604 [DOI] [PubMed] [Google Scholar]
- Sheikhlou, M., and Abbasi M. A... 2016. Genetic diversity of Iranian Lori-Bakhtiari sheep assessed by pedigree analysis. Small Ruminant Res. 141:99–105. doi: 10.1016/j.smallrumres.2016.07.009 [DOI] [Google Scholar]
- Stachowicz, K., Brito L. F., Oliveira H. R., Miller S. P., and Schenkel F. S... 2018. Assessing genetic diversity of various Canadian sheep breeds through pedigree analyses. Can. J. Anim. Sci. 98:741–749. doi: 10.1139/cjas-2017-0187 [DOI] [Google Scholar]
- Terrill, C. E. 1947. Breed crosses used in the development of Targhee sheep. J. Anim. Sci. 6:83–92. doi: 10.2527/jas1947.6183 [DOI] [PubMed] [Google Scholar]
- USTSA. 2023. U.S. Targhee Sheep Association. [accessed August 28, 2023]. https://www.targheesheepus.com/
- Welsh, C., Stewart T., Schwab C., and Blackburn H... 2010. Pedigree analysis of 5 swine breeds in the United States and the implications for genetic conservation. J. Anim. Sci. 88:1610–1618. doi: 10.2527/jas.2009-2537. [DOI] [PubMed] [Google Scholar]
- Wilson, C. S., Petersen J. L., Blackburn H. D., and Lewis R. M... 2022. Assessing population structure and genetic diversity in US Suffolk sheep to define a framework for genomic selection. J. Hered. 113:431–443. doi: 10.1093/jhered/esac026 [DOI] [PubMed] [Google Scholar]
- Wright, S. 1951. The genetical structure of populations. Ann. Eugen. 15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x [DOI] [PubMed] [Google Scholar]
- Zhang, L., Mousel M. R., Wu X., Michal J. J., Zhou X., Ding B., Dodson M. V., El-Halawany N. K., Lewis G. S., and Jiang Z... 2013. Genome-wide genetic diversity and differentially selected regions among Suffolk, Rambouillet, Columbia, Polypay, and Targhee sheep. PLoS One. 8:e65942. doi: 10.1371/journal.pone.0065942 [DOI] [PMC free article] [PubMed] [Google Scholar]